Abstract
In order to resolve the location and activity of submicroscopic viruses in living cells, viral proteins are often fused to fluorescent proteins (FPs) and visualized by microscopy. In this study, we describe the fusion of FPs to three proteins of pseudorabies virus (PRV) that allowed imaging of capsids in living cells. Included in this study are the first recombinant PRV strains expressing FP–pUL25 fusions based on a design applied to herpes simplex virus type 1 by Homa and colleagues. The properties of each reporter virus were compared in both in vitro and in vivo infection models. PRV strains expressing FP–pUL25 and FP–pUL36 preserved wild-type properties better than traditional FP–pUL35 isolates in assays of plaque size and virulence in mice. The utility of these strains in studies of axon transport, nuclear dynamics and viral particle composition are documented.
Fusion of fluorescent proteins (FPs) to viral proteins has illuminated numerous aspects of viral replication in the context of live-cell imaging. Studies of herpesvirus infections have included FP fusions to all components of the viral particle (envelope, tegument and capsid) and to non-structural proteins. These recombinant viruses have proven indispensable for understanding the dynamics of the herpesvirus particle composition and host–pathogen interactions in cells (Smith & Banfield, 2006). In particular, FP fusions to the capsid have aided the study of the entire course of the infection cycle, including: (i) capsid entry into the host cell, (ii) transport of the capsid to the nucleus, (iii) intranuclear capsid dynamics, and (iv) virion egress from host cells. While FPs have revolutionized the field of virology, viruses encoding FPs are mutants that may only approximate the behaviour of wild-type viruses. Although these reporter viruses can yield intriguing new data, assuring that observations are biologically relevant is best achieved by using reporter viruses that maintain wild-type properties and replicating findings with different fusions when possible.
VP26 (pUL35) was the first herpesvirus capsid protein fused to an FP and remains the most common approach for capsid visualization (Desai & Person, 1998; Frampton et al., 2010; Smith et al., 2001). The surface of capsids is decorated with six copies of pUL35 per hexon (900 copies per capsid) (Wingfield et al., 1997; Zhou et al., 1995). This high copy number results in bright fluorescence when pUL35 is fused to an FP. Additionally, pUL35 is the only non-essential capsid protein; while deletion of the UL35 gene from herpes simplex virus (HSV)-1 or pseudorabies virus (PRV) reduces propagation by approximately 1 log, fusion of GFP to the amino terminus of pUL35 in either virus has no impact on virus propagation kinetics (Desai & Person, 1998; Luxton et al., 2005). However, a more recently described PRV GFP–pUL35 isolate propagated similarly to a UL35-null virus and displayed reduced neuroinvasion and increased mean time to death in a mouse infection model (Krautwald et al., 2008).
Unlike pUL35, pUL25 is an essential capsid component that is necessary for stable genome encapsidation and capsid nuclear egress (McNab et al., 1998; O’Hara et al., 2010). Capsid-bound pUL25 interfaces with hexons, triplexes, portal, pUL17 and the pUL36 (VP1/2) large tegument protein (Coller et al., 2007; Conway et al., 2010; Pasdeloup et al., 2009). Along with its binding partner, pUL17, pUL25 forms a capsid-surface feature first identified on C capsids referred to as a C-capsid-specific component; however, an alternative name for this structure, the capsid-vertex-specific component (CVSC), was suggested because this structure is now recognized to surround the vertices of all capsid species (Toropova et al., 2011). The CVSC is present at 60 copies per capsid (Newcomb et al., 2006; Toropova et al., 2011; Trus et al., 2007). To tag HSV-1 pUL25, GFP was inserted in-frame between two conserved coding regions (Cockrell et al., 2009). Despite the multi-functionality of pUL25, the HSV-1 pUL25–GFP double in-frame fusion grew with wild-type kinetics and was used to identify the precise location of pUL25 on the capsid surface (Conway et al., 2010).
pUL36 is a large capsid-bound tegument protein that remains associated with the capsid after detergent extraction (Gibson & Roizman, 1972; Newcomb & Brown, 2010; Wolfstein et al., 2006). In yeast two-hybrid studies using HSV-1 or varicella-zoster virus proteins, pUL36 interacts with three capsid proteins: pUL17, pUL25 and pUL18 (VP23; triplex) (Fossum et al., 2009; Uetz et al., 2006). Additionally, pUL36 co-immunoprecipitates with the major capsid protein pUL19 from infected cells, although pUL25 is currently the only confirmed binding site for pUL36 in intact capsids (Coller et al., 2007; McNabb & Courtney, 1992). pUL36 is associated with herpesvirus capsids in virions and in the cytoplasm during both early and late stages of infection, making the protein an effective extra-nuclear capsid-associated reporter (Antinone & Smith, 2010; Gibson & Roizman, 1972; Granzow et al., 2005; Luxton et al., 2005; Newcomb & Brown, 2010). Like pUL25, pUL36 is essential for viral propagation (Desai, 2000; Fuchs et al., 2004; Knipe et al., 1981; Smith & Enquist, 1999). Viral propagation is unaffected by fusion of an FP to the amino terminus of pUL36, which has allowed for the observation of capsid movement in the absence of pUL35 (Antinone et al., 2006).
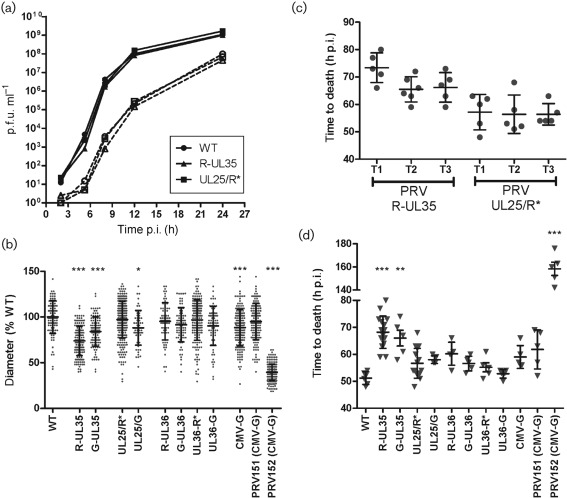
All viruses used in this study were derived from the infectious clone pBecker3 with the exception of PRV151 and PRV152 and are summarized in the Supplementary Table (available in JGV Online) (Demmin et al., 2001; Smith et al., 2000; Smith & Enquist, 2000). FP fusions are noted with letters: GFP (G), or red FPs mRFP1 (R) or mCherry (R*) are placed before or after a gene name, indicating N- or C-terminal fusion, respectively, with a slash indicating a double in-frame fusion. PRV encoding GFP or mRFP1 fused to pUL35 were described previously (Smith et al., 2001, 2004). The UL25/G and UL25/R* fusions were designed based on a previously described HSV-1 recombinant (Cockrell et al., 2009; Conway et al., 2010). All reporter viruses attained wild-type titres when grown on pig kidney epithelial (PK15) cells (Supplementary Table). Additionally, a single-step growth assay was performed on PK15 cells to compare the growth kinetics of PRV-UL25/R*, PRV-R-UL35 and PRV-WT (Fig. 1a). No kinetic defect was observed for either reporter virus in cultured cells.
Fig. 1.
In vitro and in vivo infectivity of reporter PRV. (a) Single-step growth curves comparing PRV derived from the unmodified pBecker3 infectious clone (WT) to two recombinant reporter viruses (R-UL35 and UL25/R*). Virus was harvested from adherent PK15 cells (solid lines) or infected cell supernatants (dashed lines) and titres were determined by plaque assay. (b) Plaque diameters in Vero cells at 4 days p.i. Diameters of individual plaques (dots) are expressed as a per cent of the mean plaque diameter of unmodified virus (WT). RFP (R) and GFP (G) fusions are noted based on insertion location before, after, or in the middle (noted with a slash) of the viral gene. Error bars indicate sd of plaque sizes of each reporter virus. (c and d) Virulence of reporter viruses in an intranasal model of infection in CD-1 mice. In (c), three independent trials (T1, T2 and T3) of intranasal infection by one of two reporter viruses are compared. In (d) all viruses in this study are compared. Each symbol represents one mouse. Error bars indicate sd of the mean time to death for each trial. For all panels, asterisks indicate statistically significant difference from WT (*P<0.05, **P<0.01 and ***P<0.001) as determined by Tukey’s multiple comparison test.
Plaque diameters of reporter viruses were measured to assess cell–cell spread of fluorescent viruses. Vero cells were infected in a six-well plate with 60–250 p.f.u. per well and overlaid with media containing methocel to restrict viral diffusion. Three days after infection, viable cells were stained overnight with neutral red solution. Plates were scanned and diameters (in pixels) were measured for all plaques. Plaques of PRV-R-UL35 (74 % WT) and PRV-G-UL35 (84 % WT) were both smaller than PRV-WT (Fig. 1b), consistent with observations made in PRV-Kaplan (Krautwald et al., 2008). Although we did not attempt to repair the FP–UL35 viruses by deleting the FP sequences, the similarity between different FP–UL35 fusions indicated that the plaque defect was reproducible and due to the addition of an FP to pUL35. Plaque diameters of PRV-UL25/R* were similar to PRV-WT (97 %), indicating that fusing UL25 to mCherry had little effect on the ability of the virus to spread between cells in a confluent monolayer, although PRV-UL25/G produced smaller plaques (87 %). All noted reductions in PRV-Becker plaque sizes were minimal when compared with the attenuated PRV-Bartha strain PRV152. FP fusions to either terminus of pUL36 or expression of soluble GFP in PRV-Becker strains had negligible impacts in plaque size.
A mouse CD-1 intranasal inoculation model was used to examine virulence, and allowed for comparisons with a previous study (Krautwald et al., 2008). Initially, three groups of five mice each were infected with three independent stocks of PRV-UL25/R* or PRV-R-UL35 (8–9×105 p.f.u. per nostril) and recorded to determine time of death (Fig. 1c). Variability in results obtained between replica stocks was not statistically significant. Using this same method, both PRV-R-UL35 and PRV-G-UL35 displayed reductions in virulence when compared with wild-type PRV (Fig. 1d), consistent with a previous report, although these reductions were small compared with the attenuated PRV-Bartha strain PRV152 (Krautwald et al., 2008). In contrast, FP-fusions to pUL25 or pUL36 did not significantly reduce virulence.
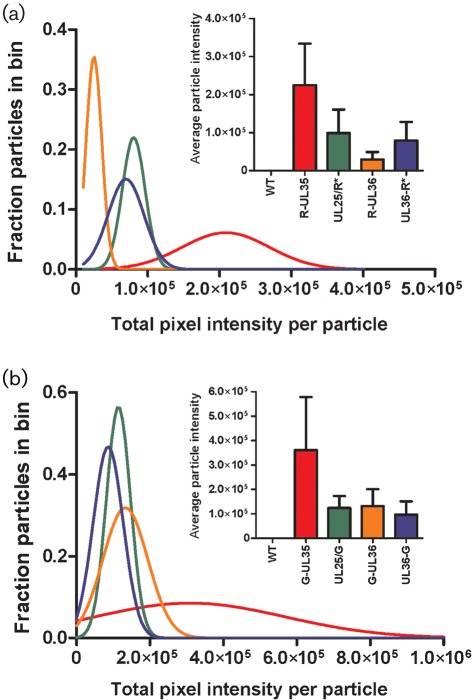
While maintenance of infectivity is an important consideration when selecting a protein for FP-fusion, the usefulness of an FP-fusion is dependent on its brightness and visibility over background. To assess relative fluorescence emission intensity, extracellular viral particles were isolated from infected PK15 cell supernatants, spotted onto coverslips and imaged by fluorescence microscopy with appropriate filters (Fig. 2; see Supplementary Methods, available in JGV Online). As expected, fluorescence emissions per particle were greatest with FP–pUL35 viruses. This was most evident when the same FP was used for each fusion; PRV-G-UL35 fluorescence was three- to fourfold higher than observed with PRV-UL25/G, PRV-G-UL36 or PRV-UL36-G (Fig. 2b). Additionally, the fluorescence intensity of extracellular viral particles from PRV-G-UL36, PRV-UL36-G and PRV-UL25/G were similar, suggesting that pUL25 and pUL36 are incorporated at 1 : 1 stoichiometry in extracellular particles, which is consistent with pUL25 serving as a binding site for pUL36 on capsids (Coller et al., 2007). Given that the CVSC is present at 60 copies per capsid we can extrapolate that there are probably 60 copies of pUL36 per PRV particle, which refines an earlier estimate of pUL36 incorporation in HSV-1 that extrapolated <150 copies of pUL36 per virion (Conway et al., 2010; Heine et al., 1974; Trus et al., 2007). The reported ratio of pUL25 : pUL35 copy number (60 : 900) was not reinforced based on the intensities from virions carrying the respective FP fusions. Because the design of the pUL25–GFP fusions allows for full occupancy on HSV-1 capsids (Conway et al., 2010), fusion of GFP to pUL35 appears to reduce pUL35 occupancy on capsid hexons. In agreement with this conclusion, the original GFP fusion to pUL35 in HSV-1 was estimated to have only 300–400 copies of GFP–pUL35 per capsid, which more closely agrees with the observed threefold difference in particle intensity (P. Desai, personal communication). The fluorescence intensities of individual viral particles were accurately modelled by Gaussian distributions, indicating that FP-fusions were incorporated into particles with normal variance. These trends were also observed in red fluorescent particles (Fig. 2a), although the difference in brightness between mRFP1 and mCherry (~25 %) prevented direct stoichiometric comparison (Shaner et al., 2004).
Fig. 2.
Characterization of fluorescence emission intensities from individual extracellular viral particles. The brightness of individual RFP-tagged (a) and GFP-tagged (b) viral particles released from PK15 cells 18–20 h p.i. was measured by summing the grey levels of all pixels from each diffraction-limited spot. Individual particles were grouped into bins according to fluorescence intensity and plotted as histograms (bin size = 10 000 grey levels for RFP viruses and bin size = 50 000 grey levels for GFP viruses; histogram bars not shown). Gaussian curves (coloured lines) were fit to histograms by non-linear regression (R2 >0.95, for all samples except G-UL36, R2 = 0.88). Insets show mean viral particle intensity with sd; bar graph colours correspond to plotted Gaussian curves.
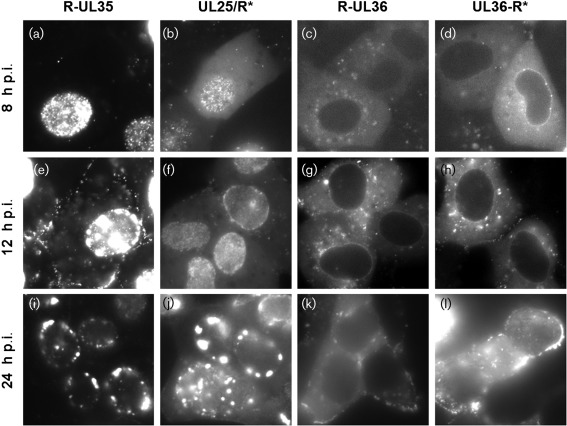
The localization of fluorescent signals was monitored in Vero cells that were infected with RFP reporter viruses at an m.o.i. of 5 (Fig. 3). At 2 and 5 h post-infection (p.i.), static fluorescent puncta were frequently observed at the nuclear membrane in PRV-R-UL35, PRV-UL25/R, PRV-R-UL36 and PRV-UL36-R* infections, indicating that these proteins remained associated with capsids after entering cells and docking at nuclear pores (not shown). At 8 h p.i., small, rapidly moving nuclear puncta consistent with single de novo assembled capsids were detected in cells infected with PRV-R-UL35 and PRV-UL25/R*, although movement of these particles may be faster than movement noted in HSV-1-infected cells (Forest et al., 2005). Due to the absence of pUL35 on procapsids and the low copy number of pUL25 on procapsids, the rapidly moving particles were probably not associated with the packaging of DNA (Sheaffer et al., 2001; Thurlow et al., 2005). However, fusion of FPs to pUL25 and pUL35 can facilitate observation of diverse processes associated with A, B and C capsid dynamics in the nucleus (Gibson & Roizman, 1972; Toropova et al., 2011). At 12 h p.i., single puncta remained predominant in PRV-UL25/R*-infected cells, but immobile aggregates (often referred to as assemblons; Ward et al., 1996) were present in nuclei of PRV-R-UL35-infected cells. At 24 h p.i., similar large aggregates in PRV-UL25/R*-infected cells were evident. Differential timing of aggregate formation suggests that tagging pUL35 with an FP exacerbates capsid aggregation in the nucleus.
Fig. 3.
Images of Vero cells infected with reporter viruses. At indicated times p.i., cells were imaged by wide-field fluorescence microscopy. All exposures are 200 ms, except (i) and (j), which are 100 ms exposures. Level settings are held constant between viruses R-UL35 and UL25/R* and between viruses R-UL36 and UL36-R* so that the apparent brightness of each image documents the true relative brightness of each sample. Image dimensions are 53 µm × 53 µm.
Prior to the onset of capsid nuclear egress, R-UL35 fluorescence was mostly confined to the nucleus, while UL25/R* displayed additional diffuse fluorescence in the cytoplasm. In contrast, fluorescent pUL36 was excluded from the nucleoplasm. Localization to the nuclear rim was observed for both termini of pUL36, although rims were apparent in PRV-UL36-R* cells at an earlier time (8 h p.i.) than in PRV-R-UL36 (12 h p.i.). In both cases, nuclear rim localization was no longer predominant by 24 h p.i. We are preparing a study that examines VP1/2 nuclear activities in greater detail.
The retrograde transport velocity of pUL35–FP PRV in axons of primary chick sensory neurons were indistinguishable from the pUL25/R* PRV (Supplementary Fig. S1, available in JGV Online). During late stages of infection with any of the FP-fusion PRV strains, capsid-like puncta were observed moving in the cytoplasm in curvilinear trajectories in the cytoplasm of Vero cells consistent with microtubule transport (data not shown) (Luxton et al., 2005).
Throughout this study, the pUL25/R* strain of PRV maintained wild-type virus properties, while allowing observation of viral capsids throughout the replication cycle. However, the R-pUL35 virus remains a useful reagent for studies of cytoplasmic capsid dynamics, including studies of microtubule transport. While G-pUL35 is the brightest tag, mutant viruses encoding FP–pUL35 fusions had defects in cell–cell spread in culture and virulence in mice, and nuclear capsids had an increased propensity to aggregate. In contrast, pUL25 and pUL36 FP-fusions performed similarly to WT viruses in assays of virulence and cell–cell spread, but the capsid tags are dimmer than pUL35, and pUL36 tags do not allow analysis of viral particles in the nucleus. Knowledge of the behaviour of these fluorescent reporters will be instrumental for more detailed studies of the dynamics of the herpesvirus infectious cycle.
Supplementary Material
Acknowledgements
We would like to acknowledge Allison Evans for excellent technical assistance and Jen Klabis and Joy Lee for help with BAC recombination. This work was funded by NIH grant R01 AI080658 to G. A. S. K. P. B. was supported by the training programme in Immunology and Molecular Pathogenesis from the National Institutes of Health (T32AI07476).
Footnotes
Supplementary material is available with the online version of this paper.
References
- Antinone S. E., Smith G. A. (2010). Retrograde axon transport of herpes simplex virus and pseudorabies virus: a live-cell comparative analysis. J Virol 84, 1504–1512 10.1128/JVI.02029-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antinone S. E., Shubeita G. T., Coller K. E., Lee J. I., Haverlock-Moyns S., Gross S. P., Smith G. A. (2006). The herpesvirus capsid surface protein, VP26, and the majority of the tegument proteins are dispensable for capsid transport toward the nucleus. J Virol 80, 5494–5498 10.1128/JVI.00026-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockrell S. K., Sanchez M. E., Erazo A., Homa F. L. (2009). Role of the UL25 protein in herpes simplex virus DNA encapsidation. J Virol 83, 47–57 10.1128/JVI.01889-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller K. E., Lee J. I., Ueda A., Smith G. A. (2007). The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J Virol 81, 11790–11797 10.1128/JVI.01113-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway J. F., Cockrell S. K., Copeland A. M., Newcomb W. W., Brown J. C., Homa F. L. (2010). Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J Mol Biol 397, 575–586 10.1016/j.jmb.2010.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmin G. L., Clase A. C., Randall J. A., Enquist L. W., Banfield B. W. (2001). Insertions in the gG gene of pseudorabies virus reduce expression of the upstream Us3 protein and inhibit cell-to-cell spread of virus infection. J Virol 75, 10856–10869 10.1128/JVI.75.22.10856-10869.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai P. J. (2000). A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J Virol 74, 11608–11618 10.1128/JVI.74.24.11608-11618.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai P., Person S. (1998). Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol 72, 7563–7568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forest T., Barnard S., Baines J. D. (2005). Active intranuclear movement of herpesvirus capsids. Nat Cell Biol 7, 429–431 10.1038/ncb1243 [DOI] [PubMed] [Google Scholar]
- Fossum E., Friedel C. C., Rajagopala S. V., Titz B., Baiker A., Schmidt T., Kraus T., Stellberger T., Rutenberg C. & other authors (2009). Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog 5, e1000570 10.1371/journal.ppat.1000570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton A. R., Jr, Uchida H., von Einem J., Goins W. F., Grandi P., Cohen J. B., Osterrieder N., Glorioso J. C. (2010). Equine herpesvirus type 1 (EHV-1) utilizes microtubules, dynein, and ROCK1 to productively infect cells. In Vet Microbiol, 141, 12–21 10.1016/j.vetmic.2009.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs W., Klupp B. G., Granzow H., Mettenleiter T. C. (2004). Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J Virol 78, 11879–11889 10.1128/JVI.78.21.11879-11889.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W., Roizman B. (1972). Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J Virol 10, 1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzow H., Klupp B. G., Mettenleiter T. C. (2005). Entry of pseudorabies virus: an immunogold-labeling study. J Virol 79, 3200–3205 10.1128/JVI.79.5.3200-3205.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine J. W., Honess R. W., Cassai E., Roizman B. (1974). Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J Virol 14, 640–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipe D. M., Batterson W., Nosal C., Roizman B., Buchan A. (1981). Molecular genetics of herpes simplex virus. VI. Characterization of a temperature-sensitive mutant defective in the expression of all early viral gene products. J Virol 38, 539–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krautwald M., Maresch C., Klupp B. G., Fuchs W., Mettenleiter T. C. (2008). Deletion or green fluorescent protein tagging of the pUL35 capsid component of pseudorabies virus impairs virus replication in cell culture and neuroinvasion in mice. J Gen Virol 89, 1346–1351 10.1099/vir.0.83652-0 [DOI] [PubMed] [Google Scholar]
- Luxton G. W., Haverlock S., Coller K. E., Antinone S. E., Pincetic A., Smith G. A. (2005). Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc Natl Acad Sci U S A 102, 5832–5837 10.1073/pnas.0500803102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab A. R., Desai P., Person S., Roof L. L., Thomsen D. R., Newcomb W. W., Brown J. C., Homa F. L. (1998). The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol 72, 1060–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb D. S., Courtney R. J. (1992). Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190, 221–232 10.1016/0042-6822(92)91208-C [DOI] [PubMed] [Google Scholar]
- Newcomb W. W., Brown J. C. (2010). Structure and capsid association of the herpesvirus large tegument protein UL36. J Virol 84, 9408–9414 10.1128/JVI.00361-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb W. W., Homa F. L., Brown J. C. (2006). Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J Virol 80, 6286–6294 10.1128/JVI.02648-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara M., Rixon F. J., Stow N. D., Murray J., Murphy M., Preston V. G. (2010). Mutational analysis of the herpes simplex virus type 1 UL25 DNA packaging protein reveals regions that are important after the viral DNA has been packaged. J Virol 84, 4252–4263 10.1128/JVI.02442-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasdeloup D., Blondel D., Isidro A. L., Rixon F. J. (2009). Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J Virol 83, 6610–6623 10.1128/JVI.02655-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N. G., Palmer A. E., Tsien R. Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22, 1567–1572 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Sheaffer A. K., Newcomb W. W., Gao M., Yu D., Weller S. K., Brown J. C., Tenney D. J. (2001). Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J Virol 75, 687–698 10.1128/JVI.75.2.687-698.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. A., Banfield B. W. (2006). The development and use of alpha-herpesviruses-expressing fluorescent proteins. In Alpha Herpesviruses: Molecular and Cell Biology, pp. 205–217 Edited by Sandri-Golden R. M. UK: Caister Academic Press [Google Scholar]
- Smith G. A., Enquist L. W. (1999). Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J Virol 73, 6405–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. A., Enquist L. W. (2000). A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc Natl Acad Sci U S A 97, 4873–4878 10.1073/pnas.080502497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B. N., Banfield B. W., Smeraski C. A., Wilcox C. L., Dudek F. E., Enquist L. W., Pickard G. E. (2000). Pseudorabies virus expressing enhanced green fluorescent protein: a tool for in vitro electrophysiological analysis of transsynaptically labeled neurons in identified central nervous system circuits. Proc Natl Acad Sci U S A 97, 9264–9269 10.1073/pnas.97.16.9264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. A., Gross S. P., Enquist L. W. (2001). Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc Natl Acad Sci U S A 98, 3466–3470 10.1073/pnas.061029798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. A., Pomeranz L., Gross S. P., Enquist L. W. (2004). Local modulation of plus-end transport targets herpesvirus entry and egress in sensory axons. Proc Natl Acad Sci U S A 101, 16034–16039 10.1073/pnas.0404686101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow J. K., Rixon F. J., Murphy M., Targett-Adams P., Hughes M., Preston V. G. (2005). The herpes simplex virus type 1 DNA packaging protein UL17 is a virion protein that is present in both the capsid and the tegument compartments. J Virol 79, 150–158 10.1128/JVI.79.1.150-158.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toropova K., Huffman J. B., Homa F. L., Conway J. F. (2011). The herpes simplex virus 1 UL17 protein is the second constituent of the capsid vertex-specific component required for DNA packaging and retention. J Virol 85, 7513–7522 10.1128/JVI.00837-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus B. L., Newcomb W. W., Cheng N., Cardone G., Marekov L., Homa F. L., Brown J. C., Steven A. C. (2007). Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol Cell 26, 479–489 10.1016/j.molcel.2007.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P., Dong Y. A., Zeretzke C., Atzler C., Baiker A., Berger B., Rajagopala S. V., Roupelieva M., Rose D. & other authors (2006). Herpesviral protein networks and their interaction with the human proteome. Science 311, 239–242 10.1126/science.1116804 [DOI] [PubMed] [Google Scholar]
- Ward P. L., Ogle W. O., Roizman B. (1996). Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J Virol 70, 4623–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield P. T., Stahl S. J., Thomsen D. R., Homa F. L., Booy F. P., Trus B. L., Steven A. C. (1997). Hexon-only binding of VP26 reflects differences between the hexon and penton conformations of VP5, the major capsid protein of herpes simplex virus. J Virol 71, 8955–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfstein A., Nagel C. H., Radtke K., Döhner K., Allan V. J., Sodeik B. (2006). The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7, 227–237 10.1111/j.1600-0854.2005.00379.x [DOI] [PubMed] [Google Scholar]
- Zhou Z. H., He J., Jakana J., Tatman J. D., Rixon F. J., Chiu W. (1995). Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol 2, 1026–1030 10.1038/nsb1195-1026 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.