Abstract
Neisseria meningitidis can utilize haem, haemoglobin and haemoglobin–haptoglobin complexes as sources of iron via two TonB-dependent phase variable haemoglobin receptors, HmbR and HpuAB. HmbR is over-represented in disease isolates, suggesting a link between haemoglobin acquisition and meningococcal disease. This study compared the distribution of HpuAB and phase variation (PV) status of both receptors in disease and carriage isolates. Meningococcal disease (n = 214) and carriage (n = 305) isolates representative of multiple clonal complexes (CCs) were investigated for the distribution, polyG tract lengths and ON/OFF status of both haemoglobin receptors, and for the deletion mechanism for HpuAB. Strains with both receptors or only hmbR were present at similar frequencies among meningococcal disease isolates as compared with carriage isolates. However, >90 % of isolates from the three CCs CC5, CC8 and CC11 with the highest disease to carriage ratios contained both receptors. Strains with an hpuAB-only phenotype were under-represented among disease isolates, suggesting selection against this receptor during systemic disease, possibly due to the receptor having a high level of immunogenicity or being inefficient in acquisition of iron during systemic spread. Absence of hpuAB resulted from either complete deletion or replacement by an insertion element. In an examination of PV status, one or both receptors were found in an ON state in 91 % of disease and 71 % of carriage isolates. We suggest that expression of a haemoglobin receptor, either HmbR or HpuAB, is of major importance for systemic spread of meningococci, and that the presence of both receptors contributes to virulence in some strains.
Introduction
Neisseria meningitidis (the meningococcus) is an obligate commensal of humans, residing in the nasopharynx of 10–30 % of individuals. Meningococci can invade host tissues and disseminate in blood, causing meningitis and septicaemia, which are leading causes of mortality among children and young adults worldwide (Pollard, 2004; Stephens, 2007). Studies in animal models have demonstrated that the virulence of pathogenic meningococci increases significantly when bacteria are injected in combination with iron complexes (Perkins-Balding et al., 2004). In humans, iron is usually sequestered in complexes with iron-binding proteins such as transferrin, lactoferrin and haemoglobin (Otto et al., 1992). Meningococci encode several surface receptors to strip and acquire iron or haem from these iron-binding proteins, including two TonB-dependent haem acquisition systems, HpuAB and HmbR (Perkins-Balding et al., 2004). HpuAB is a bipartite complex consisting of a lipoprotein, HpuA (37 kDa), and a transmembrane protein, HpuB (85 kDa), both of which are required to bind haptoglobin–haemoglobin complexes and free haemoglobin (Lewis & Dyer, 1995; Rohde & Dyer, 2004). HmbR is an 89 kDa transmembrane protein that specifically binds haemoglobin (Perkins-Balding et al., 2003). This redundancy in haemoglobin binding receptors may enable meningococci to acquire haem from a variety of sources and niches or may facilitate immune evasion.
The distribution of HpuAB and HmbR varies among meningococcal strains, with some containing both systems and others only one (Lewis et al., 1999; Richardson & Stojiljkovic, 1999). Recent studies have detected a significantly higher frequency of hmbR in disease as compared with carriage isolates, suggesting that this receptor contributes to meningococcal pathogenesis (Harrison et al., 2009). The expression of these systems is phase variable due to stretches of polyG nucleotides present in the ORFs of hpuA and hmbR (Lewis et al., 1999; Richardson & Stojiljkovic, 1999). Mutations in these tracts arising by slipped-strand mispairing during DNA replication lead to ‘ON’ and ‘OFF’ switches in the expression of these receptors. Differential expression due to changes in tract length helps bacteria to adapt to fluctuations in their microenvironments (Moxon et al., 2006), and may also contribute to escape from the adaptive immune responses within the host (Bayliss et al., 2008). Immune evasion may also be facilitated by significant levels of antigenic diversity in HmbR (Evans et al., 2010). Understanding the contributions of these haemoglobin receptors to meningococcal disease and the impact of immune responses on these surface proteins is hampered by a lack of detailed studies on the distribution, antigenic variation and phase variation (PV) status of the two receptors in epidemiological samples from disease and carriage.
Methods
Bacterial isolates, culture conditions and DNA extraction.
Isolates from four collections were analysed. The first group comprised 107 isolates assembled globally between 1937 and 1996 (Maiden et al., 1998). The second group contained 88 carriage isolates collected in 2008 from first year students at Nottingham University (Bidmos et al., 2011). The third collection included 153 isolates assembled in the Czech Republic in 1993 (Jolley et al., 2000). The fourth group contained 77 disease and 82 carriage isolates collected in England and Wales between 1999 and 2000 (from the Health Protection Agency Meningococcal Reference Unit, Manchester, UK, and the UK Carriage Study; Maiden et al., 2002). Twelve additional sequence type (ST)-41/44 isolates from the UK were also analysed (provided by I. M. Feavers). Bacteria were grown overnight, followed by either preparation of bacterial lysates or chromosomal DNA extraction using a DNeasy Blood and Tissue kit (Qiagen). Isolates were classified as disease or carriage depending on whether they were isolated from patients or carriers, respectively.
PCR amplification and sequence analysis.
The presence of hmbR was detected with primers hmbR-RF3 (5′-TGCCAACCTCTTTTACGAATGG-3′) and hmbR-RF4 (5′-GCTACTGAACACGTCGTTCC-3′), and that of hpuAB with primers hpuAC (5′-ATGCGATGAAATACAAAGCCC-3′) and hpuA350Rev (5′-GGATGAAAGGGCGTATTGCGC-3′) or hpuAC and P26.85 (5′-GGGAAACGCTTGGGCGATGG-3′). Isolates negative for hpuAB were screened with primers hpu-for1 (5′-GCAACAATGCCTTGTCATCC-3′) and hpu-rev13 (5′-TGATCGAAATGGGCGTACTC-3′), which bind on either side of the hpuAB locus. For analyses of hpuAB deletion and replacement events, hpu-for1/hpu-rev13 amplicons were sequenced with primers hpu-for1, hpu-rev13, hpuF-seq (5′-GGCAACTTTTCCACCGTCATTC-3′) and hpuF-seq2 (5′-AAACCGGCAACATCTGGAAG-3′). Complete sequences for HpuA were determined by amplification with N- and C-terminal primers, followed by sequencing of the products with both PCR primers and internal primers.
Repeat numbers for the phase-variable polyG tracts of hmbR and hpuA were determined by sequencing of the amplicons utilized for detection of these genes and enumeration of G residues in the tracts contained in this sequence data. For hpuA, a 10G tract is associated with an intact reading frame in isolates Z2491 and FAM18, and hence represents the ON status. Similarly, hmbR is in an ON state if the tract is 9G, as found in strain MC58. The alignment of sequences from test isolates with the Z2491 and MC58 sequences confirmed the ON/OFF status and showed that none of the insertion/deletion events around the repeat tracts affected the reading frame. The ON/OFF status for hpuA was also confirmed for 10 isolates by translating complete gene sequences.
The tract lengths and ON/OFF status of some isolates from the 2008 carriage study were determined by a combination of sequencing and sizing of PCR fragments. Isolates were assigned to clones on the basis of identical porA, fetA and MLST types, and then the repeat tracts of hmbR and hpuA were determined by sequencing for one or two isolates per clone. These genes were then amplified from all isolates of each clone with a 6-carboxyfluorescein (FAM)-labelled primer, hpuA-350Rev or hmbR-RF3, and the relevant non-labelled primer. PCR products were subjected to A-tailing by addition of a 4 µl reaction mix containing 0.4 µl PCR buffer (10×), 0.4 µl MgCl2 (25 mM), 0.05 µl Taq and 3.15 µl distilled H2O, followed by incubation at 72 °C for 45 min. Diluted PCR products (0.5 µl) were then mixed with 0.5 µl GeneScan 500 LIZ size standard (Applied Biosystems) and 9 µl formamide, followed by denaturation and electrophoresis on an Autosequencer (Applied Biosystems). GeneScan data were analysed using Peak Scanner v1.0 software (Applied Biosystems).
Phenotypic analysis of phase variants.
Meningococcal strains were grown overnight on brain heart infusion (BHI) plates containing Levinthal’s supplement. Suspensions of bacteria were prepared and spread onto Mueller–Hinton agar (MHA) plates containing 40 µM desferal and onto plain MHA plates. Disks were impregnated with 10 µl of either human haemoglobin (10 mg ml−1, Sigma) or transferrin (50 mg ml−1, Sigma) and placed onto a desferal-containing MHA plate along with a third disk without any added iron source. Plates were incubated overnight at 37 °C in 5 % CO2. Confluent growth was observed on the plain MHA plates but was absent on plates containing desferal and no added iron source.
Statistical analyses.
Statistical analyses were carried out using GraphPad Prism version 5. Odds ratios (ORs) and 95 % confidence intervals (CIs) were derived using a Chi-squared test and P values with a Fisher’s exact test.
Results
Sequence variation of HpuA
The variability in the amino acid sequences of the surface-exposed component of the HpuAB receptor, HpuA, has not been reported previously. Complete sequences of hpuA were generated for a serogroup B isolate (strain 8047) and isolates representing the following clonal complexes (CCs): 23, 60, 167 and 174. The nucleotide and derived amino acid sequences were aligned with those extracted from the Z2491 and FAM18 genome sequences (Supplementary Fig. S1). HpuA proteins varied in size from 326 to 342 aa due to a series of eight indels, including a large insertion of 16 aa within the N-terminal region, and exhibited significant levels of variation (76–94 % identity), with four major variable regions and 101 polymorphic sites. As nothing is known about the functional domains of this protein, it is unclear whether this variability will influence its functions or antigenicity. A similarly high level of variability was observed for TbpB, the outer membrane component of the meningococcal transferrin-binding protein, although this variability did not alter transferrin binding by this protein (Boulton et al., 1998).
Distribution of haemoglobin receptors in carriage and disease isolates
An epidemiological study of the distribution of one of the haemoglobin receptors, HmbR, has revealed a significantly higher frequency of this gene in meningococcal disease isolates compared with carriage isolates (Harrison et al., 2009). To study the combined influence of the two haemoglobin genes (hpuAB and hmbR) on meningococcal disease and carriage, the distribution of both receptors was investigated using four unrelated isolate collections containing 305 and 214 meningococcal isolates from carriage samples and disease cases, respectively. The majority (n = 422) represented nine serogroups (A, B, C, 29E, H, W-135, X, Y and Z), while 97 were non-serogroupable. A total of 33 CCs were represented, with most isolates belonging to 16 CCs.
Overall, we observed that 47 % of isolates had both genes, while 28 % had hmbR alone, and 21 % had hpuAB as their sole haemoglobin receptor. A minority (4 %) lacked both systems, and most of these (91 %) were obtained from carriers. Comparisons were made between the presence of both genes versus one gene and between hmbR only and hpuAB only (Table 1). No significant difference was observed in the frequencies of both genes versus hmbR only between disease and carriage isolates. In contrast, both genes or hmbR only occurred at significantly higher frequencies than hpuAB only in disease isolates. These differences were obtained for each isolate collection separately as well as in combination. An over-representation of hpuAB in carriage versus disease isolates was evident, although the significance was reduced on exclusion of the 2008 carriage study (Table 2), which includes multiple isolates resulting from clonal expansion of some strains (Bidmos et al., 2011).
Table 1. Distribution of haemoglobin receptors in disease and carriage isolates.
| Strain collection | Group | Both present | hmbR only | hpuAB only | Both absent | Total | Disease association OR (95 % CI) | ||
| Both vs hpuAB | Both vs hmbR | hmbR vs hpuAB | |||||||
| All | Disease | 113 (52 %) | 87 (41 %) | 12 (6 %) | 2 (1 %) | 214 | 6.83 (3.56–13.0)* | 0.58 (0.38–0.87)† | 11.88 (5.98–23.59)* |
| Carriage | 131 (43 %) | 58 (19 %) | 95 (31 %) | 21 (7 %) | 305 | ||||
| All minus 2008 | Disease | 113 (52 %) | 87 (41 %) | 12 (6 %) | 2 (1 %) | 214 | 4.85 (2.44–9.64)* | 0.67 (0.42–1.02)‡ | 7.4 (3.6–15.21)* |
| Carriage | 97 (45 %) | 49 (22 %) | 50 (23 %) | 21 (10 %) | 217 | ||||
| Czech 1993 | Disease | 26 (68 %) | 9 (24 %) | 1 (3 %) | 2 (5 %) | 38 | 10.2 (1.3–80.29) | 1.47 (0.6–3.6) | 6.92 (0.81–59.27) |
| Carriage | 51 (44 %) | 26 (23 %) | 20 (17 %) | 18 (16 %) | 115 | ||||
| 107 MLST isolates | Disease | 47 (51 %) | 33 (37 %) | 10 (12 %) | 0 | 90 | 3.92 (1.0–15.4) | 0.95 (0.25–3.63) | 4.13 (0.93–18.37) |
| Carriage | 6 (35 %) | 4 (24 %) | 5 (29 %) | 2 (12 %) | 17 | ||||
| UK 1999 | Disease | 39 (51 %) | 37 (48 %) | 1 (1 %) | 0 | 77 | 25.0 (3.23–193.8) | 0.49 (0.24–1.0) | 51.39 (6.44–410.2) |
| Carriage | 39 (48 %) | 18 (22 %) | 25 (30 %) | 0 | 82 | ||||
| ST-41/44 | Disease | 4 (9 %) | 40 (89 %) | 0 | 1 (2 %) | 45 | − | 0.47 (0.1–2.28) | − |
| Carriage | 3 (9 %) | 13 (38 %) | 0 | 18 (53 %) | 34 | ||||
P <0.0001.
P = 0.02.
Not significant.
Table 2. Distribution of hpuAB in disease and carriage isolates.
| Strain collection | Group | hpuAB-positive | hpuAB-negative | Total | Disease association OR (95 % CI) | Carriage association OR (95 % CI) |
| All | Disease | 125 (58 %) | 89 (42 %) | 214 | 0.49 (0.38–0.71) | 2.04 (1.4–2.96)* |
| Carriage | 226 (74 %) | 79 (26 %) | 305 | |||
| All minus 2008 | Disease | 125 (58 %) | 89 (42 %) | 214 | 0.67 (0.45–0.995) | 1.50 (1.01–2.22)† |
| Carriage | 147 (68 %) | 70 (32 %) | 217 |
P = 0.0002.
P = 0.047.
Serogroups A, C and 29E exhibited a high prevalence of these receptors, with 75, 87 and 57 %, respectively, of strains from these serogroups harbouring both genes. Some serogroups exhibited an under-representation of the hmbR-only phenotype; thus, none of the isolates from serogroup Y and only 2 % of 29E serogroupable strains had this receptor as the sole haemoglobin-binding protein. In contrast, hmbR only was present in 59 % of serogroup B strains, with only 12 % having an hpuAB-only phenotype. There was no significant difference between the presence of both genes versus hmbR only in disease versus carriage serogroup B isolates (OR 0.58, 0.294–1.136 at 95 % CI, P = 0.12).
A bias in the distribution of haemoglobin receptors between CCs was also evident (Table 3). One group of CCs had a high prevalence of hmbR-only isolates (e.g. 92, 90, 67 and 63 % of isolates from ST-18, ST-32, ST-41/44 and ST-269, respectively). Other CCs exhibited a high level of both receptors (e.g. 98, 92 and 90 % of isolates from ST-11, ST-5 and ST-8, respectively). In contrast, ST-167, ST-1 and ST-22 had high levels of hpuAB-only (91, 62 and 61 %, respectively) and low levels of hmbR-only isolates, while 100 % of isolates analysed from ST-174, ST-106 and ST-23 had hpuAB-only phenotypes. None of the isolates from the ST-41/44 complex had hpuAB as a sole haemoglobin receptor, while 53 % of carriage isolates from this CC lacked both receptors.
Table 3. Distribution of hpuAB deletion mechanism and haemoglobin receptors by CC.
| CC | Number of isolates | hpuAB deletion mechanism | Receptor status | Disease : carriage ratio* | ||||
| IS element | Complete deletion | Both present | hmbR only | hpuAB only | Both absent | |||
| Group 1: HmbR only in 10–100 % of isolates | ||||||||
| ST-41/44 | 79 | 69 | 3 | 7 | 53 | 0 | 19 | 1.2 |
| ST-18 | 13 | 12 | 0 | 1 | 12 | 0 | 0 | 5.5 |
| ST-213 | 8 | 6 | 0 | 2 | 6 | 0 | 0 | 0.6 |
| ST-461 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | − |
| ST-334 | 2 | 1 | 0 | 1 | 1 | 0 | 0 | − |
| ST-32 | 21 | 0 | 19 | 2 | 19 | 0 | 0 | 3.5 |
| ST-269 | 19 | 0 | 12 | 7 | 12 | 0 | 0 | 2.8 |
| ST-35 | 4 | 0 | 3 | 1 | 3 | 0 | 0 | 0.5 |
| Unspecified | 36 | 33 | 3 | 0 | 32 | 0 | 4 | − |
| Group 2: both receptors in >90 % isolates | ||||||||
| ST-11 | 59 | 1 | 0 | 58 | 1 | 0 | 0 | 6.6 |
| ST-5 | 12 | 1 | 0 | 11 | 1 | 0 | 0 | 19.5 |
| ST-4 | 11 | 0 | 0 | 11 | 0 | 0 | 0 | − |
| ST-92 | 15 | 0 | 0 | 15 | 0 | 0 | 0 | − |
| ST-8 | 10 | 0 | 1 | 9 | 1 | 0 | 0 | 24.5 |
| ST-116 | 6 | 0 | 0 | 6 | 0 | 0 | 0 | − |
| ST-231 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | − |
| ST-37 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | − |
| ST-53 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | − |
| ST-162 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | − |
| ST-292 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | − |
| ST-364 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | − |
| ST-750 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | − |
| Unspecified | 61 | 0 | 0 | 61 | 0 | 0 | 0 | − |
| Group 3: HpuAB only in 10–90 % of isolates | ||||||||
| ST-60 | 29 | 0 | 1 | 18 | 1 | 10 | 0 | 0.7 |
| ST-1157 | 10 | 0 | 0 | 8 | 0 | 2 | 0 | − |
| ST-198 | 5 | 0 | 0 | 2 | 0 | 3 | 0 | <0.1 |
| ST-1 | 13 | 0 | 1 | 4 | 1 | 8 | 0 | 5.5 |
| ST-22 | 18 | 0 | 0 | 7 | 0 | 11 | 0 | 0.6 |
| ST-103 | 5 | 1 | 0 | 2 | 1 | 2 | 0 | 1.2 |
| Group 4: HpuAB only in >90 % of isolates | ||||||||
| ST-174 | 15 | 0 | 0 | 0 | 0 | 15 | 0 | − |
| ST-106 | 11 | 0 | 0 | 0 | 0 | 11 | 0 | − |
| ST-23 | 13 | 0 | 0 | 0 | 0 | 13 | 0 | 0.8 |
| ST-167 | 11 | 0 | 0 | 1 | 0 | 10 | 0 | 0.5 |
| ST-254 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | − |
| ST-549 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | − |
| Unspecified | 20 | 0 | 0 | 0 | 0 | 20 | 0 | − |
Disease : carriage ratios were taken from Caugant & Maiden (2009).
Genetic arrangement of the hpuAB locus and deletion mechanisms
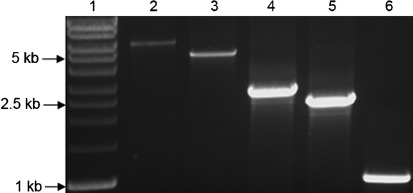
The arrangement and deletion mechanisms for the hpuAB locus were elucidated by amplification of the regions flanking this locus using primers specific for adjacent genes (encoding a hypothetical periplasmic protein and GroEL, a chaperone). These primers are predicted to produce an ~5.6 kb fragment from meningococcal strains with a full-length hpuAB system. This analysis revealed a variation in the size of PCR products from isolates that had previously tested positive for this system (Fig. 1, lanes 2 and 3), and amplicons (Fig. 1, lanes 4, 5 and 6) significantly smaller than those of the reference strain (Z2491) for hpuAB-negative isolates. A total of 264 (96 from hpuAB-positive and 168 from hpuAB-deleted strains) amplicons were generated and divided into five groups based on size and the presence (Hpu) or absence (ΔHpu) of the receptor (Fig. 1), with the following numbers of isolates in each group: Hpu1, 30; Hpu2, 66; ΔHpu3, 74; ΔHpu4, 51; and ΔHpu5, 43 (data not shown).
Fig. 1.
Variations in the size of the hpuAB locus among N. meningitidis isolates. The hpuAB locus was amplified with primers specific for flanking genes. Amplicons were grouped into five classes based on size, and representative samples are shown. Lanes: 1, size standards; 2, Hpu1, hpu+ strain (Z2491); 3, Hpu2, hpu+ strain (Z6414); 4, ΔHpu3, hpu− strain (Z4686); 5, ΔHpu4, hpu− strain (Z6427); 6, ΔHpu5, hpu− strain (Z4685).
To identify the mechanism responsible for the PCR product size variation, representative isolates of each group were sequenced and compared with the sequence of reference strain Z2491. A cluster of nine dRS3 (ATTCCCN8-GGGAAT) repetitive elements is present upstream of hpuA in the reference strain. This cluster is flanked by Correia elements and can be further subdivided into two direct repeats of 118 bp. In addition, REP elements of variable lengths (18, 22 and 25 bp) are also present. The analysis of strains positive for hpuAB (4323 and 6414), but with smaller amplicons (Hpu2) than Z2491, demonstrated a deletion of 600 bp in an upstream sequence flanked by Correia elements (Fig. 2). This deletion can be explained by a recombination event between dRS3 sequences in the upstream region.
Fig. 2.
Genetic arrangement of the hpuAB locus in N. meningitidis isolates. Directional arrows represent ORFs. Repetitive elements and the IS element are represented by different symbols. An absence of symbols indicates a region that is deleted in a particular clone. Note that the figure is not drawn to scale but is a representation of the various elements and genes present in this locus.
To explain the deletion mechanism for hpuAB, flanking regions were amplified with nested PCR primers for chromosomal walking. Subsequent sequencing of the products detected a variable number of dRS3 repeats in each of the deletion types. In addition, strains 4686 and 6427 (ΔHpu3 and ΔHpu4, respectively) contained a sequence with a high identity (95 %) to the published sequence of the transposase of IS1106A3. The complete deletion of the system in two other Hpu-negative strains, MC58 and 4685 (ΔHpu5), appears to have occurred via recombination between homologous dRS3 sequences in upstream and downstream regions, leaving a 1 kb fragment composed of repetitive elements. The presence of IS elements in non-sequenced strains with ΔHpu3 and ΔHpu4 deletion types was confirmed by performing a PCR with a flanking primer (hpu-rev13) and a primer (hpuF-seq2) specific for the IS element (data not shown).
Distribution of hpuAB deletion mechanisms within CCs
Phylogenetic analyses revealed that the majority of hpuAB-negative isolates (79 %) were restricted to 14 CCs (ST-41/44, ST-11, ST-60, ST-32, ST-269, ST-1, ST-18, ST-5, ST-8, ST-213, ST-35, ST-103, ST-334 and ST-461) (Table 3). For the 125 hpuAB-negative isolates containing the IS element, 55 % belonged to the ST41/44 complex, while 10 % were from ST-18 with high dissemination within these CCs and in ST-213 (75–92 % of isolates from each complex). Similarly, 72 % of the 43 isolates with a complete deletion of hpuAB were from two CCs, 44 % from ST-32 and 28 % from ST-269, with 75–90 % of isolates from these CCs containing the deletion.
PV status of the receptors in disease and carriage strains
The ON/OFF status of the receptors was examined in two isolate collections either by sequencing PCR products or by a combination of sizing of fluorescent PCR products (i.e GeneScan) and sequencing (Fig. 3). The 107 MLST isolates comprised strains representative of the major meningococcal clones causing disease in the latter half of the 20th century. The 2008/2009 carriage group was chosen in order to examine how expression of the haemoglobin receptors was influenced by carriage and because these isolates were subject to only one passage in vitro. Minimal passage combined with the low PV frequencies (~1×10−4) of meningococcal genes (Martin et al., 2004) reduced the chances of isolating phase variants during passage of single colonies. The observation of both ON and OFF variants amongst the carriage isolates indicated that in vitro growth did not select for particular expression states of these receptors. For disease isolates harbouring both genes (47 isolates), the majority (99 %) had either both (45 %) or one gene (54 %) in an ON state. In contrast, for carriage isolates with both receptors (41 isolates), only 22 % had both receptors ON and 34 % had both OFF. For hmbR-only strains, the receptor was ON in 94 % and 69 % of disease and carriage isolates, respectively. Surprisingly, in hpuAB-only strains, the receptor was ON in 75 % of carriage isolates but only 50 % of disease isolates, although this latter figure is based on analysis of a small number of samples. Overall, 91 % (82/90) of disease and 71 % (73/103) of carriage isolates were found to have at least one haemoglobin receptor in an ON state (OR 4.21, 95 % CI 1.82–9.77, P = 0.0005).
Fig. 3.
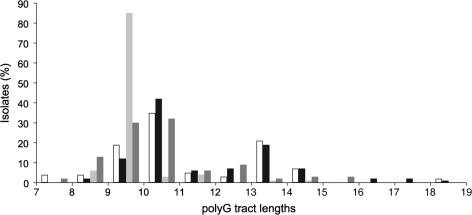
PV status of haemoglobin receptors in meningococcal disease and carriage isolates. PV status for the hmbR and hpuAB (hpu) genes was determined based on the length of the repeat tract. This tract is located within the reading frame such that alterations in these tracts switch the genes ON and OFF. Disease (n = 90) and carriage (n = 103) isolates were separated into three groups based on presence/absence of the receptors: both receptors present (n = 47 and 41 for disease and carriage, respectively); hpuAB only (n = 10 and 54); and hmbR only (n = 33 and 13). Strains with both receptors were separated into four groups depending on the ON/OFF status of the receptors. Strains with a single receptor will exhibit either an ON or OFF phenotype and so fall into two groups. Black bars, disease isolates; grey bars, carriage isolates.
The expression status of the haemoglobin receptors of a representative sample of these strains was confirmed by testing for the ability to utilize haemoglobin as the only iron source. Strain N54 had a single receptor, HpuAB, in the OFF state and only exhibited confluent growth in the presence of transferrin (Fig. 4a–c). Scattered colonies around the haemoglobin-containing disc represented a small number of ON phase variants (Fig. 4a). In contrast, strain MC58 had a single receptor, HmbR, in the ON state and exhibited confluent growth in the presence of haemoglobin and transferrin, although not without an added iron source (Fig. 4d–f). Similar results were obtained for eight further strains, indicating that the genetic screen for ON/OFF PV states correlates with phenotypic expression of the haemoglobin receptors.
Fig. 4.
Phenotypic analysis of hmbR and hpuAB phase variants. An hpuAB OFF variant of strain N54 (this strain lacks the hmbR gene) and an hmbR ON variant of strain MC58 (lacks the hpuAB genes) were tested for growth with haemoglobin or transferrin as the sole iron source. Suspensions of meningococcal strains were seeded onto MHA plates containing 40 µM desferal. Disks were inoculated with 10 µl of either human haemoglobin (10 µg ml−1) or transferrin (50 mg ml−1), or were uninoculated. Growth was recorded after overnight growth. (a) N54, with haemoglobin; (b) N54, with transferrin; (c) N54, no iron; (d) MC58, with haemoglobin; (e) MC58, with transferrin; (f) MC58, no iron.
Tract length distributions of haemoglobin receptors
The length of the homopolymer repeat tracts is one of the key factors in controlling the PV rate (De Bolle et al., 2000; Richardson et al., 2002). The sequences for hpu/hmbR were compared for variations in the polyG repeat tracts (Fig. 5). For hpuA, tract lengths in both groups ranged from 7G to 19G, with a modal repeat number of 10G, an ON number of repeats. A significant minority of isolates possessed 13G, which is also an ON state for this gene. The repeat tract of hmbR varied between 7G and 15G. The modal repeat numbers for the 107 MLST isolates and 2008/2009 carriage strains were 9G and 10G, respectively, reflecting differences in the proportion of isolates with this gene in an ON state (i.e. 9G is ON). Tract lengths longer than the modal numbers were observed at significantly higher frequencies as compared with shorter lengths for both genes (P<0.05; data not shown).
Fig. 5.
Tract distribution of hmbR and hpuAB in meningococcal disease and carriage isolates. Lengths of the polyG tracts of hmbR and hpuAB were determined by amplification with specific primers and by DNA sequencing of the products. White bars, hpuAB in disease isolates; black bars, hpuAB in carriage isolates; light-grey bars, hmbR in disease isolates; dark-grey bars, hmbR in carriage isolates. An ON PV state is produced by 9G, 12G, 15G and 18G for HmbR, and by 7G, 10G, 13G, 16G and 19G for HpuAB.
Discussion
Meningococci express a variety of iron-acquisition systems on their surfaces, enabling acquisition of iron from a range of host complexes, including transferrin, lactoferrin and haem-containing proteins (Perkins-Balding et al., 2004). The relative importance of each of these sources for invasive disease or carriage is still unclear. HmbR and HpuAB are surface receptors involved in acquisition of iron from haemoglobin or haemoglobin–haptoglobin complexes and undergo PV due to polyG repeat tracts present in the reading frame. Richardson & Stojiljkovic (1999) analysed a limited number of clinical samples and found that >50 % harbour both haemoglobin receptors, suggesting the importance of these receptors for invasion. Subsequently, HmbR was shown to be over-represented in disease isolates and to exhibit significant levels of antigenic variation, suggesting that this receptor facilitates invasion or dissemination within the body but is also subject to immune selection (Evans et al., 2010; Harrison et al., 2009). In the present study, the antigenic variation, distribution and PV of both haemoglobin receptors were assessed in four different and unrelated isolate collections. A complex relationship between presence/expression of these receptors and meningococcal disease and carriage states was revealed.
Antigenic variation of cell surface structures in bacterial pathogens is associated with escape from host defence systems and has implications for effective vaccine development. Alignment of the nucleotide sequences of the complete hpuA sequences from eight isolates and of partial sequences from several other strains detected regions of significant variation interspersed with conserved regions and an uneven distribution of single nucleotide polymorphisms. A similar pattern of variation has been observed in several other surface-exposed meningococcal proteins whose variable regions are known targets of host immune responses (Russell et al., 2004; Thompson et al., 2003). There are currently no structural or functional predictions for HpuA. Thus it is unclear whether these polymorphisms can be attributed to surface-exposed loops; however, it is likely that this variation is evidence of a high level of antigenic variation of HpuA and has arisen due to immune selection. A more detailed analysis of the diversity of both hpuA and hpuB is required to determine whether antigenic variants of this receptor are subject to CC structuring, as seen for hmbR, porA and fetA (Evans et al., 2010; Russell et al., 2004; Thompson et al., 2003).
Investigation of the distribution of genes in disease and carriage isolates is an important tool in the hunt for new virulence factors and may lead to an improved understanding of microbial pathogenesis and disease progression. A significant under-representation of an hpuAB-only phenotype (Table 1) was observed in disease isolates as compared with both receptors and an hmbR-only phenotype. This distribution may have arisen due to a combination of selection against HpuAB and a requirement for haemoglobin acquisition during invasive disease. Thus, HpuAB may be more immunogenic than HmbR, resulting in stronger selection against HpuAB-only strains than against HmbR-only strains during invasive disease and low levels of selection against strains with both receptors, as in this case switching HpuAB OFF and HmbR ON enables escape from immunity and maintenance of haemoglobin acquisition. This immunological pressure would be less severe during carriage in the nasopharynx, as HpuAB-only strains could switch the receptor OFF and acquire iron through another receptor such as the lactoferrin-binding protein. Intriguingly, all CCs with high disease : carriage ratios (>6) have both receptors (Table 3), suggesting an additional advantage associated with both receptors which may be connected with a propensity to cause disease or the rapid transmission required to compensate for low levels of carriage.
An alternative explanation is that the differing substrate specificities and affinities of these receptors influence their distribution in disease versus carriage isolates. Haemoglobin–haptoglobin complexes are likely to be a major source of iron in the nasopharynx and during initial exposure to serum, whereas free haemoglobin is likely to be present only at low levels. As haemoglobin–haptoglobin complexes can only be utilized via the HpuAB receptor, this specificity might imply that an Hpu+ phenotype would be advantageous during carriage and result in over-representation of this receptor among carriage isolates. This seems unlikely, as hmbR-only strains are prevalent in carriers, suggesting that free haemoglobin is available during carriage. It is also unclear why this specificity would have resulted in the low prevalence of an hpu-only phenotype among disease isolates, as haptoglobin–haemoglobin complexes should be readily available during systemic spread. Conversely, over-representation of hmbR in disease isolates may be due to this receptor having a higher affinity for haemoglobin than HpuAB and a greater availability of free haemoglobin during disease due to tissue destruction and cell lysis. Recently, growth of one meningococcal strain, MC58 (an hmbR-only strain), with haemoglobin but not transferrin as the sole iron source has been associated with induction of virulence genes, suggesting a direct link between haemoglobin utilization and a pathogenic phenotype (Jordan & Saunders, 2009). Thus the release of free haemoglobin by the action of haemolysins and other toxins would not only enhance growth of hmbR+ strains but also induce the expression of virulence-associated genes, leading to an increase in the likelihood of these strains causing invasive disease. If HmbR facilitates growth under these conditions, this would predict an over-representation of strains with both receptors or with an hmbR-only phenotype among disease isolates, as observed herein. Finally, the absence of both receptors in some carriage isolates implies either that acquisition of iron or haem from haemoglobin is not essential for persistence in the human nasopharynx or that an alternative haemoglobin receptor is present.
A variety and abundance of repetitive elements is evident in neisserial genomes, and repetitive elements are associated with the deletion of surface determinants such as FetA and PorA (Claus et al., 1997; Marsh et al., 2007; van der Ende et al., 1999). Deletion of hpuAB was found to be due to replacement by an IS element or recombination between repetitive elements (Fig. 2). Although present in many strains, only three types of deletion events covered all hpuAB-negative isolates (Fig. 3), and each ST was associated with a specific type of deletion event (Table 3). Stable lineages of this type suggest that deletions are infrequent, possibly because the selective advantage associated with deletion of these genes is low and may have required acquisition of hmbR as a functional replacement before dissemination of the deletion could occur.
PV of the haemoglobin receptors is likely to have a significant impact on immune evasion and niche adaptation by meningococci. Richardson et al. (2002) demonstrated that high rates of PV of these receptors were associated with epidemic spread of meningococci. This suggests that stochastic changes in expression facilitate immune evasion and transmission once the host population has developed herd immunity to circulating meningococcal strains. The presence of a significantly higher number of isolates with one or both genes in an ON state for disease (91 %) as compared with carriage (71 %) isolates reinforces the view that haemoglobin accumulation is required during invasive disease. Furthermore, the observation of a significant number of isolates with their haemoglobin receptors in the OFF state during carriage implies a less stringent requirement for haemoglobin acquisition during carriage and more frequent immune evasion. Examination of specific immune responses to these receptors will be required to determine whether PV status is influenced by adaptive immune responses.
A comment on the potential biases in analysis of PV states is appropriate at this point. As noted in Results, minimal passage, low PV frequencies (<1×10−4; Lewis et al., 1999; Richardson & Stojiljkovic, 1999), similar ON-to-OFF/OFF-to-ON switching rates and lack of selection during in vitro growth reduced the likelihood of switches occurring during the growth and analysis of isolates, such that the overall findings are likely to provide a strong reflection of the in vivo PV states of these loci. Single colony analysis, as used herein, may not reflect situations in carriers or patients who are colonized or infected with ratios of ON and OFF phase variants of <3 : 1. This situation can only be overcome by analysis of multiple colony isolates or direct analysis of bacterial DNA without growth to detect the prevalent PV state. Preliminary analyses of multiple colonies from carriers detected ratios for ON-to-OFF/OFF-to-ON variants of <3 : 1 in two of 12 individuals (S. B. Redkar and others, unpublished data), but further studies are required to ascertain how frequent such low ratios of variants are in carriers and patients. It seems unlikely, however, that we would have observed such a high prevalence of ON phenotypes if low ratios of variants were present in a significant number of carriers or patients.
Acquisition of iron is considered a prerequisite for bacterial disease. The observation of selection against an HpuAB-only phenotype in meningococcal disease isolates indicates that not all iron-acquisition systems may facilitate bacterial pathogenesis. Contrastingly, disease isolates have at least one receptor, either HpuAB or HmbR, in an ON PV state. Thus acquisition of haemoglobin appears to facilitate invasive meningococcal disease and is mediated by either a combination of haemoglobin receptors or HmbR alone. Further work is required to determine whether the high prevalence of hpuAB-negative variants and PV of these systems are driven by immune evasion, and whether expression of these receptors is required for meningococcal disease.
Supplementary Material
Acknowledgements
This work was supported by the Higher Education Commission of Pakistan, a Research Councils UK (RCUK) Fellowship and the University of Leicester, University of Nottingham and University of Oxford. M. C. J. M. is a Wellcome Trust Senior Research Fellow.
Abbreviations:
- CC
clonal complex
- CI
confidence interval
- OR
odds ratio
- PV
phase variation
- ST
sequence type
Footnotes
A supplementary figure, showing an alignment of the amino acid sequences of HpuA from carriage and disease isolates of Neisseria meningitidis, is available with the online version of this paper.
References
- Bayliss C. D., Hoe J. C., Makepeace K., Martin P., Hood D. W., Moxon E. R. (2008). Neisseria meningitidis escape from the bactericidal activity of a monoclonal antibody is mediated by phase variation of lgtG and enhanced by a mutator phenotype. Infect Immun 76, 5038–5048. 10.1128/IAI.00395-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidmos F. A., Neal K. R., Oldfield N. J., Turner D. P., Ala’aldeen D. A., Bayliss C. D. (2011). Persistence, replacement and rapid clonal expansion of meningococcal carriage isolates in a 2008 university student cohort. J Clin Microbiol 49, 506–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton I. C., Gorringe A. R., Allison N., Robinson A., Gorinsky B., Joannou C. L., Evans R. W. (1998). Transferrin-binding protein B isolated from Neisseria meningitidis discriminates between apo and diferric human transferrin. Biochem J 334, 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caugant D. A., Maiden M. C. (2009). Meningococcal carriage and disease – population biology and evolution. Vaccine 27 Suppl. 2B64–B70. 10.1016/j.vaccine.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus H., Vogel U., Mühlenhoff M., Gerardy-Schahn R., Frosch M. (1997). Molecular divergence of the sia locus in different serogroups of Neisseria meningitidis expressing polysialic acid capsules. Mol Gen Genet 257, 28–34. 10.1007/PL00008618. [DOI] [PubMed] [Google Scholar]
- De Bolle X., Bayliss C. D., Field D., van de Ven T., Saunders N. J., Hood D. W., Moxon E. R. (2000). The length of a tetranucleotide repeat tract in Haemophilus influenzae determines the phase variation rate of a gene with homology to type III DNA methyltransferases. Mol Microbiol 35, 211–222. 10.1046/j.1365-2958.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- Evans N. J., Harrison O. B., Clow K., Derrick J. P., Feavers I. M., Maiden M. C. (2010). Variation and molecular evolution of HmbR, the Neisseria meningitidis haemoglobin receptor. Microbiology 156, 1384–1393. 10.1099/mic.0.036475-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison O. B., Evans N. J., Blair J. M., Grimes H. S., Tinsley C. R., Nassif X., Kriz P., Ure R., Gray S. J., et al. (2009). Epidemiological evidence for the role of the hemoglobin receptor, HmbR, in meningococcal virulence. J Infect Dis 200, 94–98. 10.1086/599377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley K. A., Kalmusova J., Feil E. J., Gupta S., Musilek M., Kriz P., Maiden M. C. (2000). Carried meningococci in the Czech Republic: a diverse recombining population. J Clin Microbiol 38, 4492–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P. W., Saunders N. J. (2009). Host iron binding proteins acting as niche indicators for Neisseria meningitidis. PLoS ONE 4, e5198. 10.1371/journal.pone.0005198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis L. A., Dyer D. W. (1995). Identification of an iron-regulated outer membrane protein of Neisseria meningitidis involved in the utilization of hemoglobin complexed to haptoglobin. J Bacteriol 177, 1299–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis L. A., Gipson M., Hartman K., Ownbey T., Vaughn J., Dyer D. W. (1999). Phase variation of HpuAB and HmbR, two distinct haemoglobin receptors of Neisseria meningitidis DNM2. Mol Microbiol 32, 977–989. 10.1046/j.1365-2958.1999.01409.x. [DOI] [PubMed] [Google Scholar]
- Maiden M. C., Bygraves J. A., Feil E., Morelli G., Russell J. E., Urwin R., Zhang Q., Zhou J., Zurth K., et al. (1998). Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95, 3140–3145. 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden M. C. J., Stuart J. M., The UK Meningococcal Carriage Group (2002). Carriage of serogroup C meningococci 1 year after meningococcal C conjugate polysaccharide vaccination. Lancet 359, 1829–1830. 10.1128/JCM.02422-06. [DOI] [PubMed] [Google Scholar]
- Marsh J. W., O’Leary M. M., Shutt K. A., Harrison L. H. (2007). Deletion of fetA gene sequences in serogroup B and C Neisseria meningitidis isolates. J Clin Microbiol 45, 1333–1335. 10.1128/JCM.02422-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P., Sun L., Hood D. W., Moxon E. R. (2004). Involvement of genes of genome maintenance in the regulation of phase variation frequencies in Neisseria meningitidis. Microbiology 150, 3001–3012. 10.1099/mic.0.27182-0. [DOI] [PubMed] [Google Scholar]
- Moxon R., Bayliss C. D., Hood D. W. (2006). Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu Rev Genet 40, 307–333. 10.1146/annurev.genet.40.110405.090442. [DOI] [PubMed] [Google Scholar]
- Otto B. R., Verweij-van Vught A. M., MacLaren D. M. (1992). Transferrins and heme-compounds as iron sources for pathogenic bacteria. Crit Rev Microbiol 18, 217–233. 10.3109/10408419209114559. [DOI] [PubMed] [Google Scholar]
- Perkins-Balding D., Baer M. T., Stojiljkovic I. (2003). Identification of functionally important regions of a haemoglobin receptor from Neisseria meningitidis. Microbiology 149, 3423–3435. 10.1099/mic.0.26448-0. [DOI] [PubMed] [Google Scholar]
- Perkins-Balding D., Ratliff-Griffin M., Stojiljkovic I. (2004). Iron transport systems in Neisseria meningitidis. Microbiol Mol Biol Rev 68, 154–171. . 10.1128/MMBR.68.1.154-171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard A. J. (2004). Global epidemiology of meningococcal disease and vaccine efficacy. Pediatr Infect Dis J 23 Suppl. 12S274–S279. [PubMed] [Google Scholar]
- Richardson A. R., Stojiljkovic I. (1999). HmbR, a hemoglobin-binding outer membrane protein of Neisseria meningitidis, undergoes phase variation. J Bacteriol 181, 2067–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson A. R., Yu Z., Popovic T., Stojiljkovic I. (2002). Mutator clones of Neisseria meningitidis in epidemic serogroup A disease. Proc Natl Acad Sci U S A 99, 6103–6107. 10.1073/pnas.092568699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde K. H., Dyer D. W. (2004). Analysis of haptoglobin and hemoglobin-haptoglobin interactions with the Neisseria meningitidis TonB-dependent receptor HpuAB by flow cytometry. Infect Immun 72, 2494–2506. 10.1128/IAI.72.5.2494-2506.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. E., Jolley K. A., Feavers I. M., Maiden M. C., Suker J. (2004). PorA variable regions of Neisseria meningitidis. Emerg Infect Dis 10, 674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens D. S. (2007). Conquering the meningococcus. FEMS Microbiol Rev 31, 3–14. 10.1111/j.1574-6976.2006.00051.x. [DOI] [PubMed] [Google Scholar]
- Thompson E. A., Feavers I. M., Maiden M. C. (2003). Antigenic diversity of meningococcal enterobactin receptor FetA, a vaccine component. Microbiology 149, 1849–1858. 10.1099/mic.0.26131-0. [DOI] [PubMed] [Google Scholar]
- van der Ende A., Hopman C. T., Dankert J. (1999). Deletion of porA by recombination between clusters of repetitive extragenic palindromic sequences in Neisseria meningitidis. Infect Immun 67, 2928–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.