Abstract
Staphylococcus epidermidis is an opportunistic bacterium whose infections often involve the formation of a biofilm on implanted biomaterials. In S. epidermidis, the exopolysaccharide facilitating bacterial adherence in a biofilm is polysaccharide intercellular adhesin (PIA), whose synthesis requires the enzymes encoded within the intercellular adhesin operon (icaADBC). In vitro, the formation of S. epidermidis biofilms is enhanced by conditions that repress tricarboxylic acid (TCA) cycle activity, such as growth in a medium containing glucose. In many Gram-positive bacteria, repression of TCA cycle genes in response to glucose is accomplished by catabolite control protein A (CcpA). CcpA is a member of the GalR–LacI repressor family that mediates carbon catabolite repression, leading us to hypothesize that catabolite control of S. epidermidis biofilm formation is indirectly regulated by CcpA-dependent repression of the TCA cycle. To test this hypothesis, ccpA deletion mutants were constructed in strain 1457 and 1457-acnA and the effects on TCA cycle activity, biofilm formation and virulence were assessed. As anticipated, deletion of ccpA derepressed TCA cycle activity and inhibited biofilm formation; however, ccpA deletion had only a modest effect on icaADBC transcription. Surprisingly, deletion of ccpA in strain 1457-acnA, a strain whose TCA cycle is inactive and where icaADBC transcription is derepressed, strongly inhibited icaADBC transcription. These observations demonstrate that CcpA is a positive effector of biofilm formation and icaADBC transcription and a repressor of TCA cycle activity.
Introduction
Staphylococcus epidermidis is an opportunistic pathogen that primarily infects immunocompromised individuals or those with implanted biomaterials (e.g. catheters). As life expectancies and the availability of new biomaterials have increased, the number of people living with implanted biomaterials has also increased. One consequence of implanting biomaterials is that it predisposes individuals to device-related infections (Barrau et al., 2004; El-Ahdab et al., 2005; Piper et al., 2001). These infections are often difficult to treat due to the formation of a bacterial biofilm in which the bacteria are encapsulated in an exopolysaccharide matrix (Donlan, 2001; von Eiff et al., 2002). As with most virulence factors and mechanisms, the formation of biofilms and exopolysaccharide synthesis are strongly influenced by the availability of nutrients and environmental conditions (Deighton & Borland, 1993; Dobinsky et al., 2003; Vuong et al., 2005). In particular, S. epidermidis biofilm formation is enhanced during growth in media containing glucose (Dobinsky et al., 2003), suggesting a carbon catabolite-responsive regulator activates genes necessary for biofilm formation and/or represses genes that inhibit biofilm formation.
In Gram-positive bacteria, carbon catabolite repression is primarily mediated by the catabolite control protein A (CcpA) (Henkin et al., 1991; Seidl et al., 2006, 2008b). CcpA largely functions as a repressor; however, it also activates transcription of genes involved in fermentation and overflow metabolism (Shivers et al., 2006; Sonenshein, 2007). CcpA regulatory activity is controlled by interactions with the phosphorylated co-repressors, histidine-containing protein (HPr) or catabolite repression HPr (Crh) (Deutscher et al., 1994, 1995; Galinier et al., 1997; Wray et al., 1994). In addition to HPr and Crh, the activity of CcpA is regulated by the glycolytic intermediates glucose-6-phosphate and fructose-1,6-bisphosphate (Lopez & Thoms, 1977). These intermediates increase the ATP-dependent phosphorylation of HPr and/or Crh by increasing HPr kinase activity (Deutscher & Saier, 1983; Galinier et al., 1997). During growth in nutrient-rich media, CcpA is complexed with phosphorylated HPr or Crh and blocks transcription initiation by binding to catabolite responsive elements (cre) located in or near carbon catabolite repressed promoters (Kim et al., 2005; Nicholson et al., 1987). S. epidermidis has both CcpA and HPr homologues; however, it appears to lack a Crh homologue.
In Staphylococcus aureus, CcpA represses the synthesis of capsular polysaccharides and toxigenic exoproteins and promotes biofilm formation (Seidl et al., 2006, 2008a, b). On this latter point, the biofilm-promoting function of CcpA is thought to occur via its activity as a repressor of tricarboxylic acid (TCA) cycle genes (Seidl et al., 2009). Repression of TCA cycle activity derepresses synthesis of the biofilm-promoting exopolysaccharide polysaccharide intercellular adhesin (PIA) (Sadykov et al., 2008; Vuong et al., 2005). These observations and the fact that S. epidermidis biofilm formation appears to be controlled by a catabolite responsive regulator led us to inactivate the ccpA gene from S. epidermidis strain 1457 (Mack et al., 1992) and assess the effects on growth, biofilm formation and virulence.
Methods
Bacterial strains, bacteriophage, plasmids and growth conditions.
Bacterial strains, bacteriophage and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown in 2×YT broth or on 2×YT agar (Sambrook et al., 1989). S. aureus and S. epidermidis strains were grown in tryptic soy broth (TSB; BD Biosciences) or on TSB containing 1.5 % (w/v) agar. S. epidermidis cultures were inoculated 1 : 200 from overnight cultures (normalized for growth) into TSB, incubated at 37 °C, and aerated at 225 r.p.m. with a flask-to-medium ratio of 7 : 1. Bacterial growth was assessed by measuring the OD600. When used, antibiotics were used at the following concentrations: 100 µg ampicillin ml−1 for E. coli, 10 µg chloramphenicol ml−1, 10 µg erythromycin ml−1 and 10 µg tetracycline ml−1.
Table 1. Bacterial strains, bacteriophage and plasmids used in this study.
Ap, ampicillin; Cm, chloramphenicol; Em, erythromycin; Tc, tetrocycline; ts, temperature-sensitive origin of replication.
| Bacterial strain, bacteriophage or plasmid | Relevant properties | Reference or source |
| Strains | ||
| DH5α | E. coli cloning host | Invitrogen |
| RN4220 | S. aureus restriction-deficient mutant of strain 8325-4 | Novick (1991) |
| 1457 | S. epidermidis | Mack et al. (1992) |
| 1457-acnA | Strain 1457 acnA insertion mutant, TcR | Sadykov et al. (2008) |
| 1457-ΔcitZC | Strain 1457 citZC deletion mutant, EmR | Sadykov et al. (2008) |
| 1457-ΔccpA | Strain 1457 ccpA deletion mutant, EmR | This study |
| 1457-acnA-ccpA | Strain 1457 acnA and ccpA double mutant, TcR EmR | This study |
| 1457-ΔccpA(comp) | Strain 1457 with restored wild-type ccpA allele, TcR | This study |
| 1057 | Methicillin-resistant S. epidermidis (MRSE) | Nedelmann et al. (1998) |
| 1057-ΔccpA | Strain 1057 ccpA deletion mutant, EmR | This study |
| Bacteriophage | ||
| ϕ71 | S. epidermidis 1457 transducing phage | Dean et al. (1973) |
| Plasmids | ||
| pTS1-d | Shuttle vector, pE194orits, ColE1, ApR CmR | Sadykov et al. (2008) |
| pEC4 | pBluescript II KS(+) with ermB inserted into ClaI site, ApR EmR | Brückner (1997) |
| pMRS15 | Derivative of pTS1-d, with ΔccpA : : ermB | This study |
| pMRS62 | Derivative of pTS1-d, with ccpA_tetM | This study |
Construction of ccpA mutants in S. epidermidis.
All primers (Table 2) used for PCR amplification of strain 1457 genomic DNA were designed using the S. epidermidis RP62A genome sequence. The ccpA mutant was constructed by replacing a 595 bp region, which included the ccpA gene, with an erythromycin resistance gene (ermB) using the gene splicing by overlap extension (gene SOEing) technique (Horton et al., 1990). ermB was amplified from pEC4 using primers ccpA-ermB-f and ccpA-ermB-r, which contain sequences homologous to the ccpA gene. Primers BamHI-aroA-f and ermB-ccpA-r were used for amplification of a 1.1 kb region upstream of the ccpA gene. Primers ermB-ccpA-f and SacI-fhs-r were used to PCR amplify a 1.2 kb region downstream of the ccpA gene. The resulting 3.3 kb PCR product consisted of the 1 kb ermB gene with DNA flanking the ccpA gene and contained SacI and BamHI sites that were used for ligation into pTS1-d (Sadykov et al., 2008) digested with SacI and BamHI to generate plasmid pMRS15. Plasmid pMRS15 was used to construct strain 1457-ΔccpA mutant (1457-ΔccpA : : ermB) using the temperature shift protocol of Foster (1998). The ccpA mutation of strain 1457-ΔccpA was transferred into the meticillin-resistant S. epidermidis strain 1057 by phage transduction using S. epidermidis phage 71. Allelic replacement of the internal region of the ccpA genes by the ermB cassette was verified by PCR and Southern blot analysis.
Table 2. Primers used in this study.
| Primer designation | Nucleotide sequence (5′–3′) |
| BamHI-aroA-f | CCAGGATCCGATGCTGTAGCACAAGATTTGCAGG |
| ermB-ccpA-r | GACGAATCCCTCCTTCGTCGTTGTACGTTTACTTGCAAGACC |
| ccpA-ermB-f | GCAAGTAAACGTACAACGACGAAGGAGGGATTCGTCATGTTG |
| ccpA-ermB-r | CACTCCAGCATCTTGTGCACGCGACTCATAGAATTATTTCCTCCC |
| ermB-ccpA-f | GGAAATAATTCTATGAGTCGCGTGCACAAGATGCTGGAGTGAAAG |
| SacI-fhs-r | GAAGAGCTCCTAGGTACTCCGTGAGTTGTACCAC |
| tetM-ccpA-r | CGTAAGAGCATATTTGTAAAGGAATCTCCTTACTGAGTTGTCCCACGGTATTC |
| ccpA-tetM-f | AGAATACCGTGGGACAACTCAGTAAGGAGATTCCTTTACAAATATGCTCTTACG |
| fhs-tetM-r | GATTGATTCAGATACAATGAGTGAATATGACGATCTCCTCCTTTCCACTTTAATTC |
| tetM-fhs-f | GAATTAAAGTGGAAAGGAGGAGATCGTCATATTCACTCATTGTATCTGAATCAATC |
| KpnI-fhs-r | GAAGGTACCGTGCATTTTCAAAGCACGAATC |
| gcaD-f | CGATTATTCTGGCAGCAGGTAAG |
| gcaD-r | CGATTGATACGTTGTTGCAAAGC |
| icaProbeforward | GACAGTCGCTACGAAAAG |
| icaProbereverse | CCGAATAATTTGTAAATTTCC |
| pfkA-f | GAAGAAGATTGCAGTTTTAACTAGCGG |
| pfkA-r | GCTACATCCTTAATATCTGTATTGACTTCTGG |
| femC-f | GATGTTTGATGGTTCATCTATTGAAGGTTTCG |
| femC-r | GCAGTATCAGTCAATTGTAAATCACCTTCAG |
| aroA-f | GAATTACTATCGAAGCGAGGAGAATTAGC |
| fhs-r | CAACTGCTGATGGATCGAAACC |
| fhs-r1 | CAAGATTAACAAGTCCCGCTTCAAC |
| gyrB-RT-F1 | CTAATGCTGATTTACGACGCGTAA |
| gyrB-RT-D1 | TCTGTAGGACGCATTATTGTTGAAA |
| icaD-RT-F | ATGGTCAAGCCCAGACAGAG |
| icaD-RT-D | CGTGTTTTCAACATTTAATGCAA |
Construction of the S. epidermidis ccpA complementation strain.
For complementation of the ccpA mutation, a 1.9 kb PCR product containing the wild type ccpA allele and a portion of the upstream region was combined in a SOEing reaction with a 2.3 kb tetM gene and 1.4 kb of the downstream region of ccpA. The tetM gene was amplified from pWF105 using primers ccpA-tetM-f and fhs-tetM-r. Primers BamHI-aroA-f and tetM-ccpA-r were used to amplify a 1.9 kb fragment containing the intact ccpA gene. A 1.4 kb region of the ccpA downstream region was amplified using tetM-fhs-f and KpnI-fhs-r primers. The resulting 5.6 kb PCR product contained KpnI and BamHI sites that were used for ligation into pTS1-d (Sadykov et al., 2008) to generate plasmid pMRS62. Plasmid pMRS62 was used to restore the wild type ccpA allele to strain 1457-ΔccpA. Restoration of the ccpA gene was verified by PCR and Southern blot analysis.
Measurement of acetate and glucose concentrations in culture supernatants.
Acetate and glucose concentrations were determined with kits purchased from R-Biopharm and used according to the manufacturer’s protocol.
Enzymic activity assays.
Aconitase activity assays were performed as described by Sadykov et al. (2010). Isocitrate dehydrogenase enzymic activity assays were performed as described previously (Somerville et al., 2003). Protein concentrations were determined by using the modified Lowry assay (Pierce Chemical).
Transcript analysis.
Northern blot analysis was performed as described by Sadykov et al. (2008). Four micrograms of total RNA was loaded per lane. Probes for Northern blotting were generated by PCR amplification of the internal DNA fragments of the glmU, icaD, pfkA and glnA (femC) genes and labelled using the North2South random prime labelling kit (Pierce). Detection was performed using the Chemiluminescent Nucleic Acid Detection Module (Pierce). PIA biosynthesis is encoded within the icaADBC four gene operon. To assess icaADBC transcription, the polycistronic icaADBC mRNA was probed for icaD and not each individual gene.
RNA isolation and real-time RT-PCR were carried out as described by Chatterjee et al. (2005), using the icaD primer pair (Arciola et al., 2001). The level of icaD mRNA was normalized against the internal control gyrB. Transcripts amounts were expressed as the fold difference relative to the control gene (2ΔΔCT, where ΔCT represents the difference in threshold cycle between the target and control genes).
Biofilm forming ability.
Bacterial cultures were grown in TSB supplemented with 1 % glucose for 24 h at 37 °C in glass culture tubes with aeration (150 r.p.m.) or under static conditions in 24-well plates. Growth medium was inoculated with 0.5 McFarland units. Following 24 h incubation, tubes and/or wells were washed three times with PBS, air-dried, stained with 0.1 % safranin for 30 s and washed three times with distilled water. For 24-well plate cultures, the adhering dye was dissolved with 30 % acetic acid, and the absorption was measured at 530 nm.
Biofilm formation was also monitored under flow conditions using the BioFlux 200 system (Fluxion Biosciences) and a 48-well microfluidic device. Experiments were performed essentially as described by Benoit et al. (2010) with minor modifications. Microfluidic channels of the BioFlux flowthrough device were covered with TSB supplemented with 1 % glucose for 5 min at a flow rate of 5 µN cm2, and subsequently inoculated with bacteria (0.5 McFarland) at a flow rate of 2 µN cm2 from the ‘outlet’ well until the bacterial solution flooded the viewing channel of the device. Bacteria were allowed to attach for 15 min at 37 °C. Afterwards, fresh medium was pumped from the ‘inlet’ to the ‘outlet’ well through the microfluidic channels at a constant flow rate of 5 µN cm2 and 37 °C. Biofilm formation was monitored after 18 h by light microscopy using an inverted microscope (Leica DMI 4000 B; Leica Mikrosysteme). Phase-contrast images were obtained using a digital camera (Leica DFC 420 C) and the Leica application suite software V3.6.
Mouse virulence.
Non-obese diabetic female mice (NOD/LtJ; Charles River Laboratories) were kept under specific pathogen-free conditions in the Animal House of the Department of Biomedicine, University Hospital Basel, according to the regulations of the Swiss veterinary law. Female NOD mice spontaneously develop type 1 diabetes mellitus usually between 15 and 30 weeks of age (Leiter, 1993). Animals of 12 weeks of age were tested weekly for increased urinary glucose levels, and subsequently analysed for blood glucose levels. Mice with blood glucose ≥15 mmol l−1 were included as diabetic animals.
Mice were anaesthetized and sterile catheter segments were inserted subcutaneously as described by Kristian et al. (2008) and Rupp et al. (1999). Catheters were infected with 20 µl pyrogen-free saline containing 104 c.f.u. S. epidermids strain 1457 and its isogenic 1457-ΔccpA mutant, respectively, before the incisions were closed with wound clips. The diameter of swelling/oedema was measured daily using a caliper. Ten days after infection, mice were euthanized by CO2 or intraperitoneal injection of 500 mg thiopenthal kg−1, and the catheters and surrounding tissues were aseptically removed and separated. Bacteria adherent to the catheters were detached by vortexing in 0.9 % saline, 0.15 % EDTA, followed by sonication for 2 min at 250 W. Tissue samples were homogenized in 1 ml 0.9 % NaCl. Serial dilutions were plated on Mueller–Hinton agar to enumerate bacterial load in catheters and tissues.
Statistical analysis.
The significance of changes was assessed using Student’s t-test or Mann–Whitney Rank Sum Test. P-values <0.05 were considered significant.
Results
Growth characteristics of S. epidermidis strain ccpA deletion mutant
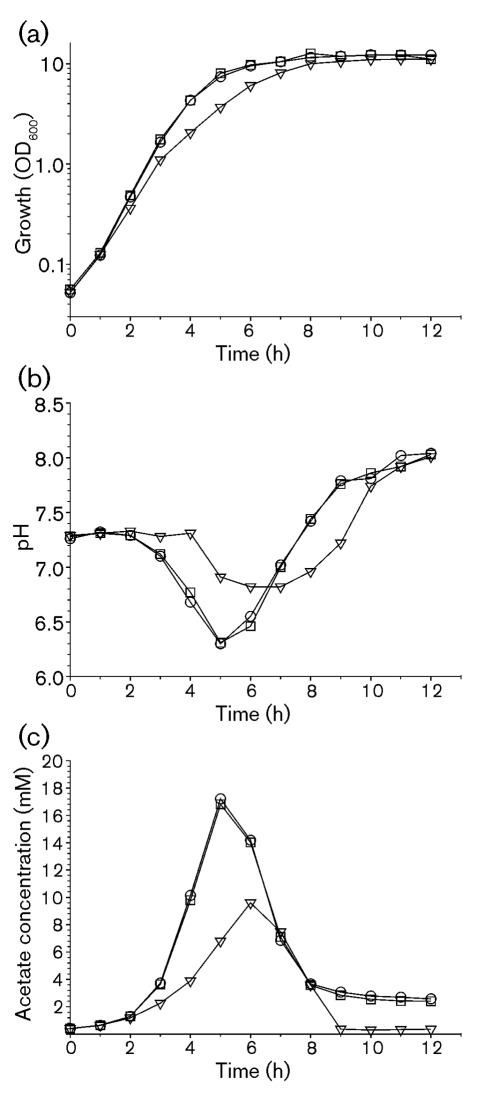
CcpA is a TCA cycle repressor and an activator of genes involved in fermentation and overflow metabolism (Shivers et al., 2006; Sonenshein, 2007). These observations suggest that deletion of ccpA would increase carbon flow into the TCA cycle and away from overflow metabolism. Because staphylococci have evolved to utilize nutrients rapidly but inefficiently when they are in abundance, redirecting carbon into the more efficient TCA cycle would likely result in a slower growth rate. Consistent with this suggestion, deletion of ccpA in strain 1457 resulted in a slightly decreased growth rate relative to strain 1457; nevertheless, the growth yields of both strains were similar (Fig. 1a). In Bacillus subtilis, deletion of ccpA impairs growth on minimal media (Ludwig et al., 2002). This growth impairment is the result of CcpA regulating glutamate and branched chain amino acid biosynthesis; hence, growth can be restored by the addition of amino acids to the minimal medium. While it is possible that amino acid auxotrophies in strain 1457-ΔccpA contribute to its slower growth rate relative to strain 1457, growth studies were conducted in the amino acid- and peptide-rich medium TSB, minimizing this possibility. Importantly, complementation of the ccpA deletion restored the growth rate to that of strain 1457. If the decreased growth rate was due to carbon being directed away from overflow metabolism and into the TCA cycle, then the accumulation of organic acids in the culture medium of strain 1457-ΔccpA will be less than in the culture medium of the wild type strain 1457. To assess the accumulation of organic acids in the culture medium, the pH and acetic acid concentrations were measured during growth (Fig. 1b, c). In the exponential growth phase, the pH of the culture medium of strain 1457-ΔccpA remained more basic than did the pH of the culture medium of the wild type strain 1457. Acidification of the culture medium was restored to wild type levels by complementing the ccpA deletion with a wild type ccpA gene. Similar to the culture medium pH profiles, strain 1457-ΔccpA accumulated less acetate in the medium than did the wild type strain or strain 1457-ΔccpA(comp). These data suggest that CcpA regulates overflow and TCA cycle metabolism in S. epidermidis.
Fig. 1.
Growth characteristics of strains 1457 (○), 1457-ΔccpA (▽) and 1457-ΔccpA(comp) (□). Growth curves (a), culture medium pH profiles (b) and acetate accumulation and depletion from the culture medium (c) of these strains are shown. The data are representative of two independent experiments.
CcpA represses TCA cycle activity
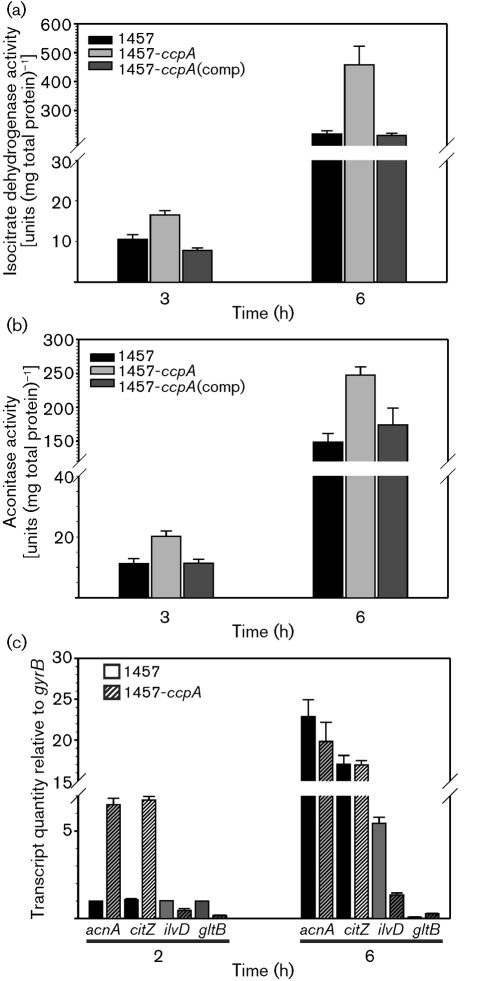
The decreased accumulation of acetate in the culture medium of strain 1457-ΔccpA relative to strain 1457 (Fig. 1c) indicated that less carbon was entering into overflow metabolism. The more likely explanation for this observation was increased carbon flow into the TCA cycle. To determine if deletion of ccpA increased TCA cycle enzymic activity, the activities of aconitase and isocitrate dehydrogenase were measured in the exponential and post-exponential phases of growth (Fig. 2a, b). As expected, isocitrate dehydrogenase and aconitase activities were significantly increased in strain 1457-ΔccpA relative to strain 1457 (P<0.005). Complementation of the ccpA deletion restored enzymic activity to the level of the wild type. Although ccpA deletion increased enzymic activity, it was less than a twofold difference, raising the possibility that ccpA deletion had an indirect effect on TCA cycle activity. In B. subtilis, CcpA regulates TCA cycle activity at the transcriptional level (Kim et al., 2002). To determine if ccpA deletion affected transcription of citZ (citrate synthase) and acnA/citB (aconitase), quantitative RT-PCR was used to determine the relative levels of citZ and acnA/citB mRNA. In addition, we assessed the effect of ccpA deletion on ilvD (dihydroxy-acid dehydratase, an indicator of branched chain amino acid biosynthesis) and gltB (the large subunit of glutamate synthase) to see if changes in transcription could account for the slower growth rate of strain 1457-ΔccpA. Similar to in B. subtilis, deletion of ccpA derepressed transcription of both citZ and acnA/citB relative to the parental strain during the exponential growth phase (Fig. 2c). During the exponential growth phase, the mRNA levels of ilvD and gltB were low, suggesting that they do not account for the difference in the growth rates between strains 1457 and 1457-ΔccpA. In addition, these data demonstrate the normal temporal induction of transcription that coincides with the depletion of glucose and the release of catabolite repression. These data confirm that CcpA functions as a regulator of the TCA cycle in S. epidermidis.
Fig. 2.
CcpA deletion increases aconitase and isocitrate dehydrogenase activity and de-represses transcription during the exponential growth phase. Isocitrate dehydrogenase (a) and aconitase (b) activity was assessed during the exponential (3 h) and post-exponential (6 h) growth phases. The data are presented as the mean±sem of two independent experiments each determined in triplicate. (c) Quantitative RT-PCR analysis of the effects of ccpA inactivation on the quantity of acnA/citB, citZ, ilvD and gltB.
CcpA regulates transcription of genes involved in PIA biosynthesis
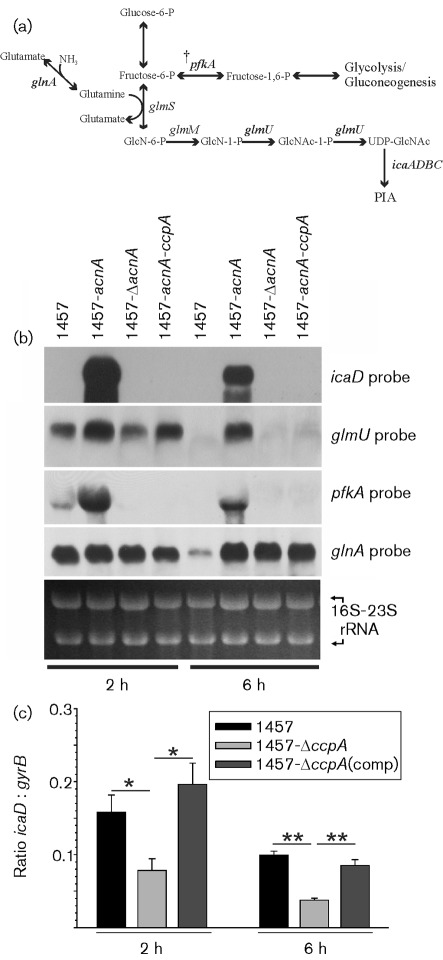
TCA cycle activity regulates synthesis of the biofilm-promoting exopolysaccharide PIA both at the transcriptional level and by altering the availability of PIA biosynthetic precursors (Sadykov et al., 2008; Vuong et al., 2005). Specifically, deletion of any of the first three genes of the TCA cycle derepresses transcription of PIA biosynthetic genes (i.e. icaADBC) and PIA biosynthesis. Although it is possible that TCA cycle enzymes directly regulate icaADBC transcription, the more likely possibility is that a regulatory protein(s) responding to TCA cycle-induced changes in the metabolic status alters transcription of icaADBC. Notably, one of the metabolic changes caused by TCA cycle stress (i.e. any external stressor that is capable of altering TCA cycle activity) is an increase in the intracellular concentration of glucose-6-phosphate (Sadykov et al., 2010). Because the regulatory activity of CcpA is enhanced by the glycolytic intermediate glucose-6-phosphate, this raised the possibility that CcpA both regulates TCA cycle activity (Fig. 2a, b) and responds to metabolic signals transduced by the TCA cycle (Brückner & Titgemeyer, 2002; Deutscher & Saier, 1983; Sonenshein, 2007; Warner & Lolkema, 2003). These observations led us to assess the effect of ccpA deletion on the transcription of a subset of genes involved in PIA biosynthesis; specifically, icaD [IcaD forms a heterodimeric UDP-N-acetylglucosamine transferase with IcaA (Gerke et al., 1998)], N-acetylglucosamine biosynthesis (glmU; glucosamine-1-phosphate N-acetyltransferase), glnA (glutamine synthetase) and pfkA (6-phosphofructokinase) (Fig. 3a). In addition, to determine whether metabolic signals transduced by the TCA cycle require CcpA, the effect of ccpA deletion on transcription of these same genes in an aconitase (acnA/citB) mutant background was investigated (Fig. 3b). Of the genes involved in PIA biosynthesis that were examined, a putative cre site was identified [using the B. subtilis cre consensus site (TGWNANCGNTNWCA) as a template (Weickert & Chambliss, 1990)] in the promoter region of pfkA (TTTTAAAACGTTTACATC) and another putative cre site was found in the 3′ portion of glmU (TGACGGCGCTAACAAA). During the exponential phase of growth when glucose was present in the medium, ccpA deletion decreased the transcription or stability of pfkA mRNA relative to strain 1457 (Fig. 3b). These data and the presence of a cre site in the promoter region of pfkA indicate that CcpA regulates pfkA transcription; however, as with B. subtilis, the effect of CcpA on glycolytic genes may be indirect (Tobisch et al., 1999). Interestingly, aconitase inactivation caused a remarkable increase in pfkA mRNA relative to strain 1457 and CcpA was required for this increased level of pfkA mRNA. Similarly, CcpA was required for the transduction of TCA cycle metabolic signals into an increased level of icaD mRNA (Fig. 3b). This observation was interesting because Northern blot analysis revealed that inactivation of ccpA had a small effect on icaD transcription and no cre sites were identified in icaADBC or its promoter region (Fig. 3b and data not shown). Since it was possible that changes in icaD transcript levels of the wild-type and ΔccpA mutant strains were below the detection limit of Northern blots, icaD transcription was assessed by real-time PCR (Fig. 3c). Using RT-PCR, we observed that inactivation of ccpA had a small but significant effect on icaD transcription (Fig. 3c). Specifically, deletion of ccpA decreased the exponential (2 h) and post-exponential growth phase (6 h) level of icaD mRNA by a factor of two to three. Complementation of the mutant with CcpA reversed this effect, suggesting this effect was CcpA-dependent. Neither glmU nor glnA transcription was affected by ccpA deletion during the exponential phase of growth; however, ccpA deletion derepressed glnA transcription during the post-exponential growth phase (Fig. 3b). Taken together, these data suggest that CcpA has a modest effect on some genes involved in PIA biosynthesis (i.e. icaD and pfkA). In addition, this suggestion is consistent with PIA immunoblot data demonstrating that ccpA inactivation had only a minor effect on PIA accumulation (Sadykov et al., 2010).
Fig. 3.
Northern blot analysis of icaD, glmU, pfkA and glnA mRNA levels in the exponential (2 h) and post-exponential (6 h) phases of growth. (a) The diagram shows the relationship of the genes chosen for Northern analysis to the synthesis of PIA. The presence of a cre site in the promoter region of pfkA is indicated by a dagger (†). -6-P indicates a 6-phosphate chain; -1,6-P indicates 1,6-bisphosphate; GlcN, glucosamine; GlcNAc, N-acetyl-glucosamine; UOP, unicline diphosphate. (b) Representative Northern blots probed with PCR products for icaD, glmU, pfkA and glnA. 23S and 16S rRNA were visualized by ethidium bromide staining and used as loading controls. The results are representative of at least two independent experiments. (c) Quantitative transcript analysis of icaD in strains 1457, 1457-ΔccpA, and 1457-ΔccpA(comp) after 2 and 6 h of growth. The data are presented as the mean±sd of four independent experiments each determined in duplicate. Statistical significance between the wild type and ccpA mutant was determined by using the Mann–Whitney test. *P<0.05; **P<0.01.
CcpA is required for biofilm formation
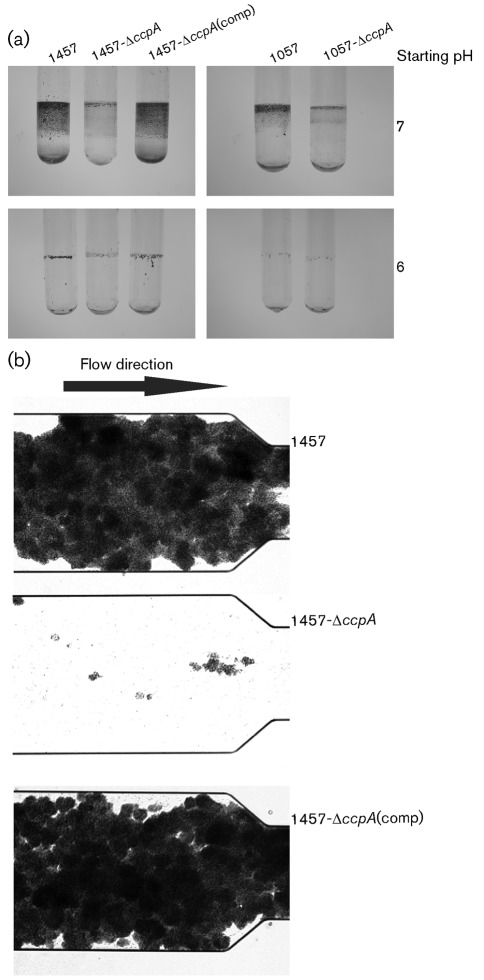
The CcpA-dependent regulation of pfkA (Fig. 3b) and the modest effect of CcpA on icaD transcription (Fig. 3c) suggested that deletion of ccpA might hinder the ability of strain 1457 to form a biofilm. To test if ccpA deletion altered the ability of S. epidermidis to form a biofilm, strains 1457, 1057, 1457-ΔccpA, 1057-ΔccpA and 1457-ΔccpA(comp) were grown in static biofilms and the biofilms were quantified using a dye retention assay (data not shown). Deletion of ccpA decreased the ability of S. epidermidis to form a biofilm, while complementation of the ccpA deletion restored the biofilm forming capacity (data not shown). Similarly, strains 1457-ΔccpA and 1057-ΔccpA grown in glass culture tubes (1 : 5 medium-to-flask ratio) with 150 r.p.m. aeration displayed a greatly diminished ability to form biofilms relative to strains 1457, 1057 and 1457-ΔccpA(comp) (Fig. 4a). The effect of ccpA deletion on biofilm formation is most apparent in a microfluidic shear flow assay (Fig. 4b), where strain 1457-ΔccpA does not form a biofilm. These data demonstrate that CcpA is critical for the ability of S. epidermidis to form a biofilm in a glucose-containing medium.
Fig. 4.
Deletion of ccpA reduces S. epidermidis biofilm forming capacity. (a) The ability of strains 1457, 1057, 1457-ΔccpA, 1057-ΔccpA and 1457-ΔccpA(comp) to form biofilms under aerobic conditions was assessed in glass culture tubes that were stained with safranin (figure is representative of three independent experiments). (b) Biofilm formation monitored under shear flow conditions using the BioFlux 200 system and a 48-well microfluidic device.
To facilitate biofilm formation in the laboratory, glucose is often added to the culture medium in abundance (Dobinsky et al., 2003; Lim et al., 2004). One outcome of the addition of excess glucose is the induction of overflow metabolism and a decrease in the extracellular pH. This decreased pH is one factor that can contribute to biofilm formation. Because ccpA deletion alters the carbon flow, the pH of the culture medium is more alkaline than it is with the wild type strain (Fig. 1), raising the possibility that the more alkaline pH is responsible for the decreased ability of strain 1457-ΔccpA to form a biofilm. To test this possibility, static and aerated cultures of strains 1457, 1057, 1457-ΔccpA and 1057-ΔccpA were grown in TSB at pH 7.0 and 6.0, and the ability of the bacteria to form a biofilm was assessed (Fig. 4a and data not shown). Surprisingly, the decreased ability of ccpA deletion strains to form a biofilm was independent of changes in the extracellular pH.
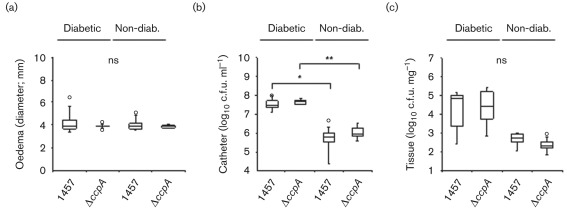
CcpA is not required for virulence in a mouse catheter/biofilm infection model
CcpA is required for biofilm formation (Fig. 4); hence, it was reasonable to hypothesize that CcpA is required for virulence in a biofilm-promoting infection model. To test this hypothesis, we assessed the effect of ccpA deletion in a mouse catheter/biofilm infection model (Fig. 5). Because the regulatory activity of CcpA is affected by glycolytic intermediates, diabetic mice (blood glucose levels ≥15 mmol l−1) were used to exacerbate any differences in virulence associated with ccpA inactivation. In contrast with our hypothesis, deletion of ccpA did not significantly alter the infectious outcomes in diabetic or non-diabetic mice in the catheter/biofilm infection model. Previously, we observed that the antiphagocytic properties of PIA were important for S. epidermidis virulence in our mouse catheter/biofilm infection model (Kristian, et al., 2008). Therefore, the absence of any significant difference between the wild type and ccpA mutant strains in our mouse catheter infection model (Fig. 4) is consistent with the modest effect CcpA has on icaADBC transcription (Fig. 3b, c) and PIA accumulation (Sadykov, et al., 2010). Alternatively, these data may indicate that biofilm formation is not regulated by the availability of carbohydrates in vivo.
Fig. 5.
The virulence of S. epidermidis strain 1457 in a catheter-related infection model is independent of CcpA. Sterile catheter segments were inserted subcutaneously into non-diabetic (non-diab.; blood glucose levels <10 mM) and diabetic (blood glucose levels >15 mM) NOD mice and subsequently infected with 104 bacteria before the incisions were closed with wound clips. On day 10 after infection, oedema end points (a) at the insertion site were determined, and the catheters (b) and the surrounding tissues (c) were aseptically removed and separated. Bacteria adherent to the catheters were detached by vortexing in saline, and tissue samples were homogenized in 1 ml 0.9 % NaCl. For c.f.u. determinations, serial dilutions were plated on Mueller–Hinton agar. Statistical significance was determined by comparing mice from the same age group using the Mann–Whitney test. *P<0.05; **P<0.01; ns, not significant. Data are presented as “box and whisker plots” showing the interquartile range (25–75 %; box), median (horizontal line), whiskers (bars) and min/max outliers (○).
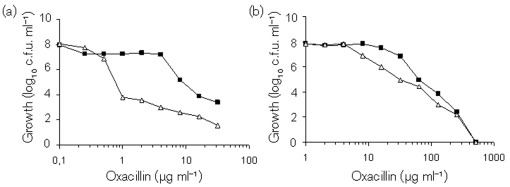
CcpA decreases oxacillin susceptibility
Inactivation of ccpA in S. aureus increases methicillin and oxacillin susceptibility (De Lencastre et al., 1999; Seidl et al., 2006), raising the possibility that deletion of ccpA in S. epidermidis would increase oxacillin susceptibility. To determine if ccpA deletion altered the susceptibility of the methicillin-resistant strain 1057 to oxacillin, strains 1057 and 1057-ΔccpA were grown for 48 h in Mueller–Hinton broth, or Mueller–Hinton broth supplemented with 2 % NaCl, with increasing concentrations of oxacillin, and c.f.u. ml−1 was determined (Fig. 6). Similar to S. aureus, deletion of ccpA in strain 1057 caused an increase in the susceptibility to oxacillin, demonstrating that oxacillin resistance in S. epidermidis requires CcpA and suggesting that CcpA-mediated oxacillin resistance may be a common resistance mechanism.
Fig. 6.
Deletion of ccpA increased the susceptibility of methicillin-resistant S. epidermidis strain 1057 to oxacillin. Population analysis profiles were established by plating appropriate dilutions of overnight cultures of 1057 (▪) and 1057-ΔccpA (▵) on Mueller–Hinton agar (a) and cation-adjusted Mueller–Hinton broth supplemented with 2 % NaCl (b) containing increasing concentrations of oxacillin. The c.f.u. was determined after 48 h of incubation at 35 °C.
Discussion
Most S. epidermidis virulence determinants are involved in attachment, adherence, and the elaboration of a biofilm; hence, the primary mode of infection for S. epidermidis is its ability to colonize implanted biomaterials (von Eiff et al., 2002). Extensive data have accumulated demonstrating that formation of S. epidermidis biofilms is dependent upon the bacterial metabolic state (Cramton et al., 2001; Sadykov et al., 2008, 2010; Schlag et al., 2007; Vuong et al., 2005). Specifically, S. epidermidis biofilms form under nutrient-rich conditions when TCA cycle activity is repressed (Cramton et al., 2001; Dobinsky et al., 2003; Mack et al., 1992; Sadykov et al., 2008; Vuong et al., 2005).
In S. epidermidis, repression of the TCA cycle de-represses ica transcription and PIA biosynthesis, which facilitates biofilm formation (Fig. 3b; Sadykov et al., 2008; Vuong et al., 2005). As stated, the more likely hypothesis to explain how TCA cycle activity regulates icaADBC transcription and PIA biosynthesis is that changes in activity are tranduced into metabolic signals that can be ‘sensed’ by regulators of icaADBC (Sadykov et al., 2010; Somerville & Proctor, 2009). Regulation of the icaADBC operon is complex, involving at least two DNA-binding proteins (IcaR and SarA) and the alternative sigma factor σB (Conlon et al., 2002; Handke et al., 2007). If changes in TCA cycle activity produce metabolic signals that can be sensed by regulators of icaADBC, then regulatory mutants (i.e. icaR, sarA and sigB) whose TCA cycle activity is blocked should be unable to de-repress icaADBC transcription and/or PIA biosynthesis. We previously observed that addition of the aconitase-specific inhibitor, fluorocitric acid, to the culture medium of S. epidermidis strains 1457-sigB and 1457-icaR increased the accumulation of PIA relative to untreated control cultures (Sadykov et al., 2008). These data suggest that TCA cycle-mediated de-repression of icaADBC transcription and PIA synthesis occur independently of IcaR and σB (Sadykov et al., 2008). In a 1457 sarA mutant, the accumulation of PIA was similar irrespective of the presence of fluorocitric acid in the culture medium (P>0.05). These data confirmed that PIA synthesis requires SarA but that there must be at least one additional regulator that is responsive to TCA cycle-associated metabolic signals.
Environmental stressors that repress TCA cycle activity increase the intracellular concentration of the glycolytic intermediate glucose-6-phosphate (Sadykov et al., 2010). Glucose-6-phosphate increases phosphorylation of the histidine-containing protein (HPr) by enhancing the activity of the HPr kinase (Deutscher & Saier, 1983), which increases its interaction with CcpA, thus activating CcpA (Brückner & Titgemeyer, 2002; Sonenshein, 2007; Warner & Lolkema, 2003). In S. aureus, CcpA is required for ica transcription, PIA accumulation and biofilm formation (Seidl et al., 2008b). These observations raised the possibility that CcpA is the additional TCA cycle-responsive regulator required for ica transcription in S. epidermidis. Surprisingly, deletion of ccpA in S. epidermidis has only a minor effect on ica transcription (Fig. 3c) and PIA accumulation (Sadykov et al., 2010); however, it inhibits biofilm formation (Fig. 4). In S. aureus, de-repression of the TCA cycle decreases PIA accumulation and delays biofilm formation (Zhu et al., 2009); hence, the simplest explanation for CcpA-dependent biofilm formation in S. epidermidis is that CcpA represses TCA cycle activity, increasing ica transcription and PIA synthesis. If this explanation is correct, then inactivation of the TCA cycle in strain 1457-ΔccpA should de-repress ica transcription and PIA accumulation. In contrast with this expectation, inactivation of aconitase in strain 1457-ΔccpA failed to increase ica transcription (Fig. 3b) or PIA accumulation (Sadykov et al., 2010). Although the mechanism by which CcpA facilitates biofilm formation has yet to be elucidated, it is clear that CcpA is required to transduce metabolic changes associated with TCA cycle stress into alterations in icaADBC transcription and PIA accumulation. Taken together, these observations demonstrate that CcpA is an essential positive effector of biofilm formation.
Acknowledgements
G. A. S. was supported by funds provided through the Hatch Act and the Institute of Agriculture and Natural Resources and by funds provided through the National Institutes of Health (AI087668). T. A. M. and M. H. were supported in part by funds provided through the University of Nebraska’s Undergraduate Creative Activities and Research Experiences. T. H., M. H. and M. B. were supported by the Deutsche Forschungsgemeinschaft (grant BI 1350/1-1). We are grateful to S. Balkow and IUL Instruments for providing the BioFlux 200 system.
Abbreviations:
- cre
catabolite responsive elements
- PIA
polysaccharide intercellular adhesin
- TCA
tricarboxylic acid
References
- Arciola C. R., Baldassarri L., Montanaro L. (2001). Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J Clin Microbiol 39, 2151–2156. 10.1128/JCM.39.6.2151-2156.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrau K., Boulamery A., Imbert G., Casalta J. P., Habib G., Messana T., Bonnet J. L., Rubinstein E., Raoult D. (2004). Causative organisms of infective endocarditis according to host status. Clin Microbiol Infect 10, 302–308. 10.1111/j.1198-743X.2004.00776.x [DOI] [PubMed] [Google Scholar]
- Benoit M. R., Conant C. G., Ionescu-Zanetti C., Schwartz M., Matin A. (2010). New device for high-throughput viability screening of flow biofilms. Appl Environ Microbiol 76, 4136–4142. 10.1128/AEM.03065-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner R. (1997). Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol Lett 151, 1–8. 10.1016/S0378-1097(97)00116-X [DOI] [PubMed] [Google Scholar]
- Brückner R., Titgemeyer F. (2002). Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol Lett 209, 141–148. 10.1016/S0378-1097(02)00559-1 [DOI] [PubMed] [Google Scholar]
- Chatterjee I., Becker P., Grundmeier M., Bischoff M., Somerville G. A., Peters G., Sinha B., Harraghy N., Proctor R. A., Herrmann M. (2005). Staphylococcus aureus ClpC is required for stress resistance, aconitase activity, growth recovery and death. J Bacteriol 187, 4488–4496. 10.1128/JB.187.13.4488-4496.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon K. M., Humphreys H., O’Gara J. P. (2002). icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J Bacteriol 184, 4400–4408. 10.1128/JB.184.16.4400-4408.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramton S. E., Ulrich M., Götz F., Döring G. (2001). Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun 69, 4079–4085. 10.1128/IAI.69.6.4079-4085.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lencastre H., Wu S. W., Pinho M. G., Ludovice A. M., Filipe S., Gardete S., Sobral R., Gill S., Chung M., Tomasz A. (1999). Antibiotic resistance as a stress response: complete sequencing of a large number of chromosomal loci in Staphylococcus aureus strain COL that impact on the expression of resistance to methicillin. Microb Drug Resist 5, 163–175. 10.1089/mdr.1999.5.163 [DOI] [PubMed] [Google Scholar]
- Dean B. A., Williams R. E., Hall F., Corse J. (1973). Phage typing of coagulase-negative staphylococci and micrococci. J Hyg (Lond) 71, 261–270. 10.1017/S0022172400022737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deighton M., Borland R. (1993). Regulation of slime production in Staphylococcus epidermidis by iron limitation. Infect Immun 61, 4473–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J., Saier M. H., Jr (1983). ATP-dependent protein kinase-catalyzed phosphorylation of a seryl residue in HPr, a phosphate carrier protein of the phosphotransferase system in Streptococcus pyogenes. Proc Natl Acad Sci U S A 80, 6790–6794. 10.1073/pnas.80.22.6790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J., Reizer J., Fischer C., Galinier A., Saier M. H., Jr, Steinmetz M. (1994). Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J Bacteriol 176, 3336–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J., Küster E., Bergstedt U., Charrier V., Hillen W. (1995). Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in Gram-positive bacteria. Mol Microbiol 15, 1049–1053. 10.1111/j.1365-2958.1995.tb02280.x [DOI] [PubMed] [Google Scholar]
- Dobinsky S., Kiel K., Rohde H., Bartscht K., Knobloch J. K., Horstkotte M. A., Mack D. (2003). Glucose-related dissociation between icaADBC transcription and biofilm expression by Staphylococcus epidermidis: evidence for an additional factor required for polysaccharide intercellular adhesin synthesis. J Bacteriol 185, 2879–2886. 10.1128/JB.185.9.2879-2886.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan R. M. (2001). Biofilms and device-associated infections. Emerg Infect Dis 7, 277–281. 10.3201/eid0702.010226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Ahdab F., Benjamin D. K., Jr, Wang A., Cabell C. H., Chu V. H., Stryjewski M. E., Corey G. R., Sexton D. J., Reller L. B., Fowler V. G., Jr (2005). Risk of endocarditis among patients with prosthetic valves and Staphylococcus aureus bacteremia. Am J Med 118, 225–229. 10.1016/j.amjmed.2004.12.017 [DOI] [PubMed] [Google Scholar]
- Foster T. J. (1998). Molecular genetic analysis of Staphylococcal virulence. In Methods in microbiology, bacterial pathogenesis, pp. 433–454. Edited by Williams P., Ketley J., Salmond G. P. C. San Diego: Academic Press; 10.1016/S0580-9517(08)70303-9 [DOI] [Google Scholar]
- Galinier A., Haiech J., Kilhoffer M. C., Jaquinod M., Stülke J., Deutscher J., Martin-Verstraete I. (1997). The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc Natl Acad Sci U S A 94, 8439–8444. 10.1073/pnas.94.16.8439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke C., Kraft A., Süssmuth R., Schweitzer O., Götz F. (1998). Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem 273, 18586–18593. 10.1074/jbc.273.29.18586 [DOI] [PubMed] [Google Scholar]
- Handke L. D., Slater S. R., Conlon K. M., O’Donnell S. T., Olson M. E., Bryant K. A., Rupp M. E., O’Gara J. P., Fey P. D. (2007). σB and SarA independently regulate polysaccharide intercellular adhesin production in Staphylococcus epidermidis. Can J Microbiol 53, 82–91. 10.1139/w06-108 [DOI] [PubMed] [Google Scholar]
- Henkin T. M., Grundy F. J., Nicholson W. L., Chambliss G. H. (1991). Catabolite repression of α-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacl and galR repressors. Mol Microbiol 5, 575–584. 10.1111/j.1365-2958.1991.tb00728.x [DOI] [PubMed] [Google Scholar]
- Horton R. M., Cai Z. L., Ho S. N., Pease L. R. (1990). Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8, 528–535. [PubMed] [Google Scholar]
- Kim H. J., Roux A., Sonenshein A. L. (2002). Direct and indirect roles of CcpA in regulation of Bacillus subtilis Krebs cycle genes. Mol Microbiol 45, 179–190. 10.1046/j.1365-2958.2002.03003.x [DOI] [PubMed] [Google Scholar]
- Kim J. H., Yang Y. K., Chambliss G. H. (2005). Evidence that Bacillus catabolite control protein CcpA interacts with RNA polymerase to inhibit transcription. Mol Microbiol 56, 155–162. 10.1111/j.1365-2958.2005.04496.x [DOI] [PubMed] [Google Scholar]
- Kristian S. A., Birkenstock T. A., Sauder U., Mack D., Götz F., Landmann R. (2008). Biofilm formation induces C3a release and protects Staphylococcus epidermidis from IgG and complement deposition and from neutrophil-dependent killing. J Infect Dis 197, 1028–1035. 10.1086/528992 [DOI] [PubMed] [Google Scholar]
- Leiter E. H. (1993). The NOD mouse: a model for analyzing the interplay between heredity and environment in development of autoimmune disease. ILAR J 35, 1–14. [Google Scholar]
- Lim Y., Jana M., Luong T. T., Lee C. Y. (2004). Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J Bacteriol 186, 722–729. 10.1128/JB.186.3.722-729.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J. M., Thoms B. (1977). Role of sugar uptake and metabolic intermediates on catabolite repression in Bacillus subtilis. J Bacteriol 129, 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig H., Meinken C., Matin A., Stülke J. (2002). Insufficient expression of the ilv–leu operon encoding enzymes of branched-chain amino acid biosynthesis limits growth of a Bacillus subtilis ccpA mutant. J Bacteriol 184, 5174–5178. 10.1128/JB.184.18.5174-5178.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D., Siemssen N., Laufs R. (1992). Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect Immun 60, 2048–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelmann M., Sabottke A., Laufs R., Mack D. (1998). Generalized transduction for genetic linkage analysis and transfer of transposon insertions in different Staphylococcus epidermidis strains. Zentralbl Bakteriol 287, 85–92. [DOI] [PubMed] [Google Scholar]
- Nicholson W. L., Park Y. K., Henkin T. M., Won M., Weickert M. J., Gaskell J. A., Chambliss G. H. (1987). Catabolite repression-resistant mutations of the Bacillus subtilis alpha-amylase promoter affect transcription levels and are in an operator-like sequence. J Mol Biol 198, 609–618. 10.1016/0022-2836(87)90204-X [DOI] [PubMed] [Google Scholar]
- Novick R. P. (1991). Genetic systems in staphylococci. In Methods in Enzymology, pp. 587–636. San Diego: Academic Press. [DOI] [PubMed] [Google Scholar]
- Piper C., Körfer R., Horstkotte D. (2001). Prosthetic valve endocarditis. Heart 85, 590–593. 10.1136/heart.85.5.590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp M. E., Ulphani J. S., Fey P. D., Bartscht K., Mack D. (1999). Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect Immun 67, 2627–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadykov M. R., Olson M. E., Halouska S., Zhu Y., Fey P. D., Powers R., Somerville G. A. (2008). Tricarboxylic acid cycle-dependent regulation of Staphylococcus epidermidis polysaccharide intercellular adhesin synthesis. J Bacteriol 190, 7621–7632. 10.1128/JB.00806-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadykov M. R., Zhang B., Halouska S., Nelson J. L., Kreimer L. W., Zhu Y., Powers R., Somerville G. A. (2010). Using NMR metabolomics to investigate tricarboxylic acid cycle-dependent signal transduction in Staphylococcus epidermidis. J Biol Chem 285, 36616–36624. 10.1074/jbc.M110.152843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: a Laboratory Manual, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Schlag S., Nerz C., Birkenstock T. A., Altenberend F., Götz F. (2007). Inhibition of staphylococcal biofilm formation by nitrite. J Bacteriol 189, 7911–7919. 10.1128/JB.00598-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl K., Stucki M., Ruegg M., Goerke C., Wolz C., Harris L., Berger-Bächi B., Bischoff M. (2006). Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob Agents Chemother 50, 1183–1194. 10.1128/AAC.50.4.1183-1194.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl K., Bischoff M., Berger-Bächi B. (2008a). CcpA mediates the catabolite repression of tst in Staphylococcus aureus. Infect Immun 76, 5093–5099. 10.1128/IAI.00724-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl K., Goerke C., Wolz C., Mack D., Berger-Bächi B., Bischoff M. (2008b). Staphylococcus aureus CcpA affects biofilm formation. Infect Immun 76, 2044–2050. 10.1128/IAI.00035-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl K., Müller S., François P., Kriebitzsch C., Schrenzel J., Engelmann S., Bischoff M., Berger-Bächi B. (2009). Effect of a glucose impulse on the CcpA regulon in Staphylococcus aureus. BMC Microbiol 9, 95. 10.1186/1471-2180-9-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivers R. P., Dineen S. S., Sonenshein A. L. (2006). Positive regulation of Bacillus subtilis ackA by CodY and CcpA: establishing a potential hierarchy in carbon flow. Mol Microbiol 62, 811–822. 10.1111/j.1365-2958.2006.05410.x [DOI] [PubMed] [Google Scholar]
- Somerville G. A., Proctor R. A. (2009). At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol Mol Biol Rev 73, 233–248. 10.1128/MMBR.00005-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville G. A., Cockayne A., Dürr M., Peschel A., Otto M., Musser J. M. (2003). Synthesis and deformylation of Staphylococcus aureus delta-toxin are linked to tricarboxylic acid cycle activity. J Bacteriol 185, 6686–6694. 10.1128/JB.185.22.6686-6694.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshein A. L. (2007). Control of key metabolic intersections in Bacillus subtilis. Nat Rev Microbiol 5, 917–927. 10.1038/nrmicro1772 [DOI] [PubMed] [Google Scholar]
- Tobisch S., Zühlke D., Bernhardt J., Stülke J., Hecker M. (1999). Role of CcpA in regulation of the central pathways of carbon catabolism in Bacillus subtilis. J Bacteriol 181, 6996–7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eiff C., Peters G., Heilmann C. (2002). Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect Dis 2, 677–685. 10.1016/S1473-3099(02)00438-3 [DOI] [PubMed] [Google Scholar]
- Vuong C., Kidder J. B., Jacobson E. R., Otto M., Proctor R. A., Somerville G. A. (2005). Staphylococcus epidermidis polysaccharide intercellular adhesin production significantly increases during tricarboxylic acid cycle stress. J Bacteriol 187, 2967–2973. 10.1128/JB.187.9.2967-2973.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J. B., Lolkema J. S. (2003). CcpA-dependent carbon catabolite repression in bacteria. Microbiol Mol Biol Rev 67, 475–490. 10.1128/MMBR.67.4.475-490.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert M. J., Chambliss G. H. (1990). Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc Natl Acad Sci U S A 87, 6238–6242. 10.1073/pnas.87.16.6238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray L. V., Jr, Pettengill F. K., Fisher S. H. (1994). Catabolite repression of the Bacillus subtilis hut operon requires a cis-acting site located downstream of the transcription initiation site. J Bacteriol 176, 1894–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Xiong Y. Q., Sadykov M. R., Fey P. D., Lei M. G., Lee C. Y., Bayer A. S., Somerville G. A. (2009). Tricarboxylic acid cycle-dependent attenuation of Staphylococcus aureus in vivo virulence by selective inhibition of amino acid transport. Infect Immun 77, 4256–4264. 10.1128/IAI.00195-09 [DOI] [PMC free article] [PubMed] [Google Scholar]