Abstract
Helicobacter pylori moves in response to environmental chemical cues using a chemotaxis two-component signal-transduction system. Autoinducer-2 (AI-2) is a quorum-sensing signal produced by the LuxS protein that accumulates in the bacterial environment in a density-dependent manner. We showed previously that a H. pylori luxS mutant was defective in motility on soft agar plates. Here we report that deletion of the luxS gene resulted in swimming behaviour with a reduced frequency of stops as compared to the wild-type strain. Stopping frequency was restored to wild-type levels by genetic complementation of the luxS mutation or by addition of synthetic 4,5-dihydroxy-2,3-pentanedione (DPD), which cyclizes to form AI-2. Synthetic DPD also increased the frequency of stops in wild-type H. pylori, similar to the behaviour induced by the known chemorepellent HCl. We found that whereas mutants lacking the chemoreceptor genes tlpA, tlpC or tlpD responded to an exogenous source of synthetic DPD, the chemoreceptor mutant tlpB was non-responsive to a gradient or uniform distribution of the chemical. Furthermore, a double mutant lacking both tlpB and luxS exhibited chemotactic behaviour similar to the tlpB single mutant, whereas a double mutant lacking both tlpB and the chemotransduction gene cheA behaved like a nonchemotactic cheA single mutant, supporting the model that tlpB functions in a signalling pathway downstream of luxS and upstream of cheA. We conclude that H. pylori perceives LuxS-produced AI-2 as a chemorepellent via the chemoreceptor TlpB.
Introduction
Directed motility in response to environmental chemical cues, a process called chemotaxis, is an important trait for many bacterial pathogens. Helicobacter pylori, a Gram-negative gastric pathogen, is proposed to situate itself within the gastric mucosa in response to specific chemical cues such as pH (Schreiber et al., 2004). Dysregulation of chemotaxis compromises the ability of H. pylori to colonize the murine and gerbil stomach, and results in its abnormal distribution and modulation of inflammation in this tissue (Foynes et al., 2000; McGee et al., 2005; Terry et al., 2005; Williams et al., 2007).
The chemotactic response, which has been studied most extensively in Escherichia coli, is mediated by chemoreceptors termed methyl-accepting chemotaxis proteins (MCPs) (Armitage, 1999). The H. pylori chemosensory machinery is similar but not identical to that of E. coli. The core signal-transduction apparatus (CheW, CheA and CheY) is present in both bacteria, but H. pylori also possesses three CheV proteins composed of both CheW and response-regulator motifs (Lowenthal et al., 2009; Pittman et al., 2001). The H. pylori genome encodes four chemoreceptors, TlpA, TlpB, TlpC and TlpD (originally called HylB or HlyB), although strain G27 does not express TlpC due to a frameshift mutation in the gene. Unlike peritrichous E. coli cells, which exhibit forward swimming and tumbling behaviour, we and others have observed that H. pylori cells swim forward, reverse and stop (Lowenthal et al., 2009; Schweinitzer et al., 2008). Once the cells stop, they usually resume movement in a different direction, thus allowing them to explore new spaces, similar to the tumbling behaviour of E. coli.
Only a handful of chemotactic signals have been demonstrated for H. pylori chemoreceptors. In strain 26695 the chemoreceptor TlpA has been shown to respond to arginine and bicarbonate (Cerda et al., 2003), and in strain SS1 the chemoreceptor TlpB has been demonstrated to respond to acid (Croxen et al., 2006). The intracellular chemoreceptor TlpD has been described as an energy sensor, although the molecular nature of the cues it perceives is not known (Schweinitzer et al., 2008). There are no known ligands for TlpC.
One environmental condition that H. pylori experiences is the endogenously produced quorum-sensing molecule autoinducer-2 (AI-2) (Forsyth & Cover, 2000; Joyce et al., 2000; Rader et al., 2007). Autoinducers (AIs) are bacterially produced extracellular signalling molecules that trigger quorum sensing, a form of bacterial cell–cell communication (Ng & Bassler, 2009). AI concentration increases as a function of bacterial cell density, and at critical concentration thresholds it initiates coordinated gene regulation. Several types of quorum-sensing systems have been characterized. Typically, Gram-positive and Gram-negative bacteria use oligopeptides and acylated homoserine lactones as AIs, respectively. Many Gram-positive and Gram-negative bacteria possess a quorum-sensing system that produces the furanone signal AI-2, which functions as a signal for interspecific communication. AI-2 is produced as a metabolic byproduct of the reaction carried out by LuxS, which cleaves S-ribosylhomocysteine, producing homocysteine and 4,5-dihydroxy-2,3-pentanedione (DPD). DPD undergoes rapid dehydration and cyclization, existing in equilibrium as several molecules collectively termed AI-2 (Schauder et al., 2001). In many bacteria, LuxS is a key enzyme in the production of the activated methyl donor, S-adenosylmethionine (SAM); however in H. pylori, the homocysteine produced by LuxS is metabolized to produce cysteine (Doherty et al., 2010).
Previous reports have shown that several H. pylori strains produce AI-2 in a luxS-dependent fashion, as detected through a Vibrio harveyi luminescence assay (Forsyth & Cover, 2000; Joyce et al., 2000; Lee et al., 2006; Rader et al., 2007; Shen et al., 2010). Mutation of the luxS gene has been associated with reduction of the flagellin flaA transcript and protein, enhanced biofilm formation, decreased motility on soft agar plates, and colonization defects in mouse and gerbil infection models (Cole et al., 2004; Lee et al., 2006; Loh et al., 2004; Osaki et al., 2006; Rader et al., 2007). We have previously reported that LuxS-produced AI-2 functions as a signal molecule that modulates transcript levels of the flagellar regulator FlhA, thereby influencing global flagellar regulation (Rader et al., 2007). Although it is clear that AI-2 signalling affects flagellar gene expression, we found that the flagella in luxS mutants are morphologically normal and thus the altered flagellar gene regulation in this strain may not explain fully its motility defect on soft agar plates. Because colonial expansion in these plates requires both motility and chemotaxis, we sought to examine how AI-2 affects the latter process. In this study we identify a novel function for AI-2 as a chemorepellent that is sensed by the chemoreceptor TlpB.
Methods
Bacterial strains and culture conditions.
H. pylori strain G27 and its isogenic mutants were used in this study and are listed in Table 1. All H. pylori strains were maintained on blood agar plates consisting of Columbia agar (Difco) and 5 % defibrinated horse blood (Hemostat) (CHBA), or CHBA plates supplemented with 0.02 mg β-cyclodextrin ml−1 (Sigma), 8 mg amphotericin B ml−1 (Sigma) and 20 µg vancomycin ml−1 (Sigma), and incubated at 37 °C in 10 % CO2. Selective plates were supplemented with 10–15 μg kanamycin ml−1 (Fisher), 18 µg metronidazole ml−1, 10 µg chloramphenicol ml−1 or 80 mM sucrose. H. pylori liquid medium (BB10) consisted of filtered sterilized Brucella broth (Difco) supplemented with 10 % fetal bovine serum (Gibco) and 20 µg vancomycin ml−1 (Sigma). Liquid cultures were grown in 50 ml conical tubes (BD Falcon) with loosened lids shaking at 37 °C in anaerobic jars (Oxoid) with CampyGen microaerobic sachets (Oxoid) or in glass test tubes (20×148 mm) shaking in a 37 °C/8 % CO2 incubator.
Table 1. Strains used in this study.
| H. pylori strain | Description | Reference |
| G27 (KG01) | Wild-type | Covacci et al. (1993) |
| luxS (BR08) | G27, luxS : : kan sacB | Rader et al. (2007) |
| ΔluxS (BR09) | G27, deletion of luxS | Rader et al. (2007) |
| luxS* (BR10) | G27, deletion of luxS and rdx : : luxS | Rader et al. (2007) |
| mG27 (KO625) | Mouse-passaged strain G27 | Castillo et al. (2008) |
| ΔtlpA (KO1002) | mG27, deletion of tlpA | This study |
| tlpB (KO1003) | mG27, tlpB : : kan-sacB | This study |
| ΔtlpB (KO1004) | mG27, deletion of tlpB | This study |
| tlpB* (BR31) | mG27, deletion of tlpB and rdx : : tlpBSS1-aphA-3 | This study |
| luxS tlpB (CW30) | mG27. tlpB : : cat and luxS : : kan sacB | This study/Williams et al. (2007) |
| tlpC (KO1005) | mG27, tlpC : : aphA-3 | This study |
| tlpD (KO1006) | mG27, tlpD : : cat (made using pL30A2cat2) | This study/Williams et al. (2007) |
| cheA (KO629) | mG27, cheA : : cat reverse (made using pKT22) | This study/Terry et al. (2005) |
| tlpB cheA (CW31) | G27, tlpB : : kan sacB and cheA : : cat reverse | This study |
Construction of H. pylori mutants.
All transformations were performed using natural transformation. PCR for cloning purposes was done with either Pfu Turbo (Stratagene) or Phusion (NEB), and all DNA manipulation enzymes were from New England Biolabs. The G27 luxS, ΔluxS, and luxS* isogenic strains (strains BR08, BR09 and BR10, respectively) were constructed as previously described (Rader et al., 2007). Throughout, we use ‘Δ’ to designate deletion mutations, in which most of a gene’s coding sequence is removed by allelic exchange, and the superscript ‘*’ to designate complemented strains in which a wild-type copy of a mutated gene is supplied at another genomic locus from the mutated gene. The luxS* strain contains a wild-type copy of the luxS gene and its promoter introduced in the rdxA locus of the ΔluxS mutant chromosome. For most chemotaxis mutants, a mouse-selected variant of G27, mG27, was used as the parent strain (Castillo et al., 2008).
The cheA mutant (strain KO629) was created by transforming H. pylori mG27 with pKT22 (Terry et al., 2005) and selecting for resistance on chloramphenicol CHBA.
ΔtlpB (strain KO1004) was made in two steps. First, mG27 was transformed to kanamycin resistance (KmR) using the plasmid pTC-B111, which bears a tlpB : : kan-sacB allele. The correct nature of this mutation was verified by PCR using primers that flank the insertion site. pTC-B111 was made starting with pTCB101, which has tlpB from strain SS1 cloned into pBluescript (McGee et al., 2005). pTCB101 was then subjected to iPCR using primers tlpB-30 and tlpB-40 (sequence in McGee et al., 2005). The kan-sacB construct was obtained from pKSFII (Copass et al., 1997) by digesting with SmaI and XhoI. The kan-sacB and tlpB flanking pieces were ligated using T4 ligase. The correct nature of the plasmid was verified by digestion. In this construct, tlpB is transcribed in the same direction as sacB (opposite to aphA-3). This strain is called KO1003. KO1003 was then transformed with a plasmid bearing a large deletion of tlpB, pTC-B113, followed by selection for sucrose resistance (SucR). pTC-B113 was created using iPCR with primers tlpB-30 and tlpB-40, followed by intramolecular ligation of the plasmid. The deletion removes all of tlpB except the first 27 and the last 17 amino acids, although the deletion is +1 out of frame, resulting in a 3′ end that encodes a different sequence with three additional amino acids. The tlpB region from SucR, kanamycin-sensitive (KmS) colonies was amplified using PCR with primers that flank tlpB. This PCR product was sequenced to verify that tlpB was deleted.
To create the tlpB* complemented strain (strain BR31), mG27 ΔtlpB mutant bacteria were naturally transformed with a construct encoding the rdxA locus with an insertion composed of the full-length tlpB gene, expressed from the cheY promoter, and cloned upstream of the non-polar aphA-3 cassette. Disruption of the rdxA locus confers metronidazole resistance, and the non-polar aphA-3 cassette contains two internal ribosome-binding sites flanking the kanamycin-resistance gene conferring that resistance. Transformants were first screened on kanamycin plates, further screened on metronidazole-supplemented plates, and verified by PCR amplification and sequencing of the genomic locus. For the complementation construct, the non-polar aphA-3 [original sequence from pUC18 K-2 (Ménard et al., 1993)] was PCR amplified from the previously described G27 fliA mutant strain (Rader et al., 2007) using M13F (5′-GTAAAACGACGGCCAGT-3′) and M13R (5′-CAGAAGACAGCTATGA-3′) primers. To create the final DNA construct, a modified form of the vector pK0140 (Terry et al., 2005) was generated called pKO140-tlpB in which the cheY coding sequence was replaced by the tlpB coding sequence from SS1, retaining the cheY promoter sequence. The aphA-3 DNA and pKO140-tlpB were digested with SalI and EcoRI and ligated together, creating the rdx : tlpBSS1-aphA-3 construct pK1337. The rdx:tlpBSS1-aphA-3 sequence was then PCR amplified from pK1337 using the RdxA1F and RdxA2R primers (Rader et al., 2007). This PCR product was then introduced into the mG27 ΔtlpB mutant through natural transformation followed by selection for KmR isolates. Transformants were further screened on metronidazole supplemented plates and verified by PCR amplification and sequencing of the genomic locus.
The tlpA mutant (strain KO1002) was made in two steps by first transforming mG27 to KmR with pTA12, which bears a tlpA : : kan-sacB allele, to create strain KO1001, followed by transformation with a plasmid that bears a ΔtlpA deletion (pTA14). pTA12 was made from pTA10, which has tlpA from strain SS1 cloned, with ~500 bp flanking sequence, by PCR amplification with primers TlpA6 (5′-ATTGAGCGCAAAAATAGGGGC-3′) and TlpA7 (5′ TTTTCTCTCGCCAAAGCTTGC-3′). This piece was phosphorylated with T4 polynucleotide kinase, and then cloned into EcoRV-digested pBluescript KS+ (Stratagene) to create pTA10. To create pTA12, iPCR was carried out with pTA10 template plus primers TlpA12 (5′-CACCCACAATATGATTTTATTACCG-3′) and TlpA13 (5′-CAAGAAATTGACAAAGTCTCTAACG-3′) to create an in-frame deletion that leaves a region coding for 14 amino acids at the 5′ end, and 19 amino acids at the 3′ end of tlpA. This iPCR product was then ligated to a kan-sacB fragment that came from pKS2. pKS2 derived from pKS1, which was made via a three-piece ligation of the following pieces: (1) the aphA3 gene (also called kan) isolated from pBS-kan (Terry et al., 2005) by cutting with SmaI+XhoI, followed by dephosphorylation; (2) the sacB gene from pKSFII (Copass et al., 1997) generated by cutting with BamHI, blunting with T4 polymerase followed by cutting with PstI; (3) the vector backbone from pBluescript KS+ cut with PstI and XhoI, followed by phosphatasing and gel purification of the 3.0 kb piece. These three pieces were ligated together to generate pKS1. pKS1 has the aphA3 promoter driving transcription of both aphA3 and the downstream sacB gene. To create pKS2, pKS1 was digested with PvuI to remove the bla gene followed by self-ligation. For cloning of the aphA3-sacB fragment from pKS2, PCR amplification using primers reverse (universal) and sacBend2 (5′-CTTTTGCGTTTTTATTTGTTAAC-3′) was carried out, to amplify aphA3-sacB without the sacB transcriptional terminator, and the resulting fragment was ligated with the iPCR fragment of pTA10 to create pTA12. In pTA12, tlpA, aphA3 and sacB are all in the same transcriptional orientation. To create pTA14, the same pTA10 iPCR fragment was self-ligated. H. pylori KO1001 was then transformed with pTA14, followed by selection for sucrose resistance and screening for kanamycin sensitivity. The tlpA allele was PCR amplified from SucR KmS colonies using primers tlpA10 (5′-TCTAAAGGTTTGAGTATCGG-3′) and tlpA11 (5′-GCTCGAATTCGAAAACTGCTTTTTATTCACATC-3′), and sequenced to verify the correct deletion. This tlpA strain is also called mG27 tlpA or KO1002.
The mG27 tlpC mutant is tlpC : : aphA3 (strain KO1005). This mutant was constructed by transforming mG27 with plasmid pKO150. This plasmid was made using iPCR of a plasmid containing tlpC from SS1 called pTC100 (Andermann et al., 2002) with primers tlpC31 (5′-TCATCACAATTTTAGAACC-3′) and tlpC40 (5′-CCTTGCAACAAGATGTGCAGG-3′). The product, 3.5 kb, was ligated with the aphA3 gene prepared by PCR from pBS-kan with primers Kanup (5′-GGCCGGATCCGATAAACCCAGCGAACC-3′) and Kandown (5′-GGCCAAGCTTTTTAGACATCTAAATC-3′). The resulting product was phosphorylated with T4 kinase, and ligated to the pTC100ΔtlpC product to create pKO150. The tlpD mutant was made by transforming mG27 to chloramphenicol resistance using plasmid pL30A2cat2 (Williams et al., 2007), which creates tlpD : : cat. Note that tlpD was previously referred to as hylB.
To generate strain CW30, with mutations in both luxS and tlpB genes, we transformed H. pylori mG27 tlpB : : catR (Williams et al., 2007) with PCR DNA generated from luxS : : kan sacB genomic DNA and selected for kanamycin resistence. This luxS : : kan-sacB DNA was amplified with primers luxSfor (5′-AACGCTGGGATTACGCATGGA-3′) and luxSrev (5′-AAGCCGCCCGTGAATGTCTGAA-3′).
To generate the bacterial strain CW31 with mutations in both cheA and tlpB genes, we first created a tlpB : : kan-sacB mutation in wild-type G27 bacteria and then added the cheA : : cat reverse mutation. To generate the DNA for the tlpB : : kan-sacB mutation, SS1 tlpB was amplified from the pK1337 plasmid using SS1TlpBF (5′-TAAGGCGTTAGAGACGCTTTGGCT-3′) and SS1TlpBR (5′-AAACACGCCGTGATCACAGAAACC-3′) primers and ligated into the pCR 2.1-TOPO plasmid using the TOPO TA Cloning kit (Invitrogen), creating pTOPO-tlpB. The kan-sacB cassette (Copass et al., 1997) was amplified from the pKSF-II plasmid using primers kansacBF (5′-CTCCATGGTCCCGGGCGAACCATTTGAGGTGA-3′) and kansacbR (5′-GACTGAGGATGCGCCCGGGTATAAGCCCATTTTCATGC-3′). The kan-sacB cassette was inserted into the HincII restriction site at base-pair position 735 of the tlpB gene, creating pTOPO-tlpB : : kansacB. The tlpB : : kan-sacB sequence was amplified from pTOPO-tlpB : : kansacB and transformed into naturally competent G27 H. pylori. Positive tlpB : : kan-sacB transformants were selected by kanamycin resistance and confirmed via PCR using primers TlpBF (5′-ACTTCAAAAGACGGGAGGACT-3′) and TlpBR (5′-TATCCCCACTCGCACGC-3′). The tlpB : : kan-sacB bacteria were transformed with PCR DNA generated from cheA : : cat reverse genomic DNA, and selection for chloramphenicol resistance was applied. The correct genotype was confirmed by PCR. The cheA : : cat reverse DNA, was amplified with primers CheA_for1 (5′-GTGCTGAAAGGGCTAAAGAAATG-3′) and CheA_rev1 (5′-GGATAATCGCTCTGTCCGTG-3′).
Bioluminescence assay and synthetic DPD.
Synthetic DPD, a kind gift from Martin Semmelhack and Bonnie Bassler (Princeton University), was prepared as described by Semmelhack et al. (2005). This DPD was synthesized with a protecting group that we removed by treatment with 15 mM H2SO4, followed by neutralization to pH 6.9 with potassium phosphate. As a mock DPD treatment, we neutralized 15 mM H2SO4 to pH 6.9 with potassium phosphate. Alternatively, we used DPD from OMM Scientific that did not contain a protecting group; BB10 medium was used to dilute this DPD and as a mock treatment. The concentration of bioluminescence-inducing activity in H. pylori cell-free supernatant from early stationary-phase cultures was approximately equivalent to 0.1 mM synthetic DPD (Rader et al., 2007).
Microscopy.
Swimming behaviour of bacterial strains was visualized using an inverted Leica DMIL phase-contrast light microscope (Leica Microsystems). G27 wild-type, ΔluxS, luxS*, tlpB and tlpB* isogenic strains were grown overnight on a shaker to an OD600 of 1.0. Aliquots (2 ml) of this overnight culture were then added to a test tube containing 500 µl fresh BB10 medium and placed stationary at 37 °C in 10 % CO2 for 2 h. These conditions promoted high levels of motility. Approximately 8 µl of the stationary culture was spotted onto a glass slide followed by the same amount of 0.1 M HCl, 0.1 mM synthetic DPD, mock DPD solution, or BB10 medium as a negative control. A coverslip was added and sealed on all sides with clear finger-nail polish. After 4–10 min, videos were recorded at 15 or 30 frames s−1 on a Cohu High Performance CCD camera using Scion Image software. The number of stops exhibited by individual bacteria was determined by examining 5 s of swimming for multiple bacterial cells per video, as described previously (Terry et al., 2006). Stops were defined as the cessation of movement in any direction for three or more frames; we did not score reversals that occurred without pauses. Individual bacterial cells were scored if they met the following criteria: they were motile at the beginning of the 5 s interval, they remained visible for the duration of the 5 s interval, and they stopped only for at most 30 frames, and resumed swimming after this period. Video scoring was performed without knowledge of the genotype or treatment condition. At least two independent trials were performed per condition and at least 15 bacterial cells were scored per condition.
To assay bacterial taxis away from a source of chemical repellent, we adopted the previously described barrier formation assay (Croxen et al., 2006). Wild-type mG27, ΔtlpA, ΔtlpB, tlpB*, tlpC, tlpD and cheA isogenic strains were grown overnight, with shaking, in BB10 to an OD600 of 1.0, followed by 2 h without shaking, as above. Samples (8 µl) of culture were spotted onto the centre of a glass slide and covered with a no. 1, 22×22 mm wide glass coverslip. Three sides of the coverslip were sealed with clear finger-nail polish. Eight microlitres of 0.1 M HCl, 0.1 mM synthetic DPD, or mock DPD solution was placed on the open side of the coverslip. After 5–10 min incubation, the slides were examined and still images were taken using Scion Image software at ×10 through a ×40 phase filter, conditions that mimic dark-field microscopy.
Western blotting.
Total cell proteins were prepared from H. pylori cultured on CHBA plates for 2 days by resuspending and lysing the cells in 2× Laemmli sample buffer. Samples were separated on a 10 % SDS-PAGE gel, transferred to Immunoblot PVDF membranes (Bio-Rad) and incubated with 1 : 5000 dilution of anti-GST_TlpA22 (Williams et al., 2007). This rabbit polyclonal antibody recognizes the conserved CheW-interacting methyl-accepting domain of all H. pylori chemoreceptors. For visualization, the blots were incubated with the secondary antibody goat anti-rabbit-HRP (Santa Cruz Biotech) at a dilution of 1 : 2000, followed by incubation with luminol, p-coumaric acid and hydrogen peroxide. Luminescent blots were visualized by exposure to Biomax Light film (Kodak).
Statistical analysis.
The frequency of stops was analysed statistically using either one-way ANOVA (www.physics.csbsju.edu/cgi-bin/stats/anova_pnp) when strains were tested or two-way ANOVA (http://faculty.vassar.edu/lowry/anova2u.htl) when both strains and chemical treatment were tested. Post-hoc multiple comparisons to identify significance within individual hypotheses were performed using t-tests, and the P-values were adjusted using Bonferroni correction to correct for an increase in type I error. P-values of <0.001 were considered significant and used for Bonferroni correction.
Results
luxS is required for normal swimming behaviour of H. pylori
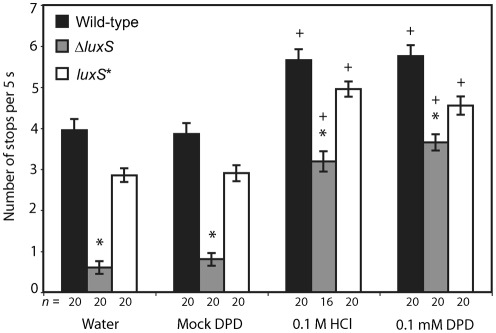
When examining the swimming behaviour of wild-type and an isogenic luxS deletion mutant (referred to as ΔluxS) of H. pylori strain G27 (Rader et al., 2007), we observed that the ΔluxS mutant displayed a propensity for swimming in a straight line (running), as opposed to the wild-type, which exhibited more typical run–stop–run behaviour. We quantified this behaviour in bacterial cultures using video microscopy to observe swimming behaviour. We found that the ΔluxS mutant exhibited significantly fewer stops per second than the wild-type strain (Fig. 1). Complementation of the luxS mutation with a wild-type copy of the luxS gene at the rdxA locus [referred to as luxS* (Rader et al., 2007)] significantly increased the number of stops per second as compared with the ΔluxS strain (Fig. 1). It should be noted that over the course of these experiments, we observed some variation between trials in the average number of stops per second exhibited by the strains, but the responses to chemicals and differences between genotypes were reproducible across experiments. We attribute these differences to factors that were difficult to control across all experiments, such as differences in the lots of serum used in the media.
Fig. 1.
LuxS is required for normal swimming behaviour of H. pylori. Swimming behaviours of G27 wild-type, ΔluxS mutant and luxS* complemented isogenic strains were observed by video microscopy, and the number of stops that individual bacteria performed during 5 s was recorded. Each strain was treated with water, mock DPD, 0.1 M HCl and 0.1 mM synthetic DPD. The number of individual bacterial cells scored (n) for each condition is indicated below the column; bars indicate se. * indicates each strain that is statistically significant different from all other strains within a treatment, + indicates chemorepellent treatments (HCl, synthetic DPD) that are significantly different from controls (water, mock DPD) within a strain (P<0.001 with Bonferroni correction).
The LuxS product, AI-2, is a chemorepellent for H. pylori
To test whether the decrease in the number of stops in the ΔluxS mutant relative to the G27 wild-type and luxS* isogenic strains was due to the loss of the AI-2 signal, we treated each strain with 0.1 mM synthetic DPD, corresponding to the approximate concentration of AI-2 in the cell-free supernatant of a early stationary-phase culture of wild-type H. pylori (Rader et al., 2007). We also treated each isogenic strain with sterile water, the DPD diluent, as a negative control. Bacterial cultures were incubated with treatment for 10 min before video microscopy. With the addition of synthetic DPD, the number of stops in the ΔluxS mutant was increased to a level similar to that seen in the wild-type strain treated with water (Fig. 1). Interestingly, in the cultures of the wild-type and the complemented strains, the number of stops also increased in the presence of synthetic DPD, suggesting that all three isogenic strains responded to changes in AI-2 concentration.
The swimming behaviours exhibited in all three isogenic strains upon addition of synthetic DPD were strikingly similar to the reported repellent response of H. pylori strain SS1 to the chemorepellent HCl (Croxen et al., 2006). To investigate whether strain G27 responded similarly to HCl and synthetic DPD, we incubated wild-type, ΔluxS and luxS* G27 with 0.1 M HCl and observed their swimming behaviour. The pH of the synthetic DPD solution was 6.9, in contrast to the pH of 1.0 of the 0.1 M HCl solutions. All three isogenic strains exhibited an increase in the number of stops when exposed to 0.1 M HCl, and this increase did not differ significantly from that in response to synthetic DPD (Fig. 1). To ensure that this similarity was not due to some component of the solution used to rehydrate and activate the synthetic DPD, we also performed the experiment with a solution identical to the synthetic DPD solution, to which the synthetic DPD was not added (mock DPD). This treatment did not alter the swimming behaviour of the strains relative to their behaviour in response to water (Fig. 1). All subsequent experiments employed mock DPD as the negative control. These experiments thus suggest that DPD functions as a chemorepellent for H. pylori, because it increases the frequency of bacterial stopping.
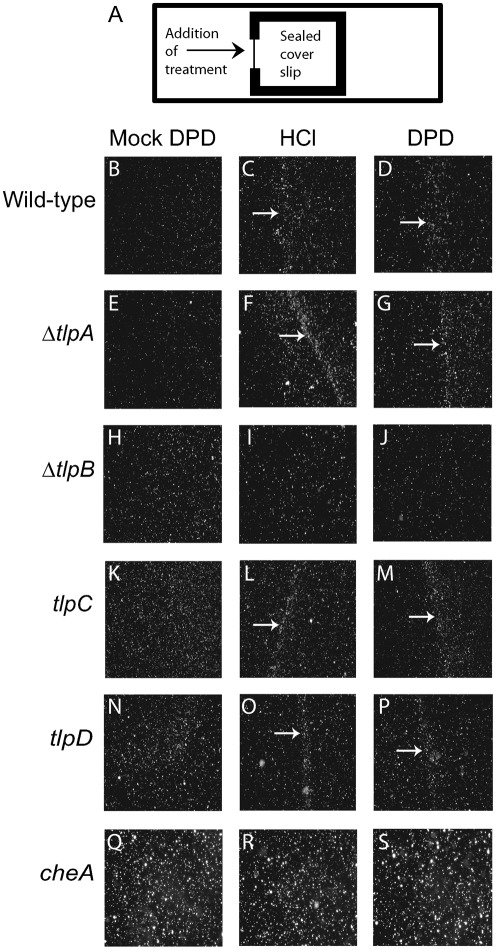
To further explore whether H. pylori uses chemotaxis to respond to synthetic DPD, we employed a previously described wet-mount assay to monitor spatial chemotactic responses (Croxen et al., 2006). Wild-type G27 bacterial suspensions in BB10 medium were prepared as described earlier, seeded onto a glass microscope slide, and covered with a coverslip that was then sealed on three sides. Synthetic DPD (0.1 mM), HCl (0.1 M), mock DPD or water was then spotted onto the part of the coverslip that remained unsealed (Fig. 2a). Movement of the bacterial population was monitored before, immediately after and 5 min after addition of the chemical stimulus. Prior to the addition of treatment the bacteria were motile and uniformly distributed below the coverslip. Immediately after addition of treatment, the bacteria were flushed away from the liquid source; however after 5 min the hydrodynamic flow had ceased, and in the case of mock DPD or water, the bacteria returned to a uniform distribution. As previously reported (Croxen et al., 2006), in response to 0.1 M HCl, the percentage of motile bacteria immediately increased, with the majority of the bacteria exhibiting an increase in the frequency of stops, resulting in net movement away from the source of the acid. This increased movement proximal to the acid source ended in the formation of a visible barrier of bacterial cells toward the centre of the coverslip that accumulated presumably at a preferred concentration of the chemical treatment (Fig. 2C). Beyond this barrier, bacterial movement resembled that of the untreated controls. These observations show that H. pylori strain G27 responds to pH similarly to strain SS1 (Croxen et al., 2006). When treated with 0.1 mM synthetic DPD, the cultures exhibited a response that was similar to that to acid: the cells showed increased frequency of stops and formed a bacterial barrier at a similar location away from the chemical source (Fig. 2D). In response to mock DPD, the percentage of motile bacteria in the population increased, but there was no obvious net directional movement, and after 5 min no bacterial barrier had formed and the bacteria were evenly distributed underneath the coverslip (Fig. 2B). Our data thus suggest that H. pylori cells respond chemotactically to synthetic DPD similarly to how they respond to HCl.
Fig. 2.
Chemotaxis response of wild-type and mutant H. pylori to HCl and synthetic DPD. A wet-mount chemotaxis assay (illustrated in A) was used to analyse chemotatic behaviour of mG27 wild-type and the indicated isogenic mutant strains in response to mock DPD, the known chemorepellent 0.1 M HCl, and 0.1 mM synthetic DPD. White arrows indicate bacterial barrier formation.
The chemotaxis receptor TlpB is required for chemotaxis away from AI-2
Bacterial tactic movement toward or away from a chemical source is generally regulated by the chemotaxis system. Key to this system are the chemoreceptors which sense environmental signals and transmit the information through a transduction cascade, eventually affecting flagellar rotation (Armitage, 1999). To test whether the response to synthetic DPD was due to detection by any of the four H. pylori chemoreceptors, we repeated the wet-mount assay described above, using mouse-passaged H. pylori mG27 isogenic strains harbouring deletions or insertional mutations in the chemoreceptor genes tlpA (strain ΔtlpA), tlpB (strain ΔtlpB), tlpC (strain tlpC) or tlpD (strain tlpD). The wild-type strain mG27 does not produce TlpC as detectable by Western blotting (Fig. 3A), but we included the engineered tlpC mutant in our analysis for completeness. Each of these strains was challenged with 0.1 mM synthetic DPD, 0.1 M HCl or mock DPD. As a negative control for chemotaxis we employed a non-chemotactic mutant with a mutation in the chemotransduction gene cheA. The ΔtlpA, tlpC and tlpD mutants all formed bacterial barriers when challenged with synthetic DPD and HCl (Fig. 2F, G, L, M, O, P). As previously reported, the ΔtlpB mutant failed to form a bacterial barrier when challenged with HCl (Fig. 2I). In addition, this mutant failed to form a bacterial barrier when challenged with synthetic DPD (Fig. 2J). As expected, the cheA mutant did not form a bacterial barrier in response to HCl or synthetic DPD (Fig. 2R, S). None of the five strains produced bacterial barriers in response to mock DPD (Fig. 2E, H, K, N, Q). These data implicated TlpB in the sensing of AI-2.
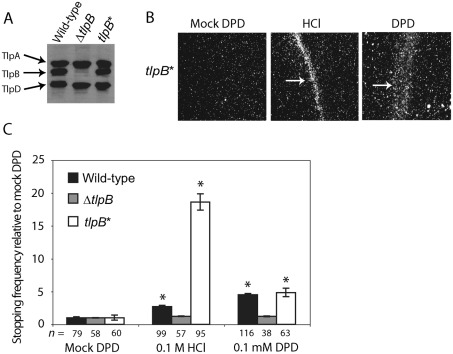
Fig. 3.
TlpB is required for chemotactic responses to HCl and synthetic DPD. (A) Western blot analysis of mG27 wild-type, ΔtlpB, and tlpB* isogenic strains with an antibody that recognizes a conserved region of all H. pylori chemoreceptors. (B) Chemotactic behaviour of the tlpB* complemented strain was assayed using the wet-mount assay in response to mock DPD, 0.1 M HCl and 0.1 mM synthetic DPD. (C) The normalized stopping frequency for mG27 wild-type, ΔtlpB and tlpB* isogenic strains treated with mock DPD, 0.1 M HCl and 0.1 mM synthetic DPD is shown, where the average number of stops for each strain in each condition is normalized to that strain’s average number of stops in mock DPD. The number of individual bacterial cells scored (n) for each condition is indicated below the column; bars indicate se. * indicates statistically significant differences in stopping frequency as compared to the mock DPD treatment of the same strain (P<0.001 with Bonferroni correction).
To verify that the failure of the ΔtlpB mutant to form a bacterial barrier in response to DPD was due to the lack of the tlpB gene product, we constructed a complemented strain (referred to as tlpB*) by placing the full-length tlpB gene in the rdxA locus of the ΔtlpB mutant chromosome. Western blot analysis verified production of the TlpB protein in the tlpB* strain (Fig. 3A). We then repeated the wet-mount assay with the tlpB* strain. When challenged with 0.1 mM synthetic DPD or 0.1 M HCl, the tlpB* strain responded by producing a bacterial barrier, in contrast to the mock DPD solution, which produced a uniform distribution of bacteria (Fig. 3B).
To further characterize TlpB as a chemoreceptor for synthetic DPD, we repeated the video taxis assay with the mG27 wild-type, ΔtlpB and tlpB* isogenic strains. The average number of stops for each genotype was recorded for each strain grown in mock DPD, 0.1 M HCl or 0.1 mM synthetic DPD. As reported previously (Croxen et al., 2006), we observed that the ΔtlpB mutant displayed an increased stopping frequency as compared to its wild-type parent in medium without added chemorepellents (see below). To best represent the relative responsiveness of the different strains to HCl and DPD, we therefore show the normalized stopping frequencies for each strain in each condition relative to the average number of stops exhibited by that strain in the mock DPD medium (Fig. 3C). In accordance with the wet-mount assay, the ΔtlpB mutant exhibited significantly reduced responsiveness to DPD and HCl as compared with the wild-type strain. The complemented tlpB* strain had restored responsiveness to DPD and greater responsiveness to acid than the wild-type strain. We do not know the reason for this strain’s hyper-responsiveness to acid, but it could be due to altered expression of the tlpB gene under acidic conditions when expressed from a different promoter and genomic locus than those of the native gene.
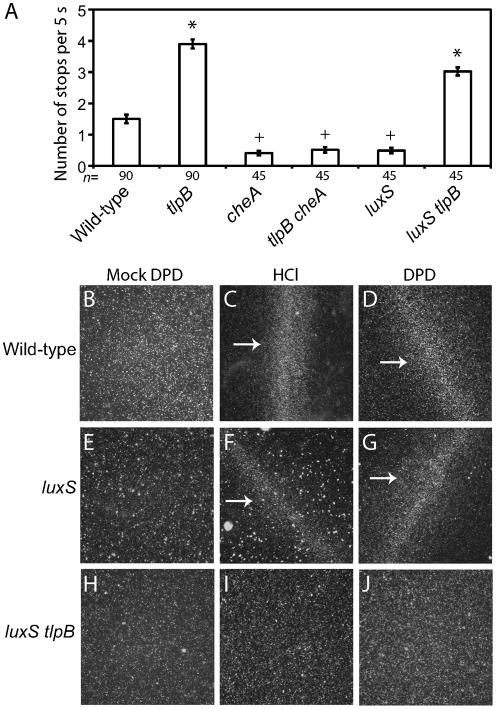
As noted above, the ΔtlpB mutant displayed an increased stopping frequency as compared to its wild-type parent when grown in medium in the absence of exogenously added chemorepellents (Fig. 4A). We wondered whether this increased frequency was due to a hyper-activation of the chemotaxis signal transduction pathway, or some other effect on swimming behaviour caused by the loss of tlpB. To distinguish these possibilities, we constructed a tlpB cheA double mutant and compared the swimming behaviour of this strain to the tlpB and cheA single mutants. As expected, the cheA single mutant, which lacks a functional chemotaxis signal tranduction pathway, exhibited fewer stops than the wild-type strain in BB10 (Fig. 4A). The tlpB cheA double mutant was indistinguishable from the cheA single mutant, indicating that the increased stopping observed with the tlpB mutant was due to increased activation of the chemotaxis pathway, possibly as a result of increased signalling from other chemoreceptors in the absence of TlpB.
Fig. 4.
tlpB functions in a signalling pathway downstream of luxS and upstream of cheA. (A) Swimming behaviours in BB10 of G27 wild-type, tlpB, cheA, tlpB cheA, luxS and luxS tlpB isogenic strains were observed by video microscopy, and the number of stops that individual bacteria performed during 5 s was recorded. The number of individual bacterial cells scored (n) for each strain is indicated below the column; bars indicate se. Symbols (*, +) indicate strains that are statistically indistinguishable from each other (P>0.001 with Bonferroni correction). (B–J) The wet-mount chemotaxis assay was used to analyse chemotatic behaviour of the indicated isogenic mutant strains in response to mock DPD, the known chemorepellent 0.1 M HCl and 0.1 mM synthetic DPD. White arrows indicate bacterial barrier formation.
TlpB functions downstream of LuxS in AI-2 responses
Our data suggest that TlpB functions as the chemoreceptor for avoidance responses to AI-2, a molecule produced by the bacteria themselves using the enzyme encoded by luxS. If this model were correct, then we would predict that cells lacking both LuxS, the enzyme that makes AI-2, and TlpB, the chemoreceptor involved in its perception, should resemble cells lacking only the receptor. We generated a luxS tlpB double mutant and compared its swimming behaviour to that of luxS and tlpB single mutants. We observed that the luxS tlpB double mutant exhibited a high stopping frequency, statistically indistinguishable from the tlpB single mutant (Fig. 4A). Importantly, whereas the luxS single mutant formed barriers to both DPD and HCl in the wet-mount assay (Fig. 4F, G), the luxS tlpB double mutant failed to form any barriers (Fig. 4I, J), similar to the tlpB single mutant (Fig. 2I, J). These data demonstrate that tlpB is epistatic to luxS, consistent with our model that TlpB perceives a molecule produced by the luxS gene product.
Discussion
In this study we have shown that the quorum-sensing signal AI-2 acts as a chemorepellent for H. pylori, and demonstrated that the chemoreceptor TlpB is required for its perception. We previously reported that elimination of the luxS gene product in H. pylori exhibited a motility defect on soft agar (Rader et al., 2007), which was complemented by exogenous AI-2, a result confirmed by others (Shen et al., 2010). This assay cannot distinguish between dysfunction of the motility apparatus and dysregulation of chemotaxis. To better characterize the chemotactic response of H. pylori to the absence or presence of AI-2, we observed both swimming behaviour of individual bacterial cells and the response of populations of bacteria to chemical gradients. We found that luxS-deficient bacterial cells exhibited a decrease in stopping frequency as compared to wild-type cells. This behaviour was restored to wild-type levels by the addition of synthetic DPD. Synthetic DPD also increased the frequency of stops in the wild-type strain, and caused populations of wild-type cells to move away from a source of the chemical, suggesting that AI-2 is a chemorepellent for H. pylori. Our ability to modulate H. pylori swimming behaviour with exogenous AI-2 argues against a metabolic requirement for the luxS gene in chemotaxis. Additionally, although many bacteria regulate the sensitivity of their chemoreceptors via methylation, a mechanism which could be influenced by function of the SAM pathway, H. pylori lacks the methyl-utilizing enzymes required for this process (Szurmant & Ordal, 2004). We further demonstrated that an H. pylori strain deficient for the chemoreceptor TlpB failed to move away from a source of synthetic DPD, and did not display increased stopping behaviour upon addition of synthetic DPD. These behaviours were restored upon genetic complementation of the tlpB gene.
TlpB and AI-2 perception
Most reported responses to AIs involve regulation of transcription. Recently, however, E. coli was shown to respond to AI-2 as a chemoattractant through a mechanism that involves both the AI-2 periplasmic binding protein LsrB and the chemoreceptor Tsr (Bansal et al., 2008; Hegde et al., 2011). Here we show that H. pylori also perceives AI-2 as a chemical cue in response to which it directs its movement, in this case by moving away from the signal. We demonstrate that the H. pylori chemoreceptor TlpB is required for perception of AI-2 and confirm its role in negative pH taxis. Bacterial chemoreceptors can often sense more than one ligand and can elicit both positive and negative behavioural responses. For example, the E. coli chemoreceptor Tsr not only senses AI-2, but also mediates taxis toward the attractants serine and related amino acids, and taxis away from weak acids, indole and leucine, and transduces oxygen and redox signals (Boyd & Simon, 1982; Rebbapragada et al., 1997). We do not yet know whether TlpB senses AI-2 directly or via other binding proteins. TlpB shares no sequence similarity with the previously identified AI-2 binding proteins, V. harveyi LuxP (Chen et al., 2002), S. typhimurium LsrB (Miller et al., 2004) and Aggregatibacter actinomycetemcomitans RbsB and LsrB (Shao et al., 2007), and the H. pylori genome lacks homologues of any of these genes. We believe that the molecular mechanism of chemotaxis from AI-2 is distinct from AI-2-mediated regulation of flhA transcription in H. pylori, because the latter does not require tlpB (B. A. R. and K. G., unpublished results). In addition, neither tlpB nor several other chemotaxis genes appear to be regulated transcriptionally by AI-2 because their transcript levels are the same in wild-type and ΔluxS mutant strains (Rader et al., 2007; B. A. R. and K. G., unpublished results).
AI-2-regulated motility is likely to be important in the gastric environment
For H. pylori, motility is essential for colonization of piglet, gerbil and mouse stomachs, where bacterial interactions with the mucosa promote gastric pathology (Eaton et al., 1992; McGee et al., 2002; O’Toole et al., 2000; Ottemann & Lowenthal, 2002). It is well established that non-chemotactic mutants (Che−) of H. pylori do not infect mice to the full wild-type level (Foynes et al., 2000; Terry et al., 2005). These mutants, lacking cheA, cheW or cheY, all engage almost exclusively in straight runs without stops or changes in direction. We found that the luxS mutant exhibits a similar swimming behaviour to the Che− mutants in broth, suggesting that LuxS is responsible for production of a significant proportion of chemorepulsive signals present in H. pylori batch culture. We have confirmed previous reports that a luxS mutant is defective in colonization of the rodent stomach; however in these experiments it was not possible to determine whether the colonization defect was due to reduced AI-2 concentrations in the mouse stomach or metabolic deficiencies in the bacteria (Lee et al., 2006; Osaki et al., 2006), (B. A. R., K. G. and K. M. O., unpublished results). Whereas the ΔtlpB mutant colonizes wild-type mice and gerbils to normal levels (McGee et al., 2005; Williams et al., 2007), it is impaired in its ability to colonize the stomachs of IL-12-deficient mice (Croxen et al., 2006). There are likely to be additional chemical cues to which H. pylori responds within a mouse stomach (Schreiber et al., 2004). Indeed, non-chemotactic mutants are less closely associated with mouse gastric epithelia and induce a diminished inflammatory response as compared to wild-type H. pylori, suggesting that chemical cues from the epithelium are important for directing H. pylori localization within the stomach (Williams et al., 2007).
The fact that gastrointestinal pathogens have chemotactic responses to AI-2 raises the question of the role of this response in host colonization. In the case of enteropathogenic E. coli, the pathogen perceives AI-2 as an attractant, possibly using it as a cue to direct itself toward the bacteria-dense colon. We hypothesize that H. pylori perceives AI-2 both as a repellent and as a cue to regulate flagellar gene expression as a means to coordinate its distribution and motility within the stomach. We imagine that at some threshold level of AI-2, the bacteria respond to this molecule as a chemorepellent and move away from the bulk bacterial population, thereby avoiding niche competition and promoting dispersal throughout the stomach. Consistent with this model, luxS mutants in several H. pylori strains have been reported to form biofilms more readily than their wild-type counterparts (Cole et al., 2004). With sustained AI-2 levels, indicative of high bacterial populations, the elevated AI-2 may alter FlhA levels to downregulate motility. Such a response might allow bacteria to avoid wasting energy on swimming and instead focus on adherence. Several bacterial species perform reciprocal regulation of motility and adherence (Holden & Gally, 2004), although this type of response has not been shown for H. pylori. Furthermore, our understanding of the gastric environment has changed in part due to 16S rRNA gene enumeration studies, which identified over 120 phylotypes of bacteria in the human stomach, many of which potentially produce AI-2 (Bik et al., 2006; Federle & Bassler, 2003). It is possible that H. pylori will move away from AI-2 produced by coincident bacteria, providing a further means of avoiding niche competition. As disease outcome is correlated with localization of H. pylori populations within the stomach (Blaser & Atherton, 2004), AI-2 may be an important environmental factor in the progression of disease in H. pylori infections. Further understanding of those factors that regulate H. pylori motility and chemotaxis within the gastric environment will undoubtedly enhance our ability to predict disease outcome and design therapies that can control H. pylori-caused chronic inflammation and subsequent progression to gastric cancer.
Acknowledgements
This work was supported by National Institutes of Health Public Health Service grants R01 AI050000 (to K .M. O.) and R01 DK075667 (to K. G.). We thank Bonnie Bassler and Martin Semmelhack for the generous gift of synthetic DPD, Tessa Andermann and Yu-Ting Chen for construction of the tlpA and tlpC mutational vectors, and Will Finch for construction of the pKO140-tlpB vector. We thank Khoosheh Gosnik, Jim Remington and members of the Guillemin lab for fruitful discussions.
Abbreviations:
- AI-2
autoinducer-2
- DPD
4,5-dihydroxy-2,3-pentanedione
- iPCR
inverse PCR
- SAM
S-adenosylmethionine
References
- Andermann T. M., Chen Y. T., Ottemann K. M. (2002). Two predicted chemoreceptors of Helicobacter pylori promote stomach infection. Infect Immun 70, 5877–5881. 10.1128/IAI.70.10.5877-5881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage J. P. (1999). Bacterial tactic responses. Adv Microb Physiol 41, 229–289. 10.1016/S0065-2911(08)60168-X. [DOI] [PubMed] [Google Scholar]
- Bansal T., Jesudhasan P., Pillai S., Wood T. K., Jayaraman A. (2008). Temporal regulation of enterohemorrhagic Escherichia coli virulence mediated by autoinducer-2. Appl Microbiol Biotechnol 78, 811–819. 10.1007/s00253-008-1359-8. [DOI] [PubMed] [Google Scholar]
- Bik E. M., Eckburg P. B., Gill S. R., Nelson K. E., Purdom E. A., Francois F., Perez-Perez G., Blaser M. J., Relman D. A. (2006). Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A 103, 732–737. 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser M. J., Atherton J. C. (2004). Helicobacter pylori persistence: biology and disease. J Clin Invest 113, 321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A., Simon M. (1982). Bacterial chemotaxis. Annu Rev Physiol 44, 501–517. 10.1146/annurev.ph.44.030182.002441. [DOI] [PubMed] [Google Scholar]
- Castillo A. R., Arevalo S. S., Woodruff A. J., Ottemann K. M. (2008). Experimental analysis of Helicobacter pylori transcriptional terminators suggests this microbe uses both intrinsic and factor-dependent termination. Mol Microbiol 67, 155–170. 10.1111/j.1365-2958.2007.06033.x. [DOI] [PubMed] [Google Scholar]
- Cerda O., Rivas A., Toledo H. (2003). Helicobacter pylori strain ATCC700392 encodes a methyl-accepting chemotaxis receptor protein (MCP) for arginine and sodium bicarbonate. FEMS Microbiol Lett 224, 175–181. 10.1016/S0378-1097(03)00423-3. [DOI] [PubMed] [Google Scholar]
- Chen X., Schauder S., Potier N., Van Dorsselaer A., Pelczer I., Bassler B. L., Hughson F. M. (2002). Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415, 545–549. 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- Cole S. P., Harwood J., Lee R., She R., Guiney D. G. (2004). Characterization of monospecies biofilm formation by Helicobacter pylori. J Bacteriol 186, 3124–3132. 10.1128/JB.186.10.3124-3132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copass M., Grandi G., Rappuoli R. (1997). Introduction of unmarked mutations in the Helicobacter pylori vacA gene with a sucrose sensitivity marker. Infect Immun 65, 1949–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covacci A., Censini S., Bugnoli M., Petracca R., Burroni D., Macchia G., Massone A., Papini E., Xiang Z., et al. (1993). Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci U S A 90, 5791–5795. 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxen M. A., Sisson G., Melano R., Hoffman P. S. (2006). The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J Bacteriol 188, 2656–2665. 10.1128/JB.188.7.2656-2665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty N. C., Shen F., Halliday N. M., Barrett D. A., Hardie K. R., Winzer K., Atherton J. C. (2010). In Helicobacter pylori, LuxS is a key enzyme in cysteine provision through a reverse transsulfuration pathway. J Bacteriol 192, 1184–1192. 10.1128/JB.01372-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton K. A., Morgan D. R., Krakowka S. (1992). Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J Med Microbiol 37, 123–127. 10.1099/00222615-37-2-123. [DOI] [PubMed] [Google Scholar]
- Federle M. J., Bassler B. L. (2003). Interspecies communication in bacteria. J Clin Invest 112, 1291–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth M. H., Cover T. L. (2000). Intercellular communication in Helicobacter pylori: luxS is essential for the production of an extracellular signaling molecule. Infect Immun 68, 3193–3199. 10.1128/IAI.68.6.3193-3199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foynes S., Dorrell N., Ward S. J., Stabler R. A., McColm A. A., Rycroft A. N., Wren B. W. (2000). Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect Immun 68, 2016–2023. 10.1128/IAI.68.4.2016-2023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde M., Englert D. L., Schrock S., Cohn W. B., Vogt C., Wood T. K., Manson M. D., Jayaraman A. (2011). Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemoreceptor and the periplasmic LsrB AI-2-binding protein. J Bacteriol 193, 768–773. 10.1128/JB.01196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden N. J., Gally D. L. (2004). Switches, cross-talk and memory in Escherichia coli adherence. J Med Microbiol 53, 585–593. 10.1099/jmm.0.05491-0. [DOI] [PubMed] [Google Scholar]
- Joyce E. A., Bassler B. L., Wright A. (2000). Evidence for a signaling system in Helicobacter pylori: detection of a luxS-encoded autoinducer. J Bacteriol 182, 3638–3643. 10.1128/JB.182.13.3638-3643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. K., Ogura K., Loh J. T., Cover T. L., Berg D. E. (2006). Quantitative effect of luxS gene inactivation on the fitness of Helicobacter pylori. Appl Environ Microbiol 72, 6615–6622. 10.1128/AEM.01291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh J. T., Forsyth M. H., Cover T. L. (2004). Growth phase regulation of flaA expression in Helicobacter pylori is luxS dependent. Infect Immun 72, 5506–5510. 10.1128/IAI.72.9.5506-5510.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenthal A. C., Simon C., Fair A. S., Mehmood K., Terry K., Anastasia S., Ottemann K. M. (2009). A fixed-time diffusion analysis method determines that the three cheV genes of Helicobacter pylori differentially affect motility. Microbiology 155, 1181–1191. 10.1099/mic.0.021857-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee D. J., Coker C., Testerman T. L., Harro J. M., Gibson S. V., Mobley H. L. (2002). The Helicobacter pylori flbA flagellar biosynthesis and regulatory gene is required for motility and virulence and modulates urease of H. pylori and Proteus mirabilis. J Med Microbiol 51, 958–970. [DOI] [PubMed] [Google Scholar]
- McGee D. J., Langford M. L., Watson E. L., Carter J. E., Chen Y. T., Ottemann K. M. (2005). Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infect Immun 73, 1820–1827. 10.1128/IAI.73.3.1820-1827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard R., Sansonetti P. J., Parsot C. (1993). Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol 175, 5899–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. T., Xavier K. B., Campagna S. R., Taga M. E., Semmelhack M. F., Bassler B. L., Hughson F. M. (2004). Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol Cell 15, 677–687. 10.1016/j.molcel.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Ng W. L., Bassler B. L. (2009). Bacterial quorum-sensing network architectures. Annu Rev Genet 43, 197–222. 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole P. W., Lane M. C., Porwollik S. (2000). Helicobacter pylori motility. Microbes Infect 2, 1207–1214. 10.1016/S1286-4579(00)01274-0. [DOI] [PubMed] [Google Scholar]
- Osaki T., Hanawa T., Manzoku T., Fukuda M., Kawakami H., Suzuki H., Yamaguchi H., Yan X., Taguchi H., et al. (2006). Mutation of luxS affects motility and infectivity of Helicobacter pylori in gastric mucosa of a Mongolian gerbil model. J Med Microbiol 55, 1477–1485. 10.1099/jmm.0.46660-0. [DOI] [PubMed] [Google Scholar]
- Ottemann K. M., Lowenthal A. C. (2002). Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect Immun 70, 1984–1990. 10.1128/IAI.70.4.1984-1990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman M. S., Goodwin M., Kelly D. J. (2001). Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for CheW and the three CheV paralogues, and evidence for CheV2 phosphorylation. Microbiology 147, 2493–2504. [DOI] [PubMed] [Google Scholar]
- Rader B. A., Campagna S. R., Semmelhack M. F., Bassler B. L., Guillemin K. (2007). The quorum-sensing molecule autoinducer 2 regulates motility and flagellar morphogenesis in Helicobacter pylori. J Bacteriol 189, 6109–6117. 10.1128/JB.00246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbapragada A., Johnson M. S., Harding G. P., Zuccarelli A. J., Fletcher H. M., Zhulin I. B., Taylor B. L. (1997). The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci U S A 94, 10541–10546. 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder S., Shokat K., Surette M. G., Bassler B. L. (2001). The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol 41, 463–476. 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- Schreiber S., Konradt M., Groll C., Scheid P., Hanauer G., Werling H. O., Josenhans C., Suerbaum S. (2004). The spatial orientation of Helicobacter pylori in the gastric mucus. Proc Natl Acad Sci U S A 101, 5024–5029. 10.1073/pnas.0308386101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinitzer T., Mizote T., Ishikawa N., Dudnik A., Inatsu S., Schreiber S., Suerbaum S., Aizawa S., Josenhans C. (2008). Functional characterization and mutagenesis of the proposed behavioral sensor TlpD of Helicobacter pylori. J Bacteriol 190, 3244–3255. 10.1128/JB.01940-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semmelhack M. F., Campagna S. R., Federle M. J., Bassler B. L. (2005). An expeditious synthesis of DPD and boron binding studies. Org Lett 7, 569–572. 10.1021/ol047695j. [DOI] [PubMed] [Google Scholar]
- Shao H., James D., Lamont R. J., Demuth D. R. (2007). Differential interaction of Aggregatibacter (Actinobacillus) actinomycetemcomitans LsrB and RbsB proteins with autoinducer 2. J Bacteriol 189, 5559–5565. 10.1128/JB.00387-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F., Hobley L., Doherty N., Loh J. T., Cover T. L., Sockett R. E., Hardie K. R., Atherton J. C. (2010). In Helicobacter pylori auto-inducer-2, but not LuxS/MccAB catalysed reverse transsulphuration, regulates motility through modulation of flagellar gene transcription. BMC Microbiol 10, 210. 10.1186/1471-2180-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H., Ordal G. W. (2004). Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev 68, 301–319. 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry K., Williams S. M., Connolly L., Ottemann K. M. (2005). Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect Immun 73, 803–811. 10.1128/IAI.73.2.803-811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry K., Go A. C., Ottemann K. M. (2006). Proteomic mapping of a suppressor of non-chemotactic cheW mutants reveals that Helicobacter pylori contains a new chemotaxis protein. Mol Microbiol 61, 871–882. 10.1111/j.1365-2958.2006.05283.x. [DOI] [PubMed] [Google Scholar]
- Williams S. M., Chen Y. T., Andermann T. M., Carter J. E., McGee D. J., Ottemann K. M. (2007). Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect Immun 75, 3747–3757. 10.1128/IAI.00082-07. [DOI] [PMC free article] [PubMed] [Google Scholar]