Abstract
The Gram-negative pathogen Acinetobacter baumannii strain M2 was found to exhibit a robust surface motility on low-percentage (0.2–0.4 %) agar plates. These patterns of motility were dramatically different depending on whether Difco or Eiken agar was used. Motility was observed in many, but not all, clinical and environmental isolates. The use of drop collapse assays to demonstrate surfactant production was unsuccessful, and the role of surfactants in A. baumannii M2 motility remains unclear. Surface motility was impaired by an insertion in pilT, encoding a gene product that is often required for retraction of the type IV pilus. Motility was also dependent on quorum sensing, as a null allele in the abaI autoinducer synthase decreased motility, and the addition of exogenous N-(3-hydroxy)-dodecanoylhomoserine lactone (3-OH C12-HSL) restored motility to the abaI mutant. Transposon mutagenesis was used to identify additional genes required for motility and revealed loci encoding various functions: non-ribosomal synthesis of a putative lipopeptide, a sensor kinase (BfmS), a lytic transglycosylase, O-antigen biosynthesis (RmlB), an outer membrane porin (OmpA) and de novo purine biosynthesis (PurK). Two of the above genes required for motility were highly activated by quorum sensing, and may explain, in part, the requirement for quorum sensing in motility.
Introduction
Bacterial motility is a complex process that employs mechanisms such as swimming, swarming, twitching, gliding and sliding, and these mechanisms can require surface appendages such as flagella, pili and nozzles (reviewed by Harshey, 2003; Henrichsen, 1972; Jarrell & McBride, 2008). Swimming and swarming are both mediated by flagella, and are distinguished by the swarming process occurring on solid surfaces, and in some cases requiring differentiation to an elongated swarmer cell (Harshey, 2003; Jarrell & McBride, 2008; Rather, 2005). Twitching motility is mediated by the extension and retraction of type IV pili (Harshey, 2003; Henrichsen, 1972; Jarrell & McBride, 2008; Mattick, 2002; Merz et al., 2000; Skerker & Berg, 2001; Wall & Kaiser, 1999). Gliding motility is associated with the extrusion of cellular substances via a cell surface nozzle, or by protein motors that move adhesins across the cell surface (Jarrell & McBride, 2008). Sliding motility is defined as the growth-promoted expansion of cells across a surface that is aided by surface properties of the cell (Henrichsen, 1972; Jarrell & McBride, 2008). There are no reports of a surface structure involved in sliding motility; however, a type-I secretion system has recently been shown to have a role in sliding motility in Legionella pneumophila, where it appears to aid in the secretion of a surfactant (Stewart et al., 2009).
Acinetobacter baumannii is a Gram-negative nosocomial pathogen that has been described as non-motile (Bergogne-Bérézin & Towner, 1996; Tomaras et al., 2003). Recently, a form of motility in A. baumannii was reported that is decreased in the presence of light (Mussi et al., 2010). Various types of motility have been reported in other species of Acinetobacter (Barker & Maxted, 1975; Henrichsen, 1984). In Acinetobacter anitratus, a form of motility termed ‘ditching’ was described by Barker and Maxted, in which cells generate furrows in the agar and migrate along these furrows (Barker & Maxted, 1975). This type of motility was proposed to represent a type of swarming, although this was later disputed and reclassified as twitching (Henrichsen, 1984). Subsequent to the above study, A. anitratus was reclassified as A. baumannii (Bouvet & Grimont, 1987), although it is unclear whether A. baumannii represented the organism used in the original study (Barker & Maxted, 1975).
Bioinformatic analysis of recently completed A. baumannii genome sequences has revealed that flagellar genes are absent, indicating that swarming is unlikely in this organism. However, a number of genes devoted to the expression and assembly of type IV pili that participate in twitching motility appear to be present in A. baumannii, including genes involved in assembly (pilA, pilB, pilC, pilF, pilM, pilN, pilO, pilP, pilQ, pilZ, pilW), twitching (pilR, pilS, pilT, pilU) and the pilin filament (pilA). Additional pili present in A. baumannii are encoded by the csu locus, which contains a chaperone–usher complex required for production and assembly of pili that are involved in surface adhesion (Tomaras et al., 2003).
In this study, a reanalysis of A. baumannii motility was conducted, including the identification of genes required for this process. A pilT mutation rendered cells significantly less motile than the wild-type. In addition, surface motility was shown to be dependent on quorum sensing. Additional genes required for motility were identified by transposon mutagenesis, and encoded functions predicted to be involved in O-antigen/LPS synthesis, two-component regulation, de novo purine biosynthesis, peptidoglycan hydrolysis, and synthesis of a non-ribosomally produced lipopeptide.
Methods
Bacterial strains and growth conditions.
A. baumannii strain M2 is a clinical isolate obtained from the Metro Health Medical Center, Cleveland, OH, USA, and has been described previously (Niu et al., 2008). Additional clinical isolates of A. baumannii were obtained from the strain collection at the Louis Stokes Cleveland VA Medical Center, Cleveland, OH. Environmental isolates of A. baumannii were obtained from Dr Judith Noble-Wang at the US Centers for Disease Control, Atlanta, GA (Table 1). Growth media consisted of modified Luria–Bertani (LB) broth containing (per litre) 10 g tryptone, 5 g yeast extract and 5 g NaCl. Agar was obtained from Difco or Eiken (Eiken Chemical, Tokyo, Japan) and was added at the indicated concentrations. Surfactin was obtained from Sigma Chemical and used at a concentration of 10 mg ml−1. Supplementation of plates with N-(3-hydroxy)-dodecanoylhomoserine lactone (3-OH C12-HSL) was done at a concentration of 500 nM.
Table 1. Bacterial strains used in this study.
| Strain | Characteristics | Source or reference |
| M2 | Clinical isolate | OH, USA |
| M5 | Clinical isolate | OH, USA |
| M8 | Clinical isolate | OH, USA |
| 1746 | Clinical isolate | Iraq |
| 1822 | Clinical isolate | Iraq |
| 1883 | Clinical isolate | Iraq |
| ATCC17978 | Sequenced strain | American Type Culture Collection; Smith et al. (2007) |
| AB0057 | Clinical isolate | Adams et al. (2008) |
| BM4454 | Clinical isolate | Paris, France |
| CDC 113-03 | Environmental isolate, strain type 10 | KY, USA |
| CDC 16-27-02 | Environmental isolate, strain type 12 | AZ, USA |
| CDC 126-16 | Environmental isolate, strain type 54 | KY, USA |
Drop collapse assays for surfactant production.
Liquid cultures of bacteria were grown to OD600 1.1 in LB medium. One millilitre of cells from each tube was pelleted by centrifugation, and 30 µl of the resulting supernatant was spotted onto the top of a polystyrene microtitre plate lid. Spreading of the liquid drop was used to assess the presence of surfactant (Jain et al., 1991). A drop that remained as a bead indicated that surfactant was not present. For harvesting media from cells grown on 0.3 % agar, cells were grown overnight on 5 ml 0.3 % Eiken agar present in the well of a six-well microtitre plate. After overnight growth, the agar/cell mixture was removed and placed in a 15 ml conical tube. This mixture was then vortexed at high speed for 2 min followed by centrifugation. The resulting supernatants were then tested as above. For drop collapse assays in the presence of oil to create a more hydrophobic surface, 1.8 µl mineral oil was spread on each well of a microtitre plate prior to assaying. As controls, Pseudomonas aeruginosa PAO1 and a lasI/rhlI mutant defective in rhamnolipid production (Whiteley et al., 1999) and known to exhibit positive and negative results in this assay were grown under identical conditions and used at the same time.
Motility assays.
Modified LB broth was used as a base medium for all motility assays, and was solidified using the indicated concentrations of agar, from either Difco or Eiken. Plates were used on the same day that they were prepared. For testing the motility of the M2 strain and various mutants, cultures were grown to early stationary phase and adjusted to the same optical density by the addition of sterile LB broth, and a 1 µl drop was placed on the centre of the plate. Plates were incubated at 37 °C except for the abaI mutant, which was incubated at 30 °C due to the apparent increased stability of 3-OH C12-HSL at lower temperatures.
Microscopic analysis of motility fronts.
A. baumannii cultures were spotted (1 µl) on 0.35 % Eiken agar present in an adhesive rubber chamber on a microscope slide. The slide was incubated in a humidified chamber for 6 h, at which time surface motility had initiated. A glass coverslip was then placed on the top of the chamber and the motility front was visualized by phase-contrast microscopy at ×1000 and photographed.
Construction of defined mutations.
The A. baumannii orthologue of csuD (A1S_2214) was inactivated by a Campbell-type insertion using the suicide vector pKNG101 (Kaniga et al., 1991). An internal segment of the csuD gene was amplified from M2 chromosomal DNA by PCR using the primers 5′-ACATGTCGACGGGGAAGCCGTTGTGGTTAC-3′ and 5′-ATGCTCTAGAATCACGGTTGGCACTTAATG-3′. For this amplification, one primer contained an XbaI site and the second primer contained a SalI site, and these enzymes were used to clone the PCR product into XbaI- and SalI-digested pKNG101 (Kaniga et al., 1991). Recombinant plasmids were initially isolated in CC118 λpir and then transferred to SM10 λpir for matings with A. baumannii M2. Introduction of each plasmid into A. baumannii M2 was done by a conjugal mating, as described previously (Niu et al., 2008), followed by plating mating mixtures on LB agar plates containing ampicillin (20 µg ml−1), to counterselect the Escherichia coli SM10 donor, and streptomycin (50 µg ml−1), to select for insertions of pKNG101 derivatives into A. baumannii. The correct disruption of csuD was confirmed by Southern blot analysis.
A null allele in the pilT gene was constructed by using PCR to amplify the pilT coding region from ATCC17978 chromosomal DNA. This PCR fragment was cloned into the EcoRV site of pBluescript KS. In vitro transposon mutagenesis was then carried out with the EZ : : TN R6K γori KAN-2 transposon (Epicentre), and insertions in the pilT coding region were identified by mini-prep and DNA sequence analysis. One insertion at a position in pilT corresponding to amino acid 217 of the 355 aa protein was chosen, and the entire pilT : : EZTN region was cloned into the SmaI site of the suicide vector pKNG101. The recombinant pKNG101 plasmid was introduced into A. baumannii M2 by electroporation, followed by selection for streptomycin (50 µg ml−1) and kanamycin resistance (20 µg ml−1). Transformants were grown for approximately 10 generations in the absence of antibiotic selection and plated on LB agar plates containing 10 % sucrose. Sucrose-resistant colonies were screened for kanamycin resistance and streptomycin sensitivity, and the correct pilT : : EZTN R6K γori KAN-2 disruption was confirmed by both PCR and Southern blot analysis.
Transposon mutagenesis.
A. baumannii M2 was mutagenized with the EZ : : TN R6K γori KAN-2 transposon (Epicentre) by electroporating cells with the transposon/transposase (transposome) complex followed by plating on LB agar plates containing 20 µg kanamycin ml−1. Colonies were then individually inoculated into the wells of a microtitre plate containing 100 µl LB and stored at −80 °C. Insertions were screened using a sterile toothpick to individually transfer media from each well of the microtitre plate to the surface of a 0.3 % agar plate. Plates were incubated overnight at 37 °C, and mutants with altered motility were rescreened to confirm the motility phenotype. The site of EZ : : TN R6K γori KAN-2 insertion in each mutant was determined by digesting chromosomal DNA with either XbaI or SalI, followed by ligation and electroporation into E. coli CC118 λpir to rescue clones with recircularized chromosomal DNA containing the R6K ori from the transposon. The site of insertion was determined by sequencing plasmids with the RP-1 primer, which reads outward from the end of the EZ : : TN transposon and into the chromosomal junction.
Construction of transcriptional lacZ fusions.
A DNA fragment containing the promoter region of the A1S_0112 gene from ATCC17978 was amplified by PCR using the primers 5′-CCGCGGATCCAGCATGGCAGAAACGTTATG-3′ and 5′-GTTCCCAAGCTTCCAAATACTCAAAAGCGATC-3′. The PCR fragments were digested with BamHI and HindIII and cloned into the promoter probe vector pQF50 (Farinha & Kropinski, 1990) cut with the same enzymes. This vector contains a promoterless lacZ gene that possesses its own ribosome-binding site and a ColE1 origin of replication. It is unable to replicate in A. baumannii and acts as a suicide vector. Recombinant pQF50 plasmids containing promoter fusions to lacZ were then electroporated into A. baumannii M2 or the isogenic abaI : : Km mutant (Niu et al., 2008), and plasmids that integrated into the chromosome by homologous recombination were selected on plates containing 600 µg ampicillin ml−1. The correct site of insertion was confirmed using an outward-reading primer from the lacZ gene and a primer to a region upstream of the cloned promoter fragment.
Complementation of the pilT mutation.
A full-length copy of the pilT gene along with its native ribosome-binding site was generated by PCR using the primers 5′-CCGCGGATCCGGTAAGCCTCGGTGAAAGCC-3′ and 5′-CCGCGGATCCCCGTTAATCTTCATAGAGGG-3′ to amplify the pilT gene from chromosomal DNA of ATCC17978. The PCR primers generated BamHI restriction sites, and these were used to clone the pilT gene into pWH1266, an E. coli–A. baumannii shuttle vector (Hunger et al., 1990).
Results
Motility of A. baumannii
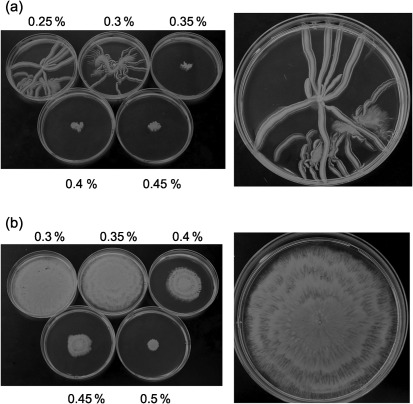
To test whether A. baumanni was capable of motility, a 1 µl drop of an overnight culture of strain M2 was placed on LB media with agar concentrations ranging from 0.25 to 0.45 %. Approximately 4–5 h after inoculation, outward motility was evident, and after 14 h of growth, strain M2 had migrated across the plate from the central point of inoculation in a manner that resembled branching tentacles. This branching pattern of motility was highly variable and could involve only a few expanding branches or multiple branches, as shown in Fig. 1(a). After a period of expansion, a migrating branch could then give rise to offshoot branches, as seen in the lower half of the enlarged plate in Fig. 1(a). This branching migration pattern was similar to that seen in A. anitratus (Barker & Maxted, 1975) and in Mycobacterium smegmatis; in the latter, motility has been described as sliding (Martínez et al., 1999). In A. baumannii, the expanding branches rarely touched each other, and when branches approached each other, they would usually curve away and then migrate in a parallel manner. In addition, despite the low agar concentration, cells never migrated into the agar and motility always occurred on the surface. This may be due to the strict oxygen requirements of A. baumannii. The migrating branches did appear to be slightly sunken into the agar surface, and it was not clear whether this resulted from the weight of the cell mass or whether they were capable of degrading the agar. The branching motility stopped at agar concentrations of 0.35 % and above (Fig. 1a).
Fig. 1.
Motility on agar plates. An overnight culture of A. baumannii M2 was grown in 3 ml LB, and a 1 µl drop was placed on the surface of an LB agar plate prepared with either Difco Bacto agar (a) or Eiken agar (b) at the various percentages indicated. The larger panels on the right-hand side are enlargements showing representative motility on 0.25 % Difco and 0.35 % Eiken agar. Cultures were incubated at 37 °C for 14 h.
Earlier motility studies with E. coli, Salmonella typhimurium and Serratia marcescens have demonstrated that agar produced by Eiken Chemical is able to promote enhanced motility relative to Difco agar (Harshey & Matsuyama, 1994; Matsuyama et al., 1995). When the motility of A. baumannii was tested on media made with 0.3–0.5 % Eiken agar, motility also initiated 4–5 h after inoculation. However, prominent differences were noted from the motility seen on plates with Difco agar, with migration occurring in an even manner from the point of inoculation, resulting in a circular expansion of cells (Fig. 1b). Eiken agar concentrations at 0.5 % were inhibitory to motility, but this was higher than for Difco agar, on which motility was inhibited at 0.35 %. Therefore, as observed with other bacteria, Eiken agar was more permissive for A. baumannii motility than Difco agar. Based on these results, Eiken agar was used for all subsequent experiments. The motility rate for the M2 strain on 0.35 % Eiken agar was calculated to be 2 µm s−1 at 37 °C. Strain M2 also exhibited motility on M9 minimal medium with either glucose or Casamino acids as a carbon source (data not shown).
Motility in clinical and environmental isolates of A. baumannii
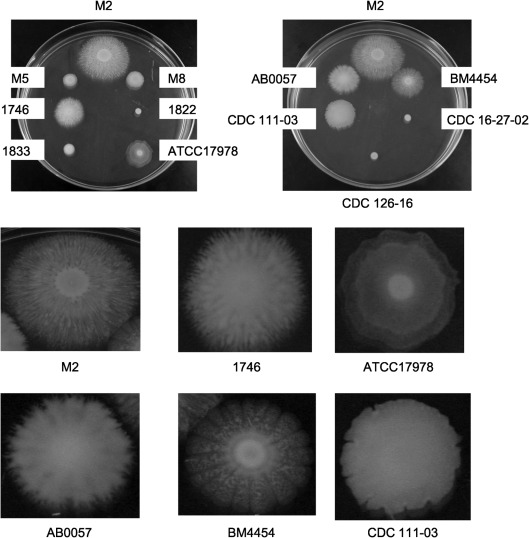
A variety of clinical and environmental isolates from distinct geographical locations (OH, KY and AZ, USA, and France and Iraq) were tested for motility on LB Eiken agar plates (0.3 %). As shown in Fig. 2, the motility exhibited by these isolates was highly variable. Interestingly, in addition to M2, strains 1746, ATCC17978, AB0057, BM4454 and CDC 111-03 exhibited highly complex patterns that ranged from the flower-like patterns of BM4454 to the concentric rings formed by ATCC17978 (Fig. 2, lower panel). The clinical isolates 1833, M5 and M8 were noticeably less motile than the above strains and did not exhibit any patterns. In addition, the clinical isolate 1822 and the environmental isolates CDC 16-27-02 and CDC 126-16 did not exhibit any signs of motility.
Fig. 2.
Motility of clinical and environmental isolates on Eiken agar. A variety of clinical and environmental isolates from diverse geographical areas were examined for motility on 150 mm LB plates with Eiken agar at 0.3 %. For each strain, cells were grown overnight in 3 ml LB broth, and all cultures were adjusted to the same optical density by the addition of fresh LB broth. A 1 µl drop was added to the plates and incubated at 37 °C for 12 h. As a reference, at the top of each plate is the M2 strain used in this study. The lower panels represent enlargements of selected strains to demonstrate the distinct and complex motility patterns.
Features associated with motility
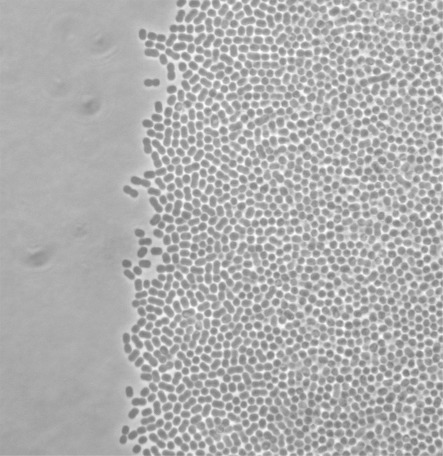
The morphology of A. baumannii M2 cells was examined during motility at the edge of the migration front and in the interior (Fig. 3). Cells at both locations exhibited similar morphology and appeared as coccobacilli. This morphology was also observed during growth in liquid medium. Therefore, there does not appear to be any change in cell shape during motility. Cells at the edge of the motility front moved in a manner that resembled twitching. This movement was independent of cell–cell contact, as the movement of individual cells was clearly observed.
Fig. 3.
Analysis of cell morphology at motility fronts. Cells of wild-type M2 were analysed by phase-contrast microscopy at the motility front on 0.35 % Eiken agar after 6 h of incubation at 37 °C.
During the migration of A. baumannii on either Difco or Eiken agar, a translucent zone was visible in the agar immediately ahead of the advancing colony. This type of zone has been reported before during the motility of other bacteria on agar surfaces, and has been proposed to represent a secreted surfactant (Stewart et al., 2009). However, when a drop of water was placed on this translucent area, it did not disperse any faster than a drop placed elsewhere on the agar, suggesting that the zone was not composed of a secreted surfactant (data not shown). To further investigate the presence of a surfactant, cell-free supernatants were prepared from overnight stationary phase cultures, filter-sterilized, and used in a drop collapse assay on polystyrene (Jain et al., 1991). As positive and negative controls for this assay, we utilized P. aeruginosa strain PAO1, which produces rhamnolipids and is positive in the drop collapse assay, and a lasI/rhlI mutant derivative that fails to produce rhamnolipids (Whiteley et al., 1999). Based on this assay, there was no evidence of surfactant activity in undiluted supernatants of A. baumannii M2 (data not shown). In contrast, supernatants from the wild-type PAO1 strain collapsed immediately and the supernatant from the lasI/rhlI double mutant failed to collapse.
To test the possibility that surfactant production only occurred when cells were grown on solid surfaces, we also tested for surfactant production on agar plates by harvesting the soft agar from wild-type A. baumannii M2 cells that had completely migrated across the plate, centrifuging out the cells and agar, and filter-sterilizing the resulting supernatant. However, the drop collapse assay under these conditions also failed to show any evidence of surfactant production (data not shown). In addition, similar results were obtained with liquid and agar supernatants when the wells were pre-coated with mineral oil (data not shown).
The pilT gene is required for full motility
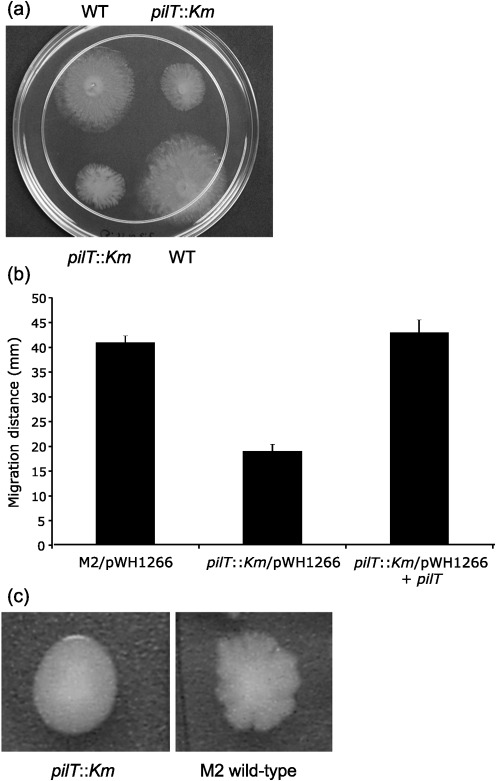
To investigate the role of twitching via type IV pili in the motility of strain M2, a null allele was constructed in the pilT gene (A1S_0897), encoding an ATPase shown to mediate pilus retraction in other bacteria (Herdendorf et al., 2002; Mattick, 2002; Merz et al., 2000). After 10 h of incubation at 37 °C, the pilT : : Km mutant exhibited a motility rate that was significantly lower than that of the wild-type, 19±1.3 mm versus 41±1.3 mm, respectively (Fig. 4a, b). To verify that the reduction in motility was due to loss of pilT, we introduced a plasmid containing the pilT gene (pWH1266+pilT) into the pilT : : Km mutant. This plasmid restored motility back to wild-type levels (43±2.5 mm) (Fig. 4b).
Fig. 4.
Role of pilT in motility. (a) Wild-type M2 and the isogenic pilT : : Km mutant were grown overnight in 2 ml LB with shaking at 200 r.p.m. at 37 °C. Cells were adjusted to the same optical density with sterile LB, and a 1 µl drop was placed on the surface of an LB plate containing 0.35 % Eiken agar. Plates were incubated for 10 h at 37 °C. (b) Complementation of the pilT : : Km mutation. Cells were grown as described for (a), except that ampicillin (800 µg ml−1) was added to LB broth. The distance migrated (diameter) is shown for each strain and represents the mean of duplicate samples from two independent experiments; error bars, sd. (c) Colony morphology of the wild-type and the isogenic pilT : : Km mutant on 1 % agar plates.
The colony morphology of pilT mutants was significantly different from that of wild-type M2 when grown on 1 % agar plates. Wild-type M2 formed very irregular colonies that had multiple short protrusions extending from each colony (Fig. 4c). In contrast, the pilT : : Km mutant formed colonies with a smooth border. The smooth colony morphology of the pilT mutant was restored to an irregular appearance by pWH1266+pilT (data not shown).
Quorum sensing mediated by the AbaI autoinducer synthase is required for motility
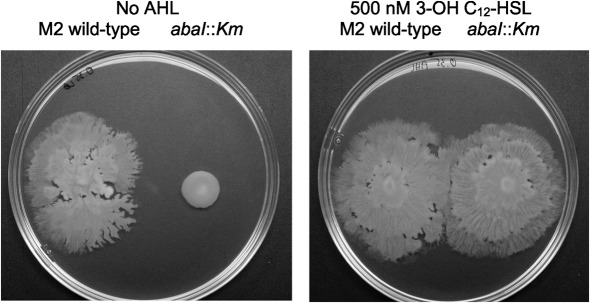
In A. baumannii, the only autoinducer synthase predicted to be encoded in the genome is AbaI, which produces 3-OH C12-HSL as the primary signal (Niu et al., 2008). To test the possible role of quorum sensing in motility, an abaI : : Km mutant was compared with the wild-type M2 parent on 0.35 % Eiken agar plates. As seen in Fig. 5, the abaI : : Km mutant was significantly less motile, with a rate reduced by 75 % when compared with the wild-type M2 parent. This motility defect was rescued when 3-OH C12-HSL (500 nM) was incorporated into the agar plate (Fig. 5), indicating that the motility defect likely resulted from the altered expression of one or more genes that were regulated by the AbaI quorum-sensing pathway (see below).
Fig. 5.
Role of quorum sensing in motility. (a) Overnight cultures of wild-type M2 and an isogenic autoinducer synthase mutant (abaI : : Km) were grown overnight in 3 ml LB broth and adjusted to the same optical density with sterile LB. A 1 µl drop was applied to an LB agar plate containing 0.35 % Eiken agar with and without 500 nM 3-OH C12-HSL (AHL) and incubated for 12 h at 30 °C.
Use of transposon mutagenesis to identify additional genes required for motility
To identify additional genetic requirements for motility, A. baumannii M2 was mutagenized with the EZ : : TN R6K γori KAN-2 transposon, and seven mutants were identified with significant motility defects (all with P values <0.05) (Fig. 6). All strains had the same growth rate as wild-type M2, with the exception of M2-9, which was slightly reduced. The individual features of each gene product are discussed below in comparison with the published sequence of the ATCC17978 strain (Smith et al., 2007).
Fig. 6.
Motility of EZ : : TN mutants. Wild-type M2 and various EZ : : TN insertion mutants were grown overnight in LB broth and adjusted to the same optical density with sterile LB broth. A 1 µl drop was placed on the surface of a 0.35 % Eiken agar plate and motility was measured after 8 h of growth at 37 °C. The values presented represent the average of two independent experiments with the ranges shown as error bars.
Mutant M2-2
The EZ : : TN Kan insertion is within A1S_0113, at a position corresponding to amino acid 481 of the putative 541 aa protein. This protein is highly similar to members of the acyl-CoA dehydrogenase family. The A1S_0113 ORF is potentially part of a large operon, in which it represents the second of eight genes (Fig. 7). The gene products of this operon appear to be related to the production of a lipopeptide, and include two polyketide synthases (A1S_0112 and A1S_0115).
Fig. 7.
Genetic organization of the chromosome in mutants M2-2 and M2-11. The possible organization of ORFs A1S_0112 to A1S_0119 as an eight-gene operon is shown, with the relative size of each gene depicted. The insertion sites of the EZ : : TN transposon are depicted above the A1S_0113 and A1S_0115 genes. The putative functions of the additional genes are as follows: A1S_0112, acyl-CoA synthetase/AMP-acid ligase, polyketide synthetase family; A1S_0113, acyl-CoA dehydrogenase; A1S_0114, acyl carrier protein; A1S_0115, polyketide synthetase; A1S_0116, RND superfamily transporter; A1S_0117, unknown function; A1S_0118, esterase–lipase superfamily; A1S_0119, phosphopantetheine protein transferase.
M2-11
The EZ : : TN Kan insertion is inserted within A1S_0115 at a position corresponding to amino acid 592 of a 1281 aa protein exhibiting significant homology to non-ribosomal peptide synthases. This gene is the fourth gene in a potentially large operon that also includes the A1S_0113 gene, which is described above and is disrupted in mutant M2-2 (Fig. 7). In particular, the protein encoded by A1S_0115 contains the following domains: (i) a condensation domain from amino acids 25 to 260 that catalyses peptide bond formation, (ii) an acyl-protein synthetase domain from amino acids 450 to 925 characteristic of LuxE-type proteins, which catalyses the formation of an acyl-protein thioester between a fatty acid and a protein, (iii) a phosphopantetheine attachment site from amino acids 950 to 1000 used for the attachment of activated fatty acid and amino acid groups, and (iv) a thioesterase domain from amino acids 1025 to 1281 that adds the last amino acid to non-ribosomally synthesized peptides.
M2-4
The EZ : : TN Kan insertion is in the A1S_3027 gene at a site corresponding to residue 250 of the 597 aa protein. This protein is similar to lytic transglycosylases that cleave the β-1,4-glycosidic bond between N-acetylmuramic acid and N-acetyl-d-glucosamine. This protein contains both lytic transglycosylase (LT) and goose egg white lysozyme (GEWL) domains. The A1S_3027 gene is upstream and in the same direction as a putative RNase, but is separated by 181 bp, suggesting that they are unlikely to be encoded within the same operon. A possible explanation for the motility defect in this mutant may be the role of this transglycosylase in the localized degradation of peptidoglycan, which may be required for the extrusion or assembly of a surface structure involved in motility. Similar enzymes in E. coli are required for the assembly of type IV pili and flagella (Vollmer et al., 2008).
M2-5
The EZ : : TN insertion is within a gene encoding a product with extensive homology to RmlB, a dTDP-glucose 4,6-dehydratase that converts dTDP-d-glucose to dTDP-4-dehydro-6-deoxy-d-glucose. This pathway is required for the biosynthesis of dTDP-l-rhamnose, a sugar present in the O-antigen of Gram-negative bacteria. Interestingly, among the A. baumannii genomes that have been sequenced to date, this gene is only present in strain AB900 (Adams et al., 2008). In AB900, the rmlB gene is upstream of ORFs that encode orthologues of rmlD, rmlA and rmlC, all of which function in the biosynthesis of dTDP-l-rhamnose. Therefore, since rmlB and the downstream genes all function in the same pathway for dTDP-l-rhamnose synthesis, polar effects would only be predicted to affect production of dTDP-l-rhamnose. The addition of 1 % l-rhamnose to 0.3 % Eiken agar restored the motility of M2-5 back to wild-type levels (data not shown). Alterations in the O-antigen have been shown to alter motility in other bacteria by a variety of mechanisms, including loss of cell surface wettability, altered flagellar synthesis or function, and increased adherence to solid surfaces (Toguchi et al., 2000).
M2-6
The EZ : : TN insertion is within A1S_2963, encoding a protein with 39 % identity and 55 % similarity to purK of E. coli, which encodes a subunit of the N5-carboxyaminoimidazole ribonucleotide synthase and is required for de novo purine synthesis. The purK gene appears to be monocistronic, and the EZ : : TN insertion is not predicted to have any polar effects. The EZ : : TN element inserted at a position corresponding to amino acid 215 of the 296 aa protein. This insertion resulted in a 70 % reduction in motility compared with wild-type M2 (Fig. 6). The basis for the motility defect of this mutant is unclear, as motility was not rescued by purine addition to the medium (data not shown).
M2-9
The EZ : : TN insertion is within A1S_2840, encoding an OmpA-like protein that has been previously described as a virulence determinant (Choi et al., 2008a, b; Gaddy et al., 2009; Gribun et al., 2003). The ompA gene appears to be monocistronic and the insertion is not predicted to have any polar effects. Based on similarities to OmpA from other Acinetobacter species, this protein may also be secreted and act as an emulsifier (Walzer et al., 2006). The swarming defect in this mutant may be due to a requirement for this protein in emulsifying hydrophobic compounds in the agar and facilitating a localized hydrophilic environment that promotes motility.
M2-10
In this mutant, the EZ : : TN transposon is inserted into A1S_0749 (BfmS), encoding a protein with a high degree of similarity to histidine sensor kinases of the two-component family of regulatory proteins. Mutations in this gene have been described before, and reported to exhibit a mild defect in biofilm formation (Tomaras et al., 2008). The EZ : : TN insertion in strain M2-10 disrupted the 530 aa BfmS protein at amino acid 168. This mutant was essentially non-motile, with an 80 % reduction in motility when compared with wild-type M2 (Fig. 6). In addition, a second independent insertion was obtained in the bfmS gene that disrupted at amino acid 330. This insertion resulted in a motility defect similar to that described for M2-10 (data not shown).
In an earlier study, in which the BfmRS two-component system was identified, it was demonstrated that a mutation in the bfmR response regulator resulted in loss of csuABCDE operon expression, which is required for Csu pili (Tomaras et al., 2008). To test whether the Csu system is required for motility, a targeted disruption of csuD, encoding an outer membrane usher protein required for fimbrial biogenesis, was created by the integration of a suicide plasmid containing an internal fragment. This mutation did not alter motility, although as reported before for csuA and csuE mutants (Tomaras et al., 2003), the csuD mutant exhibited a defect in biofilm formation (data not shown).
The A1S_0113 and A1S_0115 genes are activated by quorum sensing
In an effort to understand the role of quorum sensing in A. baumannii, we are currently using RNA-Seq to identify genes regulated by the abaI-dependent production of 3-OH C12-HSL. A detailed summary of this study will be submitted elsewhere. However, relevant to this study, the transcriptome analysis revealed that a subset of the genes most highly activated by quorum sensing included the A1S-0112 to A1S_0118 genes, with activation values of 63-, 64-, 57-, 39-, 13-, 10- and 6.6-fold, respectively. The expression data for the A1S_0119 gene were ambiguous; however, this gene encodes a phosphopantetheine protein transferase, a function that is clearly related to the products of the proximal genes in production of a lipopeptide. This suggests that the A1S_0112 to A1S_0118 (and likely A1S_0119) genes are present in an operon activated by the AbaI-dependent quorum-sensing pathway. As discussed above, two of these genes, A1S_0113 and A1S_0115, were identified in the above screen for insertions that decrease motility.
To confirm that this operon was regulated by quorum sensing, a single-copy transcriptional lacZ fusion to the promoter for the first gene of the putative operon, A1S_0112, was constructed in both a wild-type and an abaI : : Km mutant background (see Methods). In wild-type cells at mid-exponential phase (OD600 0.6), the fusion gave expression values of 8.7±0.7 Miller units in the wild-type and 2.1±0.1 Miller units in the abaI : : Km mutant (Table 2). When the A1S_0112–lacZ fusion in the abaI : : Km mutant background was grown to the same optical density as above, but in the presence of 10 µM 3-OH C12-HSL, the expression level was 15±0.5 Miller units, a 7.1-fold increase relative to the level in LB alone (Table 2). Taken together, these results indicate that the promoter upstream from the A1S_0112 gene is activated in an abaI-dependent manner by 3-OH C12-HSL.
Table 2. Role of quorum sensing in expression from the A1S_0112 promoter.
| Strain | Growth condition* | β-Galactosidase activity (Miller units) |
| M2 wild-type/A1S_0112-lacZ | LB | 8.7±0.7 |
| M2 wild-type/A1S_0112-lacZ | LB+AHL | 15.6±0.9 |
| M2 abaI : : Km/A1S_0112-lacZ | LB | 2.1±0.1 |
| M2 abaI : : Km/A1S_0112-lacZ | LB+AHL | 15.0±0.5 |
Cells were harvested at OD600 0.6. Supplementation of LB was with 10 µM 3-OH C12-HSL (AHL).
Discussion
This study has demonstrated that A. baumannii exhibits motility on LB plates with agar concentrations between 0.2 and 0.4 %. Interestingly, this motility varied significantly depending on the agar used. The use of Difco agar resulted in a branching, tentacle-like motility similar to that reported elsewhere for Acinetobacter spp. (Barker & Maxted, 1975). In contrast, Eiken agar resulted in a dramatically different pattern of motility, with an even expansion of cells from the central point of inoculation. The basis for this difference is not clear, although it has been proposed that Eiken agar may contain compounds that induce bacterial factors that promote spreading, or that it has a composition that confers greater ‘wettability’ of the agar (Harshey & Matsuyama, 1994; Toguchi et al., 2000).
The motility exhibited by A. baumannii M2 was partially dependent on a functional pilT gene. In other bacteria, PilT functions as an ATPase proposed to be required for pilus retraction, a process essential for twitching motility across surfaces (Harshey, 2003; Henrichsen, 1972; Herdendorf et al., 2002; Jarrell & McBride, 2008; Mattick, 2002; Merz et al., 2000; Skerker & Berg, 2001; Wall & Kaiser, 1999). In A. baumannii M2, the loss of pilT resulted in a 54 % reduction in motility. The residual motility in the pilT mutant background may be due to an additional gene product that can partially compensate for PilT. It is also possible that sliding motility contributes to the residual motility in the pilT mutant. Sliding motility, initially proposed by Henrichsen, is a surface translocation due to expansive forces of a growing cell population together with special properties of the cells, which reduce friction with the solid substrate (Henrichsen, 1972). Regardless of the mechanism for the residual motility, it is clear that twitching represents a significant component of the overall motility in A. baumannii, as suggested elsewhere (Henrichsen, 1984; McBride, 2010).
Transposon mutagenesis was used to identify additional genes required for motility. The insertion library used to identify these mutations was composed of approximately 2300 members and was not saturating. Therefore, we anticipate that additional genes required for motility are present and remain to be identified. An interesting class of mutants was defined by insertions in the A1S_0113 and A1S_0115 genes, which are possibly part of a large operon (A1S_0112 to A1S_0119) devoted to the non-ribosomal production of a lipopeptide (Fig. 7). It is tempting to speculate that this lipopeptide may act as a surfactant to aid motility. However, we were unable to detect any surfactant activity in culture supernatants of A. baumannii M2. The possibility that the surfactant was surface-associated was tested by cell surface hydrophobicity based on the bacterial adhesion to hydrocarbon (BATH) test using octanol (Rosenberg et al., 1980). However, both M2-2 and M2-11 exhibited cell surface hydrophobicity that was similar to that of the wild-type (data not shown). Studies are in progress to determine whether the residual motility in the pilT mutant is dependent on the A1S_0113 or A1S_0115 gene.
Motility required a functional AbaI autoinducer synthase and was rescued in an abaI mutant by the addition of 3-OH C12-HSL to plates (Fig. 5). Preliminary RNA-Seq data indicate that the A1S_0112 to A1S_0118 genes are activated by quorum sensing. A DNA fragment containing the region upstream of the A1S_0112 gene was fused to lacZ and exhibited promoter activity that was strongly activated by quorum sensing. This suggests that these genes are co-transcribed from the promoter in front of the A1S_0112 gene. The reduced expression of the A1S_0112 to A1S_0119 genes in the abaI mutant may explain, at least in part, the requirement for quorum sensing in motility. However, the loss of motility in the abaI mutant is more severe than that resulting from the insertions in the A1S_0113 and A1S_0115 genes, indicating that additional quorum-sensing-regulated genes contribute to motility. Genetic approaches are in progress to identify these genes.
Acknowledgements
We are grateful to Dr Matthew Parsek at the University of Washington for providing the lasI/rhlI mutant, to Luis Actis at Miami University for providing pWH1266, and to Dr Judith Noble-Wang at the US Centers for Disease Control for providing environmental isolates of A. baumannii. This work was supported, in part, by the Atlanta Research and Education Foundation to P. N. R., and by the National Institutes of Health (R01AI072219-01A1). P. N. R. is supported by a Research Career Scientist award from the Department of Veterans Affairs.
Abbreviations:
- 3-OH C12-HSL,
N-(3-hydroxy)-dodecanoylhomoserine lactone
References
- Adams M. D., Goglin K., Molyneaux N., Hujer K. M., Lavender H., Jamison J. J., MacDonald I. J., Martin K. M., Russo T., et al. (2008). Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol 190, 8053–8064. 10.1128/JB.00834-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker J., Maxted H. (1975). Observations on the growth and movement of Acinetobacter on semi-solid media. J Med Microbiol 8, 443–446. 10.1099/00222615-8-3-443 [DOI] [PubMed] [Google Scholar]
- Bergogne-Bérézin E., Towner K. J. (1996). Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev 9, 148–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet P. J., Grimont P. A. (1987). Identification and biotyping of clinical isolates of Acinetobacter. Ann Inst Pasteur Microbiol 138, 569–578. 10.1016/0769-2609(87)90042-1 [DOI] [PubMed] [Google Scholar]
- Choi C. H., Lee J. S., Lee Y. C., Park T. I., Lee J. C. (2008a). Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells. BMC Microbiol 8, 216. 10.1186/1471-2180-8-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C. H., Hyun S. H., Lee J. Y., Lee J. S., Lee Y. S., Kim S. A., Chae J. P., Yoo S. M., Lee J. C. (2008b). Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity. Cell Microbiol 10, 309–319. [DOI] [PubMed] [Google Scholar]
- Farinha M. A., Kropinski A. M. (1990). Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J Bacteriol 172, 3496–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddy J. A., Tomaras A. P., Actis L. A. (2009). The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun 77, 3150–3160. 10.1128/IAI.00096-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribun A., Nitzan Y., Pechatnikov I., Hershkovits G., Katcoff D. J. (2003). Molecular and structural characterization of the HMP-AB gene encoding a pore-forming protein from a clinical isolate of Acinetobacter baumannii. Curr Microbiol 47, 434–443. 10.1007/s00284-003-4050-4 [DOI] [PubMed] [Google Scholar]
- Harshey R. M. (2003). Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol 57, 249–273. 10.1146/annurev.micro.57.030502.091014 [DOI] [PubMed] [Google Scholar]
- Harshey R. M., Matsuyama T. (1994). Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagellate swarmer cells. Proc Natl Acad Sci U S A 91, 8631–8635. 10.1073/pnas.91.18.8631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichsen J. (1972). Bacterial surface translocation: a survey and a classification. Bacteriol Rev 36, 478–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichsen J. (1984). Not gliding but twitching motility of Acinetobacter calcoaceticus. J Clin Pathol 37, 102–103. 10.1136/jcp.37.1.102-b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdendorf T. J., McCaslin D. R., Forest K. T. (2002). Aquifex aeolicus PilT, homologue of a surface motility protein, is a thermostable oligomeric NTPase. J Bacteriol 184, 6465–6471. 10.1128/JB.184.23.6465-6471.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger M., Schmucker R., Kishan V., Hillen W. (1990). Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87, 45–51. 10.1016/0378-1119(90)90494-C [DOI] [PubMed] [Google Scholar]
- Jain D. K., Collins-Thompson D. L., Lee H., Trevors J. T. (1991). A drop-collapsing test for screening surfactant-producing microorganisms. J Microbiol Methods 13, 271–279. 10.1016/0167-7012(91)90064-W [DOI] [Google Scholar]
- Jarrell K. F., McBride M. J. (2008). The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol 6, 466–476. 10.1038/nrmicro1900 [DOI] [PubMed] [Google Scholar]
- Kaniga K., Delor I., Cornelis G. R. (1991). A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109, 137–141. 10.1016/0378-1119(91)90599-7 [DOI] [PubMed] [Google Scholar]
- Martínez A., Torello S., Kolter R. (1999). Sliding motility in mycobacteria. J Bacteriol 181, 7331–7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama T., Bhasin A., Harshey R. M. (1995). Mutational analysis of flagellum-independent surface spreading of Serratia marcescens 274 on a low-agar medium. J Bacteriol 177, 987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. S. (2002). Type IV pili and twitching motility. Annu Rev Microbiol 56, 289–314. 10.1146/annurev.micro.56.012302.160938 [DOI] [PubMed] [Google Scholar]
- McBride M. J. (2010). Shining a light on an opportunistic pathogen. J Bacteriol 192, 6325–6326. 10.1128/JB.01141-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz A. J., So M., Sheetz M. P. (2000). Pilus retraction powers bacterial twitching motility. Nature 407, 98–102. 10.1038/35024105 [DOI] [PubMed] [Google Scholar]
- Mussi M. A., Gaddy J. A., Cabruja M., Arivett B. A., Viale A. M., Rasia R., Actis L. A. (2010). The opportunistic human pathogen Acinetobacter baumannii senses and responds to light. J Bacteriol 192, 6336–6345. 10.1128/JB.00917-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu C., Clemmer K. M., Bonomo R. A., Rather P. N. (2008). Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J Bacteriol 190, 3386–3392. 10.1128/JB.01929-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rather P. N. (2005). Swarmer cell differentiation in Proteus mirabilis. Environ Microbiol 7, 1065–1073. 10.1111/j.1462-2920.2005.00806.x [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Gutnick D., Rosenberg E. (1980). Adherence of bacteria to hydrocarbons: a simple method for measuring cell surface hydrophobicity. FEMS Microbiol Lett 9, 29–33. 10.1111/j.1574-6968.1980.tb05599.x [DOI] [Google Scholar]
- Skerker J. M., Berg H. C. (2001). Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci U S A 98, 6901–6904. 10.1073/pnas.121171698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. G., Gianoulis T. A., Pukatzki S., Mekalanos J. J., Ornston L. N., Gerstein M., Snyder M. (2007). New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev 21, 601–614. 10.1101/gad.1510307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C. R., Rossier O., Cianciotto N. P. (2009). Surface translocation by Legionella pneumophila: a form of sliding motility that is dependent upon type II protein secretion. J Bacteriol 191, 1537–1546. 10.1128/JB.01531-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toguchi A., Siano M., Burkart M., Harshey R. M. (2000). Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J Bacteriol 182, 6308–6321. 10.1128/JB.182.22.6308-6321.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras A. P., Dorsey C. W., Edelmann R. E., Actis L. A. (2003). Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149, 3473–3484. 10.1099/mic.0.26541-0 [DOI] [PubMed] [Google Scholar]
- Tomaras A. P., Flagler M. J., Dorsey C. W., Gaddy J. A., Actis L. A. (2008). Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 154, 3398–3409. 10.1099/mic.0.2008/019471-0 [DOI] [PubMed] [Google Scholar]
- Vollmer W., Joris B., Charlier P., Foster S. (2008). Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev 32, 259–286. 10.1111/j.1574-6976.2007.00099.x [DOI] [PubMed] [Google Scholar]
- Wall D., Kaiser D. (1999). Type IV pili and cell motility. Mol Microbiol 32, 1–10. 10.1046/j.1365-2958.1999.01339.x [DOI] [PubMed] [Google Scholar]
- Walzer G., Rosenberg E., Ron E. Z. (2006). The Acinetobacter outer membrane protein A (OmpA) is a secreted emulsifier. Environ Microbiol 8, 1026–1032. 10.1111/j.1462-2920.2006.00994.x [DOI] [PubMed] [Google Scholar]
- Whiteley M., Lee K. M., Greenberg E. P. (1999). Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 96, 13904–13909. 10.1073/pnas.96.24.13904 [DOI] [PMC free article] [PubMed] [Google Scholar]