Abstract
Human cytomegalovirus encodes an alkaline nuclease, UL98, that is highly conserved among herpesviruses and has both endonuclease (endo) and exonuclease (exo) activities. This protein is thought to be important for viral replication and therefore represents a potential target for antiviral development; however, little is known about its structure or role in viral replication. Comparative structural modelling was used to build a model of UL98 based on the known structure of shutoff and exonuclease protein from Kaposi’s sarcoma-associated herpesvirus. The model predicts that UL98 residues D254, E278 and K280 represent the critical aspartic acid, glutamic acid and lysine active-site residues, respectively, while R164 and S252 correspond to residues proposed to bind the 5′ phosphate of the DNA substrate. UL98 with an amino-terminal hexahistidine tag was expressed in Escherichia coli, purified by affinity chromatography and confirmed to have exo and endo activities. Amino acid substitutions D254A, E278A, K280A and S252A virtually eliminated exo and endo activities, whereas R164A retained full endo activity but only 10 % of the exo activity compared with the wild-type enzyme. A mutant virus lacking UL98 was viable but severely attenuated for replication, while one expressing UL98(R164A) replicated normally. These results confirm the utility of the model in representing the active-site region of UL98 and suggest a mechanism for the differentiation of endonuclease and exonuclease activities. These findings could facilitate the exploration of the roles of alkaline nucleases in herpesvirus replication and the rational design of inhibitors that target their enzymic activities.
Introduction
Human cytomegalovirus (CMV) is one of eight human pathogens in the family Herpesviridae. It causes disease in immune-compromised patients, including retinitis in human immunodeficiency virus patients, pneumonitis in transplant patients and, when acquired during pregnancy, severe birth defects characterized by deafness and mental retardation (Vancíková & Dvorák, 2001). Current options for treating CMV infections are limited and suboptimal. The approved drugs ganciclovir, its prodrug valganciclovir, foscarnet and cidofovir target the viral DNA polymerase to block viral DNA synthesis. Each is associated with significant toxicity, and studies in laboratory animals suggest that these drugs may be teratogenic (Fouda et al., 1991; Henderson et al., 1993; Klug et al., 1991). Hence, none are approved for treatment of fetal CMV infections during pregnancy. Because prolonged treatment can give rise to viral strains that are resistant to one or more of these drugs (Schleiss, 2005), there is a need for the development of new CMV antivirals, particularly those that target alternative processes and are safe for use during pregnancy.
The CMV alkaline nuclease (AN), UL98, represents one such novel target. ANs are conserved throughout the family Herpesviridae. In vitro, ANs exhibit both endonuclease (endo) and 5′→3′ exonuclease (exo) activities that are optimal at alkaline pH, sensitive to high salt concentrations and require a divalent cation, with a preference for Mg2+ (Hoffmann & Cheng, 1978). Their exo activities can hydrolyse either ssDNA or dsDNA to form mono-, di- and trinucleotides (Sheaffer et al., 1997). Although DNA structural and sequence preferences have been noted (Henderson et al., 1998; Kehm et al., 1998), neither activity appears to be sequence specific.
ANs play an important, but poorly understood, role in herpesvirus replication. Herpes simplex virus type 1 (HSV-1) AN null mutants are viable, although they exhibit a growth deficit of 2–3 logs (Gao et al., 1998; Martinez et al., 1996; Patel et al., 1996; Weller et al., 1990), whereas transposon insertions that disrupt the CMV UL98 ORF are lethal (Yu et al., 2003), thus suggesting that UL98 may be essential for CMV replication.
The fact that UL98 is important for CMV replication and possesses enzymic activities that can be assayed easily makes this protein an attractive target for antiviral development. Here, we used homology-based modelling and mutagenesis to identify the active site residues of UL98 and construction of mutant viruses to determine the importance of UL98 in CMV replication. The results provide a foundation for additional studies to elucidate the role of AN in herpesvirus replication and to explore drug discovery efforts targeting UL98.
Results
Prediction of UL98 structure, DNA interactions and active site residues
Herpesvirus ANs are classified within the λ exo family of DNases, which is within the PD-(D/E)XK superfamily of DNA-modifying enzymes (Kovall & Matthews, 1998). The catalytic sites of these enzymes contain a conserved aspartic acid (D), an aspartic or glutamic acid (D/E) and a lysine (K) (Kovall & Matthews, 1998). psi-blast identified the AN of Kaposi’s sarcoma-associated herpesvirus (KSHV), known as shutoff and exonuclease protein (KSHV-SOX), as being the closest match to UL98. The catalytic residues of KSHV-SOX are D221 and E244, which coordinate Mg2+, and K246, which stabilizes the leaving group (Dahlroth et al., 2009). An adjacent 5′-phosphate-binding pocket formed by R139, S146 and S219 was suggested by the presence of a sulphate ion in the crystal structure (Dahlroth et al., 2009). clustal x version 2 was used to identify CMV AN amino acids D254, E278 and K280 of UL98 as being the corresponding catalytic residues and R164, T171 and S252 as forming the putative 5′-phosphate-binding pocket (Supplementary Fig. S1a, available in JGV Online). When ANs were compared across the family Herpesviridae, the catalytic residues were 100 % conserved, as were R164 and S252 of the phosphate-binding pocket, while the third putative phosphate-binding residue, T171, was conserved among betaherpesviruses but replaced by serine in the alpha- and gammaherpesviruses (Supplementary Fig. S1b, available in JGV Online).
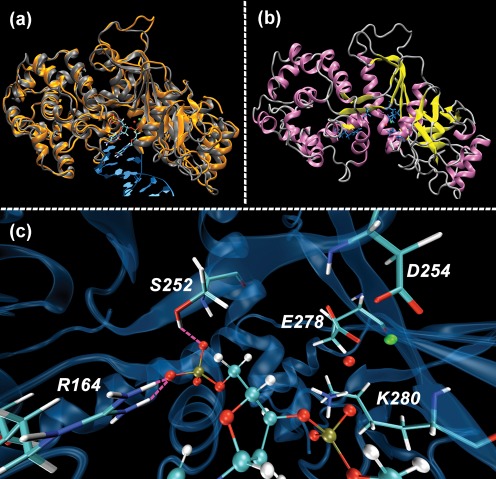
A structural model of UL98 was constructed based on the crystal structure of KSHV-SOX (Dahlroth et al., 2009) and in silico docking was used to determine the optimal binding of a dsDNA dodecamer in the predicted UL98 binding site crevice (Supplementary Materials, available in JGV Online). KSHV-SOX comprises an amino-terminal domain consisting of ten α-helices and a carboxy-terminal domain formed by five-stranded β-sheets flanked by five α-helices (Dahlroth et al., 2009). The UL98 structure has a very similar pattern and active site crevice (Fig. 1). The model again identifies catalytic site residues D254, E278 and K280, and 5′-phosphate binding residues R164 and S252, in locations precisely analogous to their counterparts in KSHV-SOX. The predicted position of DNA shows the 5′ phosphate held deep into the active-site crevice through hydrogen bonding with R164 and S252 (Fig. 1c). The scissile phosphodiester bond is appropriately positioned proximally to the metal coordination region formed by D254, E278 and K280 (Fig. 1c).
Fig. 1.
Homology-based structural model of UL98. In each panel a Mg2+ ion (green sphere) and a water molecule (red sphere) found in KSHV-SOX (PDB 3FHD) are included. (a) KSHV-SOX (3FHD) (grey) is shown overlaid with the UL98 model (orange). The 5′ end of a dsDNA (B-DNA docedamer 3BNA) is docked with UL98 with the 5′ phosphate occupying the position of the sulphate found in 3FHD. Terminal 5′ bases are shown as capped sticks while the remainder of the molecule is shown as a blue ribbon. (b) Fold structure of UL98 showing α-helices (pink), β-sheets (yellow), loops and coils (silver), and active-site residues R164, S252, D254, E278 and K280 (blue). (c) UL98 active site showing active-site residues as capped sticks and dsDNA as balls-and-sticks. Mg2+ is coordinated by the carboxylate groups of D254 and E278 and holds the water molecule at the active site. K280 is proximal to the phosphodiester group and the 5′ phosphate. Dashed magenta lines indicate strong hydrogen-bond interactions with the side chains of R164 and S252. Illustrations were prepared by using vmd (Humphrey et al., 1996; Stone, 1998).
Purification of recombinant wild-type and mutant UL98 proteins
To examine the role of these amino acids in the enzymic activity of UL98, recombinant UL98 or proteins containing single alanine substitutions (R164A, S252A, D254A, E278A or K280A) were expressed with amino-terminal hexahistidine (His6) tags. His6-tagged β-glucuronidase (GUS) was expressed as a negative control. The seven proteins were expressed and partially purified by immobilized metal affinity chromatography (IMAC) as described in Methods. With the exception of the E278A mutant, each protein was eluted in fractions 30–31 (150–200 mM imidazol), which is typical for His6-tagged proteins. The E278A mutant protein was eluted predominantly in fraction 16, suggesting that the structure of this mutant protein and/or the accessibility of the His6 tag were altered compared with the other proteins. Based on SDS-PAGE and immunoblot analysis, the His6-tagged UL98 and GUS proteins migrated at rates consistent with the calculated masses for these proteins of 68.4 and 75.9 kDa, respectively (Supplementary Fig. S2, available in JGV Online). Although the preparations contained some contaminants, these were similar across all samples. Moreover, the amounts of mutant proteins eluted were similar to the wild-type sample, with the exception of S252A.
Residues D254, E278, K280, R164 and S252 are important for exo activity
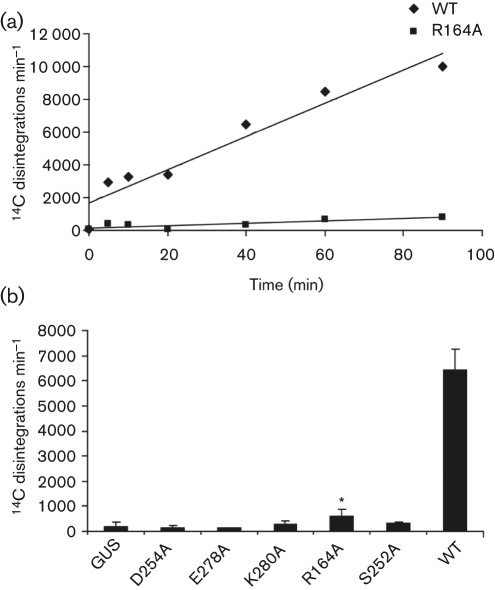
The exo activities of the IMAC-purified proteins were quantified by release of acid-soluble radioactivity from 14C-labelled DNA. Wild-type UL98 exhibited both time- and concentration-dependent exo activity (Fig. 2a and data not shown). Mutant R164A digested DNA but at a significantly reduced rate (Fig. 2a). In fact, all of the mutant proteins failed to digest substantial amounts of substrate DNA in a 1 h period (Fig. 2b). Exo activity of the R164A mutant was 10.6 % of wild-type and statistically higher than those of the other mutants and GUS (unpaired t-test, P≤0.05), whereas activities for D254A, E278A, K280A and S252A were <5 % that of wild-type and not statistically different from each other or from GUS (P>0.1).
Fig. 2.
Exo activity. (a) Exo activities were determined as acid-soluble radioactivity released during incubation of 14C-labelled DNA with 2.5 µg of IMAC-purified UL98 (WT) or R164A. (b) Exo activities of mutants were measured as above after 1 h incubation. Data are means of disintegrations per minute obtained from three independent experiments. Error bars represent means±1 sd. *, R164A differs from GUS and other mutants; unpaired t-test, P≤0.05.
Residues D254, E278, K280 and S252, but not R164, are important for endo activity
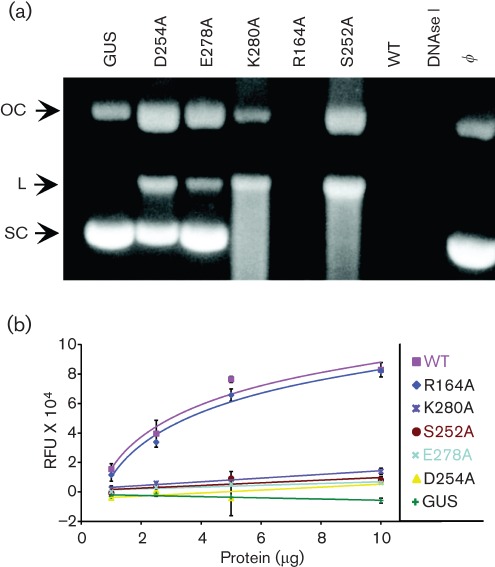
Endo activity was determined qualitatively by incubating IMAC-purified proteins with closed-circular pUC19 plasmid DNA and evaluating its conversion to open-circular and linear forms by agarose gel electrophoresis (Goldstein & Weller, 1998). Following 12 h incubation, the wild-type and R164A proteins were not only capable of nicking the supercoiled substrate, they degraded all of the DNA substrate, presumably to products too small to visualize on the gel (Fig. 3a). In contrast, the K280A and S252A mutant proteins nicked essentially all of the supercoiled DNA, although large amounts of nicked open-circular and full-length linear forms remained undigested (Fig. 3a). The smearing below the full-length linear fragment may represent the cumulative effects, over the 12 h incubation period, of endonucleolytic cleavage or low-level exo activity in these preparations. Although D254A and E278A preparations were able to convert some of the supercoiled DNA into open-circular and full-length linear DNA, the majority of the supercoiled DNA substrate remained intact after the 12 h incubation. It is possible that the low-level endo activity exhibited by the latter mutants derives from contaminating nucleases in the IMAC-purified preparations, although the similarly purified control protein, GUS, had no discernible nuclease activity (Fig. 3a).
Fig. 3.
Endo activity. (a) Supercoiled plasmid DNA (250 ng) was incubated for 12 h with buffer only (ϕ), 5 µg of IMAC-purified proteins or 1 U of DNase I. Products were analysed by agarose gel electrophoresis. Arrows indicate the positions of supercoiled (SC), open circular (OC) and linear (L) DNA. (b) A 35 nt synthetic ssDNA substrate with a fluorescent emitter at the 5′ end and a quencher at the 3′ end was incubated for 14 h with increasing amounts of each protein; fluorescence was measured as relative fluorescence units (RFU).
To quantify endo activity independent of exo activity, a fluorescence-based assay was developed in which fluorescence increases when endonucleolytic cleavage of an ssDNA substrate releases a 3′ quencher from a 5′ fluorophore. One millimole of substrate was incubated with increasing amounts of each protein for 14 h at 37 °C (Fig. 3b). No significant difference was observed between the endo activities of R164A and wild-type UL98 (P>0.05) and both wild-type and R164A endo activities were significantly different from those exhibited by GUS and the other mutant proteins (Tukey's multiple comparison test, P<0.001). The activities of S252A, D254A or E278A were not different from each other or from GUS (Tukey's multiple comparison test, P≥0.05), although the extremely low activity of K280A was statistically higher (P<0.01) than GUS. Thus, the residual endonuclease activities associated with S252A, D254A or E278A probably represent the activities of contaminating proteins.
UL98 is important but not essential for viral replication
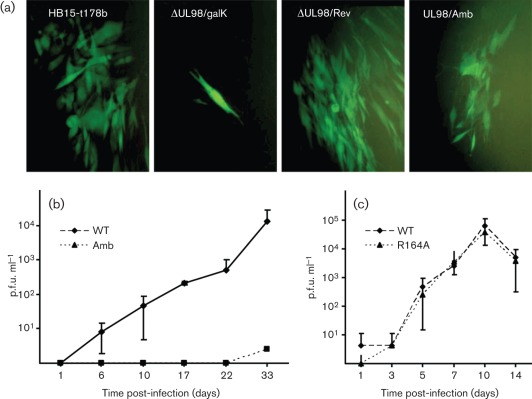
UL98 has been classified as an essential CMV gene, based on lethality of mutations that disrupt UL98 by transposon insertion (Yu et al., 2003) or gene deletion (Dunn et al., 2003). However, the mutations in both these studies had the potential to impact upon the expression of neighbouring essential genes. To definitively determine the importance of UL98 for CMV replication, we used Escherichia coli genetics to modify UL98 sequences within bacterial artificial chromosome (BAC) HB15-t178b, an infectious clone of the CMV strain AD169 genome that also contains an expression cassette for GFP (Saccoccio et al., 2011). In BAC clone ΔUL98/galK, the entire UL98 ORF was deleted and replaced by the E. coli galK gene. Consistent with the results of Dunn et al. (2003), this mutation was lethal, as transfection of MRC-5 fibroblasts with ΔUL98/galK BAC DNA resulted in isolated green cells that never expanded to form viral plaques (Fig. 4a). In contrast, both HB15-t178b and ΔUL98/Rev, a derivative of ΔUL98/galK in which galK was replaced by wild-type UL98 sequence from CMV strain Towne, gave rise to GFP+ foci that expanded over time (Fig. 4a).
Fig. 4.
Importance of UL98 for CMV replication. (a) MRC-5 cultures were transfected with the indicated BAC DNAs and photographed by UV microscopy 14 days post-transfection. (b and c) MRC-5 cultures were infected with HB15-t178b (WT) or UL98/Amb (Amb) at an m.o.i. of 0.0001 (b); or with HB15-t178b (WT) or UL98/R164A (R164A) at an m.o.i. of 0.01 (c). Titres of each virus in the culture supernatants were determined on the days indicated.
A second mutant, UL98/Amb, was constructed in which galK in ΔUL98/galK was replaced by UL98 sequences containing an amber stop codon/frameshift mutation at codon 5. Transfection of UL98/Amb DNA resulted in GFP+ foci that expanded very slowly (Fig. 4a). Production of UL98/Amb stocks required 4–6 weeks and achieved peak titres of 102 p.f.u. ml−1. A growth curve starting at a very low m.o.i. (0.0001) confirmed that UL98/Amb is extremely growth impaired – while the parental virus reached 104 p.f.u. ml−1 by day 33 post-infection, UL98/Amb was not detected in the culture supernatants until day 33, when the viral titre was <10 p.f.u. ml−1 (Fig. 4b). Although we could not detect any UL98 product in UL98/Amb-infected cells by Western blot (not shown), we cannot exclude the possibility of extremely low-level amber codon read-through or infrequent translational initiation beyond the mutation. PCR/restriction analysis detected no evidence that the mutation, which disrupts an XhoI site, had spontaneously reverted to wild-type (restoring the XhoI site) in UL98/Amb viral DNA (not shown). These results indicate that UL98 is not essential for viral replication in tissue culture; however, in the absence of UL98 viral replication is extremely attenuated.
To determine the impact of the R164A mutation on viral replication, galK in ΔUL98/galK was replaced by UL98 sequences containing the R164A mutation to yield BAC UL98/R164A. Virus UL98/R164A was readily reconstituted from this BAC and replicated in MRC-5 fibroblasts with kinetics and efficiency indistinguishable from the parental virus (Fig. 4c). PCR/restriction analysis detected no evidence that the mutation, which disrupts a BstAPI site, had spontaneously reverted to wild-type (restoring the BstAPI site) in UL98/R164A viral DNA (not shown).
Discussion
Nucleases classified within the λ exo family are encoded by a diverse range of large dsDNA viruses, including bacteriophages, insect baculoviruses and herpesviruses. While their functions remain poorly understood, the high level of conservation of these enzymes across broad groups suggests that they are important for a common replication process. The best-studied member of this family, λ exo, is thought to play a role in recombination by creating ssDNA ends that are subsequently utilized by the λ ssDNA-binding protein for strand invasion. Although in vitro the HSV-1 AN, UL12, can participate in similar strand-transfer reactions mediated by the HSV-1 ssDNA-binding protein in association with host recombination repair Mre11–Rad50–Nbs1 complex components (Balasubramanian et al., 2010; Reuven et al., 2003, 2004), the replication defect of herpesvirus AN null mutants has not as yet been linked to a defect in DNA recombination (Porter & Stow, 2004a). To date, the phenotypes of null mutants suggest a role for AN in resolving branched or abnormally structured DNA. This appears to have downstream effects on DNA packaging, capsid stability, nuclear egress and the infectivity of virions that reach the extracellular compartment (Martinez et al., 2002; Porter & Stow, 2004a, b; Shao et al., 1993; Weller et al., 1990). Although the mechanistic details remain unclear, genetic studies clearly show that herpesvirus ANs are important for viral replication. Thus, AN inhibitors are predicted to have a significant impact on viral replication.
The λ exo family belongs to a superfamily of DNA-modifying enzymes that include restriction enzymes, Holliday-junction resolvases, DNA-nicking enzymes, Vsr and the mismatch repair enzyme MutH, which all share a conserved active-site structure characterized by a PD-(D/E)XK motif (Bujnicki & Rychlewski, 2001; Venclovas et al., 1994). In λ exo four additional residues, an arginine, two serines and a glutamic acid, form a pocket that is believed to stabilize the 5′ end of the DNA substrate and position the scissile phosphodiester bond proximal to the catalytic residues (Kovall & Matthews, 1998). In the KSHV-SOX crystal structure, an analogous pocket is formed by R139, S146 and S219 and contains a sulphate ion that presumably occupies the pocket in the absence of DNA (Dahlroth et al., 2009). The UL98 model predicts that this pocket is formed by R164, T171 and S252. In a panel of seven ANs from viruses representing the three herpesvirus subfamilies, the predicted catalytic residues of UL98 (D254, E278 and K280) were fully conserved, as were two of the predicted phosphate-binding pocket residues (R164 and S252); the third (T171) was conserved as a threonine in the β subfamily and an serine in the α and γ subfamilies.
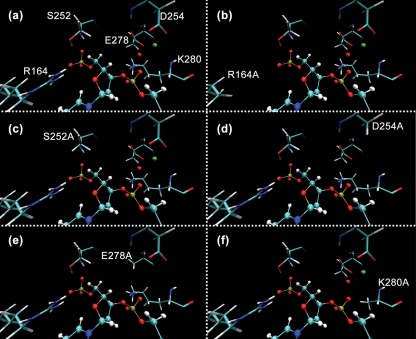
Fig. 5 shows predicted active-site structures for wild-type UL98 and each of the five mutants. The DNA is positioned in the active-site crevice through hydrogen-bond interactions with R164 and S252. Carboxylate bearing side chains of D254 and E278 coordinate the catalytically important magnesium ion, and a conserved water molecule acts as the base for hydrolysis. K280 is proximal to the scissile phosphodiester bond and serves to stabilize the leaving group. Loss of hydrogen-bonding interactions resulting from substitutions R164A or S252A may result in imprecise alignment of the DNA in the active site, resulting in improper positioning of the scissile phosphodiester bond relative to the catalytic K280 residue (Fig. 5b and 5c). This was further supported by significant changes in the predicted DNA-binding energies for R164A, S252A and K280 mutants (Supplementary Table S1, available in JGV Online). Similarly, loss of charged carboxylate residues in D254A or E278A mutants would result in failure to coordinate the magnesium ion and water molecule (Fig. 5d and 5e). Changes in predicted DNA-binding energies for mutants D254A and E278A are minimal (Supplementary Table S1), consistent with these residues interacting with Mg2+ rather than by directly interacting with the DNA. The K280A mutation would be unable to carry out the hydrolysis of the phosphodiester bond and stabilize the leaving group (Fig. 5f). This mechanistic model therefore predicts that D, E or K→A substitution mutants should lack both exo and endo activity and that R or S → A mutants may have impaired exo activity. As no 5′ phosphate is present on the substrate in endonucleolytic cleavage, endo activity might be unaffected in R or S → A mutants.
Fig. 5.
Active-site models for DNA interactions of wild-type and mutant UL98 proteins were generated from the UL98 model (see Supplementary Materials). Protein side chains are shown as capped sticks, DNA as balls-and-sticks, Mg2+ as green spheres and water as red spheres. In wild-type UL98 (a), the 5′ phosphate of DNA is positioned by R164 and S252 such that K280 is proximal to the scissile phosphodiester bond, while E278 and D254 coordinate Mg2+ and hold a water molecule in the active site. In R164A (b) and S252A (c), DNA binding or positioning may be altered by loss of one hydrogen-bond with the 5′ phosphate. In D254A (d) and E278A (e), loss of carboxylic acid-containing residues may disrupt coordination of Mg2+ at the active site. In K280A (f), loss of the lysine amino group disrupts its ability to donate a proton to stabilize the leaving group during hydrolysis.
The experimental data are generally consistent with these predictions. The D, E or K→A mutants lacked measureable exo activity. Endo activity was greatly reduced, but low-level activity was still apparent with the supercoiled dsDNA substrate. Similar findings have been reported for substitution mutants of ANs from KSHV (SOX), Epstein–Barr virus (BGLF5) and HSV-1 (UL12). In SOX mutants D221S and E244S, substitutions of the catalytic aspartic acid and glutamic acid residues inactivated exo activity (Bagnéris et al., 2011). In BGLF5 substitutions D203(E/S) or E225Q inactivated exo and endo activities, while E225D changed the divalent cation preference from Mg2+ to Mn2+ (Buisson et al., 2009; Liu et al., 2003). In UL12, substitution D340E inactivated exo activity but not endo activity (Goldstein & Weller, 1998). This was surprising given that both activities were inactivated in the BGLF5 D203E mutant (Liu et al., 2003). Of the two UL98 mutations targeting putative phosphate-binding residues, R164A behaved as predicted. Exo activity was reduced by 90 %, but there was no impact on endo activities detected with supercoiled dsDNA or end-blocked ssDNA substrates. In contrast, the S252A mutation eliminated exo activity, and reduced both ssDNA and supercoiled dsDNA endo activities to levels similar to those of the K280A mutant. While single amino acid changes in the putative phosphate-binding residues of other ANs have not been evaluated, a double substitution mutant of HSV-1 UL12 (G336A/S338A, where S338UL12 is equivalent to S252AUL98) was significantly impaired for both exo and endo activity (Goldstein & Weller, 1998). In both UL98 and UL12, the proximity of the serine-to-alanine mutation, one residue from the catalytic aspartic acid, may have perturbed the local geometry of the active site enough to impair catalytic function, and hence to have impacted not only exo but also endo activity.
The crystal structure of KSHV-SOX solved by Dahlroth et al. (2009) suggests that its active-site canyon probably accommodates four nucleotides. They successfully modelled a duplex DNA 11-mer into the pocket with the 5′ end in the active site. Consistent with these findings, our model suggests that the active-site canyon of UL98 is able to accommodate duplex DNA, and that the active-site residues show significant interactions with the scissile phosphodiester group. Bagnéris et al. (2011) have suggested that exonucleolytically processed substrates are bound as duplexes, and that the only rearrangements required for cleavage are those necessary for recognition of the 5′-phosphate group in a serine-rich pocket. Our model shows a similar potential 5′-phosphate-binding pocket, and docking of dsDNA exhibited a close approximation of the 5′ phosphate within the pocket. However, upon minimization, the dsDNA was minimally altered to conform to the hydrophobic cleft, suggesting that some conformational change of the DNA owing to steric influences may be necessary to access the 5′-phosphate-binding pocket. That active-site–dsDNA interactions are predicted mainly with phosphate groups of the 5′–3′ strand further suggests that ssDNA could bind in a manner similar to dsDNA. Consistent with this, Sheaffer et al. (1997) observed that UL98 hydrolyses ssDNA and dsDNA with similar turnover rates, affinities and rate constants.
There have been suggestions that the ‘bridge’ architecture plays an important role in exonucleolytic cleavage by inducing strand separation (Bagnéris et al., 2011; Buisson et al., 2009). The bridge region is also proposed to promote endonucleolytic processing by developing local regions of ssDNA within the substrate, which are the likely targets for endo cleavage. However, the high degree of disorder in the structure of the bridge region in both KSHV-SOX and the KSHV-SOX–DNA complex structures made it difficult to model the corresponding region in UL98. Thus, our modelling focused on the active-site region and cannot be extrapolated to the role of ‘the bridge’ in the functions of UL98.
Two previous studies classified UL98 as being essential for CMV replication (Dunn et al., 2003; Yu et al., 2003). This was surprising given that the HSV-1 UL12 mutants are viable, although significantly attenuated for growth in cell culture (Gao et al., 1998; Martinez et al., 1996; Patel et al., 1996; Weller et al., 1990). However, UL98 lies within a UL93–UL99 gene cluster that exhibits complex 5′-nested transcripts that are 3′ coterminal owing to the shared use of a polyadenylation signal downstream of UL99 (Wing & Huang, 1995). Consequently, the transposon insertions in UL98 described by Yu et al. (2003) could have had polar effects on transcripts for other genes in this cluster, most notably UL99, an essential gene that encodes the pp28 tegument protein (Silva et al., 2003). Similarly, because the deletion described by Dunn et al. (2003) removed the entire UL98 ORF, and UL98 overlaps the 5′ end of UL99 (Adam et al., 1995), this mutation must have eliminated expression of both UL98 and pp28. We therefore constructed mutant UL98/Amb to selectively inactivate UL98 expression with a minimal risk of perturbing expression of nearby genes. That UL98/Amb was able to replicate, albeit minimally, indicates that UL98, while extremely important for replication, is not essential in cell culture. However, the slow rate of replication of this mutant suggests that it might easily be cleared in vivo. Thus, UL98 remains a viable antiviral target as drugs inactivating its functions should dramatically reduce the ability of CMV to replicate.
Surprisingly, virus UL98/R164A exhibited no growth impairment in cultured fibroblasts. This suggests that CMV can tolerate a 90 % reduction in exonuclease activity without losing replication efficiency. Alternatively, given the wild-type kinetics of UL98/R164 replication, it may be the endo activity of UL98 that is important for replication. Even so, wild-type levels of exo activity may be important in vivo, as CMV infects a variety of other cell types and the importance of exo activity may be cell-type dependent.
In the absence of a crystallographic structure for UL98, these results indicate that in silico modelling can provide structural models that are sufficient to predict which residues are important for nucleolytic functions. Such models should prove valuable for continued structure/function dissection of ANs and for informing structure-based drug discovery. The ability to mutagenically target the 5′-phosphate-binding pocket to inhibit exo but not endo activity may provide a useful tool for exploring the biological roles of each activity in herpesvirus replication.
Methods
Plasmid construction.
Oligonucleotide sequences are listed in Supplementary Table S2 (available in JGV Online). The UL98 ORF was PCR-amplified from CMV strain AD169 DNA by using primers GW8-F and GW8-R and a High Fidelity EasyA PCR mixture (Bioline), cloned into vector pCR-8/GW/TOPO (Invitrogen) and moved, by using a clonase reaction, to Gateway destination vector pDest17 (Invitrogen), thus generating E. coli expression vector pD17-98(wt). Mutations were constructed by overlap extension PCR. Template DNA from pD17-98(wt) was amplified by using primer GW8-F paired with primers DR, ER, KR, RR or SR, or primer GW8-R paired with primers DF, EF, KF, RF or SF to generate overlapping products containing the desired nucleotide changes. For each mutant the two PCR products were purified, mixed and amplified again with primers GW8-F and GW8-R. The products were cloned into pCR-8/GW/TOPO and transferred as above to generate plasmids pD17-98(D), pD17-98(E), pD17-98(K), pD17-98(R) and pD17-98(S), which contain the mutations specified in Supplementary Table S2. Plasmids pGW8-98(Towne)L, pGW8-98(S5Amber)L and pGW8-98(R164A)L were constructed by similar methods, except that primers UL98-Forward and UL98-Reverse replaced primers GW8-F and GW8-R, respectively, in order to generate mutations in UL98 with flanking 500 bp homologies for use in recombinant BAC construction. For pGW8-98(Towne)L, DNA from CMV strain Towne was used as the PCR template. The entire UL98 ORF in each vector was sequenced to confirm the predicted changes and to exclude the possibility of other mutations. A control pDest17 vector expressing GUS was constructed as directed by the manufacturer’s (Invitrogen) protocol.
Expression of recombinant proteins in E. coli.
Optimal expression of soluble UL98 was obtained by using BL-21(DEQ) pLysS (Bioline) cells. Overnight culture was diluted 1 : 30 with Luria broth containing 50 µg carbenicillin ml−1 and grown to mid-exponential phase (A595 of 0.6) with shaking at 37 °C. Cultures were induced with 1 mM IPTG and incubated with shaking for an additional 3 h at 22 °C. Bacteria were harvested by centrifugation at 4000 g. for 20 min at 4 °C and cell pellets were stored at −80 °C.
IMAC purification of recombinant proteins.
Bacterial cell pellets were thawed on ice and lysed by incubation for 30 min on ice in buffer A (50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 15 % glycerol, 1 % NP-40 and 0.1 mM PMSF) containing 10 mM imidazole. Lysates were homogenized by intermittent ultrasonic disruption on ice over a 20 min period then clarified by centrifugation at 20 000 g for 20 min at 4 °C. Supernatants were applied to a 5 ml Ni–NTA agarose column equilibrated with buffer A containing 10 mM imidazole and washed extensively with the same buffer. Proteins were eluted with an 80 ml linear gradient of imidazole (10–300 mM) in buffer A. Fast protein liquid chromatography fractionation was conducted at room temperature and all fractions were collected at 0 °C. UL98-containing fractions were identified by immunoblotting with UL98-specific antibody I2 [a gift from Jay Nelson, Oregon Health and Science University, Portland, OR, USA (Adam et al., 1995)], pooled and dialysed against B-2 buffer (50 mM KH2PO4, pH 8.0, 50 mM NaCl, 15 % glycerol, 0.1 mM EDTA). Protein concentrations were determined by Bio-Rad assay as described by the manufacturer.
Quantitative exo assay.
The exo activities were quantified by release of trichloroacetic acid-soluble radioactivity from uniformly 14C-labelled E. coli DNA as previously described (Henderson et al., 1998). Reactions (200 µl final volume) were performed at 37 °C in nuclease buffer (50 mM Tris/HCl pH 9.0, 6 mM MgCl2, 1 mM DTT) and contained 3 µg 14C-labelled DNA. Reactions were initiated by the addition of protein and terminated by the addition of 25 µl 50 % trichloroacetic acid and 25 µl of salmon sperm DNA (2 mg ml−1). Undigested DNA was precipitated by incubation on ice for 5 min and centrifugation for 10 min at 3000 r.p.m. in a Beckman TJ-6 table-top centrifuge. The supernatant (200 µl) was neutralized with 75 µl of 1.2 M KOH, mixed with 7.5 ml ScintiSafe (Fisher) and the radioactivity measured by liquid scintillation spectrometry.
Qualitative endo assay.
IMAC-purified proteins (2.5 µg) were mixed with 250 ng supercoiled pUC19 plasmid DNA and incubated in nuclease buffer for 12 h at 37 °C. Reaction products were separated by electrophoresis on 1 % agarose gels, stained with ethidium bromide and visualized by UV light.
Fluorescence-based endonuclease assay.
A 35 nt ssDNA oligonucleotide substrate containing a 5′ 6-carboxyfluorescein (6-FAM) fluorophore and 3′ Iowa Black FQ (IABLFQ) quencher (6-FAM–A*GCTACGACGAACACCTCTATGTCATCAATAATC–IABLFQ) was synthesized by Integrated DNA Technologies. A thioate bond at the 5′ end (*) was added to inhibit 5′ exo activity. One millimole of DNA substrate was incubated with increasing amounts of recombinant protein in nuclease buffer for 14 h at 37 °C. Fluorescence was measured at 535 nm with a Beckman Victor2 Multilabel plate reader using an excitation wavelength of 485 nm.
Recombinant virus construction.
Construction of BAC HB15-t178b has been described (Saccoccio et al., 2011). ‘Recombineering’ was used to replace the UL98 ORF in HB15-t178b with galK as described (Warming et al., 2005). Briefly, a linear recombination fragment containing galK flanked by 50 bp homologies to the 5′ and 3′ ends of the UL98 ORF was generated by PCR using plasmid pGalK as the template for amplification with primers UL98-GalKF and UL98-GalKR (Table S2). The PCR product was electroporated into E. coli SW102 cells containing BAC HB15-t178b following induction at 42 °C for 15 min. Colonies were selected on minimal-media plates containing 0.2 % galactose and 15 µg chloramphenicol ml−1. GalK-positive colonies were confirmed by using MacConkey agar plates and then screened by HindIII restriction pattern to identify clones that both lacked 6.3 kb and 6.6 kb fragments and had gained a 12.9 kb fragment (as predicted by the anticipated ΔUL98/galK sequence). One clone was selected and designated BAC ΔUL98/galK. Correct insertion of galK in ΔUL98/galK was confirmed by sequencing across the CMV/galK breakpoints.
BACs ΔUL98/Rev, UL98/Amb and UL98/R164A were constructed from ΔUL98/galK by selection against GalK. Plasmids pGW8-98(Towne)L, pGW8-98(S5Amber)L and pGW8-98(R164A)L were digested with BsiWI to generate linear recombination fragments that were purified by agarose electrophoresis and electroporated into E. coli SW102 cells containing BAC ΔUL98/galK following induction at 42 °C for 15 min. GalK-negative colonies were selected for by growth on minimal-media plates containing 0.2 % deoxygalactose and 15 µg chloramphenicol ml−1. Candidate clones were screened for by detecting restoration of the 6.6 kb HindIII fragment and were confirmed by targeted sequencing.
Virus reconstitution, propagation and growth analysis.
BAC DNAs were purified and transfected into MRC-5 fibroblasts (ATCC CCL-171) as previously described (Sauer et al., 2010). GFP+ foci were observed by using an Olympus LX70 Inverted UV microscope 10–20 days post-transfection. At 15–30 days post-transfection culture supernatants and cells were transferred to 75 cm2 flasks of confluent MRC-5 cells and incubated until extensive cytopathic effect was observed. Viral stocks were prepared from cell-culture media that were clarified by centrifugation, adjusted to 0.2 M sucrose, aliquoted, stored at –80 °C and titrated on MRC-5 cells by limiting-dilution in 96-well plates as described (Cui et al., 2008). Growth curves were generated by infecting 75 cm2 flasks of confluent MRC-5 cells with carefully matched viral inocula. Cultures were washed 3 h post-infection and samples of culture media were removed periodically for titration as described above.
Supplementary Material
Acknowledgements
The authors thank Darrell Peterson and Tom Segar for guidance in protein expression and purification, Daniel Tenney for advice and guidance and for sharing unpublished data, and Jay Nelson for providing mAb I2. We also thank Søren Warming for pGalK and SW102 cells. This work was supported by Public Health Services grant R21AI071995 (to M. A. M.) from the National Institute of Allergy and Infectious Diseases and grants R01GM071894 (to G. E. K.) and R01GM073832 (to D. S. P.) from the National Institute of General Medical Sciences.
Footnotes
Three supplementary figures, two supplementary tables and supplementary methods are available with the online version of this paper.
References
- Adam B. L., Jervey T. Y., Kohler C. P., Wright G. L., Jr, Nelson J. A., Stenberg R. M. (1995). The human cytomegalovirus UL98 gene transcription unit overlaps with the pp28 true late gene (UL99) and encodes a 58-kilodalton early protein. J Virol 69, 5304–5310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnéris C., Briggs L. C., Savva R., Ebrahimi B., Barrett T. E. (2011). Crystal structure of a KSHV-SOX-DNA complex: insights into the molecular mechanisms underlying DNase activity and host shutoff. Nucleic Acids Res 39, 5744–5756 10.1093/nar/gkr111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian N., Bai P., Buchek G., Korza G., Weller S. K. (2010). Physical interaction between the herpes simplex virus type 1 exonuclease, UL12, and the DNA double-strand break-sensing MRN complex. J Virol 84, 12504–12514 10.1128/JVI.01506-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisson M., Géoui T., Flot D., Tarbouriech N., Ressing M. E., Wiertz E. J., Burmeister W. P. (2009). A bridge crosses the active-site canyon of the Epstein–Barr virus nuclease with DNase and RNase activities. J Mol Biol 391, 717–728 10.1016/j.jmb.2009.06.034 [DOI] [PubMed] [Google Scholar]
- Bujnicki J. M., Rychlewski L. (2001). The herpesvirus alkaline exonuclease belongs to the restriction endonuclease PD-(D/E)XK superfamily: insight from molecular modeling and phylogenetic analysis. Virus Genes 22, 219–230 10.1023/A:1008131810233 [DOI] [PubMed] [Google Scholar]
- Cui X., McGregor A., Schleiss M. R., McVoy M. A. (2008). Cloning the complete guinea pig cytomegalovirus genome as an infectious bacterial artificial chromosome with excisable origin of replication. J Virol Methods 149, 231–239 10.1016/j.jviromet.2008.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlroth S. L., Gurmu D., Schmitzberger F., Engman H., Haas J., Erlandsen H., Nordlund P. (2009). Crystal structure of the shutoff and exonuclease protein from the oncogenic Kaposi’s sarcoma-associated herpesvirus. FEBS J 276, 6636–6645 10.1111/j.1742-4658.2009.07374.x [DOI] [PubMed] [Google Scholar]
- Dunn W., Chou C., Li H., Hai R., Patterson D., Stolc V., Zhu H., Liu F. (2003). Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A 100, 14223–14228 10.1073/pnas.2334032100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouda N., Caracatsanis M., Kut I. A., Hammarström L. (1991). Mineralization disturbances of the developing rat molar induced by mono- and bisphosphonates. J Biol Buccale 19, 106–115 [PubMed] [Google Scholar]
- Gao M., Robertson B. J., McCann P. J., O’Boyle D. R., Weller S. K., Newcomb W. W., Brown J. C., Weinheimer S. P. (1998). Functional conservations of the alkaline nuclease of herpes simplex type 1 and human cytomegalovirus. Virology 249, 460–470 10.1006/viro.1998.9344 [DOI] [PubMed] [Google Scholar]
- Goldstein J. N., Weller S. K. (1998). The exonuclease activity of HSV-1 UL12 is required for in vivo function. Virology 244, 442–457 10.1006/viro.1998.9129 [DOI] [PubMed] [Google Scholar]
- Henderson G. I., Hu Z. Q., Yang Y., Perez T. B., Devi B. G., Frosto T. A., Schenker S. (1993). Ganciclovir transfer by human placenta and its effects on rat fetal cells. Am J Med Sci 306, 151–156 10.1097/00000441-199309000-00004 [DOI] [PubMed] [Google Scholar]
- Henderson J. O., Ball-Goodrich L. J., Parris D. S. (1998). Structure–function analysis of the herpes simplex virus type 1 UL12 gene: correlation of deoxyribonuclease activity in vitro with replication function. Virology 243, 247–259 10.1006/viro.1998.9054 [DOI] [PubMed] [Google Scholar]
- Hoffmann P. J., Cheng Y. C. (1978). The deoxyribonuclease induced after infection of KB cells by herpes simplex virus type 1 or type 2. I. Purification and characterization of the enzyme. J Biol Chem 253, 3557–3562 [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. (1996). vmd: visual molecular dynamics. J Mol Graph 14, 33–38, 27–28 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Kehm E., Göksu M., Bayer S., Knopf C. W. (1998). Herpes simplex virus type 1 DNase: functional analysis of the enzyme expressed by recombinant baculovirus. Intervirology 41, 110–119 10.1159/000024922 [DOI] [PubMed] [Google Scholar]
- Klug S., Lewandowski C., Merker H. J., Stahlmann R., Wildi L., Neubert D. (1991). In vitro and in vivo studies on the prenatal toxicity of five virustatic nucleoside analogues in comparison to aciclovir. Arch Toxicol 65, 283–291 10.1007/BF01968962 [DOI] [PubMed] [Google Scholar]
- Kovall R. A., Matthews B. W. (1998). Structural, functional, and evolutionary relationships between λ-exonuclease and the type II restriction endonucleases. Proc Natl Acad Sci U S A 95, 7893–7897 10.1073/pnas.95.14.7893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. T., Hu H. P., Hsu T. Y., Chen J. Y. (2003). Site-directed mutagenesis in a conserved motif of Epstein–Barr virus DNase that is homologous to the catalytic centre of type II restriction endonucleases. J Gen Virol 84, 677–686 10.1099/vir.0.18739-0 [DOI] [PubMed] [Google Scholar]
- Martinez R., Shao L., Bronstein J. C., Weber P. C., Weller S. K. (1996). The product of a 1.9-kb mRNA which overlaps the HSV-1 alkaline nuclease gene (UL12) cannot relieve the growth defects of a null mutant. Virology 215, 152–164 10.1006/viro.1996.0018 [DOI] [PubMed] [Google Scholar]
- Martinez R., Goldstein J. N., Weller S. K. (2002). The product of the UL12.5 gene of herpes simplex virus type 1 is not essential for lytic viral growth and is not specifically associated with capsids. Virology 298, 248–257 10.1006/viro.2002.1444 [DOI] [PubMed] [Google Scholar]
- Patel A. H., Subak-Sharpe J. H., Stow N. D., Marsden H. S., Maclean J. B., Dargan D. J. (1996). Suppression of amber nonsense mutations of herpes simplex virus type 1 in a tissue culture system. J Gen Virol 77, 199–209 10.1099/0022-1317-77-2-199 [DOI] [PubMed] [Google Scholar]
- Porter I. M., Stow N. D. (2004a). Replication, recombination and packaging of amplicon DNA in cells infected with the herpes simplex virus type 1 alkaline nuclease null mutant ambUL12. J Gen Virol 85, 3501–3510 10.1099/vir.0.80403-0 [DOI] [PubMed] [Google Scholar]
- Porter I. M., Stow N. D. (2004b). Virus particles produced by the herpes simplex virus type 1 alkaline nuclease null mutant ambUL12 contain abnormal genomes. J Gen Virol 85, 583–591 10.1099/vir.0.19657-0 [DOI] [PubMed] [Google Scholar]
- Reuven N. B., Staire A. E., Myers R. S., Weller S. K. (2003). The herpes simplex virus type 1 alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J Virol 77, 7425–7433 10.1128/JVI.77.13.7425-7433.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuven N. B., Willcox S., Griffith J. D., Weller S. K. (2004). Catalysis of strand exchange by the HSV-1 UL12 and ICP8 proteins: potent ICP8 recombinase activity is revealed upon resection of dsDNA substrate by nuclease. J Mol Biol 342, 57–71 10.1016/j.jmb.2004.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccoccio F. M., Sauer A. L., Cui X., Armstrong A. E., Habib S. E., Johnson D. C., Ryckman B. J., Klingelhutz A. J., Adler S. P., McVoy M. A. (2011). Peptides from cytomegalovirus UL130 and UL131 proteins induce high titer antibodies that block viral entry into mucosal epithelial cells. Vaccine 29, 2705–2711 10.1016/j.vaccine.2011.01.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer A., Wang J. B., Hahn G., McVoy M. A. (2010). A human cytomegalovirus deleted of internal repeats replicates with near wild type efficiency but fails to undergo genome isomerization. Virology 401, 90–95 10.1016/j.virol.2010.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiss M. R. (2005). Antiviral therapy of congenital cytomegalovirus infection. Semin Pediatr Infect Dis 16, 50–59 10.1053/j.spid.2004.09.012 [DOI] [PubMed] [Google Scholar]
- Shao L., Rapp L. M., Weller S. K. (1993). Herpes simplex virus 1 alkaline nuclease is required for efficient egress of capsids from the nucleus. Virology 196, 146–162 10.1006/viro.1993.1463 [DOI] [PubMed] [Google Scholar]
- Sheaffer A. K., Weinheimer S. P., Tenney D. J. (1997). The human cytomegalovirus UL98 gene encodes the conserved herpesvirus alkaline nuclease. J Gen Virol 78, 2953–2961 [DOI] [PubMed] [Google Scholar]
- Silva M. C., Yu Q. C., Enquist L., Shenk T. (2003). Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J Virol 77, 10594–10605 10.1128/JVI.77.19.10594-10605.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J. (1998). An efficient library for parallel ray tracing and animation. Rolla: University of Missouri-Rolla [Google Scholar]
- Vancíková Z., Dvorák P. (2001). Cytomegalovirus infection in immunocompetent and immunocompromised individuals – a review. Curr Drug Targets Immune Endocr Metabol Disord 1, 179–187 10.2174/1568008013341334 [DOI] [PubMed] [Google Scholar]
- Venclovas C., Timinskas A., Siksnys V. (1994). Five-stranded β-sheet sandwiched with two α-helices: a structural link between restriction endonucleases EcoRI and EcoRV. Proteins 20, 279–282 10.1002/prot.340200308 [DOI] [PubMed] [Google Scholar]
- Warming S., Costantino N., Court D. L., Jenkins N. A., Copeland N. G. (2005). Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33, e36 10.1093/nar/gni035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller S. K., Seghatoleslami M. R., Shao L., Rowse D., Carmichael E. P. (1990). The herpes simplex virus type 1 alkaline nuclease is not essential for viral DNA synthesis: isolation and characterization of a lacZ insertion mutant. J Gen Virol 71, 2941–2952 10.1099/0022-1317-71-12-2941 [DOI] [PubMed] [Google Scholar]
- Wing B. A., Huang E. S. (1995). Analysis and mapping of a family of 3′-coterminal transcripts containing coding sequences for human cytomegalovirus open reading frames UL93 through UL99. J Virol 69, 1521–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Silva M. C., Shenk T. (2003). Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc Natl Acad Sci U S A 100, 12396–12401 10.1073/pnas.1635160100 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.