Abstract
Despite the detrimental role that endogenously generated reactive oxygen species (ROS) may play in bacteria exposed to aerobic environments, very few sources of ROS have been identified in vivo. Such studies are often precluded by the presence of efficient ROS-scavenging pathways, like those found in the aerotolerant anaerobe Bacteroides fragilis. Here we demonstrate that deletion of the genes encoding catalase (Kat), alkylhydroperoxide reductase (AhpC) and thioredoxin-dependent peroxidase (Tpx) strongly inhibits H2O2 detoxification in B. fragilis, thereby allowing for the quantification of ROS production. Exogenous fumarate significantly reduced H2O2 production in a ΔahpCΔkatΔtpx B. fragilis strain, as did deletion of fumarate reductase subunit c (frdC). Deletion of frdC also increased the aerotolerance of a strain lacking superoxide dismutase, indicating that fumarate reductase is a major contributor to ROS formation in B. fragilis exposed to oxygen.
Introduction
Life in an oxygenated world presents interesting and difficult challenges to organisms from a variety of environments. Due to its small size and unpaired outer-orbital electrons, molecular oxygen (O2) can permeate cells and strip electrons from a wide variety of biomolecules (Fridovich, 1998, 1999; Imlay, 2003). These adventitious reactions result in the formation of partially reduced derivatives of O2 appropriately known as reactive oxygen species (ROS). Such ROS include superoxide (), hydrogen peroxide (H2O2) and the hydroxyl radical, and their effects on the cell include inactivation of integral proteins and damage to DNA, eventually leading to cell death (Imlay, 2003). To survive in such a climate, an organism must attempt to limit the rate of endogenous ROS production and to detoxify ROS quickly upon formation. To this end, organisms such as Escherichia coli have evolved ROS-detoxifying enzymes such as superoxide dismutase (Sod) (McCord & Fridovich, 1969), alkylhydroperoxide reductase (AhpC) (Imlay, 2008; Parsonage et al., 2008) and catalases. E. coli can easily shift between anaerobic and aerobic environments, in part due to this robust response to oxidative stress, but many bacteria do not display such metabolic flexibility and are therefore relegated to niches containing little to no oxygen. These so called ‘anaerobes’ stop growing upon the introduction of O2 and many die, but the nature of this sensitivity is still to be elucidated. One interesting and important model for studying the effects of oxygen on anaerobic bacteria is Bacteroides fragilis. Long regarded as a strict anaerobe, this mammalian commensal has been found to benefit from nanomolar concentrations of dissolved oxygen (≤0.05 % atmospheric), but cannot grow when O2 levels are increased further (Baughn & Malamy, 2004). The bacterium can maintain some viability in room air (20.9 % atmospheric, 210 µM dissolved O2 at 37 °C) even after several days of exposure, at least partially due to strong and sophisticated ROS detoxification systems. Superoxide dismutase and AhpC have been shown to protect the bacterium from O2 exposure (Privalle & Gregory, 1979; Rocha & Smith, 1999), and KatB, the cytoplasmic catalase found in B. fragilis, has been shown to detoxify millimolar concentrations of hydrogen peroxide (Rocha et al., 1996). Additionally, B. fragilis encodes two putative rubrerythrins and several peroxidases, including a thioredoxin-dependent peroxidase (Tpx) shown to contribute to defence against organic peroxides (Herren et al., 2003). Much work has been done to characterize the components of this elaborate ROS detoxification response, but there are no studies investigating ROS production. We therefore sought to quantify endogenous ROS generation in aerated B. fragilis and to begin to identify the enzyme(s) responsible for their production. In this way, we might gain insights into the molecular mechanisms underpinning this organism’s inability to grow in room air.
Methods
Reagents.
Hydrogen peroxide, horseradish peroxidase (HRP), antibiotics and fumaric acid were purchased from Sigma, and Amplex Red (AR) was obtained from Invitrogen. Restriction enzymes were purchased from New England Biolabs.
Growth conditions.
Anaerobiosis was maintained by using a Coy anaerobic chamber containing 85 % nitrogen, 10 % hydrogen and 5 % carbon dioxide. B. fragilis was grown in brain heart infusion broth supplemented with 0.5 % yeast extract and 15 µg haematin ml−1 (BHIS), or anaerobic minimal medium (AMM) containing 0.5 % glucose, as previously described (Baughn & Malamy, 2002). In some cases super anaerobic minimal medium (SAMM) plates containing 150 µg haemin ml−1 were used. Because all the strains employed are thyA mutants, thymine was added to 50 µg ml−1. Gentamicin (50 µg ml−1), rifampicin (50 µg ml−1), erythromycin (8 µg ml−1), trimethoprim (80 µg ml−1) and tetracycline (2.5 µg ml−1) were added to plates where appropriate. E. coli was grown aerobically in L broth, and chloramphenicol (25 µg ml−1) and tetracycline (10 µg ml−1) were added where appropriate.
Strains and plasmids.
Strains and plasmids used in this study are listed in Table 1. E. coli strain DH5α was used for cloning, and strain HB101/pRK231 was used for mobilization of plasmids from DH5α to B. fragilis recipient strains (Godoy et al., 1993). DH5α was made competent for transformation through the use of the RbCl method previously described (Hanahan et al., 1991).
Table 1. Bacterial strains and plasmids used in this study.
Abbreviations: CmR, TetR, TpR, chloramphenicol, tetracycline and trimethoprim resistance, respectively. AmpS, ampicillin sensitivity.
| Strain or plasmid | Relevant characteristics | Reference or source |
| B. fragilis strains | ||
| ADB77 | TM4000 ΔthyA TpR | Baughn & Malamy (2002) |
| ADB247 | ADB77 ΔfrdC247 | Baughn & Malamy (2003) |
| MBD616 | TM4000thyA2Δsod | Laboratory stock |
| ADB247/616 | ADB247Δsod | A. D. Baughn, University of Minnesota |
| BM28 | ADB77ΔahpC | This work |
| BM50 | BM28Δkat | This work |
| BM95 | ADB77Δtpx | This work |
| BM105 | BM50Δtpx | This work |
| BM112 | BM105ΔfrdC | This work |
| E. coli strains | ||
| DH5α | λ Nonlysogen | Woodcock et al. (1989) |
| HB101 | rpsL20, host strain for pRK231 | Thompson & Malamy (1990) |
| Plasmids | ||
| pRK231 | AmpS derivative of RP4, TetR Tra+ | Godoy et al. (1993) |
| pYT102 | p15A ori, CmR, RP4 oriT, B. fragilis suicide vector containing B. fragilis thyA TetR | Tang & Malamy (2000) |
| pADB242 | pYT102 derivative, 0.35 kb BamHI–HindIII fragment replaced by 18 bp BamHI–HindIII fragment from pCR2.1-TOPO (Invitrogen) CmR | Baughn & Malamy (2003) |
| pYT102SD | pYT102 containing Δsod | This work |
| pADB247 | pADB242 derivative with ΔfrdC247 allele | Baughn & Malamy (2003) |
| pADB261m7 | pADB261 frdA : : Tn1000 (frdC+) | Baughn & Malamy (2003) |
| pADB242ΔkatB | pADB242 containing ΔkatB | This work |
| pADB242ΔahpC | pADB242 containing ΔahpC | This work |
| pADB242Δtpx | pADB242 containing Δtpx | This work |
DNA manipulation.
Primers used in this study are listed in Table 2. Primers were designed using the B. fragilis NCTC9343 annotated sequence found on the Pedant3 webpage (http://pedant.gsf.de) and synthesized by IDT (Iowa City, IA). Genomic DNA was amplified using HotStarTaq Master Mix (Invitrogen). Plasmid and PCR product purifications were performed with QIAprep spin columns (Qiagen). Where indicated, DNA was digested with restriction enzymes purchased from New England Biolabs. Ligations were performed using T4 DNA Ligase from Invitrogen.
Table 2. Primers used in this study.
| Primer | Region of homology | Sequence (5′–3′) |
| 1843 | pADB242 | CCCATCGGTGATGTCGGC |
| 61RAB | pADB242 | GGCGCGCCGTAAGGAAAGTGGCTCTCAG |
| BAM56 | ahpC1 | CGATGGATCCGCAAAGGTAGGGTGAAG |
| BAM57 | ahpC2 | TGACCCATGGCCCTTTACGTCTTCGC |
| BAM58 | ahpC3 | GGCACCATGGCGAAGCAACCCTGAAAC |
| BAM59 | ahpC4 | AGGTGCGGCCGCGATGGAAGTTTCCGCAC |
| BAM60 | ahpC5 | CGACTCTTAGGTACTGG |
| BAM68 | kat1 | ATATGGATCCATCCCCCTGTGGTA |
| BAM69 | kat2 | ATATCCATGGTTAGCGCTACGCATGTT |
| BAM70 | kat3 | ATATCCATGGTTGAAGGTATCGGCTTC |
| BAM71 | kat4 | ATATGCGGCCGCCTGGGCATTTCTTTG |
| BAM72 | kat5 | CTGCACTTTACGCACTG |
| BAM216 | tpx1 | ACATGGATCCGCTTCATTAATCTGG |
| BAM217 | tpx2 | CAATCCATGGGTGGCATCGAATTTCG |
| BAM218 | tpx3 | ACATCCATGGCATGAAAGCTACCGAAG |
| BAM219 | tpx4 | CAATGCGGCCGCAAACATCGCTTTAAAG |
Strain construction.
All in-frame deletions of B. fragilis genes were created using a two-step double-crossover technique (Tang et al., 1999). In order to build a deletion construct for ahpC, an N-terminal fragment was amplified using primers BAM56 and BAM57, and a C-terminal fragment was amplified using primers BAM58 and BAM59. Purified PCR products were digested with BamHI and NcoI (N-terminal) or NotI and NcoI (C-terminal), and ligated via a three-way reaction with pADB242 that had been digested with BamHI and NotI to create pADB242ΔahpC. The insert was verified via PCR with primers BAM68 and BAM71.
The suicide plasmid pADB242ΔahpC was delivered to the recipient B. fragilis strain as previously described (Thompson & Malamy, 1990). Tetracycline-resistant colonies were screened for the appropriate cointegrant event using primers 1843 and BAM59. Isolates demonstrating recombination at the ahpC locus were grown overnight in BHIS with thymine to allow for recombination events leading to resolution of the disrupting plasmid. The presence of the thyA gene on pADB242 sensitizes cointegrants to trimethoprim, while those resolvants that have excised the plasmid are trimethoprim-resistant. For this reason, the culture was plated to SAMM containing glucose, thymine and trimethoprim. Colonies arising after 3–4 days were purified and then screened for tetracycline sensitivity on the appropriate BHIS+thymine plates. Tetracycline-sensitive colonies were then used as template in a PCR with primers BAM59 and BAM60 to identify ΔahpC clones.
The deletion construct for sod was created by PCR-amplifying a region consisting of 570 bp of sod 5′ upstream sequence and 133 bp of N-terminal coding sequence to create the upstream fragment. A fragment of 150 bp of sod C-terminal coding sequence and 539 bp of downstream sequence was amplified for the downstream fragment. These pieces were ligated into pYT102 via the BamHI and HindIII cloning sites.
The deletion construct for katB was created by PCR-amplifying an N-terminal fragment with primers BAM68 and BAM69 and a C-terminal fragment with primers BAM70 and BAM71. Fragments were again digested with BamHI and NcoI (N-terminal) or NotI and NcoI (C-terminal), and ligated to the BamHI/NotI-digested pADB242 to create pADB242Δkat. The procedure described above was used to create cointegrants, which were screened by PCR with primers 61RAB and BAM72. Resolvants were screened with primers BAM68 and BAM71.
pADB242Δtrx was created by PCR-amplifying an N-terminal fragment with primers BAM216 and BAM217 and a C-terminal fragment with BAM218 and BAM219. Digestions and ligations to pADB242 were performed as above. Cointegrants were screened with primers 61RAB and BAM216 as well as 1843 and BAM219. Resolvants were screened with primers BAM216 and BAM219.
H2O2 detection.
The protocol for measuring H2O2 scavenging was adapted from the method of Seaver & Imlay (2001). Briefly, solutions of HRP and AR reagent were made in PBS (pH 7.2) (per litre: 8 g NaCl, 0.2 g KCl, 1.44 g Na2PO4, 0.24 g KH2PO4) to concentrations of 20 and 56 µg ml−1, respectively. An H2O2 stock was made in PBS (pH 7.2) from a 30 % solution and the concentration was determined by measuring the A240 of the solution in a spectrophotometer (ϵ = 43.6 M−1 cm−1). This solution was further diluted to give a 200 µM H2O2 stock solution. Aliquots were added to 500 µl HRP mixed with 500 µl AR in 4 ml polystyrene cuvettes and diluted to a final volume of 2 ml with PBS. These samples were briefly shaken by hand and placed in an Aminco Bowman Series 2 luminescence spectrophotometer for reading with an excitation wavelength set to 563 nm (band pass = 1) and an emission wavelength set to 587 nm (band pass = 4). Fluorescence readings were recorded and used to construct a standard curve for H2O2 concentration. All experiments were conducted in the presence of oxygen, except where indicated. Optical densities were measured using a Beckman DU 640 spectrophotometer and quartz cuvettes with a 1 cm path length.
H2O2 scavenging.
Strains were first grown anaerobically to mid-exponential phase in BHIS or AMM. Cultures were adjusted to OD600 = 0.1 in BHIS or AMM lacking cysteine, placed in foam-stoppered flasks, and transferred to 37 °C under room air (21 % oxygen). Samples were shaken for 1 h at 250 r.p.m. then centrifuged for 10 min at 2000 g. Supernatants were removed, and the cell pellets were washed twice with an equal volume of PBS, as the growth medium was found to significantly reduce the sensitivity of the assay. Following the second wash, cell pellets were resuspended in 10 ml PBS and transferred to tubes for testing. Aliquots of H2O2 stock were added to the 10 ml cultures to give a starting concentration of 2.5–5 µM. Immediately after the addition of peroxide, the tubes were quickly capped and upended once to mix. A 1.1 ml volume was immediately removed to a 1.5 ml microcentrifuge tube and spun for approximately 10 s at 6000 r.p.m. in a microcentrifuge (model SD110, Clover Laboratories). A 1 ml volume of supernatant was removed to a 4 ml polystyrene cuvette (VWR), 500 µl HRP and 500 µl AR solutions were added, and the fluorescence was read. For anaerobic peroxide-scavenging experiments, cultures were transferred to conical tubes in the anaerobic chamber and sealed before removal for centrifugation. Supernatants were poured off in the anaerobic chamber, and cells were washed and ultimately resuspended in pre-reduced PBS with thymine and 0.5 % glucose. These suspensions were transferred to Hungate tubes before removal from the chamber. H2O2 additions and sample removal were performed by inserting a needle through the rubber septum to minimize oxygen exposure.
ROS generation.
Strains were grown for 2 days at 37 °C in 5 ml AMM. Aliquots of these dense cultures were then used to inoculate 10 ml AMM. Cultures were shaken anaerobically at 100 r.p.m. in 50 ml flasks and grown for several generations. When the OD600 reached approximately 0.3, cultures were transferred anaerobically to conical screw-capped tubes, sealed, and centrifuged for 5 min at 2000 g. After transferring tubes to the anaerobic chamber and decanting the supernatants, cell pellets were resuspended in 10 ml AMM lacking cysteine and transferred to 125 ml foam-stoppered flasks. These flasks were shaken at 250 r.p.m. in room air at 37 °C for 1 h. Cultures were again transferred to conical screw-capped tubes and centrifuged as before. Cell pellets were washed twice with PBS and then resuspended in 20 ml PBS+0.05 % fresh glucose and thymine to OD600 = 0.1. Flasks were shaken at 250 r.p.m. in room air at 37 °C. Samples of 1.1 ml volume were drawn at various time points as described above to determine H2O2 concentrations in the supernatants.
O2 sensitivity.
Strains were grown overnight in BHIS [with or without erythromycin (±erm)] under anaerobic conditions. Aliquots of these starter cultures were used to inoculate 10 ml BHIS, and cultures were grown for several generations. When cultures reached mid-exponential phase they were transferred to 15 ml conical tubes in the anaerobic chamber. The tubes were capped, removed from the chamber, and centrifuged for 5 min at 2000 g in a tabletop centrifuge. Supernatants were poured off, and the pellets were resuspended in 10 ml oxygenated PBS (pH 7.2)+0.5 % glucose and thymine. These suspensions were transferred to 125 ml flasks stoppered with foam, and shaken at 250 r.p.m. and 37 °C under room air. Samples were taken over time and diluted in PBS. Ten microlitre volumes of diluted samples were spotted to BHIS+thymine plates (±erm) that had been placed in the anaerobic chamber overnight to reduce. After 24–48 h, c.f.u. were enumerated under a stage microscope.
Results
A B. fragilis strain missing Kat, AhpC and Tpx is severely compromised in its ability to scavenge H2O2
Some redox-active enzymes, including flavoproteins, can adventitiously donate electrons to O2, thus generating superoxide or H2O2. The rate at which these ROS are generated depends on the accessibility of the flavin to the solvent as well as the abundance of the enzyme (Imlay, 1995; Messner & Imlay, 2002). In order to quantify the rate of this ROS formation in vivo, it is necessary to identify and inactivate the major hydrogen peroxide-scavenging enzymes. In a strain lacking such detoxifiers, H2O2 produced intracellularly will leak across the cell membrane and accumulate in the extracellular milieu, thus allowing quantification with AR (Seaver & Imlay, 2001). Unlike H2O2, superoxide cannot cross membranes at physiological pHs, but if strains encoding Sod are utilized it is converted enzymically to H2O2. In such strains, therefore, endogenous sources of both H2O2 and superoxide can be assayed using AR, thereby giving an overall picture of ROS formation.
In E. coli, deletion of ahpC and the genes encoding two catalases, katE and katG, allows for monitoring of H2O2 production by decreasing peroxide-scavenging rates to less than 5 % of wild-type (Seaver & Imlay, 2001). Because B. fragilis appears to encode only one catalase (katB), we hypothesized that a ΔahpCΔkatB mutant should permit quantification of endogenous ROS production. However, when cells were shaken under room air in rich medium and then assayed for H2O2 consumption in PBS, the ΔahpCΔkatB strain retained significant scavenging activity (Fig. 1a), potentially interfering with ROS quantification. In an attempt to identify the gene product(s) responsible for the residual scavenging activity, we deleted other putative peroxidases in the ΔahpCΔkat background. While most deletions did not appear to impair scavenging activity further, deletion of a gene encoding a putative thioredoxin-dependent peroxidase (Tpx) resulted in a strain (ΔahpCΔkatΔtpx) with approximately 20 % of the peroxide-scavenging activity of the wild-type when the cells were grown in BHIS (Fig. 1a).
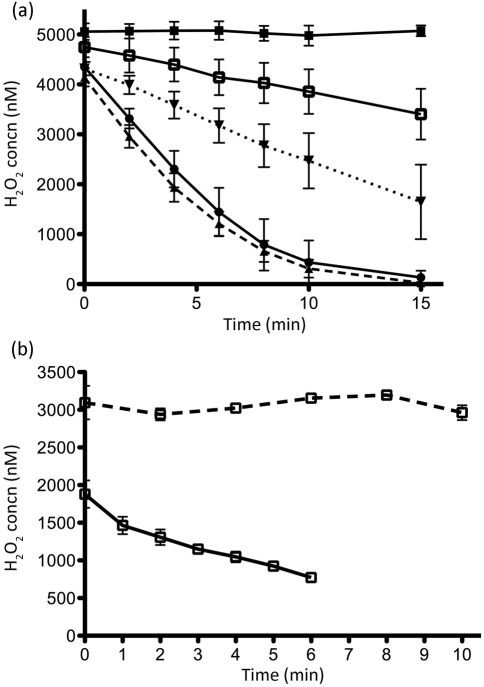
Fig. 1.
H2O2 scavenging rates for B. fragilis cells grown in rich medium (a) and minimal medium (b). Cells were washed and resuspended in PBS to OD600 0.1. Aliquots of H2O2 were added to suspensions to start the assay and samples were taken over time. After a brief centrifugation, supernatants were assayed for H2O2 concentration using the AR protocol. (a) PBS (▪), ADB77 (wild-type, ▴, dashed line), BM50 (ΔahpCΔkat, ▾, dotted line), BM95 (Δtpx, •), BM105 (ΔahpCΔkatΔtpx, □). Scavenging assays were done in the presence of oxygen. (b) BM105 was assayed in the absence (solid line) and presence (dashed line) of oxygen. Shown are the mean rates of peroxide scavenging for three experiments±sem.
We also tested the H2O2-scavenging activity of cells grown in minimal medium (AMM), and investigated the effect of oxygen on this activity. Samples of the ΔahpCΔkatΔtpx strain were grown anaerobically, washed anaerobically, and resuspended to OD600 0.1 in 10 ml pre-reduced PBS in the anaerobic chamber. An aliquot of H2O2 was added to bring the initial concentration to ~2.5 µM, while excluding oxygen. Under such conditions, these cells were capable of scavenging H2O2 at a rate of ~155 nM min −1 (Fig. 1b). However, when these cells were first shaken for 1 h under room air in AMM lacking cysteine and then suspended in aerated PBS, H2O2-scavenging activity could no longer be detected (Fig. 1b). In fact, the H2O2 concentration at the start of the assay was significantly higher in these samples than in the anaerobically prepared cells, possibly due to the production of ROS during the preparation of the sample. Exposure to oxygen was clearly interfering with H2O2 detoxification, and we reasoned that preparation of the ΔahpCΔkatΔtpx strain under these conditions might allow us to quantify endogenously generated H2O2.
Fumarate reductase is a significant contributor to ROS formation when B. fragilis is aerated
When the B. fragilis ΔahpCΔkatΔtpx strain was prepared as above, adjusted to OD600 0.1 in 10 ml PBS containing thymine and glucose, and shaken in room air, we detected the accumulation of H2O2 in the buffer at a rate of 36±6 nM min−1 (Fig. 2). Given such a rate, we would expect that this strain would experience significant oxidative stress in less than 30 min, as substantial damage to E. coli metabolism occurs even in the face of submicromolar H2O2 concentrations (Park et al., 2005).
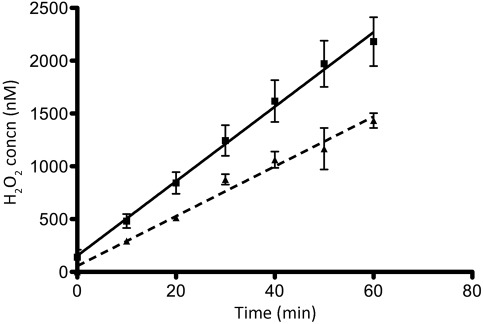
Fig. 2.
Disrupting the fumarate reductase complex reduces the rate of ROS formation in B. fragilis. Cells grown to exponential phase were centrifuged, resuspended in AMM lacking cysteine, and shaken for 1 h at 37 °C under room air. Cells were again centrifuged, washed and resuspended in PBS+thymine to OD600 0.1. Freshly prepared glucose was added to 0.05 % just before the start of assay. Suspensions were shaken at 37 °C under room air throughout the course of the assay. Samples were taken over time and cells were centrifuged. Supernatants were assayed for H2O2 using the AR protocol. BM105 (ΔahpCΔkatΔtpx, ▪), BM112 (ΔahpCΔkatΔtpxΔfrdC, ▴, dashed line). Shown are the means of three assays±sem.
Endogenous sources of ROS have been identified in E. coli (Korshunov & Imlay, 2010), and among these, fumarate reductase (Frd) has been described as a potent generator of both superoxide and H2O2 due to its solvent-exposed flavin cofactor (Imlay, 1995; Korshunov & Imlay, 2006; Messner & Imlay, 2002). With this in mind, we made a deletion of the frdC gene in the B. fragilis ΔahpCΔkatΔtpx background to test whether loss of fumarate reductase activity significantly reduced the rate of ROS formation in cells challenged under room air. As illustrated in Fig. 2, we detected the accumulation of H2O2 at a rate of 19±1 nM min−1 in strain BM112 (ΔahpCΔkatΔtpxΔfrdC), indicating that disruption of the fumarate reductase complex reduced ROS generation by approximately 47 % when B. fragilis was exposed to room air.
frdC mutants are much smaller than wild-type, and an OD600 0.1 in these experiments corresponded to ~4×107 c.f.u. ml−1 for ΔahpCΔkatΔtpx and ~2×108 c.f.u. ml−1 for ΔahpCΔkatΔtpxΔfrdC. To make sure that these discrepancies were not affecting our results, cells of both strains were sonicated and assayed for intracellular protein content. At this cell density, ΔahpCΔkatΔtpx and ΔahpCΔkatΔtpxΔfrdC gave mean protein contents of 4.7 and 5.4 µg protein ml−1, respectively, and normalizing ROS generation rates to protein content gave similar results to those reported above. Additionally, these strains were found to scavenge peroxide at indistinguishable rates in both the presence and absence of oxygen (data not shown).
Fumarate inhibits ROS formation
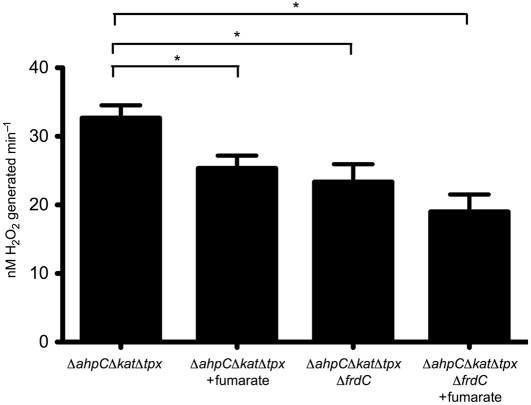
If fumarate reductase is a source of superoxide or H2O2 when cells are aerated, then addition of fumarate should reduce ROS formation by competing with oxygen for the electrons carried on the Frd flavin group. To test this, BM105 (ΔahpCΔkatΔtpx) and BM112 (ΔahpCΔkatΔtpxΔfrdC) were prepared as above for the ROS accumulation assay. Just prior to the start of the assay, cultures were split into two flasks, and fumarate was added exogenously to one flask per strain. As shown in Fig. 3, addition of fumarate to BM105 reduced ROS production to a rate similar to that of the ΔfrdC strain, indicating that fumarate was partially inhibiting the adventitious reduction of oxygen in this strain. Fumarate also appeared to reduce ROS production in BM112 slightly. While this reduction was not statistically significant, it may indicate that fumarate is inhibiting another ROS-generating enzyme.
Fig. 3.
Exogenous fumarate inhibits ROS formation. Strains BM105 (ΔahpCΔkatΔtpx) and BM112 (ΔahpCΔkatΔtpxΔfrdC) were prepared as in Fig. 2, with cell suspensions divided into two flasks each. Just prior to sampling, 50 mM fumarate was added to one flask per strain. The AR protocol was then used to determine the H2O2 concentration over time. Rates are recorded as nM H2O2 generated min−1 for cells suspended to OD600 0.1 in 10 ml PBS. Shown are the means of three assays±sem. A one-way analysis of variance (ANOVA) revealed significant differences between strains (P = 0.0154). Asterisks denote significant differences between means (P<0.05) in a Newman–Keuls multiple comparison post test.
Deleting frdC from a superoxide dismutase mutant partially restores aerotolerance
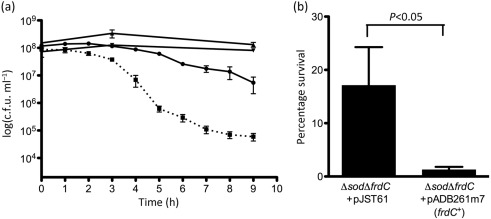
Given that the B. fragilis fumarate reductase plays such a major role in ROS production, we hypothesized that deleting frdC might increase aerotolerance. However, no significant differences in viability were seen when BM105 (ΔahpCΔkatΔtpx), BM112 (ΔahpCΔkatΔtpxΔfrdC), ADB247 (ΔfrdC) and ADB77 (wild-type) were exposed to room air for up to 9 h in PBS containing thymine and glucose (data not shown). This indicated that an inability to scavenge H2O2 did not sensitize B. fragilis to aerobic conditions during the course of these experiments. However, Fig. 4(a) shows that a Δsod mutant lost greater than three logs of viability under these conditions, suggesting that, at least initially, the major ROS generated by aerated B. fragilis is superoxide. A ΔsodΔfrdC strain (ADB247/616) showed reduced oxygen sensitivity, losing only about one log of viability during this time period, indicating that deletion of frdC restores some aerotolerance to a Δsod strain. Anaerobic controls showed that the number of c.f.u. increased slightly for the ΔsodΔfrdC strain during the course of the experiment, while the number of c.f.u. for the Δsod strain decreased slightly, resulting in an approximately threefold difference (data not shown). In separate experiments, we found that introduction of a wild-type copy of frdC in trans resensitized the ΔsodΔfrdC strain to room air (Fig. 4b), while a strain carrying an empty vector maintained significantly higher viability.
Fig. 4.
Survival of B. fragilis strains under room air. Anaerobically grown cells were suspended in PBS with 0.5 % glucose and thymine, and shaken in room air at 37 °C. Samples were taken over time, diluted, and plated to pre-reduced BHIS+thymine (a) or BHIS+thymine+erm (b). c.f.u. were enumerated after 24–48 h. (a) ADB77 (wild-type, ▾), ADB247 (ΔfrdC, ▴), MBD616 (Δsod, ▪, dotted line), ADB247/616 (ΔfrdCΔsod, •). Shown are the means of three experiments±sem. A Student’s paired t test analysing the percentages of viable c.f.u. remaining at the 6 h time point indicated a significant difference between MBD616 and ADB247/616 with P<0.5. (b) Percentage of viable c.f.u. remaining after 6 h exposure to room air for ADB247/616 (pJST61) and ADB247/616 (pADB261m7). Shown are the means of nine experiments±sem. A Student’s paired t test indicated P<0.05.
Discussion
In order to quantify ROS production rates in B. fragilis, we first constructed a strain that was incapable of scavenging any H2O2 formed when cells were aerated. We were surprised to uncover a potential role for Tpx in H2O2 scavenging, as this enzyme had previously been shown to play a role in protection against organic peroxides, but it did not appear to contribute significantly to protection against H2O2-mediated growth inhibition (Herren et al., 2003). Indeed, deletion of tpx alone did not affect the ability of B. fragilis to scavenge micromolar concentrations of H2O2 when applied exogenously. It is possible that deletion of tpx led to increased expression of Kat or AhpC, as ahpC mutants of E. coli have been shown to increase transcription of katG via activation of the transcriptional regulator OxyR (Seaver & Imlay, 2001). Alternatively, Tpx may have a lower affinity for reductant than AhpC, thus masking its activity when AhpC is present. Clearly the role of Tpx is complex and warrants further investigation.
The residual H2O2-scavenging activity exhibited by ΔahpCΔkatΔtpx in BHIS was lost when cells were shaken under room air in minimal medium lacking cysteine. By removing cysteine, which acts as a redox buffer in this medium, cells quickly become saturated by O2. Presumably, O2 is participating in reactions that consume reductant, like ROS generation or the cytochrome bd oxidase-mediated reduction of O2 to water, and thus is competing with H2O2 detoxification reactions for substrate. Under such conditions, therefore, this strain cannot effectively scavenge H2O2, thus allowing us to dissect the pathways contributing to endogenous ROS generation in B. fragilis.
Fumarate reductase has been found to produce ROS in anaerobically grown E. coli shaken in room air (Korshunov & Imlay, 2010). We demonstrate in this work that it makes a significant contribution to ROS production in B. fragilis as well, most likely by directly reducing oxygen via its flavin cofactor. The finding that deletion of frdC restored aerotolerance to a Δsod strain suggests that in vivo under aerobic conditions Frd is generating superoxide rather than H2O2. Interestingly, earlier work has shown that many of the tricarboxylic acid cycle genes of B. fragilis are upregulated five- to 28-fold when the organism is aerated (Sund et al., 2008). However, transcription of frd is downregulated approximately threefold, perhaps suggesting that an adaptive response is taking place to reduce ROS generation.
The finding that fumarate slowed ROS production slightly in the ΔahpCΔkatΔtpxΔfrdC strain suggests that another ROS-generating enzyme might also utilize fumarate as its natural substrate. One such candidate would be NadB, a key component of the NAD+ biosynthetic pathway. This enzyme has also been shown to participate in ROS formation in E. coli (Korshunov & Imlay, 2010) and to be inhibited by fumarate. Experiments are under way to test the potential contribution of NadB to ROS production in B. fragilis.
Previous work with Bacteroides thetaiotaomicron has suggested that this organism (and, by extension, B. fragilis) might generate ROS intracellularly at a faster rate than E. coli (Pan & Imlay, 2001), potentially explaining its inability to grow aerobically. We have shown that when B. fragilis was shaken in room air, the extracellular concentration of H2O2 increased at a rate of 36 nM min−1 for cells suspended to OD600 0.1. To convert this rate to reflect intracellular concentrations, we took advantage of the fact that the intracellular concentration of protein in E. coli is approximately 330 mg (ml cytoplasm)−1 (J. Imlay, personal communication). Assuming a similar concentration in B. fragilis, 1 ml of cells at OD600 0.1 contains ~5 µg cytoplasmic protein and thus represents 15 nl of cytoplasmic volume. Converting the extracellular H2O2 accumulation rates thus shows intracellular rates of ROS formation to be 40 µM s−1. An E. coli strain missing catalase and peroxidase activity (but encoding Sod) accumulates H2O2 intracellularly at a rate of 15 µM s−1 (Imlay, 2008). While a direct comparison between these organisms is complicated by differences in growth media and by the fact that E. coli is actively growing under the assay conditions while B. fragilis is merely surviving, these estimates suggest that B. fragilis is producing ROS at a rate that is only ~2.5-fold faster than E. coli, a seemingly modest increase. Such an increase in ROS generation would not be without consequence, however, as even a threefold increase in the rate of superoxide production can significantly reduce the steady-state activity of crucial dehydratases in E. coli (Gort & Imlay, 1998). This suggests that the rate at which an organism generates ROS may play an important role in its ability to cope with aerobic environments.
Acknowledgements
We are grateful to Dr Jim Imlay for helpful discussions and to Dr Carol Kumamoto for assistance with the fluorometric studies. This work was supported by The National Institute of Allergy and Infectious Disease of the National Institutes of Health (Public Health Grant AI 19497). B. M. M. was supported by Public Health Training Grant T32-GM007310.
Abbreviations:
- AR
Amplex Red
- HRP
horseradish peroxidase
- ROS
reactive oxygen species
References
- Baughn A. D., Malamy M. H. (2002). A mitochondrial-like aconitase in the bacterium Bacteroides fragilis: implications for the evolution of the mitochondrial Krebs cycle. Proc Natl Acad Sci U S A 99, 4662–4667. 10.1073/pnas.052710199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughn A. D., Malamy M. H. (2003). The essential role of fumarate reductase in haem-dependent growth stimulation of Bacteroides fragilis. Microbiology 149, 1551–1558. 10.1099/mic.0.26247-0 [DOI] [PubMed] [Google Scholar]
- Baughn A. D., Malamy M. H. (2004). The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 427, 441–444. 10.1038/nature02285 [DOI] [PubMed] [Google Scholar]
- Fridovich I. (1998). Oxygen toxicity: a radical explanation. J Exp Biol 201, 1203–1209. [DOI] [PubMed] [Google Scholar]
- Fridovich I. (1999). Fundamental aspects of reactive oxygen species, or what’s the matter with oxygen? Ann N Y Acad Sci 893, 13–18. 10.1111/j.1749-6632.1999.tb07814.x [DOI] [PubMed] [Google Scholar]
- Godoy V. G., Dallas M. M., Russo T. A., Malamy M. H. (1993). A role for Bacteroides fragilis neuraminidase in bacterial growth in two model systems. Infect Immun 61, 4415–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gort A. S., Imlay J. A. (1998). Balance between endogenous superoxide stress and antioxidant defenses. J Bacteriol 180, 1402–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Jessee J., Bloom F. R. (1991). Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol 204, 63–113. 10.1016/0076-6879(91)04006-A [DOI] [PubMed] [Google Scholar]
- Herren C. D., Rocha E. R., Smith C. J. (2003). Genetic analysis of an important oxidative stress locus in the anaerobe Bacteroides fragilis. Gene 316, 167–175. 10.1016/S0378-1119(03)00759-5 [DOI] [PubMed] [Google Scholar]
- Imlay J. A. (1995). A metabolic enzyme that rapidly produces superoxide, fumarate reductase of Escherichia coli. J Biol Chem 270, 19767–19777. [PubMed] [Google Scholar]
- Imlay J. A. (2003). Pathways of oxidative damage. Annu Rev Microbiol 57, 395–418. 10.1146/annurev.micro.57.030502.090938 [DOI] [PubMed] [Google Scholar]
- Imlay J. A. (2008). Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77, 755–776. 10.1146/annurev.biochem.77.061606.161055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunov S., Imlay J. A. (2006). Detection and quantification of superoxide formed within the periplasm of Escherichia coli. J Bacteriol 188, 6326–6334. 10.1128/JB.00554-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunov S., Imlay J. A. (2010). Two sources of endogenous hydrogen peroxide in Escherichia coli. Mol Microbiol 75, 1389–1401. 10.1111/j.1365-2958.2010.07059.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord J. M., Fridovich I. (1969). Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J Biol Chem 244, 6049–6055. [PubMed] [Google Scholar]
- Messner K. R., Imlay J. A. (2002). Mechanism of superoxide and hydrogen peroxide formation by fumarate reductase, succinate dehydrogenase, and aspartate oxidase. J Biol Chem 277, 42563–42571. 10.1074/jbc.M204958200 [DOI] [PubMed] [Google Scholar]
- Pan N., Imlay J. A. (2001). How does oxygen inhibit central metabolism in the obligate anaerobe Bacteroides thetaiotaomicron. Mol Microbiol 39, 1562–1571. 10.1046/j.1365-2958.2001.02343.x [DOI] [PubMed] [Google Scholar]
- Park S., You X., Imlay J. A. (2005). Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx– mutants of Escherichia coli. Proc Natl Acad Sci U S A 102, 9317–9322. 10.1073/pnas.0502051102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonage D., Karplus P. A., Poole L. B. (2008). Substrate specificity and redox potential of AhpC, a bacterial peroxiredoxin. Proc Natl Acad Sci U S A 105, 8209–8214. 10.1073/pnas.0708308105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalle C. T., Gregory E. M. (1979). Superoxide dismutase and O2 lethality in Bacteroides fragilis. J Bacteriol 138, 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha E. R., Smith C. J. (1999). Role of the alkyl hydroperoxide reductase (ahpCF) gene in oxidative stress defense of the obligate anaerobe Bacteroides fragilis. J Bacteriol 181, 5701–5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha E. R., Selby T., Coleman J. P., Smith C. J. (1996). Oxidative stress response in an anaerobe, Bacteroides fragilis: a role for catalase in protection against hydrogen peroxide. J Bacteriol 178, 6895–6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaver L. C., Imlay J. A. (2001). Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol 183, 7173–7181. 10.1128/JB.183.24.7173-7181.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sund C. J., Rocha E. R., Tzianabos A. O., Wells W. G., Gee J. M., Reott M. A., O’Rourke D. P., Smith C. J. (2008). The Bacteroides fragilis transcriptome response to oxygen and H2O2: the role of OxyR and its effect on survival and virulence. Mol Microbiol 67, 129–142. 10.1111/j.1365-2958.2007.06031.x [DOI] [PubMed] [Google Scholar]
- Tang Y. P., Malamy M. H. (2000). Isolation of Bacteroides fragilis mutants with in vivo growth defects by using Tn4400′, a modified Tn4400 transposition system, and a new screening method. Infect Immun 68, 415–419. 10.1128/IAI.68.1.415-419.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y. P., Dallas M. M., Malamy M. H. (1999). Characterization of the Batl (Bacteroides aerotolerance) operon in Bacteroides fragilis: isolation of a B. fragilis mutant with reduced aerotolerance and impaired growth in in vivo model systems. Mol Microbiol 32, 139–149. 10.1046/j.1365-2958.1999.01337.x [DOI] [PubMed] [Google Scholar]
- Thompson J. S., Malamy M. H. (1990). Sequencing the gene for an imipenem-cefoxitin-hydrolyzing enzyme (CfiA) from Bacteroides fragilis TAL2480 reveals strong similarity between CfiA and Bacillus cereus beta-lactamase II. J Bacteriol 172, 2584–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock D. M., Crowther P. J., Doherty J., Jefferson S., DeCruz E., Noyer-Weidner M., Smith S. S., Michael M. Z., Graham M. W. (1989). Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res 17, 3469–3478. 10.1093/nar/17.9.3469 [DOI] [PMC free article] [PubMed] [Google Scholar]