Abstract
Nutrient-limited continuous cultures in chemostats have been used to study microbial cell physiology for over 60 years. Genome instability and genetic heterogeneity are possible uncontrolled factors in continuous cultivation experiments. We investigated these issues by using high-throughput (HT) DNA sequencing to characterize samples from different phases of a glucose-limited accelerostat (A-stat) experiment with Escherichia coli K-12 MG1655 and a duration regularly used in cell physiology studies (20 generations of continuous cultivation). Seven consensus mutations from the reference sequence and five subpopulations characterized by different mutations were detected in the HT-sequenced samples. This genetic heterogeneity was confirmed to result from the stock culture by Sanger sequencing. All the subpopulations in which allele frequencies increased (betA, cspG/cspH, glyA) during the experiment were also present at the end of replicate A-stats, indicating that no new subpopulations emerged during our experiments. The fact that ~31 % of the cells in our initial cultures obtained directly from a culture stock centre were mutants raises concerns that even if cultivations are started from single colonies, there is a significant chance of picking a mutant clone with an altered phenotype. Our results show that current HT DNA sequencing technology allows accurate subpopulation analysis and demonstrates that a glucose-limited E. coli K-12 MG1655 A-stat experiment with a duration of tens of generations is suitable for studying cell physiology and collecting quantitative data for metabolic modelling without interference from new mutations.
Introduction
Nutrient-limited continuous culture has been used to study the cell physiology of micro-organisms for over 60 years (Monod, 1950; Novick & Szilard, 1950a). The main advantage of continuous cultivation methods (e.g. chemostat) over batch cultivation is the fact that in the former it is possible to maintain constant environmental conditions, and therefore to force cells to grow at desired specific growth rates and in strictly defined physiological states. This control allows the acquisition of coherent quantitative steady-state data for metabolic modelling. Accelerostats (A-stats) enable the collection of a vast amount of quantitative data within a large range of specific growth rates in a single experiment (Paalme et al., 1995). An A-stat experiment starts as a chemostat, but after reaching steady state, a smooth change of dilution rate (D) is applied (Fig. 1). A typical change in D is relatively slow (0.01 h−2), which allows cells to adapt to the changing conditions and still maintain a stable physiological state equivalent to steady state: a quasi-steady state. Chemostats (at steady state) and A-stats (at quasi-steady state) have been shown to yield quantitatively comparable results in terms of their main growth characteristics (e.g. biomass yield, substrate consumption and product accumulation rates) in experiments using Escherichia coli K-12 MG1655 (Valgepea et al., 2010) and Lactococcus lactis IL1403 (Lahtvee et al., 2011) over a range of specific growth rates (0.10–0.51 h−1).
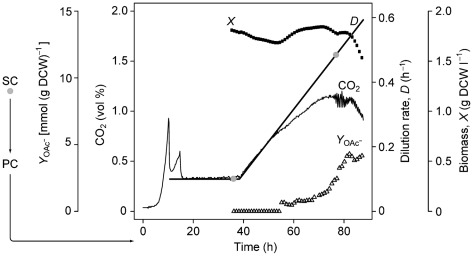
Fig. 1.
Overview of E. coli K-12 MG1655 A-stat experiment in which samples for HT DNA sequencing were taken. Grey dots denote sampling points for HT DNA sequencing: stock culture and two samples at D = 0.11 and 0.48 h−1. The numbers of generations between the sampling points were as follows: stock culture and D = 0.11 h−1, 18 generations (including four generations of continuous culture); D = 0.11 and 0.48 h−1, 16 generations. The glucose concentration remained below the detection limit (120 mg l−1) until the maximal specific growth rate was achieved. SC, stock culture; PC, pre-culture; X, dry cell weight (DCW) per litre; D, dilution rate; YOAc−, acetate production per gram dry cell weight; CO2, carbon dioxide volumetric percentage in the gas outflow. This A-stat experiment is the same as that reported in Fig. 1 of Valgepea et al. (2010).
Quantitative data collected from continuous cultivation methods are often used for metabolic modelling. Because DNA sequences encode the metabolic network of the cell, it is essential to know if and when the genomes of cells are changing during these experiments. In fact, chemostats were initially introduced to study evolution and selection in a constant environment. Fitter clones are known to rapidly arise in some cases (Helling et al., 1987; Novick & Szilard, 1950b), introducing an uncontrolled dynamic factor that reduces the quantitative reliability of steady-state data. Although Egli and colleagues have shown that evolution and selection can be reproducible at the phenotypic level in large (1011 cells) chemostat populations (Wick et al., 2001, 2002), other studies have detected significant genotypic variation among similarly evolved cultures (Notley-McRobb & Ferenci, 1999a, b; Maharjan et al., 2006). Therefore, checking the stability of the genome of the cultured strain and monitoring the genetic homogeneity of the whole population over time in continuous cultures are vital for understanding when genetic heterogeneity and evolution interfere with steady-state physiology measurements of growing cells.
Flaws in stock culture handling are one possible source of genetic heterogeneity in cell physiology studies. Indeed, heterogeneity in stab agar cultures has been reported during long-term storage of E. coli K-12 W3110 at room temperature (Jishage & Ishihama, 1997; Naas et al., 1994, 1995). Considerably fewer mutations are expected when cell cultures are stored in the frozen state, in which no metabolic activity is possible. Nevertheless, genetic differences, including loss of genes and variation in growth phenotype on various carbon sources, have been detected in E. coli K-12 MG1655 strains acquired from different stock centres and laboratories (Soupene et al., 2003). What is more, variation in motility has been observed between E. coli K-12 strain derivatives (including MG1655), whereby increased motility is found to be caused by insertion sequence (IS)-mediated activation of flhD, a regulator of flagellar gene transcription (Barker et al., 2004). Presumably, periodic subculturing contributes to this heterogeneity.
Genetic heterogeneity may also arise during continuous cultivation experiments as the result of spontaneous mutations. Information about which genes are possible targets for beneficial mutations is available from continuous cultivation experiments that are much longer than ‘routine’ cell physiology studies. Long-term evolution of E. coli in glucose-limited chemostats has been studied quite intensively during the last decade (Adams, 2004; Ferenci, 2008). As might be expected, mutations that increase glucose transport capabilities are common in glucose-limited chemostats (Helling et al., 1987; Manch et al., 1999; Wick et al., 2001, 2002). These mutations are observed in prolonged continuous cultivation experiments that exceed the number of generations necessary for cell physiology studies by at least an order of magnitude. Therefore, to study cell physiology properly in continuous culture, or any other cultivation system, it is important to find out how long a genetically uniform culture can be maintained.
Generally, pumping through five working volumes in a chemostat is assumed to be enough to reach steady state (50 h and seven generations at D = 0.1 h−1). The presence of a true steady state at this point has been questioned, based on how rapidly fitter clones may arise (Ferenci, 2008). Indeed, mutations in the stress-induced sigma factor rpoS have been detected in E. coli K-12 BW2952 cultures growing in glucose-limited chemostats at D = 0.1 h−1 after only 24–72 h (three to 10 generations) of cultivation (Notley-McRobb et al., 2003). However, the appearance of rpoS mutations appears to be strain-dependent. None were detected in E. coli K-12 MG1655 after 96 h (14 generations) at D = 0.1 h−1 (King et al., 2004), making it difficult to generalize about how long a genetically uniform culture of E. coli K-12 can be maintained. Moreover, it is plausible that MG1655 acquires mutations other than those in rpoS during glucose-limited continuous cultivation experiments.
Recent advances in DNA sequencing technologies (Mardis, 2008) have made it possible to routinely determine the full genome sequences of micro-organisms and to generate high-coverage datasets for analysing genetic diversity within communities (Barrick & Lenski, 2009). High-throughput (HT) DNA sequencing was used in the present work to examine genome stability and genetic heterogeneity during a short-term continuous cultivation experiment (A-stat, 20 generations of continuous cultivation) of E. coli K-12 MG1655. We found that E. coli K-12 MG1655 is suitable for studying cell physiology using glucose-limited A-stat experiments with a duration of up to 20 generations of continuous cultivation without interference from new, spontaneous mutations. However, we found that substantial genetic heterogeneity and mutational diversity existed in bacterial cultures from stock centres, which underscores the importance of using HT DNA sequencing to verify genome integrity in any experiment.
Methods
Strains and preparation of stock culture.
E. coli K-12 MG1655 [λ− F− rph-1 Fnr+; Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ), DSM no. 18039] was used in all A-stat experiments. The strain ordered from DSMZ was originally obtained from the Coli Genetic Stock Center (CGSC) and has the collection number CGSC 6300. This strain was used for validation of consensus mutations and subpopulations. Additionally, E. coli K-12 MG1655 strain (reference no. C 438-01), obtained from Statens Serum Institute (SSI), was used for comparison of K-12 MG1655 strains from two different stock centres. This strain also originated from CGSC; however, SSI obtained the E. coli K-12 MG1655 strain through a collaborator, and therefore it is possible that this stock is a subculture of the original CGSC sample (Karen A. Krogfelt, personal communication).
The stock culture used to start the A-stat experiment was prepared by inoculating directly from the stab agar culture obtained from DSMZ into Luria–Bertani (LB) medium and cultivating aerobically until the late exponential growth phase (around 10 generations). The cells were then washed, suspended in defined minimal medium containing 1.2 % (v/v) glycerol, divided into aliquots, lyophilized, and stored at −80 °C. A single stock aliquot was used to inoculate each A-stat cultivation experiment.
Medium and cultivation conditions.
A detailed description of the medium, cultivation conditions and growth characteristics in these A-stat experiments has been reported previously (Valgepea et al., 2010). In short, defined minimal medium with 4.5 g α-d-glucose l−1, a temperature of 37 °C, pH 7, an agitation speed of 800 r.p.m. and aerobic conditions (air flow rate 150 ml min−1) were used in all experiments. After the culture had been stabilized under chemostat conditions at D = 0.10 h−1 to achieve steady state, a continuous increase in D with an acceleration rate of 0.01 h−2 was applied.
Genomic DNA extraction.
An aliquot of the lyophilized stock culture (4.2×106 cells) and two whole-population samples (not single colonies) acquired at D = 0.11 h−1 (8.4×109 cells) and 0.48 h−1 (3.0×109 cells) from one A-stat experiment were chosen for DNA extraction and subsequent HT DNA sequencing analysis. Genomic DNA for resequencing and mutation validation was extracted using the RTP Bacteria DNA Mini kit (Invitek) following the manufacturer’s protocol.
Illumina whole-genome resequencing.
Whole-genome resequencing was performed by GATC Biotech using an Illumina Genome Analyzer II instrument. A single-end library was prepared from each genomic DNA sample and sequenced using the manufacturer’s standard protocols. FASTQ read files were generated by Sequencing Control software (version 2.6) with Real-time Analysis (version 1.6.32) and GA Pipeline (version 1.5.1). Raw read data have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (accession no. SRP006176).
Genome sequence data analysis.
Sequencing reads were compared with the E. coli K-12 MG1655 reference genome (accession no. U00096.2) using the breseq analysis pipeline (Barrick et al., 2009; Woods et al., 2011) (version 1.00rc7). Source code for breseq is freely available online (http://barricklab.org/twiki/bin/view/Lab/ToolsBacterialGenomeResequencing). The online documentation describes the methods used to predict consensus point mutations, small indels, large deletions and new sequence junctions. Specific settings for this study are provided in Supplementary Methods.
Mutation validation with Sanger sequencing.
Predicted mutations were validated by performing PCR followed by Sanger sequencing. Sanger sequencing was performed by the Estonian Biocentre using an Applied Biosystems 3730xl DNA analyser and the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems). Subpopulations with base substitutions were confirmed by Sanger sequencing PCR products from mixed population samples (Supplementary Figs S1–S5). The Supplementary Methods fully describe the PCR protocol and subpopulation analysis.
Results and Discussion
Mismatches from the E. coli K-12 MG1655 reference genome
The genome stability and genetic heterogeneity of E. coli K-12 MG1655 routinely used in our laboratory were studied during 20 generations of continuous cultivation in a glucose-limited A-stat experiment. Fig. 1 illustrates the time-course of one A-stat experiment, showing when samples were collected for HT DNA sequencing.
Seven differences from the E. coli K-12 MG1655 reference genome (GenBank accession no. U00096.2) were found at a 100 % frequency in all three sequenced samples (Table 1). Most of these consensus mutations were single nucleotide polymorphisms (SNPs). In addition, two relatively large deletions were detected. First, an IS1 was deleted from the regulatory region of flhDC (DNA-binding transcriptional dual regulator that controls gene expression of flagella genes). Second, a 111 bp deletion from the repetitive sequence region between genes gltP [glutamate and aspartate dicarboxylate/amino acid : cation symporter (DAACS)-family transporter] and yjcO (conserved protein) was found. Mutations that improve glucose uptake, for instance in ptsG of the glucose phosphotransferase system, or the stress-induced sigma factor rpoS, might be expected during continuous cultivation based on results in similar chemostat environments (Kinnersley et al., 2009; Notley-McRobb & Ferenci, 1999a, b; Notley-McRobb et al., 2003; Wick et al., 2001, 2002). However, we did not detect the emergence of either of these mutations.
Table 1. Consensus mutations in all HT-sequenced samples.
| Gene(s) affected | Genome position | Mutation | Annotation | Function(s) of related gene(s) |
| ylbE_1 | 547 694 | A→G | Pseudogene | Predicted protein |
| ylbE_1 | 547 835 | +G | Pseudogene | Predicted protein |
| flhD/uspC | Δ1 976 527–1 977 302 | IS1 deletion | Intergenic | Subunit of flagella regulator (FlhD2C2)/universal stress protein |
| rrlD | 3 422 257–3 422 259 | ATC→CAT | Non-coding | 23S rRNA |
| ppiC/yifO | 3 957 957 | C→T | Intergenic | Peptidyl-prolyl cis-trans isomerase C/conserved protein |
| gltP/yjcO | 4 294 291 | T→C | Intergenic | Glutamate and aspartate DAACS transporter/conserved protein |
| gltP/yjcO | Δ4 294 305–4 294 415 | Δ111 bp | Intergenic | Glutamate and aspartate DAACS transporter/conserved protein |
Detection of mutations in all samples could point to errors in the reference genome sequence as well as to problems of handling of cultures in stock centres. To address the latter possibility, we sequenced these regions in an E. coli K-12 MG1655 strain that was ordered from another stock centre (SSI). The presence of identical mutations in MG1655 strains of different origins would suggest errors in the reference genome, whereas variations in the mutations present would indicate problems in strain handling. We found that six out of seven mutations detected in the MG1655 strain obtained from the DSMZ collection (Table 1) were also present in the SSI strain. In addition, most of the consensus mutations found in the current study have been reported before in HT DNA sequencing studies of E. coli K-12 MG1655 strains acquired from other sources (Conrad et al., 2009; Harris et al., 2009; Lee & Palsson, 2010). Together, these observations suggest that most of the discrepancies are actually sequencing errors in the reference genome. On the other hand, our results also demonstrate that the two E. coli K-12 MG1655 strains investigated were not exactly the same in different stock centres (only the SSI strain had the IS1 insertion in the flhDC regulon). This observation highlights a problem that may arise during strain handling. The population bottleneck caused by picking a single colony before subculturing is particularly prone to fixing a new mutation in the resultant sample.
Search for evolution during the A-stat experiment
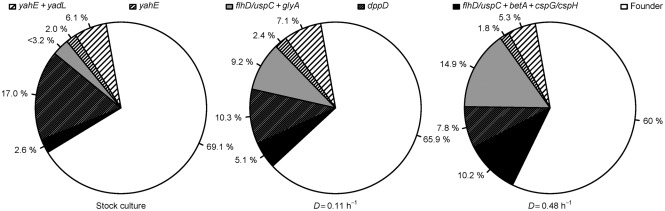
Next, we turned our attention to the dynamics of mutations that appeared to be polymorphic in the population, that is, only present in a subset of individuals. Using the HT DNA sequencing data with 111-fold average genome coverage we predicted mutations present in subpopulations (Methods, Table 2). Only a few of these appeared to be present in more than 5 % of the whole population by the end of the experiment, and all were present in fewer than 20 % of all samples (Supplementary Table S1). A problem in analysing subpopulations with a low frequency using HT DNA sequencing data is that one has to distinguish true subpopulations from false positives caused by sequencing errors with nontrivial biases. Hence, additional validation with Sanger sequencing was performed for high-scoring predictions (Methods, Supplementary Table S1). Three high-scoring subpopulations (betA, cspH/cspG, glyA) in which allele frequencies increased during the experiment were validated from the sample taken at D = 0.48 h−1 (Supplementary Figs S1–S3). Two of the three mutations (betA, cspH/cspG) were also detected in the stock culture (Fig. 2). The only subpopulation that was not detected with HT DNA sequencing from the stock culture, and therefore might have arisen during the experiment, was an SNP in glyA. However, the presence of a glyA population in the stock culture was verified by colony screening (see below), proving that all of these subpopulations were also present in the stock culture at detectable frequencies.
Table 2. Mutations present in subpopulations in the HT-sequenced samples.
Experimentally validated subpopulations are shown. See Supplementary Table S1 for complete information about all high-scoring subpopulations predicted from the HT DNA sequencing data.
| Gene(s) affected | Genome position* | Mutation | Annotation | Function(s) of related gene(s) |
| yahE | 335 361 | T→C | F71F (TTT→TTC) | Predicted protein |
| dppD | 3 701 283 | G→A | L197L (CTG→TTG) | ATP-binding component of the dipeptide ABC transporter |
| cspH/cspG | 1 050 465 | T→A | Intergenic | Stress protein, member of the CspA family/cold-shock protein |
| glyA | 2 683 035 | G→A | H165H (CAC→CAT) | Serine hydroxymethyltransferase |
| betA | 326 446 | C→T | G9D (GGT→GAT) | Choline dehydrogenase |
| yadL | 152 081–152 084 | IS5 insertion | Coding region | Gene of predicted chaperone–usher fimbrial operon |
| flhD/uspC | 1 977 510–1 977 513 | IS5 insertion | Intergenic | Subunit of flagella regulator (FlhD2C2)/universal stress protein |
Positions of IS5 insertions give the target site nucleotides that were duplicated upon insertion of the new IS copy. Both new IS5 copies were inserted in the forward (+) orientation in the genome.
Fig. 2.
Population heterogeneity in the E. coli K-12 MG1655 A-stat experiment. All the subpopulations shown here were verified with Sanger sequencing. In the stock culture, the mutation in glyA was not detected with HT DNA sequencing; therefore, the frequency for this sample was estimated from genome coverage at the position where a glyA mutation was detected in other samples. See Table 2 for details of mutations.
To further confirm that the detected heterogeneity did not arise during the cultivation experiment or result from differences between stock culture aliquots, the occurrence of subpopulations in two additional A-stat experiments (started from separate aliquots of the same stock culture) was investigated. If there was the same (reproducible) subpopulation distribution at the end of A-stat experiments, one could conclude that the mutations discovered were the consequence of selection acting on existing variation, i.e. the set of mutations already present in the stock culture at detectable levels. Indeed, it was found that all the subpopulations with SNPs that had increasing allele frequencies (betA, cspH/cspG, glyA) were reproducibly present in all three replicate A-stat experiments (Supplementary Figs S1–S3). This observation further supports the hypothesis that the above-mentioned subpopulations were already present in the stock culture. Therefore, we conclude that the current 20-generation continuous cultivation duration of E. coli K-12 MG1655 in the glucose-limited A-stat was short enough to avoid the emergence of new subpopulations, making it suitable for studying cell physiology and collecting quantitative data for metabolic modelling without interference from new spontaneous mutations.
Heterogeneity of the stock culture
After confirming that the stock culture was not uniform, we further investigated the predicted subpopulations. The following subpopulations were predicted (frequencies ranging between 6.5 and 17.0 % in stock culture) and validated: dppD, allD, recB, yahE (Supplementary Table S1). It was found that the putative mutations in allD and recB with low overall scores (see Supplementary Table S1 legend for description) were false positives (Supplementary Fig. S6), whereas high-scoring predictions in dppD and yahE were genuine (Supplementary Figs S4–S5). In addition, two IS-related mutations, namely IS5 insertions in the flhD regulon (flhD/uspC) and in yadL, were detected in the HT DNA sequencing data (Supplementary Methods) and verified to have frequencies below 10 % (data not shown).
We next investigated whether any of these mutations existed in combination in the same cells. Four mutations had increasing allele frequencies during the experiment: betA, cspH/cspG, glyA and flhD/uspC. IS elements in the regulatory region of flhD that lead to increased motility are common in E. coli (Barker et al., 2004). Therefore, we screened colonies acquired from the stock culture used in the A-stat experiments for increased motility, as described by Barker et al. (2004). Out of the 42 investigated clones, four turned out to be motile and contained the IS5 insertion described above (data not shown). All the motile colonies were screened for betA, cspH/cspG or glyA mutations. Two clones had the betA and cspH/cspG SNPs, and the other two contained the glyA mutation (data not shown). Next, we hypothesized that mutations in yahE and yadL might also exist in the same subpopulation, as the frequencies of both alleles remained constant during the course of the experiment (Supplementary Table S1). Screening of stock culture clones revealed that all but one clone isolated with the mutation in yahE also contained the IS5 insertion in yadL.
The fact that the mutation in yahE was synonymous made it seem likely that the single yahE mutant clone without yadL harboured additional mutations. Therefore, we tested it for the presence of a yhdJ/yhdU (predicted methyltransferase/predicted membrane protein) mutation, because it was the only predicted mutation with considerable reliability (high overall score) and a change in allele frequency similar to that of yahE in the A-stat experiment (Supplementary Table S1). It turned out that the yhdJ/yhdU mutation was not present in the yahE clone (data not shown). However, given its low frequency in the population, we cannot exclude the possibility that an undetected mutation may be present in this genetic background. After resolving the genetic linkage between the most common mutations in the population, we conclude that roughly 31 % of our stock culture contained at least one mutation relative to the majority genotype.
Conclusions
Our HT DNA sequencing data and detailed validation strategy enabled us to find all the mutations that appeared at appreciable frequencies during a continuous culture experiment. Sequencing of the stock culture gave, first of all, an overview of genetic heterogeneity in the stab agar received from one stock centre. We found that mutant genotypes made up roughly 31 % of our stock culture grown directly from this stab. Since the stock culture was prepared by growth through only about 10 generations in our laboratory, it is unlikely that this heterogeneity was due to new spontaneous mutations. No subpopulation emerging from a single cell could reach a detectable frequency in this short time if growth was the only competitive process. Therefore, we conclude that the mutants had to be already present in the stab agar at relatively high frequencies, due to either growth in the stab or heterogeneity in the original stock culture.
Even if our A-stat experiments had been initiated from single colonies, there would have been a large chance of picking a mutant clone. With three replicate experiments, there would have been a 67 % chance that at least one replicate would have begun with a mutation relative to the majority genotype. This surprising heterogeneity shows the importance of sequencing the genome of a culture at the start of a microbial physiology study, especially if the phenotypic data from the experiment are to be used in comparisons between different research groups or for metabolic modelling.
Taking into account the apparent differences in evolvability between E. coli K-12 MG1655 used in the current study and E. coli K-12 BW2952, where the latter apparently has mutations available in the short term that are much more beneficial in this glucose-limited environment (Notley-McRobb et al., 2003), it seems essential to evaluate the genome stability of every strain or species that is to be used for cell physiology studies in continuous cultures. Our results show that the current HT DNA sequencing technology is suitable and even necessary for accurate subpopulation analysis. We also demonstrate specifically that E. coli K-12 MG1655 has sufficient genome stability to be used in glucose-limited A-stat experiments with a duration of up to 20 generations of continuous cultivation for studying cell physiology, without mutations that occur in the course of adaptive evolution arising during the experiment.
Supplementary Material
Acknowledgements
This research was supported by EU project EU29994, Estonian targeted and science foundation projects SF0140090s08 and G8165, the US National Institutes of Health (R00 GM087550 to J. E. B.), and the US National Science Foundation BEACON Center for the Study of Evolution in Action (DBI-0939454 to J. E. B.).
Abbreviations:
- A-stat
accelerostat
- D
dilution rate
- DAACS, dicarboxylate/amino acid
cation symporter
- HT
high-throughput
- IS
insertion sequence
- SNP
single nucleotide polymorphism
Footnotes
Supplementary Methods, six supplementary figures and two supplementary tables are available with the online version of this paper.
References
- Adams J. (2004). Microbial evolution in laboratory environments. Res Microbiol 155, 311–318. 10.1016/j.resmic.2004.01.013 [DOI] [PubMed] [Google Scholar]
- Barker C. S., Prüss B. M., Matsumura P. (2004). Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon. J Bacteriol 186, 7529–7537. 10.1128/JB.186.22.7529-7537.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick J. E., Lenski R. E. (2009). Genome-wide mutational diversity in an evolving population of Escherichia coli. Cold Spring Harb Symp Quant Biol 74, 119–129. 10.1101/sqb.2009.74.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick J. E., Yu D. S., Yoon S. H., Jeong H., Oh T. K., Schneider D., Lenski R. E., Kim J. F. (2009). Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461, 1243–1247. 10.1038/nature08480 [DOI] [PubMed] [Google Scholar]
- Conrad T. M., Joyce A. R., Applebee M. K., Barrett C. L., Xie B., Gao Y., Palsson B. Ø. (2009). Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol 10, R118. 10.1186/gb-2009-10-10-r118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenci T. (2008). Bacterial physiology, regulation and mutational adaptation in a chemostat environment. Adv Microb Physiol 53, 169–229. 10.1016/S0065-2911(07)53003-1 [DOI] [PubMed] [Google Scholar]
- Harris D. R., Pollock S. V., Wood E. A., Goiffon R. J., Klingele A. J., Cabot E. L., Schackwitz W., Martin J., Eggington J., et al. (2009). Directed evolution of ionizing radiation resistance in Escherichia coli. J Bacteriol 191, 5240–5252. 10.1128/JB.00502-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling R. B., Vargas C. N., Adams J. (1987). Evolution of Escherichia coli during growth in a constant environment. Genetics 116, 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M., Ishihama A. (1997). Variation in RNA polymerase sigma subunit composition within different stocks of Escherichia coli W3110. J Bacteriol 179, 959–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King T., Ishihama A., Kori A., Ferenci T. (2004). A regulatory trade-off as a source of strain variation in the species Escherichia coli. J Bacteriol 186, 5614–5620. 10.1128/JB.186.17.5614-5620.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnersley M. A., Holben W. E., Rosenzweig F. (2009). E unibus plurum: genomic analysis of an experimentally evolved polymorphism in Escherichia coli. PLoS Genet 5, e1000713. 10.1371/journal.pgen.1000713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahtvee P.-J., Adamberg K., Arike L., Nahku R., Aller K., Vilu R. (2011). Multi-omics approach to study the growth efficiency and amino acid metabolism in Lactococcus lactis at various specific growth rates. Microb Cell Fact 10, 12. 10.1186/1475-2859-10-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.-H., Palsson B. Ø. (2010). Adaptive evolution of Escherichia coli K-12 MG1655 during growth on a nonnative carbon source, l-1,2-propanediol. Appl Environ Microbiol 76, 4158–4168. 10.1128/AEM.00373-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharjan R., Seeto S., Notley-McRobb L., Ferenci T. (2006). Clonal adaptive radiation in a constant environment. Science 313, 514–517. 10.1126/science.1129865 [DOI] [PubMed] [Google Scholar]
- Manch K., Notley-McRobb L., Ferenci T. (1999). Mutational adaptation of Escherichia coli to glucose limitation involves distinct evolutionary pathways in aerobic and oxygen-limited environments. Genetics 153, 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis E. R. (2008). Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet 9, 387–402. 10.1146/annurev.genom.9.081307.164359 [DOI] [PubMed] [Google Scholar]
- Monod J. (1950). La technique de culture continue, theorie et applications. Ann Inst Pasteur (Paris) 79, 390–410. [Google Scholar]
- Naas T., Blot M., Fitch W. M., Arber W. (1994). Insertion sequence-related genetic variation in resting Escherichia coli K-12. Genetics 136, 721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas T., Blot M., Fitch W. M., Arber W. (1995). Dynamics of IS-related genetic rearrangements in resting Escherichia coli K-12. Mol Biol Evol 12, 198–207. [DOI] [PubMed] [Google Scholar]
- Notley-McRobb L., Ferenci T. (1999a). Adaptive mgl-regulatory mutations and genetic diversity evolving in glucose-limited Escherichia coli populations. Environ Microbiol 1, 33–43. 10.1046/j.1462-2920.1999.00002.x [DOI] [PubMed] [Google Scholar]
- Notley-McRobb L., Ferenci T. (1999b). The generation of multiple co-existing mal-regulatory mutations through polygenic evolution in glucose-limited populations of Escherichia coli. Environ Microbiol 1, 45–52. 10.1046/j.1462-2920.1999.00003.x [DOI] [PubMed] [Google Scholar]
- Notley-McRobb L., Seeto S., Ferenci T. (2003). The influence of cellular physiology on the initiation of mutational pathways in Escherichia coli populations. Proc Biol Sci 270, 843–848. 10.1098/rspb.2002.2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick A., Szilard L. (1950a). Description of the chemostat. Science 112, 715–716. 10.1126/science.112.2920.715 [DOI] [PubMed] [Google Scholar]
- Novick A., Szilard L. (1950b. ). Experiments with the chemostat on spontaneous mutations of bacteria. Proc Natl Acad Sci U S A 36, 708–719. 10.1073/pnas.36.12.708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paalme T., Kahru A., Elken R., Vanatalu K., Tiismaa K., Vilu R. (1995). The computer-controlled continuous culture of Escherichia coli with smooth change of dilution rate. J Microbiol Methods 24, 145–153. 10.1016/0167-7012(95)00064-X [DOI] [Google Scholar]
- Soupene E., van Heeswijk W. C., Plumbridge J., Stewart V., Bertenthal D., Lee H., Prasad G., Paliy O., Charernnoppakul P., Kustu S. (2003). Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. J Bacteriol 185, 5611–5626. 10.1128/JB.185.18.5611-5626.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valgepea K., Adamberg K., Nahku R., Lahtvee P.-J., Arike L., Vilu R. (2010). Systems biology approach reveals that overflow metabolism of acetate in Escherichia coli is triggered by carbon catabolite repression of acetyl-CoA synthetase. BMC Syst Biol 4, 166. 10.1186/1752-0509-4-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick L. M., Quadroni M., Egli T. (2001). Short- and long-term changes in proteome composition and kinetic properties in a culture of Escherichia coli during transition from glucose-excess to glucose-limited growth conditions in continuous culture and vice versa. Environ Microbiol 3, 588–599. 10.1046/j.1462-2920.2001.00231.x [DOI] [PubMed] [Google Scholar]
- Wick L. M., Weilenmann H., Egli T. (2002). The apparent clock-like evolution of Escherichia coli in glucose-limited chemostats is reproducible at large but not at small population sizes and can be explained with Monod kinetics. Microbiology 148, 2889–2902. [DOI] [PubMed] [Google Scholar]
- Woods R. J., Barrick J. E., Cooper T. F., Shrestha U., Kauth M. R., Lenski R. E. (2011). Second-order selection for evolvability in a large Escherichia coli population. Science 331, 1433–1436. 10.1126/science.1198914 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.