Abstract
Microbial competition exists in the general environment, such as soil or aquatic habitats, upon or within unicellular or multicellular eukaryotic life forms. The molecular actions that govern microbial competition, leading to niche establishment and microbial monopolization, remain undetermined. The emerging technology of imaging mass spectrometry (IMS) enabled the observation that there is directionality in the metabolic output of the organism Bacillus subtilis when co-cultured with Staphylococcus aureus. The directionally released antibiotic alters S. aureus virulence factor production and colonization. Therefore, IMS provides insight into the largely hidden nature of competitive microbial encounters and niche establishment, and provides a paradigm for future antibiotic discovery.
Introduction
Microbial species exist in perpetual competition with one another for suitable ecological niches to support their survival and growth. Diverse niches exist in the general environment, such as soil or aquatic habitats, upon or within unicellular or multicellular eukaryotic life forms. On human skin and mucosal surfaces, the outcome of microbial competition determines our normal flora, which are essential for health and immune homeostasis. Colonization of normal flora by potential pathogens is the initial step in the pathogenesis of most infectious diseases (Grice et al., 2009; Hoffman et al., 2006). Efficiency of nutrient acquisition and strategies for surface attachment are essential factors for microbial niche survival, whilst elaboration of compounds that kill or limit the growth of competing strains or species can promote niche monopolization. The released compounds include secondary metabolite antibiotics (e.g. penicillin, chloramphenicol, tetracycline), bacteriocin peptides or low-molecular-mass toxic molecules such as hydrogen peroxide, each coupled to mechanisms for intrinsic resistance/immunity by the producing strain (Gonzalez et al., 2010; Li & Walsh, 2010; Queck et al., 2009).
An attractive hypothesis suggests that microbes regulate and optimize their production of such molecules to kill, limit the growth of or modulate the metabolism of potential niche competitors for maximal advantage. Different factors contribute to the outcome of microbial competition, such as the collection of molecules exchanged between the competing organisms, their respective cell densities and the initial spatial configuration or direction of the microbe–microbe interaction. To date, there have been limited studies addressing the events associated with microbial competition in a spatial and multiplexed fashion, due in part to the lack of available tools. The present work applies the emerging technology of imaging mass spectrometry (IMS) to the study of such interactions, using as models two well-characterized, Gram-positive bacterial species, avirulent Bacillus subtilis and Staphylococcus aureus.
B. subtilis is a bacterium found on skin, in the digestive tract, in epithelial wounds, on extremities of the human body, in livestock and in soil (Ara et al., 2006; Earl et al., 2008). Because B. subtilis is ubiquitous, it has developed adaptive strategies to subsist in diverse environments via the production and secretion of a large number of genetically encoded molecules that control the growth of neighbouring organisms (Liu et al., 2010; Stein, 2005). For this reason, B. subtilis is sold commercially as a skincare product, a food ingredient for human consumption, animal feed, fertilizer and an antibiotic substitute. The molecular mechanisms by which these products work are poorly understood. Similarly, S. aureus, a pre-eminent human pathogen causing an array of serious hospital- or community-acquired infections worldwide (Geng et al., 2010; Klevens et al., 2007; Otto, 2010), is found on human skin, digestive tracts, nares, livestock and surgical instrumentation (Iwase et al., 2010; Otto, 2010; Roberson et al., 1994). At minimum, 30 % of the world population is colonized with S. aureus, a bona fide pathogen that has developed significant resistance against a variety of antibiotics and is the cause of more fatalities in the USA than HIV/AIDS (Enright et al., 2002; Klevens et al., 2007).
Although S. aureus colonizes a large proportion of the world’s inhabitants, it only produces clinical infection in a subset of this population. One attractive hypothesis is that neighbouring organisms occupying the same environmental niche (e.g. skin) respond by secreting an array of antibiotic-type molecules to control S. aureus developmental phenotypes and thereby alter its ability to proliferate in the host. The study described herein reveals that B. subtilis, a bacterium that is nearly ubiquitous in nature, occupying many environmental niches and therefore a common but transient skin microbe (Ara et al., 2006), can inhibit the growth of an epidemic S. aureus isolate and possess the ability to directionally release a molecule with antimicrobial and metabolism-altering properties. Ultimately, the study indicates a utility for IMS in dynamic analysis of interspecies metabolic exchange and perhaps a paradigm for future discovery of novel antibiotic candidates.
Methods
Sample preparation for IMS.
ISP2 agar medium was prepared with 2 g yeast extract, 4 g malt extract, 2 g glucose and 10 g agar, by addition to 500 ml deionized water and autoclaving. Autoclaved agar medium (10 ml) was poured into sterile Petri dishes under sterile conditions. A lawn of meticillin-resistant S. aureus skin isolate ST59-MRSA-IV (Geng et al., 2010) was prepared on an ISP2 agar plate. Thereafter, a paper disc containing B. subtilis NCBI 3610 (undomesticated wild-type) was placed in the middle of the S. aureus ST59 lawn and allowed to grow overnight. For the T-shape experiment, colony growth was initiated by streaking 3 µl overnight growths of S. aureus ST59 and B. subtilis NCBI 3610 at equal cell densities on the prepared agar ISP2 plates. Bacterial colonies were allowed to grow for 48 h at 30 °C before transferring the co-culturing experiment to a Bruker MSP 96 MALDI anchor plate. Thereafter, the target plate was covered uniformly with Sigma universal matrix (α-cyano-4-hydroxycinnamic acid and 2,5-dihydroxybenzoic acid) by the use of a 50 µm sieve. Once the sample was covered uniformly with matrix, it was placed in a 37 °C oven for 3–4 h until it was deemed dried, at which point it was subjected to IMS.
IMS.
The Bruker MSP 96 anchor plate containing the sample was inserted into a Microflex Bruker Daltonics mass spectrometer outfitted with the Compass 1.2 software suite (FlexImaging 2.0, FlexControl 3.0 and FlexAnalysis 3.0; Bruker Daltonics). The sample was run in positive mode, with 200–350 µm laser intervals in xy and 50–54 % laser power. A photomicrograph of the colonies to be imaged by IMS was loaded onto the FlexImaging command window. Three teach points were selected in order to align the background image with the sample target plate. After calibration of the target plate was complete, the AutoXecute command was used to analyse the samples. The method setting under the FlexControl panel was ImagingRPpepmix, consisting of the following settings. Laser: fuzzy control, on; weight, 1.00; laser power varied between 50 and 54 %; matrix blaster, 0. Evaluation: peak selection masses from m/z 400 to 4000; mass control list, off; peak exclusion, off; peak evaluation processing method, default; smoothing, off; baseline subtraction, on; peak resolution, >100. Accumulation: parent mode, on; sum up to 20 satisfactory shots in 20 shots; dynamic termination, off. Movement: random walk, two shots at raster spot. Quit sample after two subsequent failed attempts. Processing: flex analysis method, none; Boito’s MS method, none; sample carrier, nothing; spectrometer, on; ion source, 1–19 mV, ion source 2, 16.40 mV; lens, 9.45 mV; reflector, 20.00; pulsed ion extraction, 190 ns; polarity, positive. Matrix suppression, deflection; suppress up to m/z 400; detector gain, reflector 4.1; sample rate, 2.00 GS s−1; mode, low range; electronic gain, enhanced, 100 mV; real-time smooth, off. Spectrometer: size, 81040; delay, 42968. Processing method, factory method RP_2465. Setup: mass range, low; laser frequency, 20 Hz; auto teaching, off. Instrument-specific settings: digitizer trigger level, 2000 mV; digital off linear, 127 cant; digital off reflector, 127 cant; detector gain voltage offset, linear, 1300 V; reflector, 1400 V. Laser attenuator: offset, 12 %; range, 30 %. Electronic gain button definitions: regular, 100 me (offset line), 100 mV (offset ref), 200 mV/full scale; end, 51 mV (offset line), 51 mV (offset ref), 100 mV/full scale; highest, 25 mV (offsetting), 25 mV (offset ref), 50 mV/full scale. Calibration was accomplished by using Bruker Daltonics Pepmix 4 as an external standard. Zoom range was 1.0 %; peak assignment tolerance was user-defined (500 pap).
Data processing.
After acquisition, the datasets were analysed by using the FlexImaging software. The resulting mass spectrum was filtered manually in 0.5–3.0 Ad increments, with individual colours assigned to the specific masses. Ions of interest were identified by the use of tandem mass spectrometry. IMS of over 12 co-culturing interactions between S. aureus and B. subtilis showed similar consistent phenotypes.
Visualization of membrane damage.
B. subtilis NCBI 3610 cells were harvested from a colony that was either adjacent to that of S. aureus ST59 or grown in isolation. Cells were resuspended in T-base, stained with a final concentration of 30 µg FM 4-64 ml−1, which stains the membranes red, and 2.5 µM SYTOX Green (Invitrogen), which brightly stains only cells with membrane damage. The cells were immobilized with poly-l-lysine and visualized with an Applied Precision Spectris microscope. Experiments performed on different inocula and microscopes showed consistent data.
Isolation of B. subtilis from skin.
The left eyebrow of a healthy male subject was swabbed using a sterile cotton-tipped applicator. Immediately following the collection, the cotton-tipped applicator was placed into a sterile Falcon tube labelled HR13 and sealed. Luria–Bertani (LB) broth agar was then inoculated with the cotton-tipped applicator. The inoculated Petri dish was then placed into the incubator at 37 °C overnight. Following a growth period of about 18 h, the inoculated plate was examined for bacterial growth. The Petri dish, labelled HR13, was found to contain multiple, phenotypically different bacterial species. Each of the phenotypically different microbial colonies was then excised from the Petri dish by using sterile 10 µl inoculation loops. The excised colonies were then transferred to their own Petri dish containing LB agar Miller growth medium for the purpose of raising single colonies. Each Petri dish containing a single colony was labelled as a subset of the region from which they were originally harvested from the body. More precisely, seven phenotypically different microbial colonies were excised from the initial swab plate. These plates were numbered HR13.1–HR13.7. After the plates were inoculated, they were again sealed in Parafilm and placed back into the plastic sleeve. The plates were then placed into the incubator at 37 °C for 18 h. After the growth period, the plates were removed from the incubator and examined for bacterial growth. The Petri dish labelled HR13.2 appeared to have only a single inhabitant growing on the agar. The microbe growing on the plate was then made into a cell stock in 20 % glycerol. For identification, 16S rRNA gene sequencing was performed and showed the bacterial growth to be B. subtilis. Mass spectral fingerprinting of the metabolites produced by the identified human-derived B. subtilis isolate aligned with a fingerprint obtained from B. subtilis NCBI 3610, further confirming its identity.
Inhibition of S. aureus growth on solid agar.
Purified surfactin and plipastatin were resuspended in 70 % ethanol alone or in combination and 10 µl solution was pipetted onto prewarmed agar plates and allowed to dry. Spots, prepared in triplicate, were control (70 % ethanol), surfactin (20 µg), plipastatin (20 µg) or surfactin and plipastatin together (20 µg each). S. aureus ST59 (2×105 c.f.u.) was pipetted onto the agar at the location of compounds and allowed to dry. Plates were incubated at 37 °C for 4 h. Spots were then excised and placed into 2 ml screw-cap tubes containing 1 mm silica/zirconia beads in 1 ml PBS. The samples were homogenized in a mini-BeadBeater-8 (BioSpec) for 1 min at full speed twice, placing the tubes on ice in between. Homogenized samples were diluted serially in sterile PBS and plated on Todd–Hewitt agar plates for enumeration. Inhibition of bacterial growth by purified compounds was calculated as a percentage of initial inoculum.
Nanocapillary liquid chromatography tandem mass spectrometry (LC-MS/MS).
Samples were prepared by spiking S. aureus ST59 inocula with 10 µg purified surfactin ml−1 and allowing them to grow for 16 h. Samples were then checked for cell density by the use of a spectrometer. Thereafter, samples were extracted with 1-butanol, lyophilized and resuspended for LC-MS/MS. The LC-MS/MS apparatus was prepared as follows. The nanocapillary columns were prepared by drawing 360 m outer diameter, 100 m inner diameter deactivated, fused silica tubing (Agilent) with a model P-2000 laser puller (Sutter Instruments) (heat: 330, 325, 320 °C; velocity, 45 mm min−1; delay, 125 ms) and were packed at 600 p.s.i. (4.14 MPa) to a length of 10 cm with C18 reverse-phase resin suspended in methanol. The column was equilibrated with 90 % solvent A (water, 0.1 % acetic acid) and loaded with S. aureus extracts with or without 10 µg surfactin ml−1 by flowing 90 % solvent A and 10 % solvent B (CH3CN, 0.1 % acetic acid) at 20 µl min−1 for 5 min, 15 µl min−1 for 3 min and 10 µl min−1 for 12 min. At 20 min, the flow rate was increased to 200 µl min−1 and infused into a split flow so that 200–500 nl min−1 went through the capillary column, whereas the remainder of the flow was diverted to waste. A gradient for eluting extract contents was established with a time-varying solvent mixture and electrosprayed directly into a calibrated Thermo Finnigan LTQ-XL (source voltage, 2.0 kV; capillary temperature, 200 °C). Note that, on all occasions, the LTQ mass spectrometer was tuned and calibrated to achieve a background signal NL<4.5×103.
LC-MS/MS acquisition.
Two different MS/MS acquisition methods were used to compile spectra of the surfactin-spiked S. aureus ST59 prepared extracts. Methods M1 and M2 used data-dependent acquisition with varying cut-offs on the exclusion list capacity and time, collecting fragmentation data for the first to tenth most abundant ions.
Data processing.
All collected data files (RAW) were processed with a DOS command-line version of InSpecT software (Tanner et al., 2005). Relevant input search parameters included the following: (i) post-translational modification search, +28 Da (formylation); (ii) database, modified USA300 genome, USA300 genome ‘reversed’ or ‘phony database’, and common contaminants database; (iii) PTM allowed per peptide, 3; (iv) b- and y-ion mass offset tolerance, 0.5 Da; (v) parent mass tolerance, 1.5 Da. Experiments were reproducible between multiple runs. Spectral counts were determined by taking the total number of spectra indentified for each protein. Spectral counts for each indentified protein were verified by the use of differentially prepared S. aureus growths.
In vivo (skin) inhibition of S. aureus growth.
Flanks of 8–10-week-old female CD1 mice were shaved. The following day, mice were anaesthetized with ketamine/xylazine and placed on heating pads to maintain body temperature. Purified surfactin and plipastatin were resuspended in 70 % ethanol alone or in combination and 10 µl solution was pipetted onto pre-marked spots on the mouse flank and allowed to dry. Four spots per mouse were prepared as follows: negative control (70 % ethanol), surfactin (20 µg), plipastatin (20 µg) or surfactin and plipastatin together (20 µg each). S. aureus ST59 was grown to mid-exponential phase (OD600 = 0.5), washed with sterile PBS and diluted to 2×107 c.f.u. ml−1. Ten microlitres (2×105 c.f.u.) of bacterial suspension was spotted onto agar plates and allowed to dry. Once the spots had dried, agar discs were excised using a 6 mm biopsy punch. One agar disc containing 2×105 c.f.u. S. aureus ST59 was placed onto each spot (four per mouse) and affixed to the mouse with Tegaderm transparent wound dressing. After 4 h, each spot was excised and skin, together with agar disc, was placed into 2 ml screw cap tubes containing 1 mm silica/zirconia beads in 1 ml PBS. The samples were homogenized in a mini-BeadBeater-8 (BioSpec) for 1 min at full speed twice, placing the tubes on ice in between. Homogenized samples were diluted serially in sterile PBS and plated on Todd–Hewitt agar plates for enumeration. Inhibition of bacterial growth by purified compounds was calculated as a percentage of initial inoculum.
Ethics approval.
Permission to undertake animal experiments was obtained from the animal subject ethics committee of the University of California, San Diego.
Results and Discussion
IMS of the interaction between B. subtilis and S. aureus
To initiate microbial competition, B. subtilis NCBI 3610 was spotted on top of a lawn of meticillin-resistant S. aureus skin isolate ST59-MRSA-IV (Geng et al., 2010). After the two bacteria were allowed to grow, a zone of clearing was observed on the S. aureus lawn, presumably produced by the metabolic output of B. subtilis. To provide insight into the molecular details of this interaction, the sample was subjected to IMS (Caprioli et al., 1997; Schwamborn & Caprioli, 2010; Yang et al., 2009). The imaging data identified the presence of two B. subtilis metabolites – the non-ribosomal peptide synthetase-derived lipopeptide antibiotics surfactin and plipastatin – as being localized around or within the zone of inhibition (Stein, 2005). The IMS spectrum of the S. aureus lawn displayed detectable amounts of delta-toxin (phenol-soluble modulin λ or PSMλ), a membrane-disruptive peptide produced by S. aureus (Otto, 2010) (Fig. 1a). Confirmation of the identity of each observed molecular entity was obtained by tandem mass spectrometry (Supplementary Figs S1–S4, available with the online version of this paper). Although this approach enabled the characterization of their interaction within a traditional experimental set-up and put forth potential bioactive agents against S. aureus, it is not representative of an encounter of the two organisms in nature, as the experiment lacks spatial direction.
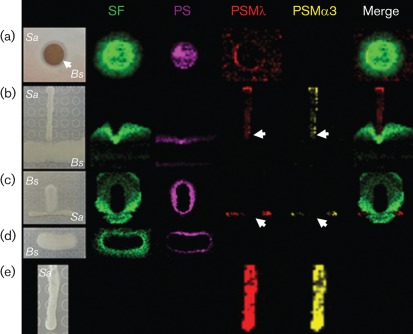
Fig. 1.
IMS of the interaction between B. subtilis (Bs) and S. aureus (Sa). (a) Zone of inhibition; (b) T-shape experiment; (c) converse of (b); (d) B. subtilis alone; (e) S. aureus alone. Ion distributions are represented by colour: surfactin (SF; green), plipastatin (PS; magenta), PSMλ (red) and PSMα3 (yellow). Arrows indicate areas of toxin suppression.
In order to provide a spatial dimension to the interacting organisms, B. subtilis was inoculated with S. aureus in a T-shaped configuration. Once again, the T-shape interaction demonstrated that B. subtilis, although minimally, could inhibit S. aureus at the bacterial interface (Fig. 1b). Mapping the chemistry associated with the observed phenotypes by IMS resulted in an unexpected observation. The IMS data suggested that the amount of surfactin produced by B. subtilis was increased at the bacterial interface (Fig. 1b, c), a directional phenomenon that did not require direct contact between the two bacterial species. The phenotype was not observed in the controls where each organism was cultured individually (Fig. 1d, e), nor was it a result of cell-membrane damage in B. subtilis (Fig. 2a). To ensure that the observed increase in surfactin production by B. subtilis was not a mass spectral artefact, known concentrations of surfactin were spotted on agar adjacent to a co-culture of B. subtilis and S. aureus (Fig. 2b). IMS was repeated on the co-cultured bacteria now including the adjacent standards of surfactin. It was observed that the highest known concentration of surfactin had the highest ion abundance (y-axis) within the mass spectrum. Accordingly, the highest ion abundance of the naturally released surfactin was observed to be at the bacterial interface. Expressing the ion abundance as a colorimetric scale, we were able to make correlations between the colorimetric scaling of the known surfactin concentration and the naturally released surfactin. Using this estimate, the concentration of the naturally released surfactin at the bacterial interface was 140 µg ml−1, a concentration that is in agreement with previous findings (Coutte et al., 2010). A similar observation of directional release of surfactin and consequent S. aureus inhibition was made with a B. subtilis isolate from human skin (Fig. 3a).
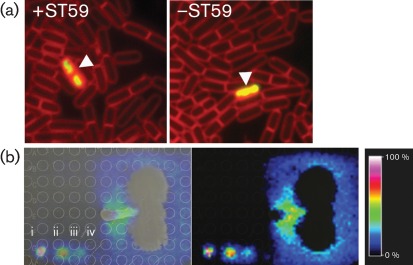
Fig. 2.
(a) Viability assessment of B. subtilis cells by fluorescence microscopy. B. subtilis on the +/−S. aureus ST59 sides was stained with SYTOX Green and FM 4-64. Viable cell counts were 97 % on the −ST59 side and 98 % on the +ST59 side. Arrowheads indicate B. subtilis membrane damage. (b) IMS of spot assays (i, 0.5 µg; ii, 0.05 µg; iii, 0.005 µg; iv, 0.0005 µg surfactin). B. subtilis is shown vertically and S. aureus horizontally. Ion-intensity colour scaling indicates that the highest naturally released surfactin is located at the bacterial interface.
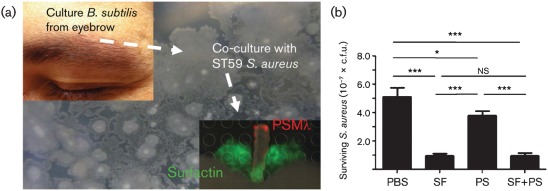
Fig. 3.
(a) B. subtilis isolated from a human eyebrow showed a similar result to wild-type B. subtilis NCBI 3610. The identity of the microbiome-derived B. subtilis isolate was validated by 16S rRNA gene sequencing. (b) Inhibition of S. aureus growth on solid agar. Purified surfactin (SF) and plipastatin (PS) were resuspended in 70 % ethanol alone or in combination and 10 µl solution was pipetted onto agar plates. S. aureus (2×105 c.f.u.) was pipetted onto agar at the location of compounds. Plates were incubated at 37 °C for 4 h. Samples were diluted serially in PBS and plated on Todd–Hewitt agar for enumeration. Inhibition of bacterial growth was calculated as a percentage of initial inoculum. *P<0.05; ***P<0.001; ns, no significant difference.
Surfactin and plipastatin bioactivity against S. aureus
Based on the ring of inhibition and T-shaped IMS data, B. subtilis has the ability to prevent growth of an antibiotic-resistant S. aureus isolate. The inhibitory phenotype may be the cumulative result of the large number of different antimicrobials that B. subtilis is known to produce (Stein, 2005). Based on the ion-localization patterns observed by IMS, the data suggest that surfactin and plipastatin play a significant role in the inhibitory phenotype. To test the inhibitory hypothesis, surfactin and plipastatin were purified individually from B. subtilis and tested for bioactivity. Both purified antibiotics suppressed S. aureus growth on solid agar; surfactin had the more pronounced effect (Fig. 3b). Moreover, the IMS analysis suggested that, beyond growth suppression, B. subtilis influenced the level of production of PSMλ and related PSMs in S. aureus (Fig. 1b, c). In the controls without B. subtilis, robust production of PSMλ and PSMα3 was observed (Fig. 1e). Community-associated meticillin-resistant S. aureus isolates have the ability to infect healthy adults and children with no predisposed risks. The emergence of highly virulent community-associated meticillin-resistant S. aureus has been attributed at least in part to high-level expression of PSMs (Wang et al., 2007). To confirm that the PSM-suppression phenotypes observed by IMS were attributable to surfactin, we spotted purified surfactin on an agar plate next to a colony of growing S. aureus; IMS of this sample confirmed that surfactin did indeed inhibit the production of PSMλ and PSMα3 (Fig. 4a).
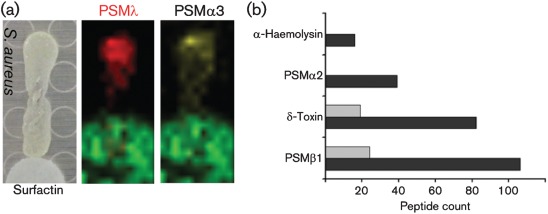
Fig. 4.
(a) S. aureus cultured adjacent to a filter disc containing surfactin showed suppression of PSMλ and PSMα3 when monitored by IMS. (b) Virulence factor suppression: peptide spectral counts of virulence factors identified in the supernatant via nanocapillary LC-MS/MS are shown. Black bars, −surfactin; grey bars, +surfactin.
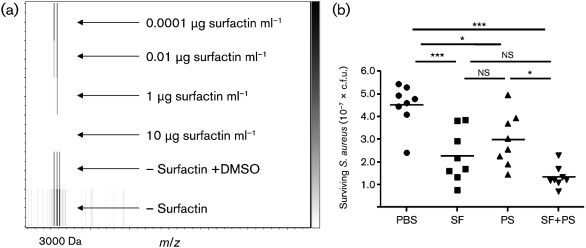
Next, S. aureus was grown in the presence and absence of surfactin, then subjected to LC-MS/MS-based spectral counting by the use of a linear-trap quadruple mass spectrometer (Carvalho et al., 2008; Liu et al., 2004). In the presence of 10 µg surfactin ml−1, a concentration that does not lyse S. aureus (Supplementary Fig. S5, available with the online version of this paper), at least a fivefold suppression of PSMλ, PSMα1, PSMβ2, and α-haemolysin (DeLeo & Chambers, 2009) was observed, even though S. aureus reached similar cell densities and c.f.u. counts to the untreated control (Fig. 4b). As a means to validate surfactin-mediated suppression of PSM, the suppression of PSMλ was monitored by the use of intact cell matrix-assisted laser desorption–ionization/time of flight (MALDI-TOF) analysis. While monitoring the PSMλ mass range (approx. 3000 Da), surfactin was titrated at 10, 1.0, 0.01, and 0.0001 µg ml−1 into inoculations of S. aureus that were allowed to grow to equal cell densities, then assessed by mass spectrometry; a reduction in the amount of PSMλ was observed. A complete loss of PSMλ was observed when surfactin reached 10 µg ml−1 (Fig. 5a).
Fig. 5.
(a) Purified surfactin was spotted next to an S. aureus intact cell. MALDI-TOF analysis of surfactin titration (monitoring detection of the PSMλ ion) is shown; the data show PSMλ suppression and restoration as surfactin is titrated. (b) In vivo growth inhibition of S. aureus on mammalian skin after 4 h. SF, Surfactin; PS, plipastatin; *P<0.05; ***P<0.001; ns, no significant difference.
Murine skin-colonization model
Given the displayed activity of growth inhibition and PSM-suppression activities observed in vitro, we tested whether bioactivity could be reconstituted in vivo in a murine skin-colonization model. The flanks of CD1 mice were shaved and treated individually with surfactin, plipastatin or surfactin/plipastatin, then inoculated with S. aureus. We found that surfactin and plipastatin could restrict growth of S. aureus on mammalian skin. The data showed that, after 4 h, a significant reduction in c.f.u. count was detected with both natural lipopeptides, consistent with their involvement in suppressing S. aureus colonization as indicated by the IMS data (Fig. 5b).
Conclusions
Because B. subtilis possesses a non-pathogenic nature in healthy individuals, strains have been explored as potential therapeutic agents of negligible consumer risk, in scenarios such as periodontitis (Tsubura et al., 2009) or Helicobacter gastritis (Park et al., 2007), or as an alternative to feed antibiotics in agricultural production of poultry (Li et al., 2006), cattle (Jenny et al., 1991), swine (Guo et al., 2006) or seafood (Liu et al., 2009). In the present study, B. subtilis inhibited S. aureus growth and virulence factor expression, at least in part, through elaboration of its natural cyclic lipopeptide antibiotics identified by IMS. We hypothesize that this strategy could be used as a platform to identify bioactive compounds against S. aureus derived from any microbe that possesses a competitive advantage. Rapid advancement in DNA-sequencing technologies is allowing metagenomic approaches to determine the composition of microbial niches. However, companion technologies that can determine the molecular interactions between organisms in a multiplexed manner are largely absent. This work highlights IMS as a hypothesis-generating tool to reveal the molecular interplay that determines the outcome of competitive microbial encounters, potentially yielding novel candidate antimicrobials or optimizing strain selection for therapeutic applications. The surprising finding of directional release of an inhibitory molecule by one bacterial strain toward a neighbouring species, in this case the human pathogen S. aureus, suggests that other secreted factors (e.g. cytotoxins, quorum-sensing factors, nutrient scavenger proteins) that play important roles in health and disease might be examined by IMS technology for more sophisticated modes of spatial deployment.
Supplementary Material
Acknowledgements
Members of the laboratory of P. C. D. appreciate the support of the NIH Haemoglobin and Blood Protein Chemistry Training Program (5T32DK007233-34), NIH grants R01 GM086283 and R01 GM094802. N. M. H. was supported by the National Institutes of Health Training Program in Marine Biotechnology (T32 GM067550) and Ruth L. Kirschstein National Research Service Award (NRSA) from National Institutes of Health grants (5 F31 GM090658-02).
Abbreviations:
- IMS
imaging mass spectrometry
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- PSM
phenol-soluble modulin
Footnotes
Five supplementary figures are available with the online version of this paper.
References
- Ara K., Hama M., Akiba S., Koike K., Okisaka K., Hagura T., Kamiya T., Tomita F. (2006). Foot odor due to microbial metabolism and its control. Can J Microbiol 52, 357–364. 10.1139/w05-130 [DOI] [PubMed] [Google Scholar]
- Caprioli R. M., Farmer T. B., Gile J. (1997). Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem 69, 4751–4760. 10.1021/ac970888i [DOI] [PubMed] [Google Scholar]
- Carvalho P. C., Hewel J., Barbosa V. C., Yates J. R., III (2008). Identifying differences in protein expression levels by spectral counting and feature selection. Genet Mol Res 7, 342–356. 10.4238/vol7-2gmr426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutte F., Leclère V., Béchet M., Guez J. S., Lecouturier D., Chollet-Imbert M., Dhulster P., Jacques P. (2010). Effect of pps disruption and constitutive expression of srfA on surfactin productivity, spreading and antagonistic properties of Bacillus subtilis 168 derivatives. J Appl Microbiol 109, 480–491. [DOI] [PubMed] [Google Scholar]
- DeLeo F. R., Chambers H. F. (2009). Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J Clin Invest 119, 2464–2474. 10.1172/JCI38226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl A. M., Losick R., Kolter R. (2008). Ecology and genomics of Bacillus subtilis. Trends Microbiol 16, 269–275. 10.1016/j.tim.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright M. C., Robinson D. A., Randle G., Feil E. J., Grundmann H., Spratt B. G. (2002). The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci U S A 99, 7687–7692. 10.1073/pnas.122108599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng W., Yang Y., Wu D., Huang G., Wang C., Deng L., Zheng Y., Fu Z., Li C., et al. (2010). Molecular characteristics of community-acquired, methicillin-resistant Staphylococcus aureus isolated from Chinese children. FEMS Immunol Med Microbiol 58, 356–362. [DOI] [PubMed] [Google Scholar]
- Gonzalez D. J., Lee S. W., Hensler M. E., Markley A. L., Dahesh S., Mitchell D. A., Bandeira N., Nizet V., Dixon J. E., Dorrestein P. C. (2010). Clostridiolysin S, a post-translationally modified biotoxin from Clostridium botulinum. J Biol Chem 285, 28220–28228. 10.1074/jbc.M110.118554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice E. A., Kong H. H., Conlan S., Deming C. B., Davis J., Young A. C., Bouffard G. G., Blakesley R. W., Murray P. R., et al. (2009). Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192. 10.1126/science.1171700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Li D., Lu W., Piao X., Chen X. (2006). Screening of Bacillus strains as potential probiotics and subsequent confirmation of the in vivo effectiveness of Bacillus subtilis MA139 in pigs. Antonie van Leeuwenhoek 90, 139–146. 10.1007/s10482-006-9067-9 [DOI] [PubMed] [Google Scholar]
- Hoffman L. R., Déziel E., D’Argenio D. A., Lépine F., Emerson J., McNamara S., Gibson R. L., Ramsey B. W., Miller S. I. (2006). Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 103, 19890–19895. 10.1073/pnas.0606756104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase T., Uehara Y., Shinji H., Tajima A., Seo H., Takada K., Agata T., Mizunoe Y. (2010). Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465, 346–349. 10.1038/nature09074 [DOI] [PubMed] [Google Scholar]
- Jenny B. F., Vandijk H. J., Collins J. A. (1991). Performance and fecal flora of calves fed a Bacillus subtilis concentrate. J Dairy Sci 74, 1968–1973. 10.3168/jds.S0022-0302(91)78364-1 [DOI] [PubMed] [Google Scholar]
- Klevens R. M., Morrison M. A., Nadle J., Petit S., Gershman K., Ray S., Harrison L. H., Lynfield R., Dumyati G., et al. (2007). Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298, 1763–1771. 10.1001/jama.298.15.1763 [DOI] [PubMed] [Google Scholar]
- Li B., Walsh C. T. (2010). Identification of the gene cluster for the dithiolopyrrolone antibiotic holomycin in Streptomyces clavuligerus. Proc Natl Acad Sci U S A 107, 19731–19735. 10.1073/pnas.1014140107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Xu C. L., Ji C., Ma Q., Hao K., Jin Z. Y., Li K. (2006). Effects of a dried Bacillus subtilis culture on egg quality. Poult Sci 85, 364–368. [DOI] [PubMed] [Google Scholar]
- Liu H., Sadygov R. G., Yates J. R., III (2004). A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 76, 4193–4201. 10.1021/ac0498563 [DOI] [PubMed] [Google Scholar]
- Liu C. H., Chiu C. S., Ho P. L., Wang S. W. (2009). Improvement in the growth performance of white shrimp, Litopenaeus vannamei, by a protease-producing probiotic, Bacillus subtilis E20, from natto. J Appl Microbiol 107, 1031–1041. 10.1111/j.1365-2672.2009.04284.x [DOI] [PubMed] [Google Scholar]
- Liu W. T., Yang Y. L., Xu Y., Lamsa A., Haste N. M., Yang J. Y., Ng J., Gonzalez D., Ellermeier C. D., et al. (2010). Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc Natl Acad Sci U S A 107, 16286–16290. 10.1073/pnas.1008368107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. (2010). Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu Rev Microbiol 64, 143–162. 10.1146/annurev.micro.112408.134309 [DOI] [PubMed] [Google Scholar]
- Park S. K., Park D. I., Choi J. S., Kang M. S., Park J. H., Kim H. J., Cho Y. K., Sohn C. I., Jeon W. K., Kim B. I. (2007). The effect of probiotics on Helicobacter pylori eradication. Hepatogastroenterology 54, 2032–2036. [PubMed] [Google Scholar]
- Queck S. Y., Khan B. A., Wang R., Bach T. H., Kretschmer D., Chen L., Kreiswirth B. N., Peschel A., Deleo F. R., Otto M. (2009). Mobile genetic element-encoded cytolysin connects virulence to methicillin resistance in MRSA. PLoS Pathog 5, e1000533. 10.1371/journal.ppat.1000533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson J. R., Fox L. K., Hancock D. D., Gay J. M., Besser T. E. (1994). Ecology of Staphylococcus aureus isolated from various sites on dairy farms. J Dairy Sci 77, 3354–3364. 10.3168/jds.S0022-0302(94)77277-5 [DOI] [PubMed] [Google Scholar]
- Schwamborn K., Caprioli R. M. (2010). Molecular imaging by mass spectrometry – looking beyond classical histology. Nat Rev Cancer 10, 639–646. 10.1038/nrc2917 [DOI] [PubMed] [Google Scholar]
- Stein T. (2005). Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol Microbiol 56, 845–857. 10.1111/j.1365-2958.2005.04587.x [DOI] [PubMed] [Google Scholar]
- Tanner S., Shu H., Frank A., Wang L. C., Zandi E., Mumby M., Pevzner P. A., Bafna V. (2005). InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal Chem 77, 4626–4639. 10.1021/ac050102d [DOI] [PubMed] [Google Scholar]
- Tsubura S., Mizunuma H., Ishikawa S., Oyake I., Okabayashi M., Katoh K., Shibata M., Iizuka T., Toda T., Iizuka T. (2009). The effect of Bacillus subtilis mouth rinsing in patients with periodontitis. Eur J Clin Microbiol Infect Dis 28, 1353–1356. 10.1007/s10096-009-0790-9 [DOI] [PubMed] [Google Scholar]
- Wang R., Braughton K. R., Kretschmer D., Bach T. H., Queck S. Y., Li M., Kennedy A. D., Dorward D. W., Klebanoff S. J., et al. (2007). Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med 13, 1510–1514. 10.1038/nm1656 [DOI] [PubMed] [Google Scholar]
- Yang Y. L., Xu Y., Straight P., Dorrestein P. C. (2009). Translating metabolic exchange with imaging mass spectrometry. Nat Chem Biol 5, 885–887. 10.1038/nchembio.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.