Abstract
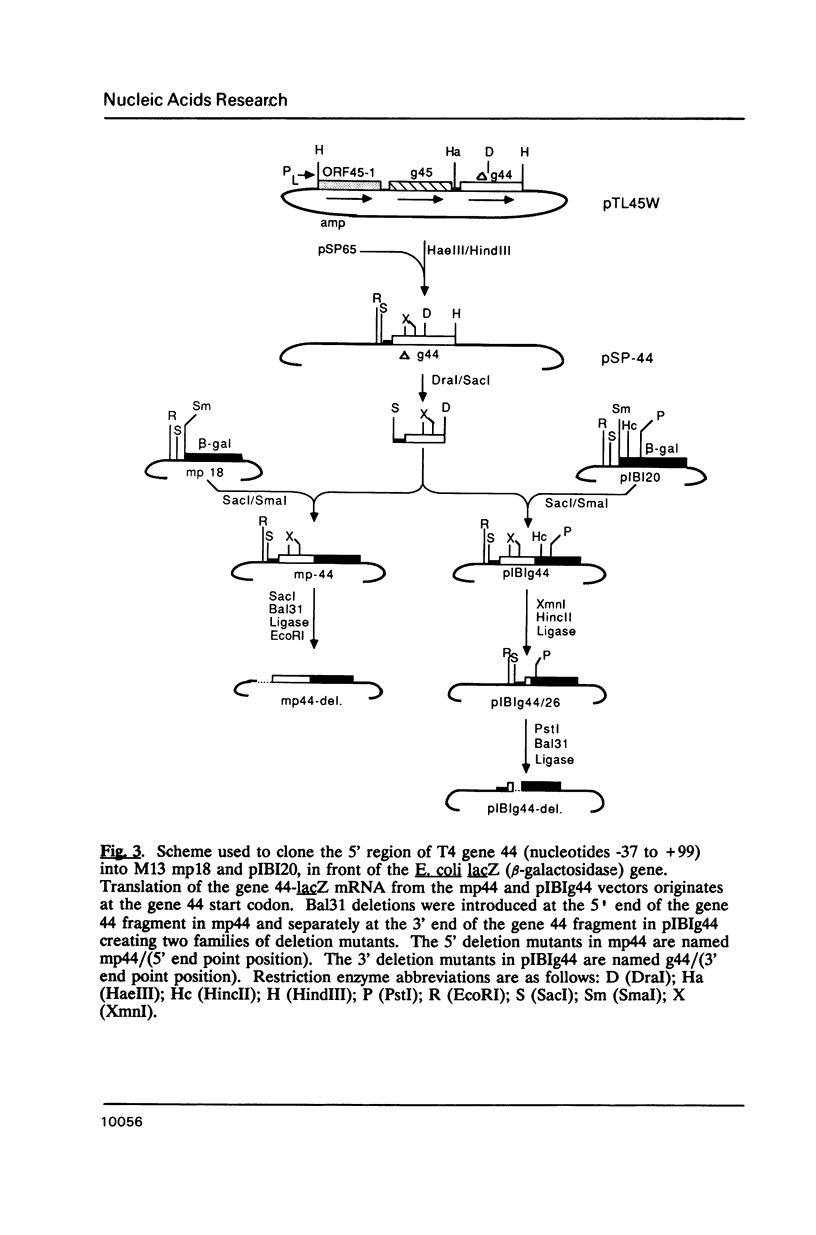
We have overproduced and purified wild type regA protein, a translational repressor encoded by bacteriophage T4. The repressor activity of the cloned regA protein has been tested on four known regA target genes (T4 genes: 44, 45, rpbA and regA) using in vitro coupled transcription-translation reactions. We have demonstrated the sensitivity of two additional T4 genes coding for alpha- and beta-glucosyltransferases to regA protein in vitro. The regA target site on the gene 44 messenger RNA has been identified through deletion analysis and RNase protection assays, using plasmids containing gene 44-lacZ fusions. The effect of regA protein on expression of 44P-beta-galactosidase fusion proteins was assayed in vitro, in coupled transcription-translation reactions. Analysis of deletion mutants of gene 44-lacZ localized the regA recognition region to between nucleotides -11 and +9 of the mRNA. RNase protection assays of g44-lacZ transcripts further defined the site of regA protein interaction to between nucleotides -10 and +2 of the mRNA. This region overlaps the gene 44 Shine-Dalgarno region and the A and U of the initiation codon.
Full text
PDF



















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adari H. Y., Rose K., Williams K. R., Konigsberg W. H., Lin T. C., Spicer E. K. Cloning, nucleotide sequence, and overexpression of the bacteriophage T4 regA gene. Proc Natl Acad Sci U S A. 1985 Apr;82(7):1901–1905. doi: 10.1073/pnas.82.7.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adari H. Y., Spicer E. K. Translational repression in vitro by the bacteriophage T4 regA protein. Proteins. 1986 Oct;1(2):116–124. doi: 10.1002/prot.340010203. [DOI] [PubMed] [Google Scholar]
- Broida J., Abelson J. Sequence organization and control of transcription in the bacteriophage T4 tRNA region. J Mol Biol. 1985 Oct 5;185(3):545–563. doi: 10.1016/0022-2836(85)90071-3. [DOI] [PubMed] [Google Scholar]
- Carey J., Cameron V., de Haseth P. L., Uhlenbeck O. C. Sequence-specific interaction of R17 coat protein with its ribonucleic acid binding site. Biochemistry. 1983 May 24;22(11):2601–2610. doi: 10.1021/bi00280a002. [DOI] [PubMed] [Google Scholar]
- Deckman I. C., Draper D. E. S4-alpha mRNA translation regulation complex. II. Secondary structures of the RNA regulatory site in the presence and absence of S4. J Mol Biol. 1987 Jul 20;196(2):323–332. doi: 10.1016/0022-2836(87)90693-0. [DOI] [PubMed] [Google Scholar]
- Derbyshire K. M., Salvo J. J., Grindley N. D. A simple and efficient procedure for saturation mutagenesis using mixed oligodeoxynucleotides. Gene. 1986;46(2-3):145–152. doi: 10.1016/0378-1119(86)90398-7. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H. Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980 Jul 25;8(14):3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulford W., Model P. Specificity of translational regulation by two DNA-binding proteins. J Mol Biol. 1984 Feb 25;173(2):211–226. doi: 10.1016/0022-2836(84)90190-6. [DOI] [PubMed] [Google Scholar]
- Gillam S., Smith M. Site-specific mutagenesis using synthetic oligodeoxyribonucleotide primers: I. Optimum conditions and minimum ologodeoxyribonucleotide length. Gene. 1979 Dec;8(1):81–97. doi: 10.1016/0378-1119(79)90009-x. [DOI] [PubMed] [Google Scholar]
- Gram H., Rüger W. Genes 55, alpha gt, 47 and 46 of bacteriophage T4: the genomic organization as deduced by sequence analysis. EMBO J. 1985 Jan;4(1):257–264. doi: 10.1002/j.1460-2075.1985.tb02344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray H. B., Jr, Ostrander D. A., Hodnett J. L., Legerski R. J., Robberson D. L. Extracellular nucleases of Pseudomonas BAL 31. I. Characterization of single strand-specific deoxyriboendonuclease and double-strand deoxyriboexonuclease activities. Nucleic Acids Res. 1975 Sep;2(9):1459–1492. doi: 10.1093/nar/2.9.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T., Wei R. X., Dawson M., Karam J. D. Identification of two new bacteriophage T4 genes that may have roles in transcription and DNA replication. J Virol. 1987 Feb;61(2):366–374. doi: 10.1128/jvi.61.2.366-374.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam J., Gold L., Singer B. S., Dawson M. Translational regulation: identification of the site on bacteriophage T4 rIIB mRNA recognized by the regA gene function. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4669–4673. doi: 10.1073/pnas.78.8.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y. M., Wei R. X., Hsu T., Alford C., Dawson M., Karam J. Autogenous regulation of the regA gene of bacteriophage T4: derepression of translation. Genetics. 1988 Aug;119(4):743–749. doi: 10.1093/genetics/119.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., Zinder N. D. Translational repression in bacteriophage f1: characterization of the gene V protein target on the gene II mRNA. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4002–4006. doi: 10.1073/pnas.86.11.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E. S., Karam J., Dawson M., Trojanowska M., Gauss P., Gold L. Translational repression: biological activity of plasmid-encoded bacteriophage T4 RegA protein. J Mol Biol. 1987 Apr 5;194(3):397–410. doi: 10.1016/0022-2836(87)90670-x. [DOI] [PubMed] [Google Scholar]
- Miller E. S., Winter R. B., Campbell K. M., Power S. D., Gold L. Bacteriophage T4 regA protein. Purification of a translational repressor. J Biol Chem. 1985 Oct 25;260(24):13053–13059. [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott J. E., Grant R. A., Ho Y. S., Platt T. Maximizing gene expression from plasmid vectors containing the lambda PL promoter: strategies for overproducing transcription termination factor rho. Proc Natl Acad Sci U S A. 1985 Jan;82(1):88–92. doi: 10.1073/pnas.82.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribnow D., Sigurdson D. C., Gold L., Singer B. S., Napoli C., Brosius J., Dull T. J., Noller H. F. rII cistrons of bacteriophage T4. DNA sequence around the intercistronic divide and positions of genetic landmarks. J Mol Biol. 1981 Jul 5;149(3):337–376. doi: 10.1016/0022-2836(81)90477-0. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Ho Y. S., Shatzman A. The use of pKc30 and its derivatives for controlled expression of genes. Methods Enzymol. 1983;101:123–138. doi: 10.1016/0076-6879(83)01009-5. [DOI] [PubMed] [Google Scholar]
- Russel M., Gold L., Morrissett H., O'Farrell P. Z. Translational, autogenous regulation of gene 32 expression during bacteriophage T4 infection. J Biol Chem. 1976 Nov 25;251(22):7263–7270. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamoo Y., Adari H., Konigsberg W. H., Williams K. R., Chase J. W. Cloning of T4 gene 32 and expression of the wild-type protein under lambda promoter PL regulation in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8844–8848. doi: 10.1073/pnas.83.23.8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer E. K., Noble J. A., Nossal N. G., Konigsberg W. H., Williams K. R. Bacteriophage T4 gene 45. Sequences of the structural gene and its protein product. J Biol Chem. 1982 Aug 10;257(15):8972–8979. [PubMed] [Google Scholar]
- Spicer E. K., Nossal N. G., Williams K. R. Bacteriophage T4 gene 44 DNA polymerase accessory protein. Sequences of gene 44 and its protein product. J Biol Chem. 1984 Dec 25;259(24):15425–15432. [PubMed] [Google Scholar]
- Tang C. K., Draper D. E. Unusual mRNA pseudoknot structure is recognized by a protein translational repressor. Cell. 1989 May 19;57(4):531–536. doi: 10.1016/0092-8674(89)90123-2. [DOI] [PubMed] [Google Scholar]
- Tomaschewski J., Gram H., Crabb J. W., Rüger W. T4-induced alpha- and beta-glucosyltransferase: cloning of the genes and a comparison of their products based on sequencing data. Nucleic Acids Res. 1985 Nov 11;13(21):7551–7568. doi: 10.1093/nar/13.21.7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojanowska M., Miller E. S., Karam J., Stormo G., Gold L. The bacteriophage T4 regA gene: primary sequence of a translational repressor. Nucleic Acids Res. 1984 Aug 10;12(15):5979–5993. doi: 10.1093/nar/12.15.5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter R. B., Morrissey L., Gauss P., Gold L., Hsu T., Karam J. Bacteriophage T4 regA protein binds to mRNAs and prevents translation initiation. Proc Natl Acad Sci U S A. 1987 Nov;84(22):7822–7826. doi: 10.1073/pnas.84.22.7822. [DOI] [PMC free article] [PubMed] [Google Scholar]