Abstract
Pseudomonas aeruginosa, which causes serious infections in immunocompromised patients, produces numerous virulence factors, including exotoxin A and the siderophore pyoverdine. As production of these virulence factors is influenced by the host environment, we examined the effect serum has on global transcription within P. aeruginosa strain PAO1 at different phases of growth in an iron-deficient medium. At early exponential phase, serum significantly enhanced expression of 138 genes, most of which are repressed by iron, including pvdS, regA and the pyoverdine synthesis genes. However, serum did not interfere with the repression of these genes by iron. Serum enhanced regA expression in a fur mutant of PAO1 but not in a pvdS mutant. The serum iron-binding protein apotransferrin, but not ferritin, enhanced regA and pvdS expression. However, in PAO1 grown in a chemically defined medium that contains no iron, serum but not apotransferrin enhanced pvdS and regA expression. While complement inactivation failed to eliminate this effect, albumin absorption reduced the effect of serum on pvdS and regA expression in the iron-deficient medium chelexed tryptic soy broth dialysate. Additionally, albumin absorption eliminated the effect of serum on pvdS and regA expression in the chemically defined medium. These results suggest that serum enhances the expression of P. aeruginosa iron-controlled genes by two mechanisms: one through apotransferrin and another one through albumin.
Introduction
Pseudomonas aeruginosa is a Gram-negative pathogen that causes acute infections in immunocompromised hosts, including severely burned patients, cancer patients undergoing chemotherapy and HIV-infected patients (Branski et al., 2009; Sadikot et al., 2005). P. aeruginosa is also a major cause of chronic lung infections in patients with cystic fibrosis (Pier & Ramphal, 2005; Sadikot et al., 2005). Damage caused by P. aeruginosa is due to the production of several cell-associated and extracellular virulence factors, such as the cell-associated flagellum, pili, lipopolysaccharide and alginate; and secreted exotoxin A (ETA), elastase, alkaline proteases and type III secretion factors (Pier & Ramphal, 2005; Sadikot et al., 2005; van Delden, 2004). The production of these factors is controlled by the environment within different infection sites.
Iron, an element essential for the growth and metabolism of bacteria, is sequestered by iron-binding proteins, such as transferrin and lactoferrin, at infection sites within the host, (González-Chávez et al., 2009; Wang & Pantopoulos, 2011). Consequently, during the infection process bacterial pathogens encounter iron-restricted conditions (Nairz et al., 2010). In response, infecting bacteria produce different iron-acquisition systems (Anzaldi & Skaar, 2010; Cornelis, 2010; Nairz et al., 2010). Iron-limited conditions trigger P. aeruginosa to produce several factors that are directly or indirectly involved in iron acquisition. These factors include the siderophores pyoverdine and pyochelin, proteases, ETA and haem utilization systems (Cornelis, 2010; Cornelis et al., 2009; Hamood et al., 2004). The production of these factors by P. aeruginosa is a complicated process that involves numerous regulators (Hamood et al., 2004). However, a key factor that regulates the activity of many of these different systems is the ferric uptake regulator (Fur) (Barton et al., 1996; Cornelis et al., 2009; Hamood et al., 2004). Upon its activation by intracellular iron, Fur represses the expression of genes that code for iron-acquisition factors (Barton et al., 1996; Cornelis et al., 2009). Genes that are directly regulated by iron-activated Fur contain specific sequences within their upstream regions to which Fur specifically binds (Fur box) (Ochsner et al., 1995, 1996; Ochsner & Vasil, 1996). Certain genes are regulated by Fur through a hierarchical system. For example, Fur represses the expression of pvdS, which codes for the extracytoplasmic function (ECF) sigma factor PvdS (Cornelis et al., 2009; Ochsner et al., 1996; Vasil & Ochsner, 1999). PvdS is, in turn, essential for the expression of the ETA gene (toxA) and the pyoverdine genes (Cornelis et al., 2009; Hamood et al., 2004; Ochsner & Vasil, 1996; Vasil & Ochsner, 1999). Similarly, Fur represses the expression of pchR, which is required for the expression of different genes involved in pyochelin production (Ochsner et al., 1995). Besides Fur, P. aeruginosa may contain other proteins that play key roles in regulating the expression of different iron-acquisition systems. However, unlike Fur, which can be activated in vitro by low-iron conditions, some of these key regulation systems may be activated in response to specific environmental cues within the host.
One host factor likely to influence the production of P. aeruginosa virulence factors is serum. During systemic infections and within infected wounds, P. aeruginosa is exposed to serum or components of serum. We recently showed that human serum and plasma interfere with the development of P. aeruginosa biofilms on intravenous catheters (Hammond et al., 2010). This hindrance appears to be related to the serum iron-binding protein transferrin (Ardehali et al., 2003; Hammond et al., 2010). Here we demonstrate that serum affects global gene expression in P. aeruginosa in the early exponential phase of growth, enhancing the expression of 154 genes, many of which are iron-controlled. The mechanism of enhancement is independent of iron and Fur, but requires functional PvdS. Serum fractionation and denaturation revealed that the enhancing activity lies within ≥50 kDa proteins. Depletion of albumin from the serum abrogated the enhancement while exogenous therapeutic human albumin solution reconstituted the effect. We propose that serum albumin plays a critical role in P. aeruginosa virulence during early phases of infection by enhancing the expression of iron-controlled genes through a Fur-independent mechanism that is not related to albumin-associated iron.
Methods
Bacterial strains and plasmids, and growth media and conditions.
Strains and plasmids utilized in this study are described in Table 1. For general growth experiments, strains were grown in the iron-containing medium Luria–Bertani (LB) broth (Miller, 1972). For all other analyses, P. aeruginosa strains were grown in either the iron-deficient medium TSB-DC (Chelex-treated trypticase soy broth dialysate) to which glycerol (1 % v/v) and monosodium glutamate (0.5 M) were added (Ohman et al., 1980) or TSB-DC containing 10 % (v/v) adult bovine serum (TSB-DC/ABS), 10 % adult human serum (TSB-DC/AHS), adult human plasma (TSB-DC/AHP), or 10 % Albuminar (CSL Behring) (TSB-DC/THA). Iron-sufficient medium was prepared by the addition of FeCl3 to TSB-DC to a final concentration of 20 µg Fe3+ ml−1 (TSB-DC/Fe) (Ohman et al., 1980). Norepinephrine (NE) was added to a concentration of 500 µM. Modified M9 medium (MM9) was prepared by the addition of glycerol (1 %, v/v) and monosodium glutamate (0.5 M) to the chemically defined medium M9 (Miller, 1972). Ferritin (Sigma-Aldrich) apotransferrin (R&D Systems) and holotransferrin (Sigma-Aldrich) were added to MM9 at concentrations ranging from 20 to 200 µg ml−1. Antibiotics were added to the growth medium as needed: 200 µg carbenicillin ml−1, 300 µg streptomycin ml−1 or 80 µg tetracycline ml−1 to maintain plasmid stability.
Table 1. Strains and plasmids used in this study.
| Strain or plasmid | Description* | Source and reference |
| Strains | ||
| PAO1 | Prototrophic PAO1 | S. E. H. West, University of Wisconsin (West et al., 1994) |
| C6 | PAO1 parent strain; Δanr, Fur mutation is Ala-10-Gly | M. L. Vasil, University of Colorado Denver (Barton et al., 1996) |
| PAO6261 | PAO1 parent strain; Δanr | M. L. Vasil (Barton et al., 1996) |
| PAO1 : : pvdS | PAO1 with 460 bp pvdS internal deletion, Gmr | I. Lamont, University of Otago, New Zealand (Cunliffe et al., 1995) |
| PW6979 : : bfrB-lacZ | bfrB-C04 : : ISlacZ/hah, Tcr | University of Washington Genome Center (Jacobs et al., 2003) |
| Plasmids | ||
| pRL88 | pSW205 carrying a regA–lacZ fusion, Cbr | D. Storey, University of Calgary, Canada (Storey et al., 1990) |
| pMP220 : : PpvdA | pMP220 carrying a pvdA promoter transcriptional lacZ fusion, Tcr | P. Visca, University ‘Roma Tre’, Italy (Ambrosi et al., 2002) |
| pMP220 : : PpvdS | pMP220 carrying a pvdS promoter transcriptional lacZ fusion, Tcr | P. Visca (Ambrosi et al., 2002) |
| pDH10 | pMP190 carrying a pchR–lacZ fusion, Smr Cmr | K. Poole, Queen’s University, Canada (Heinrichs & Poole, 1996) |
Cb, carbenicillin; Cm, chloramphenicol; Gm, gentamicin; Sm, streptomycin; Tc, tetracycline; r, resistant.
For transcriptome analysis and analysis of ETA and pyoverdine production, as well as the expression of different genes, P. aeruginosa strains were first grown overnight in LB broth at 37 °C. An aliquot of the overnight culture was pelleted, washed and resuspended in TSB-DC. The resuspended culture was used to inoculate fresh TSB-DC with or without serum, iron or other additives to an OD600 of 0.02–0.03 (4 mm cuvette pathlength; GENESYS 20, ThermoFisher Scientific). The cultures were grown at 37 °C to the indicated times with shaking at 230 r.p.m. Each experiment was conducted in triplicate (three separate flasks) with three replicates per flask.
Transcriptome analysis in the presence of serum.
Triplicate cultures of PAO1 were grown at 37 °C with shaking in either TSB-DC or TSB-DC/ABS to an OD600 of 1.11 and 0.69 (early exponential phase of growth, 4 h time point) and 4.7 and 2.5 (early stationary phase, 8 h time point), respectively. As growth levels in TSB-DC/ABS were reduced compared to growth in TSB-DC, the TSB-DC cultures were adjusted to the same OD600 as their companion cultures in TSB-DC/ABS by dilution with fresh TSB-DC prior to RNA extraction. Extraction of RNA, and cDNA synthesis, fragmentation and labelling were conducted essentially as previously described (DeRisi, 2001; Garber & Urisman, 2001; Yang et al., 2002). Labelled samples were hybridized to PAO1 genomic arrays provided by the Institute for Genomic Research (Rockville, MD; now part of the J. Craig Venter Institute) as part of the Pathogen Functional Genomics Center initiative. The hybridization, washing and drying were done using Corning hybridization chambers (Corning Life Sciences). Arrays were scanned using a GenePix 4200A array scanner (Molecular Devices). Genes were considered differentially regulated if the relative change was twofold or more between PAO1 grown in TSB-DC and TSB-DC/ABS at 4 or 8 h. Microarray data and metadata described in this study have been deposited in the GEO database (accession number GSE30698).
Assays for gene expression by β-galactosidase, ETA and pyoverdine.
Assays for β-galactosidase were performed as previously reported (Gaines et al., 2005; Miller, 1972; Stachel et al., 1985). The calculation of relative units of β-galactosidase activity takes into account the level of growth. ETA levels within the supernatant of PAO1 were determined by sandwich ELISA as previously described (Gaines et al., 2005). Values were standardized by dividing the amount of ETA in pg ml−1 by the OD600 of the culture from which the fraction was obtained. Pyoverdine levels within the supernatant fractions of PAO1 were quantified spectrophotometrically as A405 according to Stintzi et al. (1996). Normalization of pyoverdine levels for growth was obtained by dividing A405 by OD600.
Collection and preparation of adult human serum and human plasma.
Fifty millilitres of whole blood was obtained by venipuncture from each of five healthy volunteers under a protocol approved by the Institutional Review Board at Texas Tech University Health Sciences Center. The blood was drawn into three 10 ml glass tubes without additives for collection of serum and two 10 ml glass tubes containing 143 USP units of freeze-dried sodium heparin for collection of plasma (Becton Dickinson). For the separation of either serum or plasma from the cellular portion of the blood, the tubes were incubated at room temperature for 2 h. To achieve cell-free plasma (AHP) or serum (AHS), the tubes were then centrifuged at 3000 r.p.m. for 15 min at room temperature. Sera from the five donors were pooled, aliquoted and stored at −20 °C until use; the plasma samples were handled in the same manner.
Fractionation and treatment of serum.
Using molecular mass cut-off membrane filters (Vivaspin) we separated ABS and AHS into different fractions (≥3/<3 kDa and ≥50/<50 kDa). Albumin was absorbed from ABS or AHS using DEAE Affi-Gel Blue gel columns (Bio-Rad) according to the manufacturer’s recommendations. To determine effects of protein degradation, serum was heated to 70 °C for 3 or 5 h and cooled to room temperature; or treated with 3 mg trypsin ml−1 (Sigma-Aldrich) for 1 h 37 °C and the reaction was terminated by addition of 3 mg trypsin inhibitor ml−1 (Sigma-Aldrich). Trypsin treatment was confirmed by comparing the protein profile of treated and non-treated samples by SDS-PAGE analysis (data not shown). Treated serum was utilized in the expression experiments as described above.
Immunoblotting experiments.
These experiments were conducted as previously described (Schaber et al., 2004) using a commercially available rabbit anti-human transferrin antibody (Rockland Immunochemicals) that detects both apo- and holotransferrin. Albumin absorbed from AHS was eluted from the DEAE Affi-Gel Blue gel column according to the manufacturer’s instructions. Purified human apotransferrin and human holotransferrin were used as controls.
Results
Serum alters the expression of numerous P. aeruginosa PAO1 genes during early exponential growth
To examine the effect of serum on P. aeruginosa gene expression, we selected the P. aeruginosa prototrophic strain PAO1, which has been extensively utilized in numerous in vitro and in vivo virulence studies (Peluso et al., 2010; Schaber et al., 2004; Smith & Iglewski, 2003). To establish the time points at which serum would affect gene expression, PAO1 was grown in the iron-limited medium TSB-DC with or without 10 % (v/v) ABS (TSB-DC/ABS) for 10 h. Samples were obtained every 2 h and the growth index was determined. The presence of 10 % ABS significantly reduced the growth of PAO1 at time points tested (Fig. 1). Despite this reduction, PAO1 grown in ABS reached early exponential phase at 4 h and early stationary phase at 8 h in both media (Fig. 1). Accordingly, we determined the PAO1 transcriptome in both media at these two specific time points. However, to standardize the comparison, we adjusted the cultures to the same OD600 (at each time point) by adding fresh TSB-DC to the TSB-DC cultures. Total RNA was extracted and reverse transcribed, and the transcriptome analysis was conducted as described in Methods. We identified 178 PAO1 genes whose expression was significantly altered (at least a twofold change) by the presence of serum only at the early exponential phase (4 h) of growth (see Supplementary Table S1, available with the online version of this paper). Of these genes, the expression of 138 was significantly enhanced while that of 40 genes was significantly reduced (Table S1). In contrast, the expression of only a few genes was significantly altered (reduced) at the early stationary (8 h) phase of growth (Table S1).
Fig. 1.
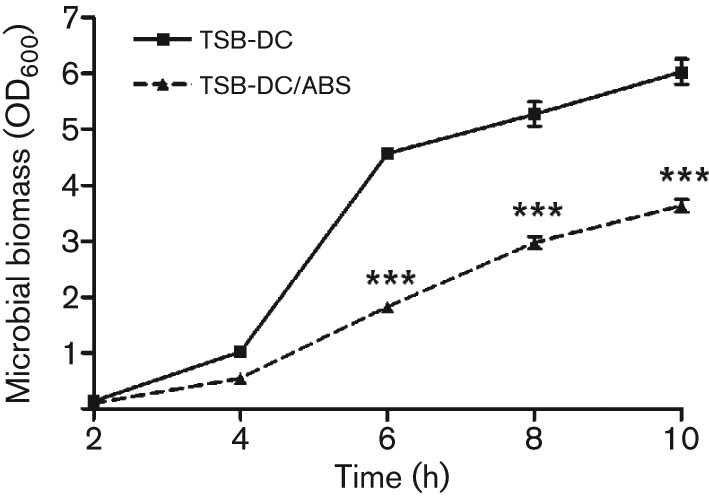
Effect of ABS on P. aeruginosa growth. One-millilitre aliquots of PAO1 overnight culture in LB broth were pelleted, washed twice with TSB-DC, resuspended in TSB-DC and inoculated into fresh TSB-DC or TSB-DC containing 10 % ABS (TSB-DC/ABS) to a starting OD600 of 0.02–0.03. Cultures were incubated at 37 °C with shaking at 230 r.p.m. Samples were obtained at 2 h intervals and the OD600 determined. Values represent the mean±sem of three independent experiments. Statistical significance was determined by one-way ANOVA with Tukey’s post test. ***, P<0.001.
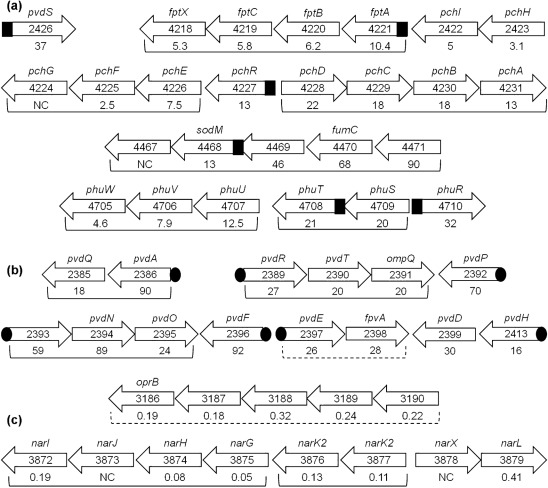
Results of the transcriptome analysis revealed that many of the genes whose expression is significantly altered at 4 h are iron-regulated genes (Table 2). However, the effect of ABS on the expression of these genes was opposite to that of iron, i.e. ABS increased the expression of iron-repressed genes and reduced the expression of iron-induced genes (Table 2). The increase in the expression of iron-repressed genes by ABS varied between eightfold (pchE) and 92-fold (pvdF) (Table 2). Several of these genes have been previously characterized while others code for hypothetical proteins (Table 2). Additionally, many of the genes are clustered in operons, including the pvd, phu, pch and nar operons (Fig. 2). While ABS enhanced the expression of every gene within certain operons, it enhanced the expression of only one or two genes within other operons (Fig. 2). ABS significantly repressed the expression of one iron-induced gene (the bacterioferritin gene, bfrB) (Table 2). Previously characterized P. aeruginosa iron-regulated genes contain specific sequences within their upstream regions to which the iron-regulatory protein Fur or the ECF sigma factor PvdS bind: the Fur box or the iron-starvation box (IS box), respectively (Cornelis et al., 2009; Hamood et al., 2004; Ochsner & Vasil, 1996; Vasil & Ochsner, 1999). A computer search of the upstream regions of the ABS-regulated, iron-controlled genes identified the presence of the Fur box or the IS box within the upstream regions of several of these genes (Table 2, Fig. 2).
Table 2. PAO1 iron-controlled genes influenced by serum in the early exponential phase of growth.
| ORF* | Gene* | Function/class* | Fold change† | Fur box (FB)GATAATGATAATCATTATC‡Iron starvation box (ISB)TAAAT-N16-CGT§ |
| PA0707 | toxR/regA | Transcriptional regulator ToxR/RegA | 14 | |
| PA1317 | cyoA | Cytochrome o ubiquinol oxidase subunit II | 11 | |
| PA2384 | nn|| | Probable DNA-binding protein | 27 | |
| Pyoverdine synthesis and transport | ||||
| PA2385 | pvdQ | PvdQ | 18 | |
| PA2386 | pvdA | l-Ornithine N5-oxygenase | 90 | ISB |
| PA2389 | pvdR | Protein of unknown function | 27 | ISB |
| PA2390 | pvdT | ABC transporter | 20 | |
| PA2391 | opmQ | Probable outer-membrane protein | 20 | |
| PA2392 | pvdP | PvdP | 70 | ISB |
| PA2393 | nn | Probable dipeptidase precursor | 59 | ISB |
| PA2394 | pvdN | PvdN | 89 | |
| PA2395 | pvdO | PvdO | 24 | |
| PA2396 | pvdF | Pyoverdine synthetase F | 92 | ISB |
| PA2397 | pvdE | Pyoverdine biosynthesis protein PvdE | 26 | ISB |
| PA2398 | fpvA | Ferripyoverdine receptor | 28 | |
| PA2399 | pvdD | Pyoverdine synthetase D | 30 | |
| PA2413 | pvdH | Diaminobutyrate-2-oxoglutarate aminotransferase | 17 | ISB |
| PA2425 | pvdG | PvdG | 51 | |
| PA2426 | pvdS | Sigma factor PvdS | 37 | FB (GtaAtTGAcAATCATTATC) |
| PA3531 | bfrB | Bacterioferritin | 0.09 | Fur-like binding sequence |
| PA4159 | fepB | Ferrienterobactin-binding periplasmic protein precursor | 9 | |
| Pyochelin synthesis and transport | ||||
| PA4168 | fpvB | Second ferric pyoverdine receptor FpvB | 10 | |
| PA4218 | fptX | Putative permease, AmpG paralogue | 5.3 | |
| PA4219 | fptC | Hypothetical protein | 5.8 | |
| PA4220 | fptB | Hypothetical protein | 6.2 | |
| PA4221 | fptA | Fe(III)-pyochelin outer-membrane receptor precursor | 10 | FB (cATAATGATAAgCATTATC) |
| PA4222 | pchI | Probable ATP-binding component of ABC transporter | 5.65 | |
| PA4223 | pchH | Probable ATP-binding component of ABC transporter | 3.12 | |
| PA4225 | pchF | Pyochelin synthetase | 2.5 | |
| PA4226 | pchE | Dihydroaeruginoic acid synthetase | 8 | |
| PA4227 | pchR | Transcriptional regulator PchR | 13 | FB (GgaAATGAgAtTtATTATC) |
| PA4228 | pchD | Pyochelin biosynthesis protein D | 22 | |
| PA4229 | pchC | Pyochelin biosynthesis protein C | 18 | |
| PA4230 | pchB | Salicylate biosynthesis protein PchB | 18 | |
| PA4231 | pchA | Salicylate biosynthesis isochorismate synthase | 9 | |
| PA4359 | feoA | Ferrous iron transport protein A | 11.5 | |
| PA4468 | sodM | Superoxide dismutase | 13 | FB (GATAATGAgAtTgATTATt) |
| PA4469 | unc¶ | Hypothetical protein | 46 | |
| PA4470 | fumC1 | Fumarate hydratase | 68 | |
| PA4471 | unc | Hypothetical protein | 90 | |
| Haem transport and utilization | ||||
| PA4705 | phuW | Hypothetical protein | 4.6 | |
| PA4706 | phuV | Haemin importer ATP binding subunit | 7.9 | |
| PA4707 | phuU | Permease of ABC transporter | 12.5 | |
| PA4708 | phuT | Haem-transport protein | 21 | FB (GcTAATGcaAATaATTATC) |
| PA4709 | phuS | Haemin degrading factor | 20 | |
| PA4710 | phuR | Haem/haemoglobin uptake outer membrane receptor precursor | 32 | FB (GATAATtATttgCATTAgC) |
| PA5531 | tonB1 | TonB1 protein | 6 | FB (ctgAATGATAATaATTATC) |
Information obtained from the Pseudomonas Genome DatabaseV2 (http://www.pseudomonas.com/) (Winsor et al., 2009).
PAO1 grown in TSB-DC/ABS compared to PAO1 grown in TSB-DC.
Consensus sequence for Fur box (Ochsner et al., 2002).
Consensus sequence for iron starvation box (Wilson et al., 2001).
Not named.
Unclassified.
Fig. 2.
Diagram of PAO1 iron-controlled genes enhanced or repressed by serum. The arrows with gene number indicate the orientation of the gene on the PAO1 chromosome (Pseudomonas Genome DatabaseV2; http://www.Pseudomonas.com; Winsor et al., 2009). Gene names, if known, are above the arrow, and the fold change is below the arrow. Brackets indicate that genes lie in operons; dashed brackets indicate putative operons. (a) Genes/operons enhanced by serum and directly or indirectly regulated by Fur; black square indicates Fur box (Table 2). (b) Genes/operons enhanced by serum and directly or indirectly regulated by PvdS; black oval indicates iron starvation box (Table 2). (c) Genes/operons repressed by serum. The upstream region of bfrB contains a Fur box-like sequence.
Serum enhances the expression of iron-controlled P. aeruginosa genes
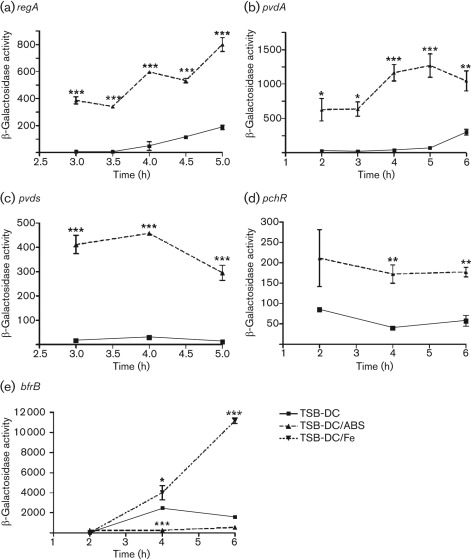
To validate the results of the transcriptome analysis, we utilized plasmids carrying lacZ transcriptional or translational fusions or PAO1 strains containing chromosomal transcriptional fusions with the iron-repressed genes regA, pvdA, pvdS and pchR, and the iron-induced gene bfrB (Table 2) (Hamood et al., 2004; Ma et al., 1999; Vasil & Ochsner, 1999; Wilderman et al., 2004). Since the significant effect of ABS on the expression of these genes was detected in the early exponential phase of growth (Fig. 1), we determined the expression of these genes over 2–6 h of growth. To compensate for the growth-related bias (Fig. 1), the growth index (OD600) was incorporated into the formula for calculating the units of β-galactosidase activity (Gaines et al., 2005; Stachel et al., 1985). Compared with PAO1, the presence of ABS significantly enhanced the expression of pvdA and regA at the indicated time points (Fig. 3a, b). In conjunction with its effect on expression of the pyoverdine genes, ABS significantly enhanced pyoverdine production by PAO1 in the mid-exponential and early stationary phases of growth (Fig. 4a), a reflection of the time required for translating and processing the pyoverdine synthesis proteins. Similarly, ETA production was also enhanced (Fig. 4b). As expression of toxA, regA and pyoverdine requires functional PvdS (Cornelis, 2010; Hamood et al., 2004; Vasil & Ochsner, 1999), we confirmed that ABS significantly enhanced the expression of pvdS (Fig. 3c). Furthermore, we showed that ABS enhances the expression of the pyochelin regulatory gene pchR (Fig. 3d), which is regulated by Fur. In contrast, ABS significantly represses the expression of the iron-inducible bfrB gene at 4 and 6 h (Fig. 3e). As expected, addition of iron increased bfrB expression significantly at these time points (Fig. 3e).
Fig. 3.
Serum enhances the expression of regA, pvdA, pvdS and pchR but represses the expression of bfrB in the early exponential phase of growth. PAO1 containing transcriptional or translational lacZ fusion plasmids was grown in TSB-DC or TSB-DC/ABS as described in Fig. 1. Samples were collected at the time points indicated on the graphs and the β-galactosidase activity was determined and normalized for growth (see Methods). Plasmids pRL88, pMP220 : : PpvdA, pMP220 : : PpvdS and pDH10 were utilized to examine the expression of regA (a), pvdA (b), pvdS (c) and pchR (d), respectively. Strain PW6979 : : bfrB-lacZ was used to examine bfrB expression (e). Iron was added to TSB-DC at 5 µg ml−1 (TSB-DC/Fe) to induce bfrB expression as a control. Values represent the means±sem of three independent experiments. ***, P<0.001; **, P<0.01; *, P<0.05.
Fig. 4.
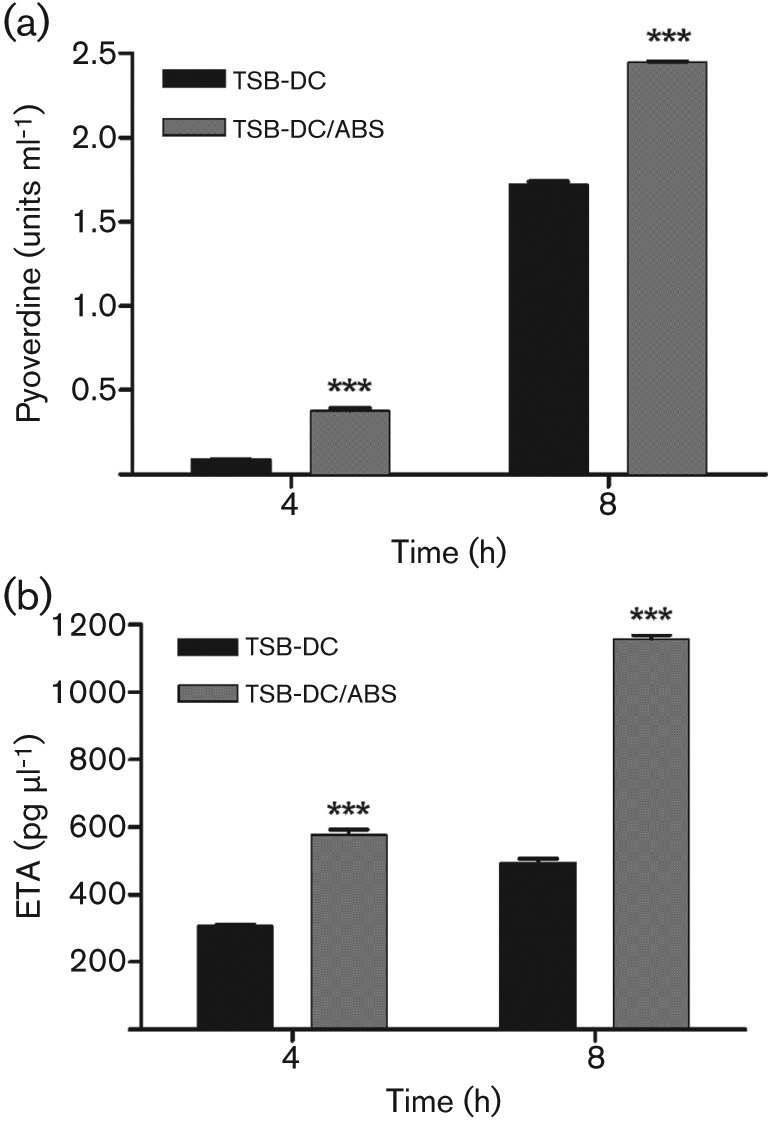
Serum enhances synthesis of pyoverdine and exotoxin A (ETA). (a) To assess pyoverdine production, PAO1 was grown in TSB-DC and TSB-DC/ABS at 37 °C. Samples were obtained at 4 and 8 h, and the level of pyoverdine within the supernatant fraction was quantified spectrophotometrically as A405. Pyoverdine levels were normalized for growth by dividing A405 by the OD600 of the cultures. (b) To analyse ETA synthesis, PAO1 was grown in TSB-DC and TSB-DC/ABS at 32 °C. Samples were obtained at 4 and 8 h, and the level of ETA within the supernatant fraction was determined by sandwich ELISA. Levels of ETA were standardized by dividing the amount of ETA in pg ml−1 by the OD600 of the culture from which the fraction was obtained. Values represent the means±sem of three independent experiments. ***, P<0.001.
Serum enhancement of target genes does not occur through iron
One possible explanation for the above results is that a serum factor interferes with the regulation of PAO1 genes by iron. As a result, iron-induced genes would be repressed and vice versa. In our experiments, cells within the initial inocula of PAO1 in TSB-DC or TSB-DC/ABS contained a residual pool of iron. Prior to their inoculation into the iron-deficient TSB-DC, PAO1 cells were grown overnight in the iron-containing medium LB broth. The intracellular pool of iron in these cells would be partially depleted at the 4 h time point and completely depleted at the 8 h time point. Therefore, in the early exponential phase of growth, ABS would increase the expression of these genes by interfering with the repressive effect of the residual pool of iron. However, by the early stationary phase of growth, the intracellular pool of iron is depleted and the target genes would no longer be regulated by either iron or the serum factor.
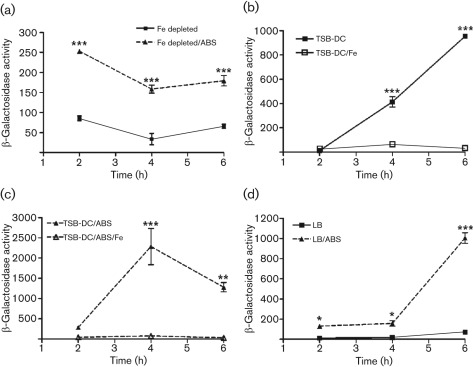
To examine this possibility, we tried two approaches. Firstly, we depleted the intracellular pool of iron in PAO1 by subculturing the cells twice in TSB-DC prior to examining the effect of ABS on gene expression. If the above scenario is correct, we would no longer detect the effect of ABS on gene expression in the early exponential phase of growth. Alternatively, if the effect of ABS is iron independent, the genes would be regulated. Since pvdS regulates the expression of a number of iron-regulated genes, we examined pvdS expression as representative of the ABS-upregulated genes. The addition of 10 % ABS to the second TSB-DC subculture of PAO1/pPMP220 : : PpvdS still significantly enhanced regA expression in the early exponential phase of growth (Fig. 5a). Secondly, we determined whether exogenously added iron would override the serum enhancement in pvdS expression in the early exponential phase of growth (two independent regulatory mechanisms). We grew PAO1/pPMP220 : : PpvdS overnight in LB broth and subcultured it into TSB-DC, TSB-DC/ABS, TSB-DC/Fe and TSB-DC/ABS/Fe. Preliminary experiments showed that iron completely represses pvdS expression at this concentration (20 µg Fe ml−1; data not shown). Control experiments showed that, when added separately, iron repressed pvdS expression (Fig. 5b). However, ABS did not enhance pvdS expression in TSB-DC containing iron (Fig. 5c). We obtained similar results when we used the two approaches to examine pvdA and regA expression (data not shown). Next, we examined the effect of serum on pvdS expression in a growth medium in which the level of iron is not sufficient to completely repress the expression of different genes (LB broth). The addition of ABS significantly enhanced pvdS expression in LB broth at 4 and 6 h (Fig. 5d). These results support the possibility that ABS regulates the expression of its target genes by an iron-independent mechanism.
Fig. 5.
Serum does not interfere with the repression of pvdS expression by iron. PAO1/pPMP220 : : PpvdS was grown in the relevant media, and samples were obtained at the indicated time points and the level of β-galactosidase activity was determined. Values represent the means±sem of three independent experiments. ***, P<0.001; **, P<0.01; *, P<0.05. (a) Serum enhances pvdS expression when the intracellular pool of iron is depleted. The intracellular pool of iron was depleted by subculturing cells twice in TSB-DC and then adding ABS or continuing the cultures in TSB-DC alone. (b) Iron exogenously added to TSB-DC (high level, 20 µg ml−1) represses pvdS expression. (c) High levels of exogenous iron override serum enhancement in pvdS expression. (d) Serum enhances pvdS expression in PAO1 grown in LB broth, a medium that contains low levels of iron.
We previously showed that the addition of norepinephrine (NE) to cultures of PAO1 in ABS significantly reduced the expression of the iron-regulated genes pvdS, regA and toxA (Li et al., 2009). Based on this and other experiments, we suggested that NE functions as a siderophore by removing iron from an iron-binding protein in the serum, such as transferrin, and transferring it to PAO1 (Li et al., 2009). As a result, the intracellular pool of iron increases, which in turn represses the expression of pvdS, regA and toxA. To further confirm that the observed influence of ABS on these genes is not directly related to iron, we examined the effect of exogenously added NE (500 µM) on pvdS expression in PAO1 in the early exponential phase of growth. While ABS enhanced pvdS expression as previously observed, this enhancement was eliminated at 2 h upon the addition of NE (Supplementary Fig. S1).
Seven of the serum-regulated genes contain a Fur box within their upstream regions (Table 2, Fig. 2), raising the possibility that Fur plays a role in the regulation of some of the serum-targeted genes. However, the above results also suggest that serum affects the expression of these genes independently of the Fur cofactor, iron (Fig. 5). Therefore, to explore a possible link between serum and Fur in regulating PAO1 genes, we utilized the PAO1 Fur-deficient mutant C6 (Barton et al., 1996). C6, which was derived from the PAO1 anr mutant strain PAO6261 (Ye et al., 1995), carries mutations in both the anr and fur-6 genes (Table 1) (Barton et al., 1996). Barton et al. (1996) previously demonstrated that C6 is the only relatively stable PAO1-fur mutant; with the exception of C6, all other Fur-deficient mutants generated directly from PAO1 reverted to the wild-type phenotype. Within C6, ETA production is deregulated with respect to iron (Barton et al., 1996). Thus, a possible role for Fur in our observed results would be indicated if ABS failed to regulate its target genes in C6. We determined the level of regA expression in PAO6261 and C6 containing pRL88 grown in either TSB-DC or TSB-DC/ABS. At 4 and 5 h, ABS significantly enhanced regA expression in both strains, suggesting that the absence of a functional Fur may not interfere with the regulation of PAO1 genes by serum (Fig. 6). However, since the C6 fur mutant is only relatively stable, a definitive conclusion regarding the role of Fur in this phenomenon cannot be made at this time.
Fig. 6.
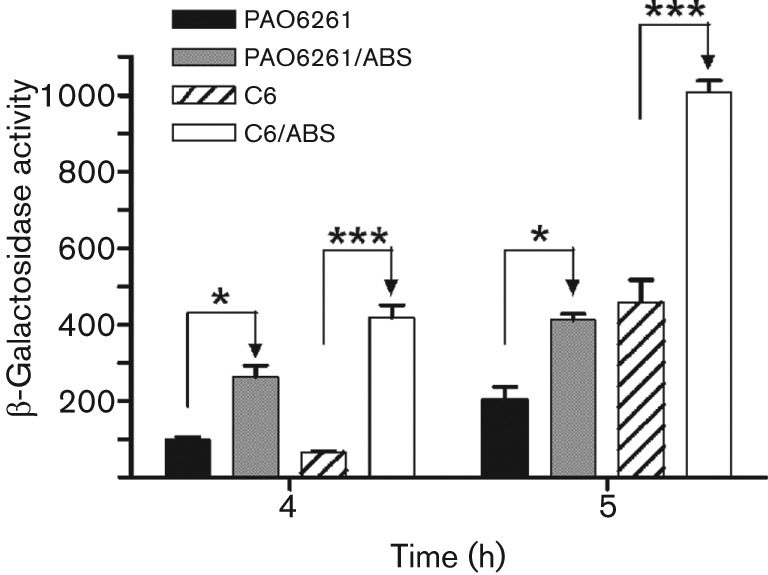
The effect of serum does not require Fur. The PAO1 fur mutant C6 and its parent strain PAO6261 carrying pRL88 were grown in TSB-DC or TSB-DC/ABS and regA expression was determined by the levels of β-galactosidase activity produced at 4 and 5 h. Values represent the means±sem of three independent experiments. ***, P<0.001; *, P<0.05.
Based on the above results, serum may regulate the expression of toxA, regA and the pyoverdine genes individually by direct regulation. Alternatively, serum may regulate these genes indirectly by increasing the expression of their main regulator, pvdS. To examine this possibility, we determined whether ABS increases regA expression in PAO1ΔpvdS, which carries an internal deletion in pvdS. As expected, the lack of functional PvdS reduced regA expression significantly; however, comparable levels of regA expression were seen in TSB-DC and TSB-DC/ABS (data not shown). Similar results were obtained when we examined toxA and pvdD expression in PAO1ΔpvdS (data not shown), suggesting that serum regulates regA and the pyoverdine genes through pvdS.
Bovine serum albumin enhances the expression of PAO1 iron-repressed genes
The above results suggest that serum influences the expression of PAO1 genes through a unique mechanism that is PvdS-dependent but iron- and Fur-independent. To further understand this mechanism, we attempted to identify the potential serum factor(s) that affect(s) the expression of these genes. We first addressed the possibility that the serum factor is a small molecule that enters the PAO1 cytoplasm (passively or actively) and programmes the expression of different genes. We membrane-fractionated ABS using a 3 kDa MWCO membrane (Vivaspin). The fraction that passed through the membrane contains small molecules and possibly peptides. Strain PAO1/pPMP220: : PpvdS was grown using the filtrate and retentate in place of serum. At both the 2 h and 4 h time points, the serum-induced enhancement in pvdS expression was eliminated from the filtrate (<3 kDa) but preserved within the retenate (≥3 kDa) (data not shown). We then repeated the process using a 50 kDa MWCO membrane. Again, enhancement of pvdS expression was detected with the retentate but not the filtrate at both time points (Supplementary Fig. S2). To confirm that the potential factor is a protein, we denatured the serum proteins by either heat or trypsin treatment. Heat treatment eliminated the effect of serum on regA expression (1400 units of β-galactosidase activity were produced by PAO1/pRL88 in untreated serum and 14 units in heat-treated serum, a 101-fold reduction in activity). We obtained similar results with trypsin treatment (units of β-galactosidase activity produced by PAO1/pRL88 were reduced eightfold in trypsin-treated serum). We eliminated the possibility that the complement system is involved in the observed effect of serum by heating the serum at 65 °C for 20 min to inactivate complement. Complement-inactivated serum still increased regA expression in PAO/pRL88 at the 2 and 4 h time points (data not shown).
Albumin, a protein of approximately 65 kDa in its monomeric form, is a major component of serum (Foster, 1977; Peters, 1996; Rondeau & Bourdon, 2011). Previous studies suggested that serum albumin affects PAO1 virulence (Foster, 1977; Peters, 1996; Rondeau & Bourdon, 2011). Thus, to determine if serum albumin is the serum factor that affects the expression of PAO1 genes, we depleted the albumin from ABS using DEAE Affi-Gel Blue gel affinity columns. Compared with the effect of intact serum, albumin depletion eliminated the ABS-induced enhancement in pvdS expression in PAO1/pPMP220 : : PpvdS at both 2 h and 4 h (Fig. 7a), suggesting that serum albumin plays a major role in ABS enhancement of PAO1 gene expression.
Fig. 7.
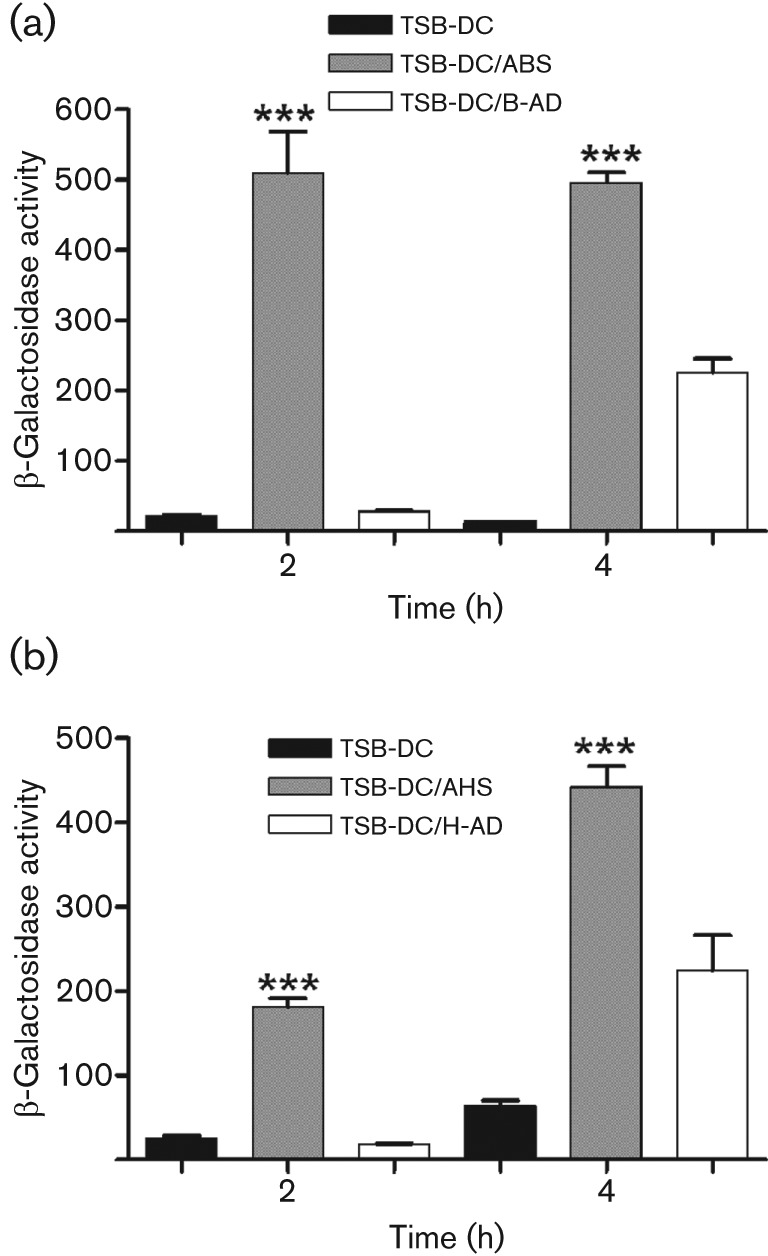
Albumin depletion abrogates serum enhancement of PAO1 gene expression. DEAE Affi-Gel Blue gel affinity columns (Bio-Rad) were used to deplete ABS (B-AD) and AHS (H-AD) of albumin. PAO1/pPMP220 : : PpvdS was grown in (a) TSB-DC, TSB-DC/ABS or TSB-DC/B-AD; or (b) TSB-DC, TSB-DC/AHS or TSB-DC/H-AD. Samples were obtained at 2 and 4 h and the level of β-galactosidase activity was determined. Values represent the means±sem of three independent experiments. ***, P<0.001.
Human serum and serum albumin produce the same effect on regA and pvdS expression
We then determined whether similar results would be obtained using adult human serum (AHS) and adult human plasma (AHP). Blood samples were collected from healthy volunteers; the serum or plasma fractions were separated and pooled, and tested as described in the above sections. Both AHS and AHP significantly enhanced pvdS and regA expression at both the 2 h and 4 h time points (Supplementary Fig. S3a, b). We then depleted the albumin from AHS using Affi-Gel Blue gel affinity columns. Compared with AHS, albumin depletion abrogated the effect on pvdS (Fig. 7b) and regA expression (data not shown) at both 2 h and 4 h. To further confirm the role of serum albumin, we examined the effect of the therapeutic human albumin (THA) solution Albuminar, which contains 10 % human albumin in normal saline. The THA enhanced expression of both pvdS and regA (Fig. 8a, b). These results strongly suggest that at early stages of growth of PAO1, human serum albumin is the serum factor that significantly increases the expression of several of P. aeruginosa iron-repressed genes.
Fig. 8.
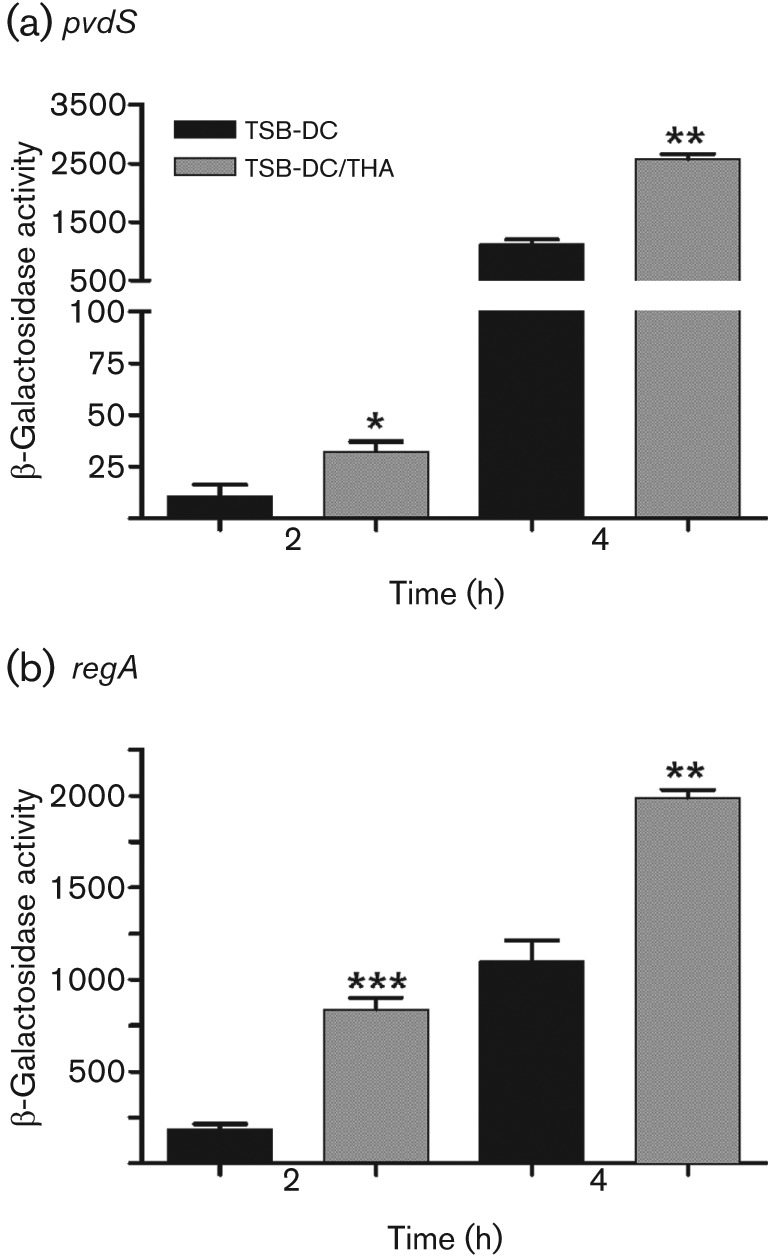
Therapeutic human albumin (THA) saline solution (Albuminar) enhances pvdS expression. PAO1/pPMP220 : : PpvdS (a) and PAO1/pRL88 (b) were grown in either TSB-DC or TSB-DC/THA. Samples were obtained at 2 and 4 h and the level of β-galactosidase activity was determined. Values represent the means±sem of three independent experiments. ***, P<0.001; **, P<0.01; *, P<0.05.
Human albumin and apotransferrin each enhance the expression of PAO1 iron-controlled genes
Serum contains many iron-binding proteins, including transferrin, ferritin and albumin. Although TSB-DC is iron-deficient (Ohman et al., 1980), our recent analysis revealed that the medium contains a trace amount of iron (0.21µg ml−1). Serum iron-binding proteins may chelate this trace amount of iron and enhance pvdS and regA expression. Therefore, we examined the effect of purified human ferritin, holotransferrin and apotransferrin on PvdS expression. At 20 µg ml−1, none of the proteins enhanced pvdS expression (data not shown), while at 80 µg ml−1 and higher, only apotransferrin enhanced pvdS expression, by about fourfold (322 units of β-galactosidase activity for PAO1/pPMP220 : : PpvdS grown in TSB-DC vs 1307 units for PAO1/pPMP220 : : PpvdS grown in TSB-DC/apoTF). To determine if serum enhances pvdS expression independently of apotransferrin, we grew PAO1/pPMP220 : : PpvdS in MM9, which contains no iron. Even at 200 µg ml−1, apotransferrin did not enhance pvdS expression (Fig. 9). However, serum significantly enhanced pvdS expression (Fig. 9). Serum also significantly enhanced regA expression PAO1/pRL88 grown in MM9 (data not shown). More importantly, albumin-deficient human serum failed to enhance pvdS expression in MM9 (Fig. 9), indicating that the effect of albumin is not directly related to iron. To exclude the possibility that passage of serum through the DEAE Affi-Gel Blue gel column eliminated both albumin and apotransferrin, we eluted the absorbed albumin-rich fraction and examined it for the presence of apotransferrin by immunoblotting. The albumin-rich fraction contained neither apo- nor holotransferrin (Fig. 10). Using the same approach, we also showed Albuminar contains neither form of transferrin (Fig. 10). These results suggest that the observed enhancement in pvdS/regA expression, and possibly the expression of other iron-controlled genes, by serum occurs through two separate mechanisms: one is apotransferrin-dependent (iron chelation) while the other is albumin-dependent.
Fig. 9.
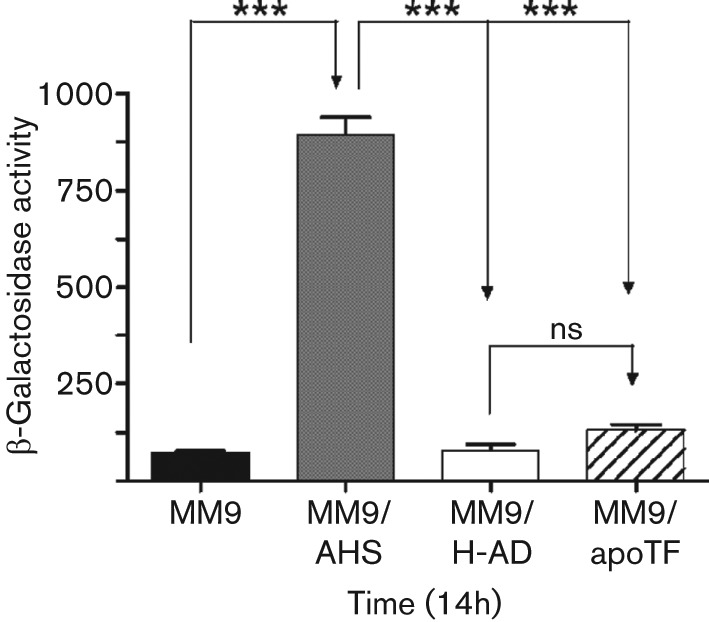
Serum enhances expression of pvdS in medium lacking iron. Cells were grown as described in Fig. 1 and inoculated into MM9 (chemically defined totally iron-deficient medium), MM9/AHS, MM9/H-AD or MM9/apoTF (200 µg apotransferrin ml−1). Cultures were incubated at 37 °C with shaking at 230 r.p.m. for 14 h and the level of β-galactosidase activity was determined. Values represent the means±sem of three independent experiments. Statistical significance was determined by one-way ANOVA with Tukey’s post test. ***, P<0.001; ns, no significant difference.
Fig. 10.
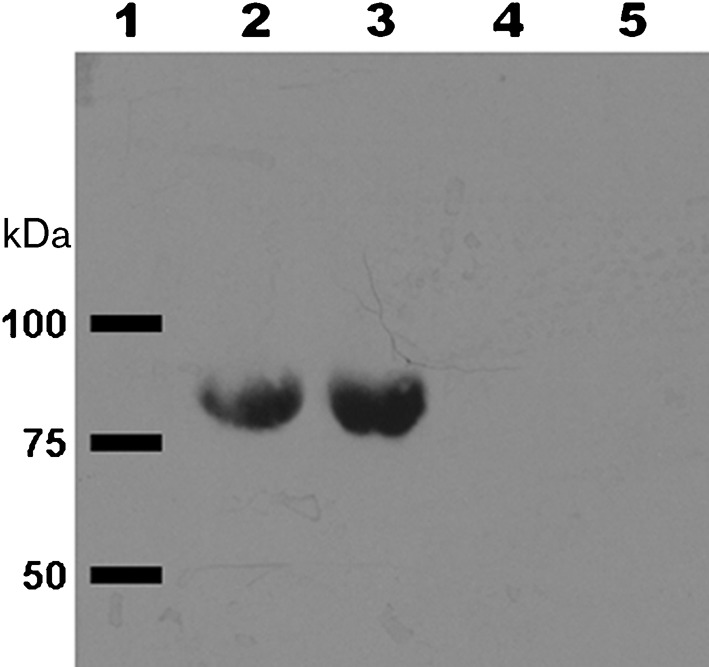
Transferrin is not present in the albumin-rich serum fraction or in Albuminar. Albumin absorbed from AHS was eluted from the DEAE Affi-Gel Blue gel column. Proteins from each sample were separated by SDS-PAGE, transferred to membrane, and probed with rabbit anti-human transferrin antibody. Lanes: 1, molecular mass standards (band positions indicated by black bars); 2, 30 µg holotransferrin; 3, 30 µg apotransferrin; 4, 50 µg albumin-rich fraction from AHS; 5, 50 µg Albuminar.
Discussion
Results presented in this study suggest a novel mechanism through which serum regulates the expression of different P. aeruginosa genes. The essential feature of this mechanism is its occurrence at early stages of growth and under iron-limited conditions. As shown by the results of the microarray analysis and confirmed by the transcriptional and translational fusion analyses, many of the serum-regulated genes are involved in either iron acquisition, iron transport or iron storage (Table 2, Fig. 2). However, although several of these serum-regulated genes also contain a Fur box within their upstream regions, their regulation by serum is opposite to their regulation by iron (Table 2). Several of these serum-regulated genes also contain a Fur box within their upstream regions (Table 2). Many genes that do not contain the Fur box contain an IS box and are regulated by PvdS, which carries a Fur box. Despite this, their regulation by serum does not appear to be directly related to iron. Our results showed that the phenomenon did not occur in response to the depletion of the intracellular pool of iron (Fig. 5a). Similarly, serum did not interfere with the repression of these genes by exogenous iron (Fig. 5c). Additionally, serum did not interfere with the repression of these two genes by NE (Fig. S1). Part of the observed effect is produced by the serum iron-binding protein apotransferrin, which chelated the trace amount of iron present in TSB-DC. However, serum significantly induced pvdS and regA expression even when PAO1 was grown in the chemically defined medium MM9, which contains no iron (Fig. 9). Therefore, it is unlikely that the observed serum-induced regulation of these genes is due only to the iron-chelating activity of apotransferritin within the serum. With respect to Fur, our results using the C6 fur mutant showed that the loss of functional Fur did not interfere with the regulation of PAO1 pvdS or regA by serum (Fig. 6). However, this conclusion must be tempered by the relative instability of this mutant.
Our experimental evidence strongly suggests that serum albumin regulates the expression of different P. aeruginosa genes. Through initial fractionation experiments, we ruled out the possibility that the mechanism involves a low-molecular-mass protein (<3 kDa) or an unbound peptide that is actively or passively internalized in PAO1 (data not shown). Additional experiments revealed that the potential factor(s) is a high-molecular-mass protein (≥50 kDa) (Fig. S2). Further analyses indicated that serum albumin contributes substantially to the observed phenomenon, as depletion of albumin from serum eliminated most of the serum-induced enhancement in pvdS and regA expression (Fig. 7). Similarly, the addition of the therapeutic human serum albumin Albuminar enhanced the expression of both pvdS and regA at early stages of growth of PAO1 (Fig. 8). Albumin is a complex molecule that contains several cofactors or associated peptides (Foster, 1977; Peters, 1996; Rondeau & Bourdon, 2011). Therefore, the observed effect on P. aeruginosa genes may be due to one ore more of these peptides or cofactors. The influence of serum albumin on the production of P. aeruginosa virulence factors has been previously reported (Hammond et al., 2010; Kim et al., 2005). Kim et al. (2005) showed that serum albumin facilitated the secretion of P. aeruginosa type III effector molecules. Analogous to the repression of different P. aeruginosa genes by iron, the expression of type III secretion genes, as well as the production of type III effectors, is repressed by calcium (Frank, 1997; Hauser, 2009). Upon its growth in a calcium-deficient medium, P. aeruginosa produces maximum levels of type III secretion effectors (Frank, 1997; Hauser, 2009). Kim et al. (2005) showed that the addition of serum albumin to a calcium-deficient medium triggered the secretion of intracellularly accumulated type III effector molecules; however, the expression of type III secretion genes was not affected. Serum produced this effect by functioning as a low-affinity calcium-binding protein (Kim et al., 2005). The addition of other low-affinity calcium-binding proteins, including casein, α-lactoalbumin and calreticulin, to the calcium-deficient medium produced a similar phenomenon (Kim et al., 2005). Besides being a low-affinity calcium-binding protein, serum albumin is a low-affinity iron-binding protein (Hider et al., 2010; Silva & Hider, 2009). Therefore, serum albumin may affect the expression of PAO1 genes within its capacity as an iron-binding protein. In serum, iron binds with high affinity to transferrin, a protein which transports iron to different tissues within the body (Hider et al., 2010; Silva & Hider, 2009). Iron may also be transported in serum via ferritin and albumin; however, iron binds to these proteins with less affinity than it binds to transferrin. Additionally, our results clearly rule out the possibility that albumin enhances the expression of different PAO1 genes through its capacity as a low-affinity iron binding protein. Albumin-deficient human serum failed to enhance pvdS expression in MM9 (Fig. 9). Further experiments will be conducted to determine if albumin or albumin-associated factor(s) influences the expression of P. aeruginosa genes besides pvdS and regA.
Although the results of our analyses strongly suggest that the influence of serum on the expression of P. aeruginosa iron-controlled genes is not directly related to Fur, the specific PAO1 gene or regulatory system through which this regulation is accomplished is not known. It is clear that the effect still requires part of the hierarchical system of iron-regulated genes. In the absence of functional PvdS, which regulates the expression of regA, toxA and the pyoverdine genes, serum failed to enhance regA expression (data not shown). At this time, little is known regarding the relationship between serum and iron-regulated genes in P. aeruginosa. One potential link is the previously described virulence and quorum-sensing regulator vqsR (Juhas et al., 2004): it was shown by microarray analysis that, in the presence of serum, 113 genes were differentially regulated in the vqsR transposon insertion mutant compared with its PAO1 parent strain (Cornelis & Aendekerk, 2004; Juhas et al., 2004).
Among the genes whose expression was significantly downregulated in the vqsR mutant are 24 genes known to be induced by iron limitation (Juhas et al., 2004). In our study, the expression of 18 of those 24 genes was significantly increased by serum at early stages of growth of PAO1, including genes involved in pyoverdine and pyochelin biosynthesis (Fig. 2, Table 2, Table S1). However, our results also suggest that the observed effect of serum in our study does not occur through vqsR. Juhas et al. (2004) reported that vqsR significantly affects pyocyanin production by PAO1; however, our microarray analysis revealed that serum does not significantly alter the expression of phenazine genes at early- or mid-exponential or late phases of growth (Table S1 and data not shown). We also showed that serum does not affect pyocyanin production by PAO1 (data not shown). Furthermore, our transcriptional analysis revealed that vqsR is not among the genes whose expression is significantly affected by serum (Table S1). In contrast, Juhas et al. (2004) showed that the addition of serum to a standard medium increased the accumulation of vqsR mRNA, suggesting that serum enhances vqsR expression. Differences in the findings between our study and that of Juhas et al. (2004) are likely due to the different experimental conditions utilized in these studies. Whereas we analysed the effect of serum on PAO1 cultures that were grown to early exponential and early stationary phases in TSB-DC containing 10 % serum, Juhas et al. (2004) examined the expression in PAO1 cells from stationary-phase cultures that were grown in RPMI medium containing 10 % serum.
Another potential candidate gene through which serum may accomplish its effect is PA2384, which has neither a Fur box nor an IS box in its upstream region, and is less likely to be regulated by Fur (Zheng et al., 2007). The amino-terminal region of the calculated protein encoded by PA2384 has a weak similarity to the DNA-binding domain of Fur (Zheng et al., 2007). Compared to PAO1, the expression of 71 iron-dependent genes was reduced in a PA2384 mutant, including 42 related to pyoverdine or pyochelin synthesis and transport, and haem uptake and utilization (Zheng et al., 2007). In addition to PA2384, 37 of these same 42 genes were upregulated by the presence of serum in our study (Fig. 2, Table 2, Table S1). Preliminary analyses revealed that, at early phases of growth, serum still enhances pyoverdine production by the PAO1-PA2384 mutant, while quantitative RT-PCR analysis showed that serum enhanced pvdS and regA expression in both PAO1 and its PA2384 mutant (data not shown). Finally, whereas we detect the effect of serum at early phases of growth, most of the PA2384 target genes are regulated during the stationary phase of growth of PAO1 (Zheng et al., 2007). Thus, serum is unlikely to regulate the expression of PAO1 genes through PA2384.
Our recent DNA/gel-shift experiments suggest that, in the early exponential phase, serum enhances the synthesis and/or the activity of a PAO1 DNA-binding protein that specifically binds to the upstream region of pvdS (data not shown). Compared with the lysate from PAO1 that was grown in TSB-DC, the lysate from PAO1 that was grown in TSB-DC/ABS produced a more intense specific gel-shift band when it was incubated with the pvdS upstream region (data not shown).
Thus, the most likely scenario to explain our results is that, at early phases of growth, serum regulates the expression of PAO1 genes through a potential regulator that functions similarly to PA2384 (i.e. is not regulated by Fur but most of its target genes carry a Fur-binding site). This candidate regulator may be one of the already identified serum-regulated hypothetical transcriptional activators or probable sigma factors (Table 2, Table S1). However, unlike PA2384, the expression of this potential regulator may not be affected by the level of iron in the growth medium.
Acknowledgements
The authors thank Iain Lamont, Keith Poole, Douglas Storey, Michael Vasil, Paolo Visca and Susan West for the strains and plasmids; and Joanna Swickard for critical reading of the manuscript. Strain PW6979 was made available through NIH grant no. P30 DK089507.
Abbreviations:
- ABS
adult bovine serum
- AHS
adult human serum
- AHP
adult human plasma
- ECF
extracytoplasmic function
- ETA
exotoxin A
- IS box
iron-starvation box
- NE
norepinephrine
- THA
therapeutic human albumin (10 % Albuminar)
- TSB-DC
chelexed tryptic soy broth dialysate
Footnotes
Supplementary material is available with the online version of this paper.
References
- Ambrosi C., Leoni L., Visca P. (2002). Different responses of pyoverdine genes to autoinduction in Pseudomonas aeruginosa and the group Pseudomonas fluorescens–Pseudomonas putida. Appl Environ Microbiol 68, 4122–4126. 10.1128/AEM.68.8.4122-4126.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzaldi L. L., Skaar E. P. (2010). Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infect Immun 78, 4977–4989. 10.1128/IAI.00613-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardehali R., Shi L., Janatova J., Mohammad S. F., Burns G. L. (2003). The inhibitory activity of serum to prevent bacterial adhesion is mainly due to apo-transferrin. J Biomed Mater Res A 66, 21–28. 10.1002/jbm.a.10493 [DOI] [PubMed] [Google Scholar]
- Barton H. A., Johnson Z., Cox C. D., Vasil A. I., Vasil M. L. (1996). Ferric uptake regulator mutants of Pseudomonas aeruginosa with distinct alterations in the iron-dependent repression of exotoxin A and siderophores in aerobic and microaerobic environments. Mol Microbiol 21, 1001–1017. 10.1046/j.1365-2958.1996.381426.x [DOI] [PubMed] [Google Scholar]
- Branski L. K., Al-Mousawi A., Rivero H., Jeschke M. G., Sanford A. P., Herndon D. N. (2009). Emerging infections in burns. Surg Infect (Larchmt) 10, 389–397. 10.1089/sur.2009.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis P. (2010). Iron uptake and metabolism in pseudomonads. Appl Microbiol Biotechnol 86, 1637–1645. 10.1007/s00253-010-2550-2 [DOI] [PubMed] [Google Scholar]
- Cornelis P., Aendekerk S. (2004). A new regulator linking quorum sensing and iron uptake in Pseudomonas aeruginosa. Microbiology 150, 752–756. 10.1099/mic.0.27086-0 [DOI] [PubMed] [Google Scholar]
- Cornelis P., Matthijs S., Van Oeffelen L. (2009). Iron uptake regulation in Pseudomonas aeruginosa. Biometals 22, 15–22. 10.1007/s10534-008-9193-0 [DOI] [PubMed] [Google Scholar]
- Cunliffe H. E., Merriman T. R., Lamont I. L. (1995). Cloning and characterization of pvdS, a gene required for pyoverdine synthesis in Pseudomonas aeruginosa: PvdS is probably an alternative sigma factor. J Bacteriol 177, 2744–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi J. (2001). Overview of nucleic acid arrays. In Current Protocols in Molecular Biology, 22.1.1–22.1.7. 10.1002/0471142727.mb2201s49 10.1002/0471142727.mb2201s49 [DOI] [PubMed] [Google Scholar]
- Foster J. F. (1977). Binding properties of albumin. In Albumin Structure, Function and Uses, pp. 53–84. Edited by Rosenoer V. M., Oratz M., Rothschild M. A. Oxford: Pergamon. [Google Scholar]
- Frank D. W. (1997). The exoenzyme S regulon of Pseudomonas aeruginosa. Mol Microbiol 26, 621–629. 10.1046/j.1365-2958.1997.6251991.x [DOI] [PubMed] [Google Scholar]
- Gaines J. M., Carty N. L., Colmer-Hamood J. A., Hamood A. N. (2005). Effect of static growth and different levels of environmental oxygen on toxA and ptxR expression in the Pseudomonas aeruginosa strain PAO1. Microbiology 151, 2263–2275. 10.1099/mic.0.27754-0 [DOI] [PubMed] [Google Scholar]
- Garber M., Urisman A. (2001). Protocol for reverse transcription and amino-allyl coupling of RNA. Stanford, CA: Stanford Functional Genomics Facility; http://www.microarray.org/sfgf/docView.do?type=2; accessed 23 June 2011. [Google Scholar]
- González-Chávez S. A., Arévalo-Gallegos S., Rascón-Cruz Q. (2009). Lactoferrin: structure, function and applications. Int J Antimicrob Agents 33, e301–e308. 10.1016/j.ijantimicag.2008.07.020 [DOI] [PubMed] [Google Scholar]
- Hammond A., Dertien J., Colmer-Hamood J. A., Griswold J. A., Hamood A. N. (2010). Serum inhibits P. aeruginosa biofilm formation on plastic surfaces and intravenous catheters. J Surg Res 159, 735–746. 10.1016/j.jss.2008.09.003 [DOI] [PubMed] [Google Scholar]
- Hamood A. N., Colmer-Hamood J. A., Carty N. L. (2004). Regulation of Pseudomonas aeruginosa exotoxin A synthesis. In Pseudomonas: Virulence and Gene Regulation, pp. 389–423. Edited by Ramos J.-L. New York, NY: Kluwer Academic/Plenum Publishers; 10.1007/978-1-4419-9084-6_14 [DOI] [Google Scholar]
- Hauser A. R. (2009). The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol 7, 654–665. 10.1038/nrmicro2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs D. E., Poole K. (1996). PchR, a regulator of ferripyochelin receptor gene (fptA) expression in Pseudomonas aeruginosa, functions both as an activator and as a repressor. J Bacteriol 178, 2586–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hider R. C., Silva A. M., Podinovskaia M., Ma Y. (2010). Monitoring the efficiency of iron chelation therapy: the potential of nontransferrin-bound iron. Ann N Y Acad Sci 1202, 94–99. 10.1111/j.1749-6632.2010.05573.x [DOI] [PubMed] [Google Scholar]
- Jacobs M. A., Alwood A., Thaipisuttikul I., Spencer D., Haugen E., Ernst S., Will O., Kaul R., Raymond C. & other authors (2003). Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100, 14339–14344. 10.1073/pnas.2036282100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M., Wiehlmann L., Huber B., Jordan D., Lauber J., Salunkhe P., Limpert A. S., von Götz F., Steinmetz I. & other authors (2004). Global regulation of quorum sensing and virulence by VqsR in Pseudomonas aeruginosa. Microbiology 150, 831–841. 10.1099/mic.0.26906-0 [DOI] [PubMed] [Google Scholar]
- Kim J., Ahn K., Min S., Jia J., Ha U., Wu D., Jin S. (2005). Factors triggering type III secretion in Pseudomonas aeruginosa. Microbiology 151, 3575–3587. 10.1099/mic.0.28277-0 [DOI] [PubMed] [Google Scholar]
- Li W., Lyte M., Freestone P. P., Ajmal A., Colmer-Hamood J. A., Hamood A. N. (2009). Norepinephrine represses the expression of toxA and the siderophore genes in Pseudomonas aeruginosa. FEMS Microbiol Lett 299, 100–109. 10.1111/j.1574-6968.2009.01739.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. F., Ochsner U. A., Klotz M. G., Nanayakkara V. K., Howell M. L., Johnson Z., Posey J. E., Vasil M. L., Monaco J. J., Hassett D. J. (1999). Bacterioferritin A modulates catalase A (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa. J Bacteriol 181, 3730–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Nairz M., Schroll A., Sonnweber T., Weiss G. (2010). The struggle for iron – a metal at the host–pathogen interface. Cell Microbiol 12, 1691–1702. 10.1111/j.1462-5822.2010.01529.x [DOI] [PubMed] [Google Scholar]
- Ochsner U. A., Vasil M. L. (1996). Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc Natl Acad Sci U S A 93, 4409–4414. 10.1073/pnas.93.9.4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner U. A., Vasil A. I., Vasil M. L. (1995). Role of the ferric uptake regulator of Pseudomonas aeruginosa in the regulation of siderophores and exotoxin A expression: purification and activity on iron-regulated promoters. J Bacteriol 177, 7194–7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner U. A., Johnson Z., Lamont I. L., Cunliffe H. E., Vasil M. L. (1996). Exotoxin A production in Pseudomonas aeruginosa requires the iron-regulated pvdS gene encoding an alternative sigma factor. Mol Microbiol 21, 1019–1028. 10.1046/j.1365-2958.1996.481425.x [DOI] [PubMed] [Google Scholar]
- Ochsner U. A., Wilderman P. J., Vasil A. I., Vasil M. L. (2002). GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol Microbiol 45, 1277–1287. 10.1046/j.1365-2958.2002.03084.x [DOI] [PubMed] [Google Scholar]
- Ohman D. E., Sadoff J. C., Iglewski B. H. (1980). Toxin A-deficient mutants of Pseudomonas aeruginosa PA103: isolation and characterization. Infect Immun 28, 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso L., de Luca C., Bozza S., Leonardi A., Giovannini G., Lavorgna A., De Rosa G., Mascolo M., Ortega De Luna L. & other authors (2010). Protection against Pseudomonas aeruginosa lung infection in mice by recombinant OprF-pulsed dendritic cell immunization. BMC Microbiol 10, 9. 10.1186/1471-2180-10-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters T., Jr (1996). The albumin molecule: its structure and chemical properties. In All About Albumin: Biochemistry, Genetics and Medical Applications, pp. 9–75. Edited by Peters T. San Diego: Academic Press. [Google Scholar]
- Pier G. B., Ramphal R. (2005). Pseudomonas aeruginosa. In Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases, pp. 2587–2615. Edited by Mandel G. L., Bennett J. E., Dolin R. Philadelphia: Elsevier/Churchill Livingstone. [Google Scholar]
- Rondeau P., Bourdon E. (2011). The glycation of albumin: structural and functional impacts. Biochimie 93, 645–658. 10.1016/j.biochi.2010.12.003 [DOI] [PubMed] [Google Scholar]
- Sadikot R. T., Blackwell T. S., Christman J. W., Prince A. S. (2005). Pathogen–host interactions in Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med 171, 1209–1223. 10.1164/rccm.200408-1044SO [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaber J. A., Carty N. L., McDonald N. A., Graham E. D., Cheluvappa R., Griswold J. A., Hamood A. N. (2004). Analysis of quorum sensing-deficient clinical isolates of Pseudomonas aeruginosa. J Med Microbiol 53, 841–853. 10.1099/jmm.0.45617-0 [DOI] [PubMed] [Google Scholar]
- Silva A. M., Hider R. C. (2009). Influence of non-enzymatic post-translation modifications on the ability of human serum albumin to bind iron. Implications for non-transferrin-bound iron speciation. Biochim Biophys Acta 1794, 1449–1458. [DOI] [PubMed] [Google Scholar]
- Smith R. S., Iglewski B. H. (2003). P. aeruginosa quorum-sensing systems and virulence. Curr Opin Microbiol 6, 56–60. 10.1016/S1369-5274(03)00008-0 [DOI] [PubMed] [Google Scholar]
- Stachel S. E., An G., Flores C., Nester E. W. (1985). A Tn3 lacZ transposon for the random generation of beta-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J 4, 891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A., Cornelis P., Hohnadel D., Meyer J. M., Dean C., Poole K., Kourambas S., Krishnapillai V. (1996). Novel pyoverdine biosynthesis gene(s) of Pseudomonas aeruginosa PAO. Microbiology 142, 1181–1190. 10.1099/13500872-142-5-1181 [DOI] [PubMed] [Google Scholar]
- Storey D. G., Frank D. W., Farinha M. A., Kropinski A. M., Iglewski B. H. (1990). Multiple promoters control the regulation of the Pseudomonas aeruginosa regA gene. Mol Microbiol 4, 499–503. 10.1111/j.1365-2958.1990.tb00616.x [DOI] [PubMed] [Google Scholar]
- van Delden C. (2004). Virulence factors in Pseudomonas aeruginosa. In Pseudomonas: Virulence and Gene Regulation, pp. 3–45. Edited by Ramos J.-L. New York: Kluwer Academic/Plenum Publishers. [Google Scholar]
- Vasil M. L., Ochsner U. A. (1999). The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol Microbiol 34, 399–413. 10.1046/j.1365-2958.1999.01586.x [DOI] [PubMed] [Google Scholar]
- Wang J., Pantopoulos K. (2011). Regulation of cellular iron metabolism. Biochem J 434, 365–381. 10.1042/BJ20101825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. E., Sample A. K., Runyen-Janecky L. J. (1994). The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J Bacteriol 176, 7532–7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman P. J., Sowa N. A., FitzGerald D. J., FitzGerald P. C., Gottesman S., Ochsner U. A., Vasil M. L. (2004). Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci U S A 101, 9792–9797. 10.1073/pnas.0403423101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. J., McMorran B. J., Lamont I. L. (2001). Analysis of promoters recognized by PvdS, an extracytoplasmic-function sigma factor protein from Pseudomonas aeruginosa. J Bacteriol 183, 2151–2155. 10.1128/JB.183.6.2151-2155.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor G. L., Van Rossum T., Lo R., Khaira B., Whiteside M. D., Hancock R. E., Brinkman F. S. (2009). Pseudomonas Genome Database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res 37 (Database issue), D483–D488. 10.1093/nar/gkn861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang I. V., Chen E., Hasseman J. P., Liang W., Frank B. C., Wang S., Sharov V., Saeed A. I., White J. & other authors (2002). Within the fold: assessing differential expression measures and reproducibility in microarray assays. Genome Biol 3, h0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R. W., Haas D., Ka J. O., Krishnapillai V., Zimmermann A., Baird C., Tiedje J. M. (1995). Anaerobic activation of the entire denitrification pathway in Pseudomonas aeruginosa requires Anr, an analog of Fnr. J Bacteriol 177, 3606–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P., Sun J., Geffers R., Zeng A. P. (2007). Functional characterization of the gene PA2384 in large-scale gene regulation in response to iron starvation in Pseudomonas aeruginosa. J Biotechnol 132, 342–352. 10.1016/j.jbiotec.2007.08.013 [DOI] [PubMed] [Google Scholar]