Abstract
Actinomycete strain RB72T was isolated from woodland bluff soil in northern Alabama, USA, and shown to produce a broad spectrum bacteriocin. Based on morphological and chemotaxonomic characteristics, the strain was determined to belong to the genus Streptomyces. Phylogenetic analysis of the near-complete 16S rRNA gene sequence indicated that it differed from those of the described streptomycetes available in public databases. The distinctive white aerial hyphae and lack of sporulation suggest a deficiency in the whi pathway of the organism. A combination of substrate utilization patterns, morphological and chemotaxonomic characteristics and DNA–DNA hybridization results supported the affiliation of strain RB72T to the genus Streptomyces and enabled the genotypic and phenotypic differentiation of strain RB72T from closely related reference strains. Strain RB72T therefore represents a novel species of the genus Streptomyces, for which the name Streptomyces scopuliridis sp. nov. is proposed. The type strain is RB72T ( = DSM 41917T = NRRL B-24574T).
The genus Streptomyces, with more than 500 species with validly published names, contains the largest number of species of any genus in the domain Bacteria (Hain et al., 1997). The genus, first proposed by Waksman & Henrici (1943), includes aerobic, Gram-positive, high G+C content (69–78 mol%) bacteria. Most members of the genus Streptomyces possess ll-diaminopimelic acid in the ultrastructure of their peptidoglycan cell wall and produce extensively branching networks of substrate mycelia that give rise to the vertical projection of branching aerial hyphae (Williams et al., 1983; Embley & Stackebrandt, 1994). Maturity of the aerial hyphae typically culminates in a sporulation event, resulting in the formation of chains of uninucleoidal spores from the multinucleoidal, filamentous hyphae (Kwak & Kendrick, 1996). The erection of aerial hyphae generally requires a minimum of 48 h of substrate mycelium growth, while the maturation of the spores can take an additional 2 to 4 days (Lawlor et al., 1987; Willey et al., 1991; Kieser et al., 2000). Mutations in the regulatory genes guiding this process can result in alterations of phenotype. Mutations in the bld cascade and/or the proposed sky pathway cause early termination of aerial hyphae production with the differentiation of the colony arrested at the substrate mycelium growth stage (Claessen et al., 2006). Mutation within the whi cascade results in the production of an aerial mycelium that does not generate mature spores and remains white in colour (Willey et al., 1991; Chater, 2001). Bacteriocin production within the genus Streptomyces has been previously reported, with bactericidal spectra described as species-specific (Zhang et al., 2003) or genus-specific (Roelants & Naudts, 1964).
In the present study, we isolated strain RB72T from a soil sample collected at Rainbow Bluff, a woodland bluff in Lynn, Alabama. Soil-extract medium, developed from a cold-water extraction of the native soil of the organism supplemented with 10 µg cycloheximide ml−1, 20 µg nalidixic acid ml−1 and 100 U catalase ml−1, was seeded with a soil sample suspension and incubated at 25 °C for 14 days (Farris & Olson, 2007). Strain RB72T was selected for its appearance as a characteristic streptomycete colony producing a leathery substrate mycelium and developing aerial hyphae with colony maturity. Colour production within the substrate mycelium and aerial hyphae was evaluated according to the Colour Harmony Manual as described by Tresner & Backus (1963) and Shirling & Gottlieb (1966). The isolate was maintained on nutrient agar slants at 25 °C and as suspensions in nutrient broth (Difco) with glycerol (20 %, v/v) at −20 °C. Biomass for the chemotaxonomic and molecular systematic studies was prepared as described previously (Li et al., 2002). Mannitol soya flour agar (Hobbs et al., 1989) was used for maintenance growth, and nutrient broth with 0.4 % glucose (w/v) was used for biomass growth.
The morphological characteristics of strain RB72T were examined using light and scanning electron microscopy of colonies grown on mannitol soya flour agar, nutrient agar with 0.4 % glucose (w/v), yeast extract-malt extract agar [International Streptomyces Project (ISP) medium 2; Shirling & Gottlieb, 1966] and oatmeal agar (ISP medium 3) after 7, 14 and 21 days at 25 °C. The coverslip method of Hopwood (1960) was used to observe hyphal characters by phase-contrast light microscopy with a Nikon Eclipse E600 microscope equipped with a Spot RT Colour imaging system (version 3.4 imaging software; Diagnostic Instruments). For high-resolution scanning electron microscopy, agar blocks containing mycelium were fixed with osmium tetroxide (1 %, w/v, in 0.1 M cacodylate buffer, pH 7.2) for 2 h, passed through increased concentrations of acetone (25, 50, 75, 90 and 100 %) and dried to critical point with a Denton DCP-1 critical point drying apparatus. The dried samples were mounted on graphite-coated aluminium stubs, coated with gold/palladium alloy by a Technics Hummer sputter coater, and examined with a Hitachi S2500 scanning electron microscope.
Colony morphology of strain RB72T was observed on several standard media [ISP2, ISP3, inorganic salts-starch agar (ISP4), glycerol-asparagine agar (ISP5)] after 14 days of incubation at 25 °C. Examination of strain RB72T for a range of biochemical and physiological characters was as described by Shirling & Gottlieb (1966), Williams et al. (1983) and Kämpfer et al. (1991). Tolerance to salt, temperature and pH was tested on nutrient agar with 0.4 % (w/v) glucose plates incubated for 7–14 days.
Liquid cultures of strain RB72T, Streptomyces hachijoensis NRRL B-3106T and Streptomyces kentuckensis NRRL B-1831T were grown under identical conditions (nutrient broth with 0.4 %, w/v, glucose, 225 r.p.m., 30 °C) until late exponential phase (8 days), washed, lyophilized and whole-cell fatty acid profiles determined for triplicate samples following standard protocols (Sasser, 2001) except that fatty acids were identified by co-elution with known standards and mass spectral analysis of their methyl and picolinyl esters (Christie, 1998).
Genomic DNA was extracted from biomass of actively growing cultures on nutrient agar supplemented with 0.4 % glucose (w/v) as described by Olson et al. (2002). PCR amplification using universal primers 24f and 1492r was performed as described by Farris & Olson (2007). Amplified fragments were ligated into pCR2.1 cloning vector (TA cloning kit; Invitrogen) and used to transform Escherichia coli DH10B (Invitrogen) according to the manufacturer’s instructions. Plasmids with inserts of the correct size were sequenced at the Macrogen (Korea) sequencing facility. Genomic DNA isolated from strain RB72T using the method of Bollet et al. (1991) was sent to the HudsonAlpha Genomic Services Lab (Huntsville, AL) for Illumina Genome Analyser IIx sequencing.
16S rRNA gene sequence data were aligned using Sequencher version 4.5 (Gene Codes) and relatedness to gene sequences of type strains of characterized species of the genus Streptomyces was determined via NCBI blast searches (Altschul et al., 1997). The reference sequences and strain RB72T sequence (GenBank accession number EF657884) were aligned in BioEdit Sequence Alignment Editor, version 7.0.5.3 (Hall, 1999), using clustal w (Thompson et al., 1994). The neighbour-joining (Saitou & Nei, 1987) and maximum-parsimony algorithms of paup* version 4.0b 10 (Swofford, 2002) were used to infer the phylogenetic relatedness of the sequences. The method of Kimura (1980) was used to generate evolutionary distance matrices for the neighbour-joining algorithm. Tree topologies were calculated by bootstrap analyses based on 1000 resamplings.
DNA–DNA relatedness experiments were performed between strain RB72T and two closely related strains, Streptomyces hachijoensis NRRL B-3106T and Streptomyces kentuckensis NRRL B-1831T ( = Streptomyces netropsis; Hatano et al. 2003), using the fluorometric method described by Gonzalez & Saiz-Jimenez (2005). Briefly, strains RB72T, Streptomyces hachijoensis NRRL B-3106T and Streptomyces kentuckensis NRRL B-1831T were grown in either nutrient broth (Difco) or SYZ (15 g soluble starch, 2 g yeast extract, 4 g NZ amine, 2 g glucose, 1 l deionized H2O; pH 6.2) medium. Genomic DNA was isolated from the above strains using the method of Bollet et al. (1991). The purified genomic DNA samples possessed A260/A280 ratios between 1.8 and 2.0. Homoduplex and heteroduplex DNA–DNA hybridizations were performed as described by Gonzalez & Saiz-Jimenez (2005) using a Tor of 82.7 °C. Thermal denaturation experiments contained 0.2 µg duplex DNA µl−1, 0.1× SSC (pH 8.0) and SYBR Green nucleic acid stain diluted 1 : 100 000. Melting curve analysis was performed using a MyiQ Real-time PCR Detection System (Bio-Rad). Tm values for homoduplex and heteroduplex genomic DNA solutions were calculated as the temperatures corresponding to a 50 % decrease in fluorescence. ΔTm values were calculated as the difference between the Tm of the heteroduplex genomic DNA solution and the Tm of the reference strain homoduplex genomic DNA solution.
The organism exhibited a range of chemotaxonomic and phenotypic characters typical of the members of the genus Streptomyces (Table 1 and Supplementary Table S1, available in IJSEM Online). Strain RB72T formed an extensively branched substrate mycelium and aerial hyphae on several standard growth media (Supplementary Figs S1 and S2). The organism produced white aerial hyphae with no spores and a golden brown substrate mycelium on all standard morphological media tested with the exception of ISP2, on which the extent of the aerial hyphae formation was reduced and the substrate mycelium did not produce pigment. Sporulation of the aerial hyphae was not detected after 14 days, and the aerial hyphae remained white in colour, typical of other Streptomyces strains that do not sporulate (Hopwood et al., 1970; Chater, 1972, 1993; Aínsa et al., 2000; Gehring et al., 2000). Interestingly, analysis of the genomic sequencing failed to identify highly conserved (within the genus Streptomyces) primers for the bacterial signal recognition particle receptor FtsY, which has been shown to regulate sporulation in Streptomyces coelicolor through interaction with whiH (Shen et al., 2008). These results suggest a deficiency (or silent transcription) in the whi pathway of the organism.
Table 1. Comparison of morphological, cultural and physiological characteristics of strain RB72T and related species of the genus Streptomyces.
Strains: 1, RB72T; 2, S. hachijoensis NRRL B-3106T; 3, S. kentuckensis NRRL B-1831T. All data were determined in the laboratory under the same growth conditions. nd, Not determined; d, variable; +, positive; −, negative. All strains were positive for growth on d-glucose and myo-inositol and negative for growth on sucrose.
| Characteristic | 1 | 2 | 3 |
| Morphology and pigmentation | |||
| Aerial mass on oatmeal agar | White | Beige | Red–white |
| Spore-chain arrangement | − | nd | nd |
| Spore surface | − | nd | nd |
| Melanin production | − | − | + |
| Production of diffusible pigments | − | − | − |
| Growth on sole carbon sources (1 %, w/v) | |||
| l-Arabinose | + | − | − |
| d-Fructose | − | d | − |
| d-Galactose | + | − | + |
| d-Mannitol | − | − | − |
| d-Raffinose | + | − | − |
| l-Rhamnose | + | − | − |
| d-Xylose | + | − | − |
| Sorbitol | − | + | + |
| Cellobiose | + | − | + |
| Melibiose | + | − | + |
| l-Sorbose | − | − | − |
| Maltose | + | + | + |
| Adonitol | + | + | + |
| Lactose | + | − | + |
| d-Mannose | + | + | + |
| Dextrin | + | + | + |
| Inulin | − | + | + |
In addition to the characters in Table 1, strain RB72T reduced nitrate to nitrite and growth occurred at sodium chloride concentrations of 4 and 7 %, but not at 10 or 13 % (w/v). Growth occurred at pH 6.0–11.0 (optimum, pH 7.0) and 15–37 °C (optimum, 25 °C). Strain RB72T hydrolysed adenine, casein, aesculin, gelatin, hypoxanthine, l-tyrosine, starch and xanthine but not cellulose.
The whole-cell fatty acid profiles of all of the species of the genus Streptomyces analysed were mainly comprised of iso-branched, even- and odd-chain, saturated and monounsaturated fatty acids (Supplementary Table S1). Strain RB72T was unique in that the iso-branched, odd-chain, unsaturated fatty acid iso-C17 : 1ω8 comprised greater than 10 % of its total fatty acids.
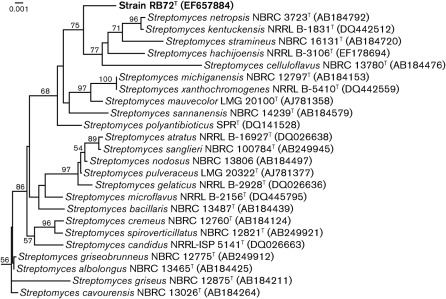
A near-complete 16S rRNA gene sequence (1376 nt) was determined for strain RB72T. Comparison of the sequence with sequences of reference micro-organisms confirmed that the unknown isolate was closely related to species of the genus Streptomyces. Phylogenetic analysis showed that strain RB72T formed a sister grouping with Streptomyces kentuckensis NRRL B-1831T (98.47 % similarity), Streptomyces netropsis NBRC 12893T (98.45 %), Streptomyces stramineus NBRC 16131T (98.45 %), Streptomyces hachijoensis NRRL B-3106T (98.61 %) and Streptomyces celluloflavus NBRC 13780T (98.59 %) (Fig. 1). Streptomyces kentuckensis NRRL B-1831T is considered a subjective synonym of Streptomyces netropsis (Labeda, 1996; Hatano et al., 2003). Genomic sequencing revealed 70.3 mol% DNA G+C content for strain RB72T.
Fig. 1.
Neighbour-joining phylogenetic tree based on near-complete 16S rRNA gene sequences showing the relationship between strain RB72T and 24 species of the genus Streptomyces. Numbers at nodes indicate levels of bootstrap support (%) based on analysis of 1000 resampled datasets; only values above 50 % are given. NCBI accession numbers for each sequence are in parentheses. Bar, 1 substitution per 1000 nt.
Analysis of DNA–DNA relatedness was performed between strain RB72T and the closely related strains Streptomyces hachijoensis NRRL B-3106T and Streptomyces kentuckensis NRRL B-1831T using the fluorometric method described by Gonzalez & Saiz-Jimenez (2005). This method measures the difference in the thermal denaturation midpoints of homoduplex versus heteroduplex genomic DNA (ΔTm) and has been used successfully in the characterization of other species of the genus Streptomyces (Kumar & Goodfellow, 2008). Distinct genomic species have a ΔTm equal to or greater than 5 °C (e.g. Wayne et al., 1987; Stackebrandt & Goebel, 1994; Rosselló-Mora & Amann, 2001). For each species–species comparison, two independent experiments were performed. The following temperature differences represent the means and single standard deviations (given in parentheses) of these experimental sets. We found a difference in melting temperature of Streptomyces hachijoensis homoduplex genomic DNA versus RB72T– Streptomyces hachijoensis heteroduplex genomic DNA of 12.2 °C (1.0 °C), confirming a definite species delineation. Likewise, the melting temperature difference between Streptomyces kentuckensis homoduplex genomic DNA and RB72T–Streptomyces kentuckensis heteroduplex genomic DNA was 7.6 °C (1.9 °C), also corroborating species delineation.
Strain RB72T demonstrated a broad spectrum of bacteriolytic activity. The purified bacteriocin (data not shown) was active against the Gram-positive bacteria Streptomyces avermitilis MA-4680T, Streptomyces coelicolor A3(2), ‘Streptomyces lividans’ 66, Streptomyces venezuelae NRRL-ISP 5230T, Nocardia salmonicida NRRL B-2778T, Nocardia vaccinii NRRL WC-3500T, Rhodococcus marinonascens DSM 43752T, Bacillus megaterium ATCC 14581T, Bacillus subtilis 168, Staphylococcus aureus FDA209, Streptococcus pyogenes ATCC 14289, Enterococcus faecalis ATCC 29212 and Micrococcus luteus strain 85W0996, and the Gram-negative bacteria Escherichia coli DH10B and Klebsiella pneumoniae ATCC 13883T.
Strain RB72T warrants classification as the type strain of a novel species of the genus Streptomyces based on comparison of its 16S rRNA gene sequence with other known species of the genus Streptomyces and the phenotypic characters of sole carbon source utilization, genomic DNA hybridization/thermal denaturation experiments, chemotaxonomic characters, broad spectrum bacteriocin production and lack of sporulation that set it apart from other described species of the genus Streptomyces. For strain RB72T, we propose the name Streptomyces scopuliridis sp. nov.
Description of Streptomyces scopuliridis sp. nov.
Streptomyces scopuliridis (scop.ul.i′rid.is. L. masc. n. scopulus cliff, bluff, crag; L. gen. n. Iridis of or belonging to the goddess of the rainbow; N.L. gen. n. scopuliridis from a rainbow cliff, referring to the location of isolation, Rainbow Bluff, a woodland bluff in Lynn, Alabama).
Aerobic, Gram-positive, non-motile, non-spore-forming actinomycete. Substrate and aerial mycelia are produced; however, the aerial hyphae fail to undergo the sporulation process. The substrate and aerial hyphae branch extensively, and the aerial hyphae remain white upon maturation. The reverse side of the substrate mycelium produces a golden brown pigment on ISP3, ISP4 and ISP5 media. In addition to the characters described in Table 1, nitrate is reduced to nitrite and growth occurs at sodium chloride concentrations of 4–7 % (w/v), at pH 6.0–11.0 and at temperatures of 15–37 °C. Hydrolyses adenine, casein, aesculin, gelatin, hypoxanthine, l-tyrosine, starch and xanthine, but not cellulose. The four most abundant fatty acids are iso-C16 : 0, iso-C17 : 1ω8, iso-C15 : 0 and anteiso-C15 : 0 and the G+C content of the genomic DNA of the type strain is 70.3 mol%. Produces a broad spectrum bacteriocin with activity against Gram-positive and Gram-negative bacteria.
The type strain, RB72T ( = DSM 41917T = NRRL B-24574T), was isolated from a soil sample collected from Rainbow Bluff, a woodland bluff in Lynn, Alabama.
Acknowledgements
Funding for this study was partially provided by a National Institutes of Health grant (1R15GM069402-01) to J. B. O.
Abbreviation:
- ISP
International Streptomyces Project
Footnotes
Two supplementary figures and a supplementary table are available with the online version of this paper.
Supplementary Material
Whole-cell fatty acid composition of strain RB72T and related speciesof the genus Streptomyces
Phase-contrast micrographs of strain RB72T. Growth of strain RB72T on nutrient medium with 0.4% glucose at 25 °C for 14 days shows extensively branching aerial hyphae with a lack of sporulation. Images (a) and (b) are ×400 magnification. Images (c), (d) and (e) are ×1000 magnification.
Scanning electron micrographs of strain RB72TT on oatmeal medium at 25 °C for 14 days shows extensively branching aerial hyphae with a lack of sporulation. Bars, 6.0 μm.
References
- Aínsa J. A., Ryding N. J., Hartley N., Findlay K. C., Bruton C. J., Chater K. F. (2000). WhiA, a protein of unknown function conserved among Gram-positive bacteria, is essential for sporulation in Streptomyces coelicolor A3(2). J Bacteriol 182, 5470–5478 10.1128/JB.182.19.5470-5478.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997). Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollet C., Gevaudan M. J., de Lamballerie X., Zandotti C., de Micco P. (1991). A simple method for the isolation of chromosomal DNA from gram positive or acid-fast bacteria. Nucleic Acids Res 19, 1955 10.1093/nar/19.8.1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater K. F. (1972). A morphological and genetic mapping study of white colony mutants of Streptomyces coelicolor. J Gen Microbiol 72, 9–28 [DOI] [PubMed] [Google Scholar]
- Chater K. F. (1993). Genetics of differentiation in Streptomyces. Annu Rev Microbiol 47, 685–711 10.1146/annurev.mi.47.100193.003345 [DOI] [PubMed] [Google Scholar]
- Chater K. F. (2001). Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr Opin Microbiol 4, 667–673 10.1016/S1369-5274(01)00267-3 [DOI] [PubMed] [Google Scholar]
- Christie W. W. (1998). Gas chromatography-mass spectrometry methods for structural analysis of fatty acids. Lipids 33, 343–353 10.1007/s11745-998-0214-x [DOI] [PubMed] [Google Scholar]
- Claessen D., de Jong W., Dijkhuizen L., Wösten H. A. (2006). Regulation of Streptomyces development: reach for the sky! Trends Microbiol 14, 313–319 10.1016/j.tim.2006.05.008 [DOI] [PubMed] [Google Scholar]
- Embley T. M., Stackebrandt E. (1994). The molecular phylogeny and systematics of the actinomycetes. Annu Rev Microbiol 48, 257–289 10.1146/annurev.mi.48.100194.001353 [DOI] [PubMed] [Google Scholar]
- Farris M. H., Olson J. B. (2007). Detection of Actinobacteria cultivated from environmental samples reveals bias in universal primers. Lett Appl Microbiol 45, 376–381 10.1111/j.1472-765X.2007.02198.x [DOI] [PubMed] [Google Scholar]
- Gehring A. M., Nodwell J. R., Beverley S. M., Losick R. (2000). Genomewide insertional mutagenesis in Streptomyces coelicolor reveals additional genes involved in morphological differentiation. Proc Natl Acad Sci U S A 97, 9642–9647 10.1073/pnas.170059797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J. M., Saiz-Jimenez C. (2005). A simple fluorimetric method for the estimation of DNA–DNA relatedness between closely related microorganisms by thermal denaturation temperatures. Extremophiles 9, 75–79 10.1007/s00792-004-0417-0 [DOI] [PubMed] [Google Scholar]
- Hain T., Ward-Rainey N., Kroppenstedt R. M., Stackebrandt E., Rainey F. A. (1997). Discrimination of Streptomyces albidoflavus strains based on the size and number of 16S–23S ribosomal DNA intergenic spacers. Int J Syst Bacteriol 47, 202–206 10.1099/00207713-47-1-202 [DOI] [PubMed] [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41, 95–98 [Google Scholar]
- Hatano K., Nishii T., Kasai H. (2003). Taxonomic re-evaluation of whorl-forming Streptomyces (formerly Streptoverticillium) species by using phenotypes, DNA–DNA hybridization and sequences of gyrB, and proposal of Streptomyces luteireticuli (ex Katoh and Arai 1957) corrig., sp. nov., nom. rev. Int J Syst Evol Microbiol 53, 1519–1529 10.1099/ijs.0.02238-0 [DOI] [PubMed] [Google Scholar]
- Hobbs G., Frazer C. M., Gardner D. C. J., Cullum J. A., Oliver S. G. (1989). Dispersed growth of Streptomyces in liquid culture. Appl Microbiol Biotechnol 31, 272–277 10.1007/BF00258408 [DOI] [Google Scholar]
- Hopwood D. A. (1960). Phase-contrast observations on Streptomyces coelicolor. J Gen Microbiol 22, 295–302 [DOI] [PubMed] [Google Scholar]
- Hopwood D. A., Wildermuth H., Palmer H. M. (1970). Mutants of Streptomyces coelicolor defective in sporulation. J Gen Microbiol 61, 397–408 [DOI] [PubMed] [Google Scholar]
- Kämpfer P., Kroppenstedt R. M., Dott W. (1991). A numerical classification of the genera Streptomyces and Streptoverticillium using miniaturized physiological tests. J Gen Microbiol 137, 1831–1891 [Google Scholar]
- Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. (2000). General introduction to actinomycete biology, In Practical Streptomyces Genetics, pp. 2–42 Norwich: The John Innes Foundation [Google Scholar]
- Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16, 111–120 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- Kumar Y., Goodfellow M. (2008). Five new members of the Streptomyces violaceusniger 16S rRNA gene clade: Streptomyces castelarensis sp. nov., comb. nov., Streptomyces himastatinicus sp. nov., Streptomyces mordarskii sp. nov., Streptomyces rapamycinicus sp. nov. and Streptomyces ruanii sp. nov. Int J Syst Evol Microbiol 58, 1369–1378 10.1099/ijs.0.65408-0 [DOI] [PubMed] [Google Scholar]
- Kwak J., Kendrick K. E. (1996). Bald mutants of Streptomyces griseus that prematurely undergo key events of sporulation. J Bacteriol 178, 4643–4650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeda D. P. (1996). DNA relatedness among verticil-forming Streptomyces species (formerly Streptoverticillium species). Int J Syst Bacteriol 46, 699–703 10.1099/00207713-46-3-699 [DOI] [Google Scholar]
- Lawlor E. J., Baylis H. A., Chater K. F. (1987). Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2). Genes Dev 1, 1305–1310 10.1101/gad.1.10.1305 [DOI] [PubMed] [Google Scholar]
- Li W., Lanoot B., Zhang Y., Vancanneyt M., Swings J., Liu Z. (2002). Streptomyces scopiformis sp. nov., a novel streptomycete with fastigiate spore chains. Int J Syst Evol Microbiol 52, 1629–1633 10.1099/ijs.0.02130-0 [DOI] [PubMed] [Google Scholar]
- Olson J. B., Harmody D. K., McCarthy P. J. (2002). Alpha-proteobacteria cultivated from marine sponges display branching rod morphology. FEMS Microbiol Lett 211, 169–173 [DOI] [PubMed] [Google Scholar]
- Roelants P., Naudts F. (1964). Properties of a bacteriocin-like substance produced by Streptomyces virginiae. Antonie van Leeuwenhoek 30, 45–53 10.1007/BF02046699 [DOI] [PubMed] [Google Scholar]
- Rosselló-Mora R., Amann R. (2001). The species concept for prokaryotes. FEMS Microbiol Rev 25, 39–67 10.1016/S0168-6445(00)00040-1 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4, 406–425 [DOI] [PubMed] [Google Scholar]
- Sasser M. (2001). Bacterial identification by gas chromatographic analysis of fatty acids methyl esters (GC-FAME). http://www.midi-inc.com/pdf/MIS_Technote_101.pdf.
- Shen X.-L., Dong H.-J., Hou X.-P., Guan W.-J., Li Y.-Q. (2008). FtsY affects sporulation and antibiotic production by whiH in Streptomyces coelicolor. Curr Microbiol 56, 61–65 10.1007/s00284-007-9039-y [DOI] [PubMed] [Google Scholar]
- Shirling E. B., Gottlieb D. (1966). Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16, 313–340 10.1099/00207713-16-3-313 [DOI] [Google Scholar]
- Stackebrandt E., Goebel B. M. (1994). Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44, 846–849 10.1099/00207713-44-4-846 [DOI] [Google Scholar]
- Swofford D. L. (2002). paup*: Phylogenetic analysis using parsimony (*and other methods), version 4. Sunderland, MA: Sinauer Associates.
- Thompson J. D., Higgins D. G., Gibson T. J. (1994). clustal w: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22, 4673–4680 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresner H. D., Backus E. J. (1963). System of color wheels for streptomycete taxonomy. Appl Microbiol 11, 335–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman S. A., Henrici A. T. (1943). The nomenclature and classification of the actinomycetes. J Bacteriol 46, 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne L. G., Brenner D. J., Colwell R. R., Grimont P. A. D., Kandler O., Krichevsky M. I., Moore L. H., Moore W. E. C., Murray R. G. E., et al. (1987). Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37, 463–464 10.1099/00207713-37-4-463 [DOI] [Google Scholar]
- Willey J., Santamaria R., Guijarro J., Geistlich M., Losick R. (1991). Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by S. coelicolor. Cell 65, 641–650 10.1016/0092-8674(91)90096-H [DOI] [PubMed] [Google Scholar]
- Williams S. T., Goodfellow M., Alderson G., Wellington E. M., Sneath P. H., Sackin M. J. (1983). Numerical classification of Streptomyces and related genera. J Gen Microbiol 129, 1743–1813 [DOI] [PubMed] [Google Scholar]
- Zhang X., Clark C. A., Pettis G. S. (2003). Interstrain inhibition in the sweet potato pathogen Streptomyces ipomoeae: purification and characterization of a highly specific bacteriocin and cloning of its structural gene. Appl Environ Microbiol 69, 2201–2208 10.1128/AEM.69.4.2201-2208.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Whole-cell fatty acid composition of strain RB72T and related speciesof the genus Streptomyces
Phase-contrast micrographs of strain RB72T. Growth of strain RB72T on nutrient medium with 0.4% glucose at 25 °C for 14 days shows extensively branching aerial hyphae with a lack of sporulation. Images (a) and (b) are ×400 magnification. Images (c), (d) and (e) are ×1000 magnification.
Scanning electron micrographs of strain RB72TT on oatmeal medium at 25 °C for 14 days shows extensively branching aerial hyphae with a lack of sporulation. Bars, 6.0 μm.