Abstract
Type 1 fimbriae produced by serovars of Salmonella are characterized by their ability to agglutinate guinea pig erythrocytes in the absence of d-mannose but not in its presence. The FimH protein is the adhesin that mediates this reaction; it is distinct from the major fimbrial protei.n (FimA) that composes the fimbrial shaft. Avian-adapted serovars of Salmonella produce non-haemagglutinating fimbriae that have been reported to mediate adherence to avian cells. A single amino acid substitution is present in the FimH adhesin of these strains compared to that of a Typhimurium isolate. Also, previous studies have shown that single nucleotide polymorphisms in two strains of the Typhimurium fimH alter the binding specificity. We therefore investigated the allelic variation of fimH from a range of serotypes (both host-adapted and non-host-adapted) and isolates of Salmonella. Most FimH adhesins mediated the mannose-sensitive haemagglutination of guinea pig erythrocytes, but many did not facilitate adherence to HEp-2 cells. A small number of isolates also produced fimbriae but did not mediate adherence to either cell type. Transformants possessing cloned fimH genes exhibited a number of different substitutions within the predicted amino acid sequence of the FimH polypeptide. No identical FimH amino sequence was found between strains that adhere to erythrocytes and/or HEp-2 cells and those produced by non-adherent strains. FimH-mediated adherence to HEp-2 cells was invariably associated with the ability to form biofilms on mannosylated bovine serum albumin.
Introduction
Salmonella enterica serovar Typhimurium (S. Typhimurium) expresses type 1 fimbriae that enable the bacteria to bind to eukaryotic cells and have been implicated in mediating colonization in vivo of host tissues (Althouse et al., 2003; Boddicker et al., 2002; De Buck et al., 2004; Thorns, 1995). Type 1 fimbriae characteristically exhibit a mannose-sensitive binding phenotype: soluble mannose and mannose derivatives inhibit attachment to mannose glycoconjugates on host cells (Clegg et al., 1985; Kisiela et al., 2005b). The type 1 fimbriae are encoded by the chromosomally borne fim gene cluster (Swenson et al., 1991; Yeh et al., 1995). The fimA gene encodes the major fimbrial subunit whereas the assembly periplasmic chaperone and scaffolding proteins are encoded by fimC and fimD respectively. We have previously demonstrated that three additional genes, fimZ, fimY and fimW, are involved in mediating the regulation of fimbrial expression (Saini et al., 2009; Tinker & Clegg, 2000; Tinker et al., 2001). The fim gene cluster also includes three additional genes. The gene encoding FimI is suggested to be a minor fimbrial protein of unknown function and the location of this gene immediately adjacent to fimA is similar to that of the fimI gene of the Escherichia coli fimbrial gene cluster (Boyd & Hartl, 1999). However, there is limited amino acid sequence relatedness between the two FimI proteins. Similarly, fimF encodes a protein that could be an adaptor polypepetide but also bears little amino acid sequence relatedness to the E. coli FimF adaptor protein. FimH is believed to be the fimbrial adhesin, and like FimI and FimF it shows little primary amino acid sequence similarity to the E. coli FimH adhesin (Madison et al., 1994; Stahlhut et al., 2009b). We have previously shown that FimH mutants of Salmonella are unable to adhere to eukaryotic cells, demonstrating that FimH is critical for the ability of type 1 fimbriae to specifically bind to host cells (Hancox et al., 1997).
The fimH gene in E. coli functions to encode the adhesin of type 1 fimbriae (Stahlhut et al., 2009b). Previous investigations of the E. coli FimH adhesin have demonstrated that allelic variation of fimH results in detectable binding differences in vitro among E. coli isolates (Stahlhut et al., 2009a). Changes in the amino acid sequence of the various FimH proteins were determined and correlated with binding to mannan. Because of the critical role of bacterial adherence in pathogenesis, changes in the FimH protein among strains of E. coli have been proposed to determine the success of specific bacterial populations to colonize different environments, a phenomenon termed pathoadaptive evolution (Chattopadhyay et al., 2009; Sokurenko et al., 1999, 2004; Weissman et al., 2007).
Our group found that two serovar Typhimurium strains, LB5010 and SL1344, have differing abilities to adhere to HEp-2 cells (Boddicker et al., 2002). The amino acid sequences of the fimH gene product of these two strains differ at only four sites. In addition, it has been shown that serovars Gallinarum and Pullorum, which do not produce a functional mannose-sensitive adhesin but do bind to avian cells, also possess a FimH polypeptide. Comparison of the adhesins from serovars Gallinarum and Typhimurium indicated only single amino acid differences in the FimH molecules (Guo et al., 2009; Kisiela et al., 2005a, b).
The genus Salmonella comprises more than 1000 distinct serovars, some of which are host-adapted, such as serovars Typhi and Paratyphi, and some of which exhibit a broader host range, such as Typhimurium. Most isolates of Salmonella produce type 1 fimbriae and exhibit the mannose-sensitive haemagglutinating phenotype associated with the production of these appendages (Duguid et al., 1966; Duguid & Campbell, 1969). However, it is currently unknown how extensive is the allelic variation of fimH among many of these serovars. We therefore examined the fimH alleles from 13 different Salmonella serovars and examined their ability to adhere to HEp-2 cells and form biofilms in vitro on a mannosylated protein. The nucleotide sequences of these alleles were determined to assess whether amino acid differences were associated with these biological functions.
Methods
Strains, culture conditions, and detection of type 1 fimbriae.
A total of 45 strains representing 14 serovars were used in this study; all non-Typhimurium serovars were obtained from the collection described by Duguid and colleagues (Duguid et al., 1966; Duguid & Campbell, 1969). The strains and serovars are shown in Table 1. As previously described, to detect optimal expression of type 1 fimbriae, bacteria were serially incubated without shaking in tubes containing 10 ml LB broth for 48 h periods at 37 °C (Old & Duguid, 1970). The production of type 1 fimbriae was detected using monospecific serum raised against purified fimbriae as described elsewhere (Clegg et al., 1996; Clegg & Hughes, 2002). Strains were considered phenotypically non-fimbriate if they did not react with the serum even after eight 48 h subcultures. Mannose-sensitive haemagglutination was detected using a 3 % suspension of guinea pig erythrocytes (Duguid, 1959; Duguid et al., 1966).
Table 1. Fimbria production and adherence of transformants.
| Source of cloned fim genes* | Reactivity with fimbrial antiserum† | MSHA‡ | No. of bacteria binding to HEp-2 cells |
| S. Typm LB5000 | ++++ | ++++ | >20 |
| S. Typm 4250 | ++++ | ++++ | >20 |
| S. Typm 6704 | ++++ | ++++ | >20 |
| S. Newport | ++++ | ++++ | >20 |
| S. Typm SL1344 | ++++ | ++++ | <5 |
| S. Typm S1098 | ++++ | ++++ | <5 |
| S. Typm DMSO 01 | ++++ | ++++ | <5 |
| S. Typm DMSO 08 | ++ | – | <5 |
| S. Typm DMSO 12 | ++++ | ++++ | <5 |
| S. Typm DMSO 22 | ++++ | ++++ | <5 |
| S. Typm DMSO 38 | ++++ | ++ | <5 |
| S. Typm DMSO 46 | ++++ | ++++ | <5 |
| S. Typm DMSO 49 | ++++ | ++ | <5 |
| S. Typm DMSO 54 | ++++ | ++ | <5 |
| S. Typm DMSO 55 | ++++ | ++++ | <5 |
| S. Typm DMSO 60 | ++++ | ++++ | <5 |
| S. Typm DMSO 67 | ++++ | – | <5 |
| S. Typm DMSO 72 | ++ | ++ | <5 |
| S. Typm DMSO 76 | ++ | – | <5 |
| S. Anatum | ++++ | ++ | <5 |
| S. Mississippi | ++++ | ++++ | <5 |
| S. Pomena | ++++ | ++++ | <5 |
| S. Heidelberg | ++++ | ++++ | <5 |
| S. Pullorum | ++++ | – | <5 |
| S. Agona | ++++ | ++++ | <5 |
| S. Dublin | ++++ | ++++ | <5 |
| S. ParaB 1076 | ++++ | ++++ | <5 |
| S. Infantis | – | – | <5 |
| S. Rubislaw | – | – | <5 |
| S. Adelaide | – | – | <5 |
| S. Alachua | – | – | <5 |
| S. ParaB 511 | – | – | <5 |
| S. ParaB S66 | – | – | <5 |
| S. ParaB 957 | – | – | <5 |
| S. Typm DMSO 13 | – | – | <5 |
| S. Typm DMSO 27 | – | – | <5 |
| S. Typm DMSO 30 | – | – | <5 |
| S. Typm DMSO 34 | – | – | <5 |
| S. Typm DMSO 37 | – | – | <5 |
| S. Typm DMSO 18 | – | – | <5 |
| S. Typm 2231 | – | – | <5 |
| S. Typm SCC100 | – | – | <5 |
| S. Typm 56631 | – | – | <5 |
| S. Typm S7471 | – | – | <5 |
| S. Typm DMSO 33 | – | – | <5 |
S. Typm, S. enterica serovar Typhimurium; S. ParaB, S. enterica serovar Paratyphi B.
++++, agglutination within 60 s; ++, agglutination within 3 min; –, no reactivity.
MSHA, mannose-sensitive haemagglutination (scores as for † footnote).
Cloning of Salmonella fim genes.
DNA primers used in this study are shown in Table 2. The forward primer HF and reverse primer WR were used to clone the intact fimHFZYW genes from Salmonella isolates, by conventional PCR procedures, into the cloning vector pGEM-T Easy (Promega). Genomic DNA was prepared using the DNeasy kit (Qiagen) according to the manufacturer’s instructions and was used as a template for the PCRs. Recombinant plasmids were introduced into the FimH− mutant S. enterica serovar Typhimurium SL1344 H3 following transformation of this strain by standard techniques. All transformants were cultured in LB broth supplemented with ampicillin at a final concentration of 100 µg ml−1. The construction and characterization of the serovar Typhimurium SL1344 H3 FimH− mutant has been described by us elsewhere (Hancox et al., 1997). The construction of recombinant plasmids possessing single fimH and fimF genes was performed using the forward and reverse primers HF and HR, and FF and FR, respectively (Table 2).
Table 2. Oligonucleotides used in this study.
| Oligo | Sequence (5′–3′) |
| HF | CGGAGGATAGCCTGAAGCAGGCGATTA |
| HR | CTTCGCCCAGAGATGAGTTGGCCTGA |
| WR | CACCATGATTCACCTGCCGTGTAGGA |
| FF | TATTCGCGCCTGGCCAATCA |
| FR | GGCCTTCACTCTATCGTTGAGCT |
| ZF | CGCGGATGCGACCTTCCTGATCAATTA |
| ZR | CTTTAAGGTGTCTGCACG |
Nucleotide sequencing of all fim genes was performed by the University of Iowa DNA sequencing facility. Overlapping fragments were sequenced as well as both strands of DNA to confirm all nucleotide sequences reported below.
Adherence to HEp-2 cells.
Adherence assays were performed as previously reported by our group (Boddicker et al., 2002). All assays were performed in triplicate and the means of three independent experiments are reported. All statistical analyses were compared by Student’s t-test for unpaired samples.
Biofilm formation.
The ability of strains to form biofilms on mannosylated bovine serum albumin (ManBSA; Sigma) was determined using a modification of the assay described by Merritt et al. (2005). The wells of 24-well microtitre plates were coated with 1 ml ManBSA (200 µg ml−1 in carbonate buffer) for 18 h at 4 °C. After removal and washing with PBS, 1 ml LB broth was added to each well and inoculated with 1×108 bacteria. Following overnight incubation with shaking at 37 °C the ability of bacteria to form a biofilm on ManBSA was determined using crystal violet as described elsewhere (Merritt et al., 2005). All microtitre plates had three wells coated with ManBSA and incubated with sterile medium. Biofilm assays are reported as A595 values following subtraction of the readings from these control wells. All assays were performed in duplicate and the mean of two independent experiments was calculated.
Some strains, transformed with a plasmid encoding GFP, were examined for their ability to form a biofilm on a monolayer of HEp-2 cells in once-flow-through chambers coated with these cells. This procedure has been described by us in detail elsewhere (Boddicker et al., 2002). Briefly, HEp-2 cells were cultured as monolayers on the surface of glass microscope slides treated to facilitate growth of the cells. All biofilms were examined by confocal microscopy after 24 h incubation and biofilm production was analysed by compilation of a z-series of sections.
Results
Production of type 1 fimbriae by transformants
The ability of transformants of the FimH− mutant, possessing cloned fimH alleles, to produce surface-associated fimbriae is summarized in Table 1. The production of surface-assembled fimbriae was detected using specific antiserum, and the mannose-sensitive adhesin was detected using guinea pig erythrocytes. Of the 45 transformants tested, 27 produced type 1 fimbriae whereas the mutant possessing the cloning vector alone was consistently negative. The presence of fimbriae on the surfaces of the positive transformants also correlated with the ability of the wild-type strains from which the genes were cloned to produce these appendages.
Transformants possessing the fim genes from 18 isolates were non-fimbriate even after repeated subculture in static liquid medium. The parental serovars and strains from which these genes were cloned are also phenotypically non-fimbriate. None of these strains were observed to mediate haemagglutination of guinea pig erythrocytes. As previously reported (Guo et al., 2009; Kisiela et al., 2005b, 2006; Wilson et al., 2000) the cloned fim genes from serovar Pullorum facilitated the production of non-haemagglutinating fimbriae on the surface of transformants. Similarly, the wild-type Pullorum isolate produces fimbriae that react with the serum but do not mediate mannose-sensitive haemagglutination. Also, three serovar Typhimurium strains, DMSO 08, 67 and 76, were found to be fimbriate but did not exhibit any haemagglutinating activity (Table 1).
Adherence to HEp-2 cells
None of the phenotypically non-fimbriate transformants were able to adhere in vitro to cultured HEp-2 cells (Table 1). Also, the wild-type strains from which these genes were cloned did not exhibit any binding activity to the cells. Only four transformants of the serovar Typhimrium FimH− mutant demonstrated an ability to bind to HEp-2 cells (Table 1); one of these was the LB5000 strain that we have previously described as binding to these cells (Boddicker et al., 2002). These four transformants represented two serovars of Salmonella: Typhimurium (three transformants) and Newport (one transformant). The wild-type strains from which the fim genes were cloned also showed binding to HEp-2 cells.
A total of 41 transformants exhibited poor (<5 bacteria per cell) binding to the HEp-2 cells. Nineteen of these did not produce demonstrable fimbrial appendages on their surfaces and all the wild-type strains from which the fim genes were cloned are also phenotypically non-fimbriate. Twenty-three transformants that were shown to be strongly fimbriate as evidenced by their reactivity with fimbria-specific antiserum also exhibited poor or no binding to the HEp-2 cells. Nineteen of these transformants were shown to produce a mannose-sensitive adhesin as indicated by their ability to agglutinate guinea pig erythrocytes. Binding of representative isolates to HEp-2 cells is shown in Fig. 1.
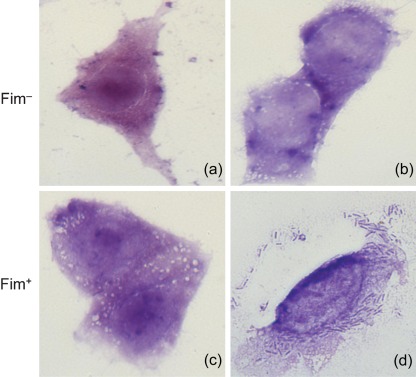
Fig. 1.
Adherence of Salmonella strains to HEp-2 cells in vitro. (a) S. Typhimurium SL1344 H3 (Fim−). (b) S. Typhimurium SL1344 H3 transformed with empty cloning vector pGEM (Fim−). (c) S. Typhimurium SL1344 H3 transformed with the fimH gene cloned from S. Typhimurium DMSO 01 (Fim+). (d) S. Typhimurium SL1344 H3 transformed with the fimH gene from S. Typhimurium 4250 (Fim+).
Biofilm formation on ManBSA and HEp-2 cells
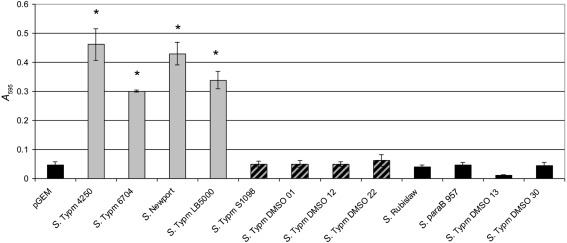
The ability of type 1 fimbriae to facilitate biofilm formation in vitro on ManBSA was determined; the results are summarized in Fig. 2. The four transformants that exhibited significant binding to the HEp-2 cells were also the only four isolates that grew as biofilms. All the remaining transformants did not grow on the ManBSA regardless of their ability to produce fimbriae. All transformants grew equally well when cultured as planktonic bacteria in shaking broth cultures.
Fig. 2.
Biofilm formation by representative strains on ManBSA. The fimbrial phenotype of the strains as determined by seroreactivity is as indicated by the shading of the columns: Grey, Fim+ and binds HEp-2 cells; striped, Fim+ and does not bind HEp-2 cells; black, Fim− and does not bind HEp-2 cells. S. Typhimurium SL1344 H3 transformed with the empty cloning vector is designated pGEM and is Fim−. *, P <0.00005.
A small number of transformants were also examined for their ability to form biofilms on HEp-2 cells in biofilm chambers (Fig. 3). As for the assay using ManBSA, the only strains that formed a mature biofilm after 24 h incubation were those that bound to the HEp-2 cells in the binding assay.
Fig. 3.
Biofilm formation by two representative strains on cultured HEp-2 cells in flow-through chambers after 24 h incubation at 37 °C. (a) S. Typhimurium SL1344 H3 transformed with the fimH gene from S. Typhimurium 6704. (Fim+ MSHA+ and binds to HEp-2 cells). (b) S. Typhimurium SL1344 H3 transformed with the fimH gene from S. Anatum (Fim+ MSHA+ but does not bind to HEp-2 cells).
Amino acid sequences of FimH
The predicted amino acid sequences derived from cloning the fimH alleles from 45 strains of Salmonella representing 14 serovars were determined and are shown in Table 3. For all serovars, except Paratyphi B, intact open reading frames encoding a product of 332 amino acids were found, with no strains encoding truncated gene products. The fimH gene product of all four strains of Paratyphi B possesses an additional asparagine residue at position 289 that is not present in other strains.
Table 3. Salmonella serovar FimH variant.
The most commonly found amino acids at variable locations in the primary sequences of FimH from the majority of serovars are indicated by the term ‘Common’. Specific substitutions in various serotypes and strains at these sites are as indicated.
| No. of changes from common | Serovar | |||||||||||||||||||||||||||||
| Location: | 1 | 2 | 9 | 12 | 13 | 20 | 32 | 46 | 48 | 61 | 63 | 66 | 70 | 71 | 74 | 78 | 80 | 84 | 89 | 94 | 101 | 111 | 117 | 118 | 126 | 127 | 128 | 131 | 137 | |
| Common: | M | K | L | T | A | A | A | G | N | G | V | N | P | A | T | T | R | S | Q | N | N | T | V | F | R | M | G | S | M | |
| 6 | S. Typm LB5000 | A | L | Y | K | |||||||||||||||||||||||||
| 5 | S. Typm 4250 | A | L | Y | K | |||||||||||||||||||||||||
| 6 | S. Typm 6704 | A | L | Y | K | |||||||||||||||||||||||||
| 2 | S. Newport | R | ||||||||||||||||||||||||||||
| 0 | S. Typm SL 1344 | A | ||||||||||||||||||||||||||||
| 6 | S. Typm S1098 | S | L | Y | K | |||||||||||||||||||||||||
| 3 | S. Typm DMSO 01 | |||||||||||||||||||||||||||||
| 5 | S. Typm DMSO 08 | L | Y | K | ||||||||||||||||||||||||||
| 4 | S. Typm DMSO 12 | R | ||||||||||||||||||||||||||||
| 1 | S. Typm DMSO 22 | R | ||||||||||||||||||||||||||||
| 3 | S. Typm DMSO 38 | L | R | |||||||||||||||||||||||||||
| 2 | S. Typm DMSO 46 | K | ||||||||||||||||||||||||||||
| 4 | S. Typm DMSO 49 | S | L | K | ||||||||||||||||||||||||||
| 0 | S. Typm DMSO 54 | |||||||||||||||||||||||||||||
| 4 | S. Typm DMSO 55 | R | ||||||||||||||||||||||||||||
| 3 | S. Typm DMSO 60 | P | R | |||||||||||||||||||||||||||
| 2 | S. Typm DMSO 67 | |||||||||||||||||||||||||||||
| 3 | S. Typm DMSO 72 | R | K | |||||||||||||||||||||||||||
| 5 | S. Typm DMSO 76 | T | R | K | ||||||||||||||||||||||||||
| 5 | S. Anatum | E | M | R | ||||||||||||||||||||||||||
| 5 | S. Mississippi | L | Y | K | ||||||||||||||||||||||||||
| 3 | S. Pomena | V | I | R | ||||||||||||||||||||||||||
| 4 | S. Heidelberg | L | R | |||||||||||||||||||||||||||
| 1 | S. Pullorum | I | ||||||||||||||||||||||||||||
| 1 | S. Agona | V | ||||||||||||||||||||||||||||
| 3 | S. Dublin | V | K | |||||||||||||||||||||||||||
| 3 | S. ParaB 1076 | K | ||||||||||||||||||||||||||||
| 4 | S. Infantis | L | P | K | ||||||||||||||||||||||||||
| 1 | S. Rubislaw | P | ||||||||||||||||||||||||||||
| 3 | S. Adelaide | |||||||||||||||||||||||||||||
| 4 | S. Alachua | S | R | |||||||||||||||||||||||||||
| 6 | S. ParaB 511 | S | S | I | ||||||||||||||||||||||||||
| 3 | S. ParaB S66 | I | ||||||||||||||||||||||||||||
| 3 | S. ParaB 957 | L | ||||||||||||||||||||||||||||
| 4 | S. Typm DMSO 13 | E | M | R | ||||||||||||||||||||||||||
| 2 | S. Typm DMSO 27 | D | ||||||||||||||||||||||||||||
| 3 | S. Typm DMSO 30 | S | I | |||||||||||||||||||||||||||
| 2 | S. Typm DMSO 34 | |||||||||||||||||||||||||||||
| 2 | S. Typm DMSO 37 | V | I | |||||||||||||||||||||||||||
| 6 | S. Typm DMSO 18 | E | M | R | ||||||||||||||||||||||||||
| 4 | S. Typm 2231 | L | Y | K | ||||||||||||||||||||||||||
| 7 | S. Typm SCC100 | A | R | L | Y | K | ||||||||||||||||||||||||
| 4 | S. Typm 56631 | R | L | Y | K | |||||||||||||||||||||||||
| 4 | S. Typm S7471 | L | Y | K | ||||||||||||||||||||||||||
| 5 | S. Typm DMSO 33 | V | R | K |
None of the predicted amino acid sequences of the FimH polypetides encoded by the 26 cloned fimH genes that enabled appendages to be assembled on the surface of the host were identical. However, strains DMSO 54 and SL1344 only differed at a single position (position 12: T and A, respectively) that is predicted to be within the signal peptide region of FimH and therefore would not be present in the assembled adhesins. Since S. Typhimurium SL1344 has been extensively used to investigate the virulence of this serovar, all of the FimH sequences reported in this study were compared to this strain. Compared to the FimH sequence of the gene product cloned from serovar Typhimurium strain SL1344 four strains, representing the serovars Agona, Pullorum, Typhimurium and Newport exhibited a single amino acid substitution. Two different strains of serovar Typhimurium possessed two substitutions whereas six strains (four Typhimurium, one Paratyphi B, and one Pomona) had three changes in FimH sequences. Four changes were found in four strains (three Typhimurium and one Heidelberg), and five changes in six strains (four Typhimurium, one Anatum and one Mississippi). Three strains (two Typhimurium and one Paratyphi B) had six changes in their FimH sequences. The sequences of all these serovars are shown in Table 3.
In strains with no demonstrable production of surface-associated fimbriae, intact fimH determinants were also found. In no case, however, was an identical sequence found in the FimH protein of a phenotypically fimbriate transformant compared to a non-fimbriate strain. One strain representing serovar Rubislaw differed in only a single amino acid change within the region of the assembled adhesin compared to the FimH of S. Typhimurium SL1344. A total of four strains all belonging to serovar Typhimurium possessed two differences in amino sequence compared to SL1344 but in each case the differences were not identical to each other. Twelve strains representing six distinct serovars exhibited three or more changes in their FimH sequences when compared to SL1344. Three changes were found in four strains, four changes in six strains, five changes in one strain, and six changes in two strains. The greatest number of changes was seven, observed in one strain (Typhimurium). These results are summarized in Table 3.
Amino acid sequences of FimF
The plasmids used to transform the FimH mutant of strain SL1344 in these studies also possessed the fimF gene. Although the precise function of FimF in Salmonella type 1 fimbrial production is unknown the location of fimF in the fim gene cluster may suggest that it could be an adaptor protein connecting FimH and FimA in the appendage. Genes encoding two adaptor proteins are located in similar regions of the E. coli fim gene cluster whereas only one is found in the Salmonella system. Therefore, we sequenced the fimF genes from representative phenotypes of some of the serovars described herein. Sequences were determined for three transformants that assembled surface-associated fimbriae and formed good biofilms on HEp-2 cells, one transformant that was fimbriate but did not bind to HEp-2 cells, and two transformants that were phenotypically non-fimbriate. In all but one case, the predicted amino acid sequences of the FimF proteins were found to be identical. The exception was the FimF protein from Typhimurium strain 6704, which differed by a T142I substitution. However, stains that exhibited fimbriate and non-fimbriate phenotypes possessed identical FimF polypeptides.
Discussion
The ability of Salmonella type 1 fimbriae to mediate adherence to the cells of their hosts has been investigated by many groups (Bäumler et al., 1996; Duguid et al., 1976; Misselwitz et al., 2011; van Asten & van Dijk, 2005). In addition, previous studies have indicated that the FimH adhesin protein of these fimbriae is necessary for binding to guinea pig erythrocytes in a mannose-sensitive manner and is responsible for the defining phenotype of this class of fimbriae. Also, it has been demonstrated that the non-haemagglutinating fimbriae, the so-called type 2 fimbriae, produced by the avian-adapted strains Pullorum and Gallinarum differ by one amino acid substitution in their FimH protein compared to the adhesin produced by a strain of Typhimurium that agglutinated erythrocytes (Guo et al., 2009; Kisiela et al., 2006). However, the FimH adhesin of the avian-adapted pathogen does function as an adhesin since it has been shown to mediate binding to chicken leukocytes (Guo et al., 2009). In addition, Kisiela et al. (2006) demonstrated that the mannose-binding activity of the FimH adhesin could be restored by mutation of the single isoleucine in the avian FimH adhesin to a threonine in the Typhimurium FimH strain. Thus a single amino acid change in the FimH adhesin can result in distinct differences in type 1 fimbria-mediated adherence to eukaryotic cells. In addition, we have previously shown that although two strains of S. Typhimurium can exhibit the canonical mannose-sensitive haemagglutination of guinea pig erythrocytes due to production of type 1 fimbriae, the fimbria-mediated binding to HEp-2 cells differs between the two strains (Boddicker et al., 2002). This difference in binding was shown to be due to a small number of changes in the amino acid sequences of the FimH adhesins produced by the strains. Therefore, we have examined the allelic variation in the fimH genes of numerous serovars of Salmonella and also distinct strains of serovar Typhimurium.
All of the strains examined by us possessed a complete fimH gene encoding a polypeptide that was either 332 or 333 amino acids in length. We decided to use the sequence of the FimH adhesin of S. Typhimurium strain SL1344 for comparison with all the other strains examined in this study since this is one of the sequences that is widely available from the online databases and SL1344 has been used extensively to investigate Salmonella pathogenesis. Only one of the isolates encoded a FimH polypeptide that was identical to the processed amino acid sequence of strain SL1344. However, the maximum number of differences observed among all the strains examined by us was seven. This would suggest that only a limited number of substitutions within the FimH polypeptide have been selected during the molecular evolution of the fim gene cluster among Salmonella strains.
The ability of different fimH alleles to mediate fimbria production, adherence to HEp-2 cells and biofilm formation was determined in a common non-fimbriate Typhimurium background host to eliminate the possibility of non-fimbrial and uncharacterized genes from each serovar affecting these phenotypes. In these assays we also used recombinant plasmids that encoded the fimbrial regulators FimZ, Y and W. S. Typhimurium transformants that carry plasmids with these genes produce large amounts of surface-assembled fimbriae (Tinker et al., 2001; Yeh et al., 2002). Consequently, any observed differences in phenotype of fimbriate transformants are not likely due to significant differences in the amount of fimbriae produced by the bacteria. Indeed, most transformants were found to be strongly fimbriate as determined by their reactivity with fimbria-specific immune serum.
Transformation of the FimH− mutant with fimH-bearing recombinant plasmids from some serovars of Salmonella did not enable the bacteria to produce detectable levels of type 1 fimbriae. Interestingly, all of these transformants possessed genes cloned from parental strains that were also phenotypically non-fimbriate. As detected by RT-PCR (data not shown), the fimH genes from these strains were expressed in transformants and therefore the inability to produce fimbriae is not likely due to the absence of FimH. However, the inability to assemble type 1 fimbriae by these strains may be a property of their FimH proteins. None of these strains possesses a FimH polypeptide with an identical amino acid sequence to any strains that assemble fimbriae on their surfaces. Also, the inability to express type 1 fimbriae was not associated with any one specific FimH amino acid sequence, and the adhesins from these strains showed a range, from one to seven, in the number of substitutions compared to the FimH of strain SL1344. In addition, the substitutions within the FimH molecules from these strains were found throughout FimH among the isolates examined and were not localized to any one specific region. Since the plasmids we used in this study also possess the fimF gene, encoding a protein that could act as an adaptor for fimbrial assembly, it is possible that the non-fimbriate strains possess a non-functional FimF protein. We sequenced the fimF genes from both fimbriate and non-fimbriate strains and found no correlation between the FimH amino acid sequence and fimbriation. Representative strains from both groups possessed identical FimF polypetides. Similarly, the sequences of the regulatory proteins (FimZ and Y) from a small number of strains from both groups were also identical (data not shown) and no association between the fimbrial phenotype and this property could be found. The reason why these strains, and transformants possessing fim genes from these strains, are non-fimbriate is unclear but must reflect the presence of specific amino acids in the FimH adhesins of these strains which prevent assembly of appendages. Currently, we are investigating this by constructing site-directed mutations of specific locations within these fimH genes.
The presence of fimbriae on the surface of bacteria correlates with the ability to mediate mannose-sensitive agglutination of guinea pig erythrocytes, and these cells are routinely used to detect type 1 fimbriae (Duguid, 1959; Duguid & Campbell, 1969; Old & Duguid, 1970; Old & Adegbola, 1982). The notable exception to this is the previously characterized Fim adhesin produced by the avian-adapted serovars Pullorum and Gallinarum. Prior investigations have demonstrated that a single substitution of an isoleucine for a threonine at position 78 in these strains results in the production of a non-haemagglutinating adhesin. Three strains of serovar Typhimurium in our studies also produced fimbriae that did not mediate haemagglutination. These fimbriae would be termed type 2 fimbriae using the classification originally developed by Duguid and colleagues (Duguid et al., 1966; Old & Payne, 1971) but they do not possess the same substitution as the Pullorum/Gallinarum fimbriae. Whether these fimbriae function as adhesins to any other cell types remains to be determined but type 2 fimbriae have occasionally been reported in strains of serovar Typhimurium (Old & Payne, 1971).
In haemagglutinating strains the presence of fimbriae is not frequently associated with adherence to other cell types since we found that many strongly fimbriate strains adhere poorly or not at all to HEp-2 cells. When adherence did occur it was associated with the ability of bacteria to form biofilms. The ability to adhere to HEp-2 cells was not associated with a single fimH allele, and in three of four adherent strains there were five or more amino acid substitutions in the FimH adhesin of these strains compared to strain LB5000, which we have previously described as adhering to HEp-2 cells (Boddicker et al., 2002). The results of the adherence assay are consistent with our previous report using two Typhimurium strains that also exhibited differential binding to HEp-2 cells. It is likely that the FimH proteins from haemagglutinating strains that did not bind to HEp-2 cells do represent functional adhesins and are also likely to mediate binding to specific host cells during the infective process. One possibility is that the allelic variants have evolved to optimize binding to various sites and tissues during infection. For example all four strains of the human-adapted serovar Paratyphi B that we examined, which were isolated from different outbreaks, possess a unique asparagine at position 289 in the FimH polypeptide. This amino acid is absent from all the other adhesins and increases the length of the Paratyphi B FimH polypetide by a single residue compared to all other serovars. However, this amino acid is not predicted to be present in the FimH adhesins of two human-adapted strains (serovars Typhi and Paratyphi B) for which sequences are currently available online.
Table 3. cont.
| No. of changes from common | Serovar | ||||||||||||||||||||||||
| Location: | 158 | 177 | 190 | 196 | 197 | 224 | 234 | 235 | 248 | 249 | 251 | 265 | 266 | 267 | 278 | 280 | 285 | 289 | 291 | 300 | 310 | 317 | 321 | 333 | |
| Common: | N | G | S | P | Q | A | Q | T | Q | A | L | M | V | S | N | N | T | NA | S | N | W | N | A | V | |
| 6 | S. Typm LB5000 | S | I | ||||||||||||||||||||||
| 5 | S. Typm 4250 | I | |||||||||||||||||||||||
| 6 | S. Typm 6704 | I | A | ||||||||||||||||||||||
| 2 | S. Newport | V | |||||||||||||||||||||||
| 0 | S. Typm SL 1344 | ||||||||||||||||||||||||
| 6 | S. Typm S1098 | S | S | ||||||||||||||||||||||
| 3 | S. Typm DMSO 01 | H | H | I | |||||||||||||||||||||
| 5 | S. Typm DMSO 08 | S | A | ||||||||||||||||||||||
| 4 | S. Typm DMSO 12 | H | H | V | |||||||||||||||||||||
| 1 | S. Typm DMSO 22 | ||||||||||||||||||||||||
| 3 | S. Typm DMSO 38 | V | |||||||||||||||||||||||
| 2 | S. Typm DMSO 46 | H | |||||||||||||||||||||||
| 4 | S. Typm DMSO 49 | V | |||||||||||||||||||||||
| 0 | S. Typm DMSO 54 | ||||||||||||||||||||||||
| 4 | S. Typm DMSO 55 | H | A | V | |||||||||||||||||||||
| 3 | S. Typm DMSO 60 | P | |||||||||||||||||||||||
| 2 | S. Typm DMSO 67 | H | F | ||||||||||||||||||||||
| 3 | S. Typm DMSO 72 | H | |||||||||||||||||||||||
| 5 | S. Typm DMSO 76 | V | A | D | |||||||||||||||||||||
| 5 | S. Anatum | H | I | ||||||||||||||||||||||
| 5 | S. Mississippi | A | S | ||||||||||||||||||||||
| 3 | S. Pomena | ||||||||||||||||||||||||
| 4 | S. Heidelberg | H | V | ||||||||||||||||||||||
| 1 | S. Pullorum | ||||||||||||||||||||||||
| 1 | S. Agona | ||||||||||||||||||||||||
| 3 | S. Dublin | V | |||||||||||||||||||||||
| 3 | S. ParaB 1076 | N | I | ||||||||||||||||||||||
| 4 | S. Infantis | V | |||||||||||||||||||||||
| 1 | S. Rubislaw | S | |||||||||||||||||||||||
| 3 | S. Adelaide | I | S | ||||||||||||||||||||||
| 4 | S. Alachua | R | V | ||||||||||||||||||||||
| 6 | S. ParaB 511 | G | N | I | |||||||||||||||||||||
| 3 | S. ParaB S66 | N | I | ||||||||||||||||||||||
| 3 | S. ParaB 957 | N | I | ||||||||||||||||||||||
| 4 | S. Typm DMSO 13 | I | |||||||||||||||||||||||
| 2 | S. Typm DMSO 27 | I | |||||||||||||||||||||||
| 3 | S. Typm DMSO 30 | I | |||||||||||||||||||||||
| 2 | S. Typm DMSO 34 | H | H | ||||||||||||||||||||||
| 2 | S. Typm DMSO 37 | ||||||||||||||||||||||||
| 6 | S. Typm DMSO 18 | H | I | ||||||||||||||||||||||
| 4 | S. Typm 2231 | I | |||||||||||||||||||||||
| 7 | S. Typm SCC100 | H | H | I | |||||||||||||||||||||
| 4 | S. Typm 56631 | I | |||||||||||||||||||||||
| 4 | S. Typm S7471 | I | |||||||||||||||||||||||
| 5 | S. Typm DMSO 33 | G | V |
As indicated above, all type 1 fimbriate strains mediate agglutination of guinea pig erythrocytes and therefore this cell type must present a mannose-containing glycoconjugate that is recognized by the fimbrial adhesin. The identical receptor must be absent or inaccessible on HEp-2 cells, which presumably produce a receptor recognized by only a few of the adhesins. FimH-mediated binding to receptor molecules involves catch-bond interactions and our results indicate that the nature of this bond will vary between FimH adhesins and target cells. Interestingly the binding to, and subsequent biofilm formation on, both ManBSA and HEp-2 cells was demonstrated by the same fimH alleles. The presentation of the mannose receptor on this molecule and cell type must be recognized by these adhesins but not by others.
In summary, the Salmonella fimH gene is represented by several allelic variants due to single nucleotide polypmorphisms in this gene. Based upon the predicted amino acid sequences of these variants there is no common sequence restricted to haemagglutinating, adherent or biofilm-forming strains. Also, the sequence of a FimH protein can influence the ability of bacteria to assemble type 1 fimbriae since the difference between several non-fimbriate and fimbriate transformants can only be attributable to differences in the primary amino acid sequence of their adhesins. An examination of the predicted amino acid sequences of the major fimbrial subunits (FimA) from a wide range of Salmonella serovars has not been reported so it is not known if this degree of variation also occurs in this structural component of the type 1 fimbriae. There have been reports that in other enterobacteria both the fimbrial shaft and the adhesin influence the binding properties of type 1 fimbriae (Madison et al., 1994). Consequently the interaction of these two proteins could influence receptor-binding specificity. However, in our studies a common FimA subunit protein makes up the shaft of the fimbrial transformants. Also, based upon our results the additional structural protein, FimF, which is proposed to be an adaptor protein in the Salmonella fimbriae, does not demonstrate the variability observed in FimH. Therefore, Salmonella FimH adhesin variability plays a major role in binding to target molecules.
Acknowledgements
This work was supported the by National Institutes of Health, Bethesda, MD, USA, grants RO1 GM084318 (E. V. S. and S. C.) and RO1 AI0746993 (S. C.).
Abbreviations:
- ManBSA
mannosylated bovine serum albumin
References
- Althouse C., Patterson S., Fedorka-Cray P., Isaacson R. E. (2003). Type 1 fimbriae of Salmonella enterica serovar Typhimurium bind to enterocytes and contribute to colonization of swine in vivo. Infect Immun 71, 6446–6452. 10.1128/IAI.71.11.6446-6452.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumler A. J., Tsolis R. M., Heffron F. (1996). Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect Immun 64, 1862–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddicker J. D., Ledeboer N. A., Jagnow J., Jones B. D., Clegg S. (2002). Differential binding to and biofilm formation on Hep-2 cells by Salmonella enterica serovar Typhimurium is dependent upon allelic variation in the fimH gene of the fim gene cluster. Mol Microbiol 45, 1255–1265. 10.1046/j.1365-2958.2002.03121.x [DOI] [PubMed] [Google Scholar]
- Boyd E. F., Hartl D. L. (1999). Analysis of the type 1 pilin gene cluster fim in Salmonella: its distinct evolutionary histories in the 5′ and 3′ regions. J Bacteriol 181, 1301–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S., Weissman S. J., Minin V. N., Russo T. A., Dykhuizen D. E., Sokurenko E. V. (2009). High frequency of hotspot mutations in core genes of Escherichia coli due to short-term positive selection. Proc Natl Acad Sci U S A 106, 12412–12417. 10.1073/pnas.0906217106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg S., Hughes K. T. (2002). FimZ is a molecular link between sticking and swimming in Salmonella enterica serovar Typhimurium. J Bacteriol 184, 1209–1213. 10.1128/jb.184.4.1209-1213.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg S., Hull S., Hull R., Pruckler J. (1985). Construction and comparison of recombinant plasmids encoding type 1 fimbriae of members of the family Enterobacteriaceae. Infect Immun 48, 275–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg S., Hancox L. S., Yeh K. S. (1996). Salmonella typhimurium fimbrial phase variation and FimA expression. J Bacteriol 178, 542–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Buck J., Van Immerseel F., Haesebrouck F., Ducatelle R. (2004). Effect of type 1 fimbriae of Salmonella enterica serotype Enteritidis on bacteraemia and reproductive tract infection in laying hens. Avian Pathol 33, 314–320. 10.1080/0307945042000220561 [DOI] [PubMed] [Google Scholar]
- Duguid J. P. (1959). Fimbriae and adhesive properties in Klebsiella strains. J Gen Microbiol 21, 271–286. [DOI] [PubMed] [Google Scholar]
- Duguid J. P., Campbell I. (1969). Antigens of the type-1 fimbriae of salmonellae and other enterobacteria. J Med Microbiol 2, 535–553. 10.1099/00222615-2-4-535 [DOI] [PubMed] [Google Scholar]
- Duguid J. P., Anderson E. S., Campbell I. (1966). Fimbriae and adhesive properties in salmonellae. J Pathol Bacteriol 92, 107–138. 10.1002/path.1700920113 [DOI] [PubMed] [Google Scholar]
- Duguid J. P., Darekar M. R., Wheater D. W. (1976). Fimbriae and infectivity in Salmonella typhimurium. J Med Microbiol 9, 459–473. 10.1099/00222615-9-4-459 [DOI] [PubMed] [Google Scholar]
- Guo A., Cao S., Tu L., Chen P., Zhang C., Jia A., Yang W., Liu Z., Chen H., Schifferli D. M. (2009). FimH alleles direct preferential binding of Salmonella to distinct mammalian cells or to avian cells. Microbiology 155, 1623–1633. 10.1099/mic.0.026286-0 [DOI] [PubMed] [Google Scholar]
- Hancox L. S., Yeh K. S., Clegg S. (1997). Construction and characterization of type 1 non-fimbriate and non-adhesive mutants of Salmonella typhimurium. FEMS Immunol Med Microbiol 19, 289–296. 10.1016/S0928-8244(97)00095-3 [DOI] [PubMed] [Google Scholar]
- Kisiela D., Kuczkowski M., Kiczak L., Wieliczko A., Ugorski M. (2005a). Differentiation of Salmonella Gallinarum biovar Gallinarum from Salmonella Gallinarum biovar Pullorum by PCR-RFLP of the fimH gene. J Vet Med B Infect Dis Vet Public Health 52, 214–218. [DOI] [PubMed] [Google Scholar]
- Kisiela D., Sapeta A., Kuczkowski M., Stefaniak T., Wieliczko A., Ugorski M. (2005b). Characterization of FimH adhesins expressed by Salmonella enterica serovar Gallinarum biovars Gallinarum and Pullorum: reconstitution of mannose-binding properties by single amino acid substitution. Infect Immun 73, 6187–6190. 10.1128/IAI.73.9.6187-6190.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisiela D., Laskowska A., Sapeta A., Kuczkowski M., Wieliczko A., Ugorski M. (2006). Functional characterization of the FimH adhesin from Salmonella enterica serovar Enteritidis. Microbiology 152, 1337–1346. 10.1099/mic.0.28588-0 [DOI] [PubMed] [Google Scholar]
- Madison B., Ofek I., Clegg S., Abraham S. N. (1994). Type 1 fimbrial shafts of Escherichia coli and Klebsiella pneumoniae influence sugar-binding specificities of their FimH adhesins. Infect Immun 62, 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J. H., Kadouri D. E., O’Toole G. A. (2005). Growing and analyzing static biofilms. Curr Protoc Microbiol Chapter 1, 1B–, 1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misselwitz B., Kreibich S. K., Rout S., Stecher B., Periaswamy B., Hardt W. D. (2011). Salmonella enterica serovar Typhimurium binds to HeLa cells via Fim-mediated reversible adhesion and irreversible type three secretion system 1-mediated docking. Infect Immun 79, 330–341. 10.1128/IAI.00581-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Old D. C., Adegbola R. A. (1982). Haemagglutinins and fimbriae of Morganella, Proteus and Providencia. J Med Microbiol 15, 551–564. 10.1099/00222615-15-4-551 [DOI] [PubMed] [Google Scholar]
- Old D. C., Duguid J. P. (1970). Selective outgrowth of fimbriate bacteria in static liquid medium. J Bacteriol 103, 447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Old D. C., Payne S. B. (1971). Antigens of the type-2 fimbriae of salmonellae: “cross-reacting material” (CRM) of type-1 fimbriae. J Med Microbiol 4, 215–225. 10.1099/00222615-4-2-215 [DOI] [PubMed] [Google Scholar]
- Saini S., Pearl J. A., Rao C. V. (2009). Role of FimW, FimY, and FimZ in regulating the expression of type i fimbriae in Salmonella enterica serovar Typhimurium. J Bacteriol 191, 3003–3010. 10.1128/JB.01694-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokurenko E. V., Hasty D. L., Dykhuizen D. E. (1999). Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol 7, 191–195. 10.1016/S0966-842X(99)01493-6 [DOI] [PubMed] [Google Scholar]
- Sokurenko E. V., Feldgarden M., Trintchina E., Weissman S. J., Avagyan S., Chattopadhyay S., Johnson J. R., Dykhuizen D. E. (2004). Selection footprint in the FimH adhesin shows pathoadaptive niche differentiation in Escherichia coli. Mol Biol Evol 21, 1373–1383. 10.1093/molbev/msh136 [DOI] [PubMed] [Google Scholar]
- Stahlhut S. G., Chattopadhyay S., Struve C., Weissman S. J., Aprikian P., Libby S. J., Fang F. C., Krogfelt K. A., Sokurenko E. V. (2009a). Population variability of the FimH type 1 fimbrial adhesin in Klebsiella pneumoniae. J Bacteriol 191, 1941–1950. 10.1128/JB.00601-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlhut S. G., Tchesnokova V., Struve C., Weissman S. J., Chattopadhyay S., Yakovenko O., Aprikian P., Sokurenko E. V., Krogfelt K. A. (2009b). Comparative structure-function analysis of mannose-specific FimH adhesins from Klebsiella pneumoniae and Escherichia coli. J Bacteriol 191, 6592–6601. 10.1128/JB.00786-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson D. L., Clegg S., Old D. C. (1991). The frequency of fim genes among Salmonella serovars. Microb Pathog 10, 487–492. 10.1016/0882-4010(91)90115-Q [DOI] [PubMed] [Google Scholar]
- Thorns C. J. (1995). Salmonella fimbriae: novel antigens in the detection and control of salmonella infections. Br Vet J 151, 643–658. 10.1016/S0007-1935(95)80146-4 [DOI] [PubMed] [Google Scholar]
- Tinker J. K., Clegg S. (2000). Characterization of FimY as a coactivator of type 1 fimbrial expression in Salmonella enterica serovar Typhimurium. Infect Immun 68, 3305–3313. 10.1128/IAI.68.6.3305-3313.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker J. K., Hancox L. S., Clegg S. (2001). FimW is a negative regulator affecting type 1 fimbrial expression in Salmonella enterica serovar Typhimurium. J Bacteriol 183, 435–442. 10.1128/JB.183.2.435-442.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Asten A. J., van Dijk J. E. (2005). Distribution of “classic” virulence factors among Salmonella spp. FEMS Immunol Med Microbiol 44, 251–259. 10.1016/j.femsim.2005.02.002 [DOI] [PubMed] [Google Scholar]
- Weissman S. J., Beskhlebnaya V., Chesnokova V., Chattopadhyay S., Stamm W. E., Hooton T. M., Sokurenko E. V. (2007). Differential stability and trade-off effects of pathoadaptive mutations in the Escherichia coli FimH adhesin. Infect Immun 75, 3548–3555. 10.1128/IAI.01963-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. L., Elthon J., Clegg S., Jones B. D. (2000). Salmonella enterica serovars Gallinarum and Pullorum expressing Salmonella enterica serovar Typhimurium type 1 fimbriae exhibit increased invasiveness for mammalian cells. Infect Immun 68, 4782–4785. 10.1128/IAI.68.8.4782-4785.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K. S., Hancox L. S., Clegg S. (1995). Construction and characterization of a fimZ mutant of Salmonella typhimurium. J Bacteriol 177, 6861–6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K. S., Tinker J. K., Clegg S. (2002). FimZ binds the Salmonella typhimurium fimA promoter region and may regulate its own expression with FimY. Microbiol Immunol 46, 1–10. [DOI] [PubMed] [Google Scholar]