Abstract
The Francisella pathogenicity island (FPI) encodes proteins thought to compose a type VI secretion system (T6SS) that is required for the intracellular growth of Francisella novicida. In this work we used deletion mutagenesis and genetic complementation to determine that the intracellular growth of F. novicida was dependent on 14 of the 18 genes in the FPI. The products of the iglABCD operon were localized by the biochemical fractionation of F. novicida, and Francisella tularensis LVS. Sucrose gradient separation of water-insoluble material showed that the FPI-encoded proteins IglA, IglB and IglC were found in multiple fractions, especially in a fraction that did not correspond to a known membrane fraction. We interpreted these data to suggest that IglA, IglB and IglC are part of a macromolecular structure. Analysis of published structural data suggested that IglC is an analogue of Hcp, which is thought to form long nano-tubes. Thus the fractionation properties of IglA, IglB and IglC are consistent with the current model of the T6SS apparatus, which supposes that IglA and IglB homologues form an outer tube structure that surrounds an inner tube composed of Hcp (IglC) subunits. Fractionation of F. novicida expressing FLAG-tagged DotU (IcmH homologue) and PdpB (IcmF homologue) showed that these proteins localize to the inner membrane. Deletion of dotU led to the cleavage of PdpB, suggesting an interaction of these two proteins that is consistent with results obtained with other T6SSs. Our results may provide a mechanistic basis for many of the studies that have examined the virulence properties of Francisella mutants in FPI genes, namely that the observed phenotypes of the mutants are the result of the disruption of the FPI-encoded T6SS structure.
Introduction
The facultative intracellular bacterium Francisella tularensis is the aetiological agent of the zoonotic disease tularaemia. The intracellular lifestyle of F. tularensis differs from that of other pathogens studied to date. After phagocytosis, the F. tularensis-laden phagosome acquires early and late endosomal markers such as EEA-1, Rab5, Rab7 and LAMP-1, but fails to acquire lysosomal markers such as cathepsin D (Clemens et al., 2004; Santic et al., 2005). Within 1 h of uptake by a macrophage, the phagosomal membrane enclosing F. tularensis is degraded, allowing the bacterium to gain access to the cytosol (Checroun et al., 2006; Clemens et al., 2004; Golovliov et al., 2003). This is followed by robust replication, leading to eventual host cell lysis. In some studies, late in the infectious cycle, Francisella has been observed to enter a double membranous compartment with the characteristics of an autophagosome (Checroun et al., 2006). The significance of the delay in phagosomal maturation and the role of autophagy are not clear; however, phagosomal escape is critical to the intracellular growth and virulence of this pathogen.
Transposon mutagenesis studies have been useful in identifying a number of genes that are required for F. tularensis to replicate intracellularly (Gray et al., 2002; Tempel et al., 2006). Several of these genes are located in an approximately 30 kb genomic region named the Francisella pathogenicity island (FPI) (Nano et al., 2004). The FPI is found in all Francisella species and strains, and is duplicated in all of the human-virulent biovars of F. tularensis. The closely related bacterium Francisella novicida, frequently referred to as ‘F. tularensis subsp. novicida’, has only one copy of the FPI, and this feature facilitates the genetic analysis of the FPI in this strain.
The FPI gene iglC is one of the most highly induced genes during intracellular growth (Golovliov et al., 1997). Deletion of iglC causes a defect in the ability of Francisella to escape the phagosome (Lindgren et al., 2004), as well as a failure to induce apoptosis and downregulate host cell signalling (Lai et al., 2004; Telepnev et al., 2005). The role of IglC in phagosomal escape is not known, but the crystal structure of IglC (Sun et al., 2007) may soon shed light on its function. Indeed, most FPI proteins do not show similarity to any other protein by blast analysis, making predictions regarding their function difficult, especially in the absence of structural information. Overall, it is clear that the FPI encodes important virulence factors, although the function of FPI proteins has remained elusive.
Recently, two analyses (Bingle et al., 2008; Boyer et al., 2009) of sequenced bacterial genomes have revealed the existence of more than 100 type VI secretion system (T6SS)-like loci, and these studies suggest that the FPI-encoded secretion system is perhaps an outlier among the T6SSs. We previously identified the FPI-encoded proteins IglA, IglB, VgrG, PdpB (IcmF) and DotU as having deduced amino acid sequence similarity to T6SS components found in other bacteria (de Bruin et al., 2007), although these five proteins fall far short of comprising the core of at least 13 proteins thought to be part of a T6SS (Zheng & Leung, 2007). Substantial evidence that the FPI encodes a secretion system comes from the finding that IglI and VgrG are secreted to the cytosol of macrophages (Barker et al., 2009). However, recent work has failed to confirm the FPI-dependent secretion of IglI (Bröms et al., 2011), and no FPI-encoded secreted effector protein has been identified.
Methods
Strains, plasmids and growth conditions.
Strains and plasmids used in this study are listed in Table 1 and described in Supplementary Table S2 (deletion mutants). The wild-type strain of F. novicida was the parent strain of all of the deletion mutants. F. novicida and F. tularensis LVS were cultured in trypticase soy broth or agar medium supplemented with 0.1 % (w/v) cysteine (TSBC or TSAC, respectively). All of the experiments were done with F. novicida strains, except for the cell fractionation studies shown in Fig. 3, for which F. tularensis LVS was used. The LVS strain expresses higher levels of the FPI-encoded proteins, presumably because it has two chromosomal copies of the FPI. Erythromycin (15 µg ml−1), kanamycin (15 µg ml−1) or 10 % (w/v) sucrose was added as needed. Escherichia coli strains were grown in Luria–Bertani broth and supplemented with kanamycin (30 µg ml−1) or ampicillin (200 µg ml−1) as needed.
Table 1. Strains and plasmids used in this study.
| Strain or plasmid | Genotype or properties | Source or reference |
| E. coli strain | ||
| DH5α | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 (ara-leu) 7697 galU galK rpsL (StrR) endA1 nupG | Invitrogen |
| F. novicida strains | ||
| Wild-type | Wild-type prototype strain, U112 | ATCC |
| LVS | F. tularensis live vaccine strain | ATCC |
| Plasmids | ||
| pJL-SKX | F. novicida integrating vector; KmR | Ludu et al. (2008b) |
| pFNLTP6-groE-GFP | F. tularensis–E. coli shuttle vector with gfp gene driven by groEL promoter; KmR | Maier et al. (2004) |
| pKH3 | pFNLTP6-groEgfp with sopB-3×FLAG replacing gfp gene. Allows cloning of DNA to replace sopB and generate in-frame fusion to 3×FLAG as EcoRI, Kpn or NdeI–NcoI fragment. NcoI site in KmR marker of pFNLTP6 removed by site-directed mutagenesis. See Supplementary Fig. S2, Supplementary Table S3 | Maier et al. (2004); this study |
Fig. 3.
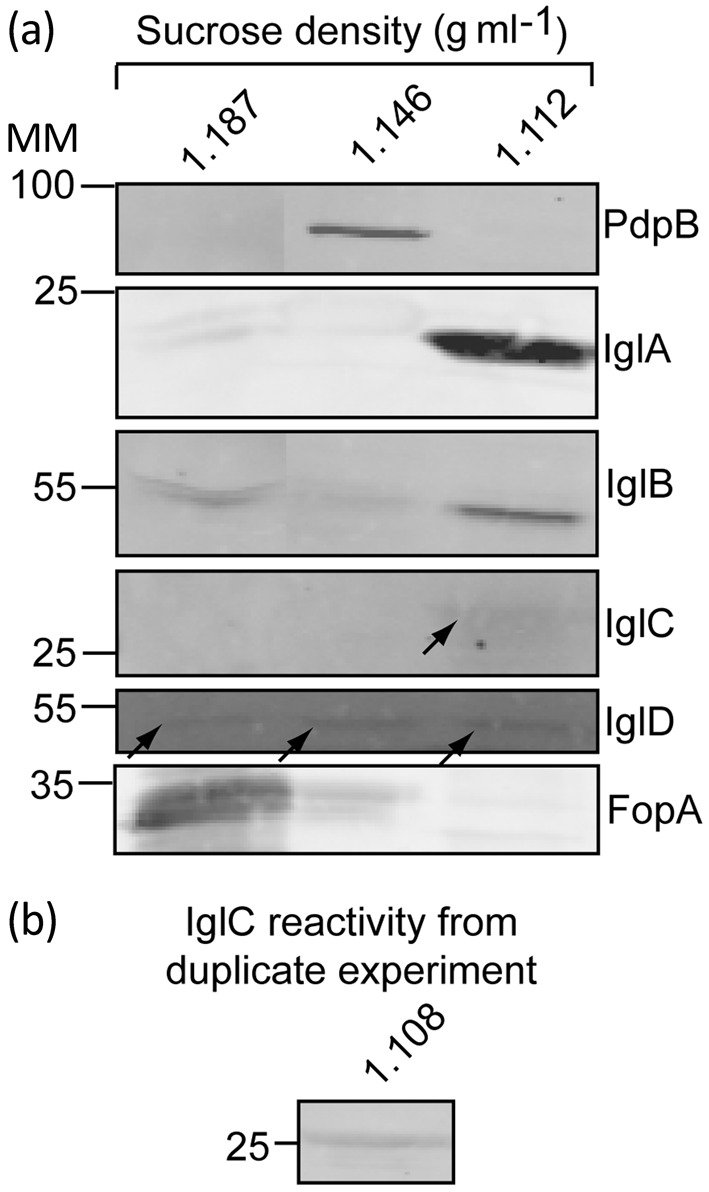
Localization of IglA, IglB, IglC and IglD following osmotic lysis of F. tularensis LVS cells. (a) Immunoblot analysis of fractions from sucrose gradient separation of insoluble material from a bacterial extract generated by osmotic lysis of cells. (b) Immunoblot of IglC from a similar sucrose fraction from a duplicate experiment of that shown in (a). MM, molecular mass (kDa).
Construction of deletion strains and strains with complementing plasmids.
Unmarked F. novicida deletion mutants of FPI genes were constructed using the sacB counter-selection method, as previously described (de Bruin et al., 2007). The approach to deletion mutagenesis is shown in Supplementary Fig. S1, and the primers used to make each deletion and the precise location of deletions in each gene are listed in Supplementary Tables S1 and S2, respectively. The entire collection of F. novicida FPI deletion mutants has been deposited with the American Type Culture Collection (ATCC) Biodefense and Emerging Infections Resources (BEI) program. To construct complementing plasmids expressing FPI-encoded proteins, targeted genes were PCR-amplified and ligated into pKH3, a pFNLTP derivative (Maier et al., 2004) with a 3×FLAG insert engineered into the plasmid to create C-terminal fusions to recombinant inserts. The primers used to amplify the FPI genes for insertion into pKH3 are listed in Supplementary Table S3. Further details of plasmid construction and complementation are presented in the Supplementary Methods.
Subcellular localization of Francisella proteins.
Two previously described methods, one that uses detergent-based separation of inner and outer membranes and one that uses sucrose gradient centrifugation separation, were used to separate FPI-encoded proteins into different biochemical fractions. Details of these methods are provided in the Supplementary Methods.
Intracellular growth assay.
Macrophage infection assays were performed as previously described (Schmerk et al., 2009a). In brief, J774A.1 mouse macrophage-like cells were seeded in 96-well cell culture plates at a density of 5×104 cells per well and allowed to adhere overnight. F. novicida strains were added to the wells at an m.o.i. of 50 : 1. The infected monolayers were incubated at 37 °C in an atmosphere of 5 % CO2 for 1 h to allow uptake of the bacteria. Sterile gentamicin sulfate was added to a final concentration of 10 µg ml−1 for 1 h. The monolayers were washed three times to remove external bacteria and gentamicin. At various times post-infection, the infected J774 cells were lysed by the addition of 0.1 % (w/v) deoxycholic acid. Lysates were serially diluted in PBS containing 0.1 % (w/v) gelatin and plated on TSAC for viable count determination.
Western immunoblot analysis.
SDS-PAGE and Western blotting were performed according to standard techniques, as previously described (de Bruin et al., 2007). The sources and dilution of the antibodies are described in the Supplementary Methods.
Results and Discussion
Deletion mutagenesis and genetic complementation reveal a requirement for individual FPI genes in intramacrophage growth
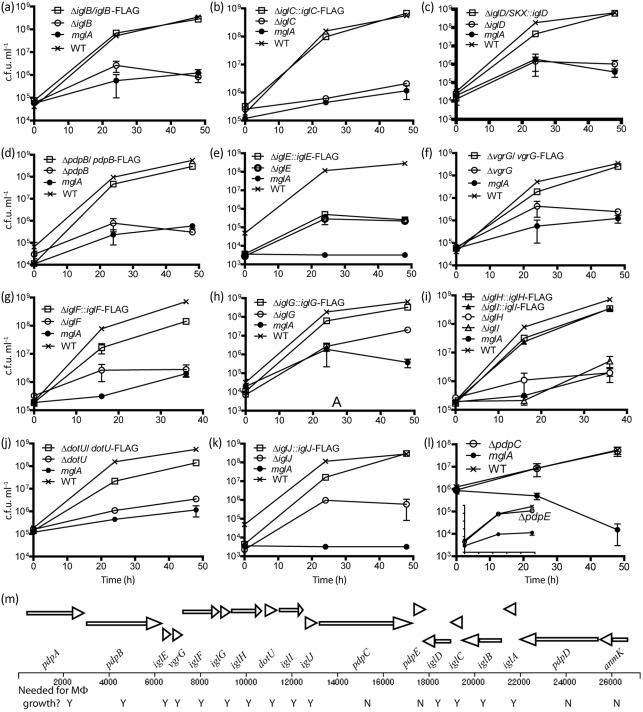
Several studies have examined the phenotypes of insertion mutants in FPI genes, and most mutants have been found to be defective for intracellular growth (Gray et al., 2002; Maier et al., 2007; Nano et al., 2004; Tempel et al., 2006). Since we now know that many FPI genes are needed for intracellular growth, and that many of the insertion mutations affect the expression of downstream genes (Chong et al., 2008; de Bruin et al., 2007; Maier et al., 2007), a thorough analysis of the role of individual FPI genes in intracellular growth was needed. To help determine which FPI-encoded gene products are required for intracellular growth we constructed an in-frame, markerless deletion mutant strain for each FPI gene (see Supplementary Fig. S1, Supplementary Table S2). Each deletion mutation was also analysed by creating strains that contained genetic complements, usually in the form of plasmid-borne copies of the cognate wild-type gene along with a C-terminal-encoded FLAG-tag (see Supplementary Fig. S2, Supplementary Table S3). As shown in Fig. 1, deletion mutants of iglBCD, pdpB, vgrG, dotU and iglEFGHIJ were defective for intramacrophage growth, and genetic complementation of each mutation restored intracellular growth, except in the case of the ΔiglE mutant. Multiple approaches to complement the ΔiglE mutant failed, possibly because both insufficient and excessive expression of IglE contribute to avirulence. Deletion of pdpC (Fig. 1l) or pdpE (Fig. 1l, insert) did not affect intracellular growth. We have previously analysed markerless deletion mutants and their complements for iglA, pdpA, pdpD and anmK, and we found that iglA and pdpA are required for intracellular growth, whereas anmK and pdpD are not required for intracellular growth but are required for virulence (de Bruin et al., 2007; Ludu et al., 2008a; Schmerk et al., 2009a). Hence, we conclude that pdpA, pdpB, dotU, vgrG and iglABCDEFGHIJ are required for intracellular growth, whereas pdpCDE and anmK are not. Interestingly, the number of FPI genes required for intracellular growth (14) is close to the minimal number of genes (13) thought to be required to encode a T6SS (Zheng & Leung, 2007).
Fig. 1.
Intracellular growth phenoptype of FPI deletion mutants. (a–l) Growth of the FPI gene mutants and their complements in J774 mouse macrophage-like cells. The growth of ΔpdpE, which is essentially the same as the wild-type (WT), is presented as an insert in (l). The mglA mutant lacks expression of a global virulence regulator that affects all FPI genes, and it is defective for intracellular growth. The mglA mutant is included as a negative control for extracellular growth in the tissue culture medium. All of the genetic complements expressed the cognate FPI gene as a 3 × FLAG-tagged version from a plasmid vector, except for the iglD complement, which carried iglD on a chromosomally integrated vector, pJL-SKX. The graphs are representative of three repetitions, which all gave similar results. The P values generated by two-way analysis of variance (ANOVA) for the differences between the mutant strain and WT or the cognate complemented strain were all <0.0001, except for the following: ΔiglA vs WT, 0.0002; ΔiglJ vs ΔiglJ/iglJ–FLAG, 0.004; ΔdotU vs WT, 0.0002; ΔiglE vs ΔiglE/iglE–FLAG, 0.6; ΔpdpC vs WT, 0.3; ΔpdpE vs WT, 0.4. A variety of strategies were attempted to complement the ΔiglE mutant strain and none succeeded in restoring WT intracellular growth. Analysis of the ΔiglE mutant showed that the product of the downstream gene, vgrG, produced WT amounts of the protein product, suggesting that the ΔiglE mutant did not have polar effects on transcription/translation coupling (see Supplementary Fig. S1c). Error bars, which are too small to be visible for some data points, show sd. (m) FPI gene organization and requirement for intramacrophage growth. FPI genes are represented by arrows, and ‘Y/N’ indicates the need of each FPI gene for intracellular growth (Y = yes, N = no).
Localization of IglA, IglB, IglC and IglD
The dominant model of the role of T6SS proteins posits that IglA and IglB homologues form a tube structure that spans the inner and outer membranes (Bönemann et al., 2009, 2010). Our analysis (see below) of the structure of IglC suggests that it is the major subunit of a tube structure that lies inside the IglAB tube. However, published studies present differing, and perhaps conflicting, data as to the biochemical properties of IglA, IglB and IglC. Studies of IglA (de Bruin et al., 2007) and IglC (Bröms et al., 2011; Golovliov et al., 1997) suggest that they are cytoplasmic proteins. Separate studies have provided evidence that IglA, IglB and IglC are surface-exposed (Ludu et al., 2008a; Melillo et al., 2006), since they can be biotinylated in intact F. novicida (IglABC) and F. tularensis LVS (IglA) cells. In an attempt to resolve these discrepancies, we examined the association of IglABCD with different biochemical fractions of the bacterial cell, and we took two different approaches to examining the localization pattern of the proteins.
A commonly used method to separate inner- and outer-membrane proteins is to treat membrane fractions with the detergent Sarkosyl. Inner-membrane proteins are usually solubilized by Sarkosyl, and are thus said to associate with the ‘Sarkosyl-soluble’ fraction of the membrane, whereas outer-membrane proteins are not, and are thus said to associate with the ‘Sarkosyl-insoluble’ fraction of the membrane. When this biochemical fractionation was applied to F. novicida, the FPI-encoded protein IglB was found in all fractions, but most heavily in the Sarkosyl-insoluble fraction (Fig. 2b). IglA and IglC were found predominantly in the Sarkosyl-insoluble fraction, with lesser amounts in the osmotic shock fraction, and a very small amount in the soluble and Sarkosyl-soluble fractions (Fig. 2a, c). IglD was found predominantly in the soluble fraction and in the osmotic shock fraction, which represents primarily periplasmic proteins (Fig. 2d). It could be argued that the appearance of IglA, IglB, IglC and IglD in the soluble fraction represents protein that localizes to the periplasm. The localization of IglA, IglB and IglC in the Sarkosyl-insoluble fraction may reflect a true localization to the outer membrane or may simply indicate that these three proteins are in a complex that is large enough to be pelleted by ultracentrifugation and are therefore not solubilized by Sarkosyl. The gene regulatory protein MglB localized, as expected, to the cytoplasm (Fig. 2e). PdpB was found in the Sarkosyl-soluble fraction (Fig. 2f). This protein has previously been shown to co-localize with the inner-membrane marker NADH oxidase (de Bruin et al., 2007; Schmerk et al., 2009b), and its localization to the Sarkosyl-soluble fraction is consistent with these previous studies. The outer-membrane protein FopA was found predominantly associated with the Sarkosyl-insoluble fraction (Fig. 2g). The IglA, IglB and IglC proteins all lack an N-terminal signal that is needed for export via the Sec pathway or the twin arginine translocation pathway, and thus any true localization to the outer membrane would require a mechanism that is not readily apparent.
Fig. 2.
Localization of IglA, IglB, IglC and IglD in different biochemical fractions using detergent-based fractionation. (a–d) Immunoblots with antibodies reactive with IglA, IglB, IglC and IglD following detergent-based separation of F. novicida extracts. The amount of proteins applied to SDS-PAGE was normalized to 5 µg for each lane. The samples in the Sarkosyl-insoluble (outer-membrane) lane were distorted by the large amount of LPS present. The samples in the osmotic shock preparation (periplasm) were concentrated by filtration through a 10 kDa selective filter. (e–g) Immunoblots to detect control proteins following SDS-PAGE. MglB is a gene regulatory protein found in the cytoplasm. PdpB had previously been shown to be associated almost exclusively with the Sarkosyl-soluble fraction and the inner-membrane marker NADH oxidase (Schmerk et al., 2009b). FopA is an outer-membrane protein (Nano, 1988).
In a second approach to bacterial cell fractionation we used a gentle osmotic lysis technique (Huntley et al., 2007), followed by sucrose gradient centrifugation to separate the insoluble fractions of Francisella. Using this approach we found that detection of FPI-encoded proteins was difficult with F. novicida cells, and was better with F. tularensis LVS cells; the latter strain has two copies of the FPI and produces higher levels of the proteins.
When osmotic lysis and sucrose density centrifugation were applied to separating insoluble fractions of F. tularensis LVS we found three major bands of protein, at sucrose densities centred at approximately 1.19, 1.15 and 1.11 g ml−1 (Fig. 3a). The precise density of the protein banding varied slightly among experiments, but always followed the same pattern. The band at 1.19 g ml−1 contained the bulk of the FopA outer-membrane protein present in the insoluble material. The major anti-PdpB reactivity was found at about 1.15 g ml−1. We had previously shown (Schmerk et al., 2009b) that PdpB co-localizes with the bulk of the NADH oxidase activity, a marker for the inner membrane. The sucrose gradient fractions in which we found FopA and PdpB correspond well with the sucrose gradient densities found by Huntley and co-workers that contained outer-membrane proteins (1.18 g ml−1) and inner-membrane proteins (1.14 g ml−1), respectively (Huntley et al., 2007).
A remarkable feature of the sucrose gradient separation was the appearance of anti-IglA, anti-IglB and anti-IglC reactivity that was found primarily in sucrose gradient fractions corresponding to 1.11 g ml−1, a region of the gradient that is far separated from those that contain membrane fractions as identified by reactivity with anti-FopA or anti-PdpB antibody (Fig. 3a, b). Note that all of the material in the sucrose gradient was present by virtue of the fact that it was part of a pellet formed after lysed cells were subjected to high-speed centrifugation. Material found in the sucrose gradient, but not associated with a clearly identifiable membrane fraction, is presumably associated with a macromolecular structure that is large enough to be pelleted by high-speed centrifugation. Thus, the presence of IglA, IglB and IglC at the 1.11 g ml−1 position in the sucrose gradient is consistent with the notion of their association with a macromolecular structure that is distinct from the inner and outer membranes.
Our observation that IglA and IglC associate with an insoluble fraction would appear to contradict the previous conclusion that these proteins are water soluble, and presumably located in the bacterial cytoplasm. The hypothesis that IglA and IglC are major components of the tube structure of an FPI-encoded T6SS provides an explanation for the apparently conflicting data. During gentle osmotic lysis of Francisella cells the tube structure may remain largely intact, and thus insoluble. However, techniques that use pressure or sonic disruption to lyse cells may insert sufficient energy to disrupt the tube structures and release IglA and IglC as water-soluble monomers.
IglC as a possible Hcp analogue
Our cell fractionation results showed that IglA, IglB and IglC can be pelleted by high-speed centrifugation and migrate together during a sucrose density separation of bacterial structures. In most gene clusters encoding T6SSs the Hcp homologue is encoded by a gene that lies immediately upstream or downstream of homologues of iglAB (Shrivastava & Mande, 2008). In the FPI, the gene encoding IglC follows those encoding IglA and IglB, suggesting the possibility that IglC is an Hcp analogue. IglA and IglB are clear homologues of VipA and VipB, respectively, and the dominant model of T6SS apparatuses posits that VipAB form an outer tube structure that surrounds an inner tube composed of Hcp units (Bönemann et al., 2010). Thus our biochemical results, together with the current model of the T6SS, suggested to us that IglC may play the role in the FPI-encoded T6SS that Hcp plays in other T6SSs.
Although matches of the amino acid sequence of IglC to sequences in protein databases fail to show any similarity to Hcp homologues, an analysis of its tertiary structure does suggest a similarity to Hcp homologues and similarities to bacteriophage components. When the structure of IglC was published, the authors found that the VAST algorithm did not detect highly significant structural similarities to proteins in the Protein Data Bank (PDB) databases (Sun et al., 2007). However, recent updates to the database have revealed structural similarities between IglC and Hcp and presumed secreted analogues of Hcp in other T6SSs; IglC has also been found to be similar to bacteriophage tail proteins. Using the DALI server, we found that the IglC crystal structure (2qwu) showed similarity to a baseplate protein of a Lactobacillus lactis bacteriophage [2wzp, root mean square deviation (RMSD) score 3.2], and Hcp3 (3he1, RMSD score 4.0) and Hcp1 (1y12, RMSD score 4.3) from Pseudomonas aeruginosa.
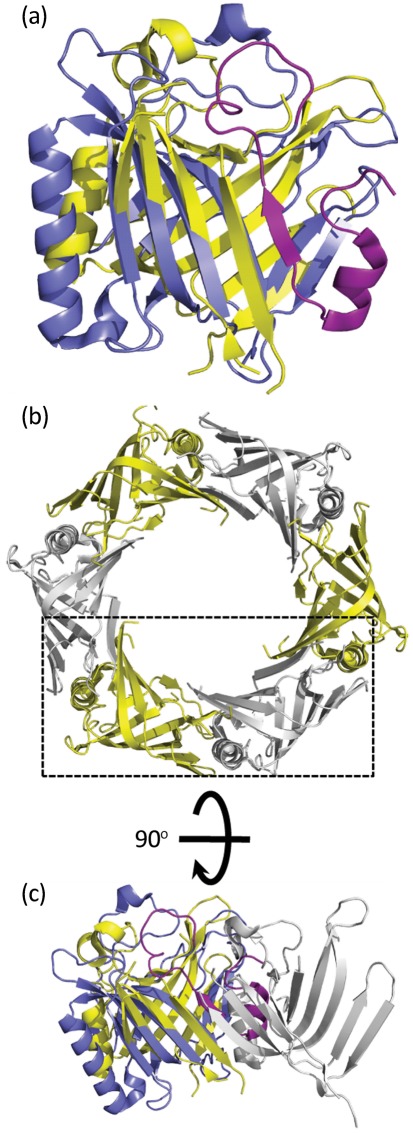
A structural overlay of IglC with Hcp3 from P. aeruginosa reveals the structural identity of the two proteins (RMSD of 4.0 Å over 128 matched Cα atoms) (Fig. 4a). This suggests that IglC may have the capacity to adopt a similar hexameric structure to Hcp3 (Osipiuk et al., 2011) (Fig. 4b). However, there are about 30 aa at the N terminus of IglC that, in this form, would appear to prevent the formation of a hexamer with identical packing to Hcp3 (Fig. 4c). There is evidence that IglC may have different isoforms (Pavkova et al., 2006; Twine et al., 2006), but at present there is no evidence that part of the N terminus is removed. Nevertheless, the clear structural similarity of IglC to Hcp proteins gives a strong indication of analogous functional roles, although possibly with IglC packing into hexamers slightly differently from Hcp analogues.
Fig. 4.
Structural comparison of IglC with the canonical Hcp from P. aeruginosa (PDB code 3he1). (a) An overlap of Hcp (yellow) with IglC (purple; RMSD of 4.0 Å over 128 matched Cα atoms). The IglC region shown in magenta indicates an ~32 aa N-terminal extension that is not structurally present in Hcp. (b) The hexameric arrangement of Hcp monomers. (c) The Hcp dimer indicated by the box in (b) rotated 90° around the horizontal axis and overlapped with IglC.
Effect of DotU loss on the stability of PdpB
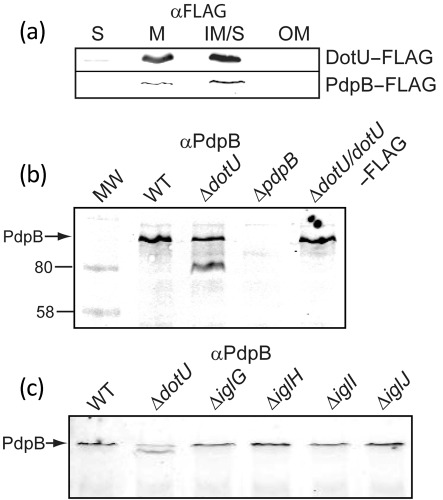
The localization that we found previously and in this study for PdpB to the inner membrane is consistent with its role as an IcmF homologue. Since IcmF homologues are known to interact with DotU (IcmH) homologues in the inner membrane, we examined the localization of the FPI-encoded DotU using a FLAG-tagged recombinant. As expected, DotU–FLAG localized to the inner membrane (Fig. 5a). In a Legionella pneumophila mutant with deletions of icmF and dotU, the Dot/Icm complex is destabilized (Sexton et al., 2004). As an approach to testing whether this phenomenon holds true in F. novicida we examined the stability of PdpB in a dotU mutant. In extracts of a ΔdotU mutant we found that anti-PdpB antibody detected two reactive bands, including one of a lower relative molecular mass than the normal PdpB band (Fig. 5b), suggesting that PdpB was being proteolytically cleaved. This phenotype was reversed by the expression of DotU–FLAG (Fig. 5b). To determine whether the stability of PdpB is dependent on other FPI gene products, we analysed whole-cell lysates of strains deleted in two genes upstream and two genes downstream of dotU and found that only the deletion of dotU affected PdpB stability (Fig. 5c). We have not shown that DotU directly interacts with PdpB, but cumulatively our data indicate that DotU expression is required to stabilize PdpB, as has been shown with IcmF and DotU orthologues in type IV secretion systems and one T6SS (Ma et al., 2009; Sexton et al., 2004).
Fig. 5.
PdpB degradation in the absence of DotU. (a) Anti-FLAG (αFLAG) immunoblot showing that DotU–FLAG and PdpB–FLAG localize in a similar fashion to the inner membrane. Columns S and M represent an experiment in which bacterial extracts were separated between a soluble and an insoluble (membrane) fraction, respectively. Columns IM/S and OM represent a separate experiment in which bacterial extracts were separated into soluble together with a Sarkosyl-soluble (inner-membrane, IM/S) fraction and a Sarkosyl-insoluble fraction (OM). (b) Immunoblot with anti-PdpB monoclonal antibody (αPdpB) against wild-type, ΔdotU and a genetically complemented ΔdotU strain, showing a difference in the number of anti-PdpB reactive bands. (c) Anti-PdpB immunoblot of cell lysates from strains deleted in genes upstream (iglG, iglH) and downstream (iglI, iglJ) of dotU.
Model of an FPI-encoded T6SS
If we assume that IglC plays the role of Hcp homologues, and that PdpB (IcmF) and DotU have similar functions to their homologues in other bacteria, then it is simple to adopt the model of the T6SS apparatus that has been proposed by Bönemann et al. (2010) for the proteins encoded by the FPI (Fig. 6). In our adaptation of the Bönemann model the IglA and IglB proteins form an outer tube, with IglC being the dominant component of the inner tube. Importantly, IglA and IglB are known to interact (Bröms et al., 2009; de Bruin et al., 2007). The localization of DotU was shown in this work to be in the inner membrane, and experiments presented here and elsewhere show that PdpB also localizes to the inner membrane (Bröms et al., 2011; Schmerk et al., 2009b). The results of Barker and colleagues suggest that IglI and VgrG are both secreted from F. novicida (Barker et al., 2009), although contradictory results have been obtained with F. tularensis LVS (Bröms et al., 2011). Although there are many issues to be resolved, an image of the FPI-encoded T6SS is beginning to emerge.
Fig. 6.
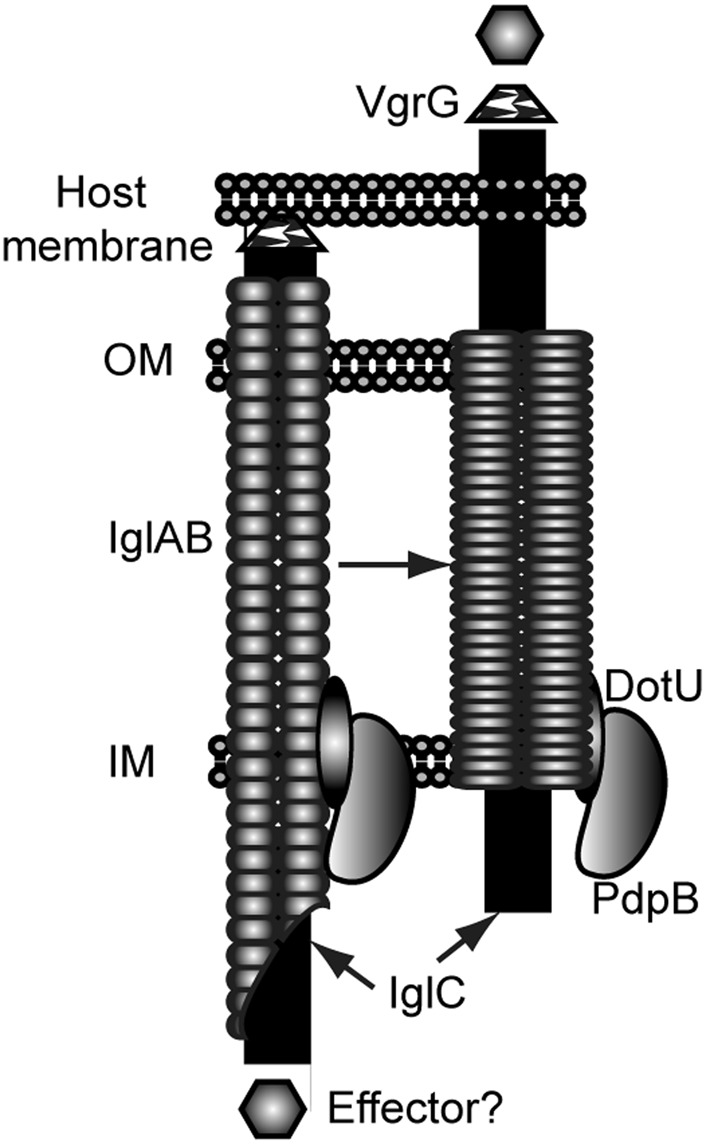
Model of the FPI-encoded T6SS-like apparatus. Polymers of IglAB are drawn as a supramolecular complex that forms a tube that spans the inner (IM) and outer (OM) membranes of F. novicida. The inner tube is proposed to be composed of a polymer of IglC. Contraction of the IglAB tube is proposed to drive the IglC inner tube through the host cell membrane, and the penetration of the membrane is aided by VgrG. DotU and PdpB are shown as interacting with each other and associated with the inner membrane. The model shows secretion of a presumed effector protein.
Supplementary Material
Acknowledgements
This work was supported by a grant from the Canadian Institutes of Health Research (MOP 89812) and by a grant from the National Institute of Allergy and Infectious Diseases (5R01 AI056212-02) to F. E. N. R. F. H. and K. H. were supported through National Center for Research Resources (NCRR) grant RR016466.
Abbreviations:
- FPI
Francisella pathogenicity island
- RMSD
root mean square deviation
- T6SS
type VI secretion system
Footnotes
Supplementary methods, two supplementary figures and three supplementary tables are available with the online version of this paper.
References
- Barker J. R., Chong A., Wehrly T. D., Yu J. J., Rodriguez S. A., Liu J., Celli J., Arulanandam B. P., Klose K. E. (2009). The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol Microbiol 74, 1459–1470. 10.1111/j.1365-2958.2009.06947.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingle L. E., Bailey C. M., Pallen M. J. (2008). Type VI secretion: a beginner’s guide. Curr Opin Microbiol 11, 3–8. 10.1016/j.mib.2008.01.006 [DOI] [PubMed] [Google Scholar]
- Bönemann G., Pietrosiuk A., Diemand A., Zentgraf H., Mogk A. (2009). Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J 28, 315–325. 10.1038/emboj.2008.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bönemann G., Pietrosiuk A., Mogk A. (2010). Tubules and donuts: a type VI secretion story. Mol Microbiol 76, 815–821. 10.1111/j.1365-2958.2010.07171.x [DOI] [PubMed] [Google Scholar]
- Boyer F., Fichant G., Berthod J., Vandenbrouck Y., Attree I. (2009). Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10, 104. 10.1186/1471-2164-10-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröms J. E., Lavander M., Sjöstedt A. (2009). A conserved α-helix essential for a type VI secretion-like system of Francisella tularensis. J Bacteriol 191, 2431–2446. 10.1128/JB.01759-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröms J. E., Lavander M., Meyer L., Sjöstedt A. (2011). IglG and IglI of the Francisella pathogenicity island are important virulence determinants of Francisella tularensis LVS. Infect Immun 79, 3683–3696. 10.1128/IAI.01344-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checroun C., Wehrly T. D., Fischer E. R., Hayes S. F., Celli J. (2006). Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci U S A 103, 14578–14583. 10.1073/pnas.0601838103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong A., Wehrly T. D., Nair V., Fischer E. R., Barker J. R., Klose K. E., Celli J. (2008). The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect Immun 76, 5488–5499. 10.1128/IAI.00682-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens D. L., Lee B. Y., Horwitz M. A. (2004). Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect Immun 72, 3204–3217. 10.1128/IAI.72.6.3204-3217.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin O. M., Ludu J. S., Nano F. E. (2007). The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol 7, 1. 10.1186/1471-2180-7-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovliov I., Ericsson M., Sandström G., Tärnvik A., Sjöstedt A. (1997). Identification of proteins of Francisella tularensis induced during growth in macrophages and cloning of the gene encoding a prominently induced 23-kilodalton protein. Infect Immun 65, 2183–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovliov I., Baranov V., Krocova Z., Kovarova H., Sjöstedt A. (2003). An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect Immun 71, 5940–5950. 10.1128/IAI.71.10.5940-5950.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray C. G., Cowley S. C., Cheung K. K., Nano F. E. (2002). The identification of five genetic loci of Francisella novicida associated with intracellular growth. FEMS Microbiol Lett 215, 53–56. 10.1111/j.1574-6968.2002.tb11369.x [DOI] [PubMed] [Google Scholar]
- Huntley J. F., Conley P. G., Hagman K. E., Norgard M. V. (2007). Characterization of Francisella tularensis outer membrane proteins. J Bacteriol 189, 561–574. 10.1128/JB.01505-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai X. H., Golovliov I., Sjöstedt A. (2004). Expression of IglC is necessary for intracellular growth and induction of apoptosis in murine macrophages by Francisella tularensis. Microb Pathog 37, 225–230. 10.1016/j.micpath.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Lindgren H., Golovliov I., Baranov V., Ernst R. K., Telepnev M., Sjöstedt A. (2004). Factors affecting the escape of Francisella tularensis from the phagolysosome. J Med Microbiol 53, 953–958. 10.1099/jmm.0.45685-0 [DOI] [PubMed] [Google Scholar]
- Ludu J. S., de Bruin O. M., Duplantis B. N., Schmerk C. L., Chou A. Y., Elkins K. L., Nano F. E. (2008a). The Francisella pathogenicity island protein PdpD is required for full virulence and associates with homologues of the type VI secretion system. J Bacteriol 190, 4584–4595. 10.1128/JB.00198-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludu J. S., Nix E. B., Duplantis B. N., de Bruin O. M., Gallagher L. A., Hawley L. M., Nano F. E. (2008b). Genetic elements for selection, deletion mutagenesis and complementation in Francisella spp. FEMS Microbiol Lett 278, 86–93. 10.1111/j.1574-6968.2007.00979.x [DOI] [PubMed] [Google Scholar]
- Ma L. S., Lin J. S., Lai E. M. (2009). An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its Walker A motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J Bacteriol 191, 4316–4329. 10.1128/JB.00029-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T. M., Havig A., Casey M., Nano F. E., Frank D. W., Zahrt T. C. (2004). Construction and characterization of a highly efficient Francisella shuttle plasmid. Appl Environ Microbiol 70, 7511–7519. 10.1128/AEM.70.12.7511-7519.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T. M., Casey M. S., Becker R. H., Dorsey C. W., Glass E. M., Maltsev N., Zahrt T. C., Frank D. W. (2007). Identification of Francisella tularensis Himar1-based transposon mutants defective for replication in macrophages. Infect Immun 75, 5376–5389. 10.1128/IAI.00238-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melillo A., Sledjeski D. D., Lipski S., Wooten R. M., Basrur V., Lafontaine E. R. (2006). Identification of a Francisella tularensis LVS outer membrane protein that confers adherence to A549 human lung cells. FEMS Microbiol Lett 263, 102–108. 10.1111/j.1574-6968.2006.00413.x [DOI] [PubMed] [Google Scholar]
- Nano F. E. (1988). Identification of a heat-modifiable protein of Francisella tularensis and molecular cloning of the encoding gene. Microb Pathog 5, 109–119. 10.1016/0882-4010(88)90013-7 [DOI] [PubMed] [Google Scholar]
- Nano F. E., Zhang N., Cowley S. C., Klose K. E., Cheung K. K., Roberts M. J., Ludu J. S., Letendre G. W., Meierovics A. I., et al. & other authors (2004). A Francisella tularensis pathogenicity island required for intramacrophage growth. J Bacteriol 186, 6430–6436. 10.1128/JB.186.19.6430-6436.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipiuk J., Xu X., Cui H., Savchenko A., Edwards A., Joachimiak A. (2011). Crystal structure of secretory protein Hcp3 from Pseudomonas aeruginosa. J Struct Funct Genomics 12, 21–26. 10.1007/s10969-011-9107-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavkova I., Reichelova M., Larsson P., Hubalek M., Vackova J., Forsberg A., Stulik J. (2006). Comparative proteome analysis of fractions enriched for membrane-associated proteins from Francisella tularensis subsp. tularensis and F. tularensis subsp. holarctica strains. J Proteome Res 5, 3125–3134. 10.1021/pr0601887 [DOI] [PubMed] [Google Scholar]
- Santic M., Molmeret M., Abu Kwaik Y. (2005). Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-γ. Cell Microbiol 7, 957–967. 10.1111/j.1462-5822.2005.00529.x [DOI] [PubMed] [Google Scholar]
- Schmerk C. L., Duplantis B. N., Howard P. L., Nano F. E. (2009a). A Francisella novicida pdpA mutant exhibits limited intracellular replication and remains associated with the lysosomal marker LAMP-1. Microbiology 155, 1498–1504. 10.1099/mic.0.025445-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmerk C. L., Duplantis B. N., Wang D., Burke R. D., Chou A. Y., Elkins K. L., Ludu J. S., Nano F. E. (2009b). Characterization of the pathogenicity island protein PdpA and its role in the virulence of Francisella novicida. Microbiology 155, 1489–1497. 10.1099/mic.0.025379-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton J. A., Miller J. L., Yoneda A., Kehl-Fie T. E., Vogel J. P. (2004). Legionella pneumophila DotU and IcmF are required for stability of the Dot/Icm complex. Infect Immun 72, 5983–5992. 10.1128/IAI.72.10.5983-5992.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava S., Mande S. S. (2008). Identification and functional characterization of gene components of type VI secretion system in bacterial genomes. PLoS ONE 3, e2955. 10.1371/journal.pone.0002955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P., Austin B. P., Schubot F. D., Waugh D. S. (2007). New protein fold revealed by a 1.65 Å resolution crystal structure of Francisella tularensis pathogenicity island protein IglC. Protein Sci 16, 2560–2563. 10.1110/ps.073177307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telepnev M., Golovliov I., Sjöstedt A. (2005). Francisella tularensis LVS initially activates but subsequently down-regulates intracellular signaling and cytokine secretion in mouse monocytic and human peripheral blood mononuclear cells. Microb Pathog 38, 239–247. 10.1016/j.micpath.2005.02.003 [DOI] [PubMed] [Google Scholar]
- Tempel R., Lai X. H., Crosa L., Kozlowicz B., Heffron F. (2006). Attenuated Francisella novicida transposon mutants protect mice against wild-type challenge. Infect Immun 74, 5095–5105. 10.1128/IAI.00598-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twine S. M., Mykytczuk N. C., Petit M. D., Shen H., Sjöstedt A., Wayne Conlan J., Kelly J. F. (2006). In vivo proteomic analysis of the intracellular bacterial pathogen, Francisella tularensis, isolated from mouse spleen. Biochem Biophys Res Commun 345, 1621–1633. 10.1016/j.bbrc.2006.05.070 [DOI] [PubMed] [Google Scholar]
- Zheng J., Leung K. Y. (2007). Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol 66, 1192–1206. 10.1111/j.1365-2958.2007.05993.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.