Abstract
Despite utilizing the same avian hosts and mosquito vectors, St Louis encephalitis virus (SLEV) and West Nile virus (WNV) display dissimilar vector-infectivity and vertebrate-pathogenic phenotypes. SLEV exhibits a low oral infection threshold for Culex mosquito vectors and is avirulent in avian hosts, producing low-magnitude viraemias. In contrast, WNV is less orally infective to mosquitoes and elicits high-magnitude viraemias in a wide range of avian species. In order to identify the genetic determinants of these different phenotypes and to assess the utility of mosquito and vertebrate cell lines for recapitulating in vivo differences observed between these viruses, reciprocal WNV and SLEV pre-membrane and envelope protein (prME) chimeric viruses were generated and growth of these mutant viruses was characterized in mammalian (Vero), avian (duck) and mosquito [Aedes (C6/36) and Culex (CT)] cells. In both vertebrate lines, WNV grew to 100-fold higher titres than SLEV, and growth and cytopathogenicity phenotypes, determined by chimeric phenotypes, were modulated by genetic elements outside the prME gene region. Both chimeras exhibited distinctive growth patterns from those of SLEV in C6/36 cells, indicating the role of both structural and non-structural gene regions for growth in this cell line. In contrast, growth of chimeric viruses was indistinguishable from that of virus containing homologous prME genes in CT cells, indicating that structural genetic elements could specifically dictate growth differences of these viruses in relevant vectors. These data provide genetic insight into divergent enzootic maintenance strategies that could also be useful for the assessment of emergence mechanisms of closely related flaviviruses.
Introduction
West Nile virus and St Louis encephalitis virus (WNV and SLEV, respectively; family Flaviviridae) are closely related members of the Japanese encephalitis serocomplex and are both transmitted between Culex mosquitoes and avian hosts. Both viruses have the capacity for eliciting encephalitis in humans and have been associated with large epidemics. Despite utilizing the same avian and mosquito species for their enzootic maintenance, marked epidemiological differences exist between the viruses. For instance, since its introduction into North America in 1999, WNV has been associated with large epiornitics, whereas SLEV has never been associated with widespread mortality in avian hosts. Interestingly, SLEV also has been documented to be poorly cytopathogenic in cell culture, requiring up to 7 days to generate plaques on Vero cells. Additionally, SLEV has demonstrated greater oral infectivity for Culex mosquitoes than WNV (Reisen et al., 2008a). These differences suggest that the two viruses have adopted dissimilar evolutionary strategies for enzootic maintenance, with WNV relying on high avian replication rates to assure transmission and SLEV utilizing high mosquito oral susceptibility for its propagation. As SLEV is a Western hemisphere-endemic virus, this could reflect long-term co-adaptation of SLEV with North American Culex mosquitoes. This would be in direct contrast to WNV, a recently introduced virus that has not adapted to North American Culex vectors, but is capable of generalized high-level replication in a broad range of avian hosts, some of which, such as the house sparrow (Passer domesticus), have been introduced from the Old World (Robbins, 1973). The emergence and proliferation of a new North American genotype with increased dissemination capacity suggests a potential subsequent evolutionary adaptation of WNV in North American mosquitoes (Moudy et al., 2007).
Culex tarsalis mosquitoes are a major vector of WNV and SLEV in western North America, based on field isolations and laboratory vector-competence studies (Goddard et al., 2002; Reisen et al., 2006). This species has previously been demonstrated to be infected at lower oral titres with SLEV than WNV (Reisen et al., 2005). House sparrows and house finches (Carpodacus mexicanus) are considered to be major amplification hosts of WNV and SLEV in North America due to their abundance, susceptibility to infection, development of high-magnitude viraemias in laboratory studies and rapid population-replacement rates (Komar et al., 2003; Langevin et al., 2005; Mahmood et al., 2004; Reisen et al., 2004).
WNV and SLEV both possess positive-sense, ssRNA genomes of approximately 11 kb in length, expressed as a single ORF that is cleaved co- and post-translationally to produce three structural [capsid (C), pre-membrane/membrane (prM/M) and envelope (E)] and seven non-structural (NS) (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) proteins. Previous reports have indicated that prME structural proteins are important for modulation of mosquito infectivity (McElroy et al., 2006b), presumably as a direct result of altered efficiency of virus–cell receptor binding. Alternatively, WNV NS genetic elements have been associated with altered growth in avian hosts (Brault et al., 2007) that could be dictated by enhanced intracellular replication in key target cells or altered replicative capacity in febrile avian hosts (Kinney et al., 2006). Therefore, structural genetic elements (prME) of SLEV could dictate increased oral infectivity of Culex vectors, whereas the NS proteins could modulate growth within avian hosts and produce the higher cytopathogenicity observed following WNV infection. To test this association, chimeric viruses were generated in which the structural components of WNV and SLEV were interchanged. The resulting mutant viruses generated from these chimeric constructs, WNV-prME/SLEV.IC and SLEV-prME/WNV.IC, were assayed in avian, mammalian and mosquito in vitro systems to determine virus-growth and cytopathogenic phenotypes.
Results
Characterization of the recombinant SLEV and prME chimeric viruses
Recombinant WNV from an infectious cDNA based on the NY99 genotype (strain 382-99) was generated and used as a surrogate for the parental strain, based on previous studies demonstrating the viruses to be phenotypically identical (Kinney et al., 2006). Infectious virus was generated successfully from the parental SLEV infectious clone-derived virus (SLEV.IC) following transfection of C6/36 cells, representing the first generation of a purely recombinant SLEV. Sequence analysis performed on the rescued SLEV.IC virus demonstrated complete identity with the genomic sequence of the original IMP115 SLEV isolate, GenBank accession no. JF460774. Chimeric viruses, SLEV-prME/WNV.IC and WNV-prME/SLEV.IC, were also rescued successfully following transfection in C6/36 cells. Sequences for all predicted amino acid substitutions were maintained following transfection in C6/36 cells for both chimeric viruses. Each chimeric virus shared complete sequence identity within the prME-encoding region for the heterologous flavivirus, containing a total of 191 amino acid substitutions (58 prM and 133 E substitutions; Table 1). WNV and SLEV shared predicted N-linked motifs for glycosylation at prM-15/prM-17, respectively, as well as at E-154.
Table 1. Amino acid identity and similarity between WNV and SLEV prME gene regions incorporated into chimeric viruses.
| Gene region | Identity | Similarity |
| Pre-membrane | ||
| Identity/similarity (%) | 65.7 | 80.5 |
| No. of amino acid differences | 58 | 33 |
| Envelope | ||
| Identity/similarity (%) | 73.5 | 85.2 |
| No. of amino acid differences | 133 | 74 |
| Total | ||
| Identity/similarity (%) | 71.5 | 84.0 |
| No. of amino acid differences | 191 | 107 |
Indirect immunofluorescence assay (IFA)
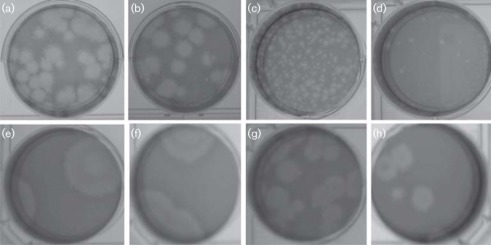
To ensure the retention of antigenic specificity of the structural elements after chimerization, IFA was performed using WNV-specific (3.67) and SLEV-specific (4A4C-4) mAbs that target the envelope protein. mAb 3.67 reacted positively with Vero cells inoculated with WNV and WNV-prME/SLEV.IC; however, this antibody failed to detect cells inoculated with SLEV.IC or SLEV-prME/WNV.IC (Fig. 1a, c, e, g). Conversely, mAb 4A4C-4 was reactive with cells inoculated with SLEV.IC or SLEV-prME/WNV.IC, but not with WNV or WNV-prME/SLEV.IC viruses (Fig. 1b, d, f, h), thereby indicating the conservation of antigenic specificity of the two chimeric viruses.
Fig. 1.
IFA performed with WNV-specific (3.67; left panels) and SLEV-specific (4A4C-4; right panels) mAbs on Vero cells inoculated with WNV, SLEV and WNV/SLEV chimeric viruses. (a, b) WNV; (c, d) WNV-prME/SLEV.IC; (e, f) SLEV-prME/WNV.IC; (g, h) SLEV.IC.
Plaque morphology
Plaques of the WNV-NY99 isolate (strain 382-99) and the WNV infectious clone-derived virus (WNV.IC) averaged approximately 2 mm in diameter at 72 h post-infection (p.i.), similar to previous reports (Kinney et al., 2006). Plaques of the IMP115 SLEV strain and the SLEV.IC averaged 6 mm in diameter at 168 h p.i. (Fig. 2) and were statistically indistinguishable (P>0.05). Comparatively, WNV.IC plaques were clearly visible (2 mm) at 72 h p.i., whereas SLEV.IC plaques were <1 mm at this time point (Figs 2 and 3). The SLEV-prME/WNV.IC chimeric virus exhibited plaques of 2 mm at 72 h p.i. and ≥20 mm by 168 h p.i., consistent with the plaque morphology of WNV.IC (Figs 2 and 3). In contrast, WNV-prME/SLEV.IC plaques were statistically indistinguishable in size from those of SLEV.IC (P>0.05), measuring <1 mm at 72 h p.i. and <6 mm at 168 h p.i.
Fig. 2.
Diameters of plaques (mean±sd, in mm) exhibited by WNV/NY99, WNV.IC, SLEV-prME/WNV.IC, SLEV/IMP115, SLEV.IC and WNV-prME/SLEV.IC (order as presented) viruses on Vero cells over time (h p.i.). Bars representing viruses containing the NS gene elements of SLEV are outlined.
Fig. 3.
Plaques after plating of viruses on Vero cells at 72 and 168 h p.i. (a, e) WNV.IC at 72 and 168 h p.i., respectively; (b, f) SLEV-prME/WNV.IC at 72 and 168 h p.i., respectively; (c, g) SLEV.IC at 72 and 168 h p.i., respectively; (d, h) WNV-prME/SLEV.IC at 72 and 168 h p.i., respectively.
Cell viability
Changes in Vero cell-culture viability based on electrical impedance (Fang et al., 2011) supported the plaque-morphology results. Overall, WNV/NY99, WNV.IC and SLEV-prME/WNV.IC viruses demonstrated statistically similar (P>0.05) cell-viability patterns, initiating at a mean cell index of 9.7±0.2 and decreasing to 5.4±0.4 by 72 h p.i. and to 0.2±0.1 by 120 h p.i. (Fig. 4). However, the SLEV/IMP115, SLEV.IC and WNV-prME/SLEV.IC viruses exhibited a different cell-viability pattern, starting with a cell index value of 9.4±0.1 and reaching a mean cell index of 5.7±0.8 by 120 h p.i., similar to the pattern of the uninfected-control cells until 96 h p.i., at which point there was a more pronounced drop in cell index values for SLEV-infected cells (Fig. 4). The mean cell index did not differ significantly (P>0.05) among SLEV/IMP115, SLEV.IC and WNV-prME/SLEV.IC viruses, but did differ significantly (P<0.05) from WNV-NY99, WNV.IC and SLEV-prME/WNV.IC viruses (Fig. 4).
Fig. 4.
Impedance data (mean±sd) demonstrating quantitative cell viability over time (48–120 h p.i.) of WNV, SLEV and WNV/SLEV chimeric viruses on Vero cells.
In vitro vertebrate cell growth
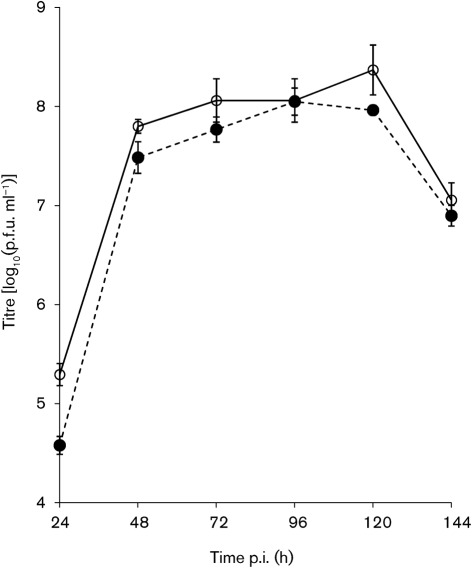
Previous work has demonstrated high growth rates of WNV in avian hosts and a rapid induction of cytopathic effect (CPE) in vertebrate cells. The validity of using vertebrate cells for assessing the genetic determinants associated with altered vertebrate replication and CPE was evaluated in vitro using growth profiles of the parental strains and chimeric viruses in avian [duck embryonic fibroblast; DEF (ATCC CCL-141)] and mammalian (Vero) cell lines. Preliminarily, the SLEV clone-derived virus was compared with the IMP115 strain in DEF cells and the growth patterns were indistinguishable (Fig. 5); SLEV.IC was therefore used as a surrogate for IMP115 for all subsequent in vitro growth comparisons.
Fig. 5.
Geometric mean titres (±sd) for growth of the parental SLEV strain IMP115 (○) and the SLEV infectious clone-derived virus, SLEV.IC (•), in DEF cells. The limit of detection was 1.7 log10(p.f.u. ml−1).
DEF cells.
The WNV.IC virus grew to an approximately 100-fold higher peak titre than the SLEV.IC virus in DEF cells. The WNV.IC virus titre peaked at 144 h p.i. at 8.5±0.2 log10(p.f.u. ml−1), significantly greater (P<0.05) than the peak SLEV.IC titre of 7.3±0.5 p.f.u. ml−1 occurring at 72 h p.i. (Fig. 6a). The mean peak titre for WNV.IC virus was not statistically different (P>0.05) from the mean peak titre for SLEV-prME/WNV.IC. Similarly, the peak titre of 7.2±0.6 log10(p.f.u. ml−1) for the WNV-prME/SLEV.IC chimeric virus at 72 h p.i. was indistinguishable (P>0.05) from that of the SLEV.IC virus. Growth patterns for both viruses containing the SLEV NS proteins plateaued at 72 h p.i., whereas the viruses that expressed the WNV NS proteins exhibited elevated growth until 96–144 h p.i. (Fig. 6a). A repeated-measures ANOVA for viruses over all time points followed by a Tukey–Kramer multiple comparison test demonstrated significant differences (P<0.05) in overall mean titres between the SLEV.IC and SLEV-prME/WNV.IC viruses, but failed to identify differences between the SLEV.IC and the chimeric virus with the homologous NS gene region, WNV-prME/SLEV.IC.
Fig. 6.
Geometric mean titres (±sd) of virus growth in vertebrate cell lines: (a) DEF; (b) Vero cells. Viruses used were WNV.IC (▴), SLEV-prME/WNV.IC (○), SLEV.IC (▪) and WNV-prME/SLEV.IC (◊). The limit of detection was 1.7 log10(p.f.u. ml−1).
Vero cells.
WNV.IC virus grew to a peak titre of 9.4±0.1 log10(p.f.u. ml−1) by 48 h p.i., which was significantly greater (P<0.05) than the SLEV-prME/WNV.IC virus titre of 8.8±0.1 log10(p.f.u. ml−1) that also peaked at 48 h p.i. (Fig. 6b). In contrast, the SLEV.IC and WNV-prME/SLEV.IC viruses demonstrated a lag in growth, both peaking at 96 h p.i. with statistically similar (P>0.05) titres of 8.2±0.1 log10(p.f.u. ml−1) and 8.7±0.1log10(p.f.u. ml−1), respectively (Fig. 6b). A repeated-measures ANOVA for viruses over all time points followed by a Tukey–Kramer multiple comparison test confirmed that there were statistically significant differences in mean titres among all viruses (P<0.05) and that the temporal pattern of virus growth varied among viruses because the virus×time interaction term was highly significant (P<0.0001). Nevertheless, the trend for the overall mean viraemia as a function of time indicated that the overall mean titres for viruses with the NS genetic elements of SLEV were significantly lower than those for viruses containing the NS genes of WNV.
In vitro invertebrate cell growth
Low oral-infection thresholds have been described previously for SLEV in Culex mosquitoes (Hardy & Reeves, 1990). To assess whether vector-competence differences between SLEV and WNV could be recapitulated in vitro and used to gauge potential genetic determinants of mosquito infection, growth phenotypes of the parental and chimeric viruses were characterized in C6/36 (Aedes albopictus) and more biologically relevant CT (Chao & Ball, 1976; Main et al., 1977) cell lines.
C6/36 cells.
Overall, the mean peak titre for the WNV.IC virus was significantly higher (P<0.05) than that of the SLEV.IC virus (Fig. 7a) in C6/36 cells. The chimeric WNV-prME/SLEV.IC virus, peaking at 8.0±0.2 log10(p.f.u. ml−1) at 144 h p.i., grew to statistically indistinguishable peak titres (P>0.05) from both SLEV.IC and WNV.IC. The SLEV.IC virus peaked at 6.9±0.1 log10(p.f.u. ml−1) at 144 h p.i., statistically lower (P<0.05) than SLEV-prME/WNV.IC, which produced a peak titre of 9.0±0.1 log10(p.f.u. ml−1) at the same time point (Fig. 7a). In the repeated-measures ANOVA, overall mean titre for the SLEV.IC virus was significantly lower (P<0.05) than those for the other three viruses, and no differences were observed between overall means among WNV.IC, WNV-prME/SLEV.IC and SLEV-prME/WNV.IC viruses.
Fig. 7.
Geometric mean titres (±sd) of virus growth in mosquito cell lines: (a) C6/36 cells; (b) CT cells. Viruses used were WNV.IC (▴), SLEV-prME/WNV.IC (○), SLEV.IC (▪) and WNV-prME/SLEV.IC (◊). The limit of detection was 1.7 log10(p.f.u. ml−1).
CT cells.
No statistically significant differences were observed for the mean peak titres among the four viruses. The WNV.IC virus peaked at 8.2±0.2 log10(p.f.u. ml−1) by 168 h p.i. (Fig. 7b), whilst the WNV-prME/SLEV.IC chimeric virus peaked at 8.1±0.2 log10(p.f.u. ml−1) at 168 h p.i. The SLEV-prME/WNV.IC and SLEV.IC viruses exhibited peak titres of 8.2±0.2 log10(p.f.u. ml−1) and 8.0±0.2 log10(p.f.u. ml−1) at 120 h p.i., respectively (Fig. 7b). Using a repeated-measures ANOVA followed by a Tukey–Kramer multiple comparison test, overall mean titres of WNV.IC and WNV-prME/SLEV.IC viruses were statistically indistinguishable from one another and were significantly greater (P<0.05) than those for SLEV.IC and SLEV-prME/WNV.IC; these means varied differently over time, as indicated by the significant (P<0.001) virus by time interaction term.
Discussion
WNV and SLEV serve as an evolutionary model for two viruses that have developed contrasting adaptive strategies for maintenance in similar enzootic transmission cycles. WNV, eliciting high viraemia in large numbers of avian hosts (Komar et al., 2003), relies on elevated viraemia in the imbibed bloodmeal for infection of mosquito midgut epithelium. Ingestion of a 1 µl bloodmeal containing 108 p.f.u. (ml blood)−1 would expose each epithelial cell to approximately 10 virus particles, based on an estimated 104 mesenteric cells in an average mosquito midgut (Houk et al., 1990), potentially reducing selective pressure for maintenance of high oral susceptibility and allowing infection of a wide variety of marginally competent vectors. Interestingly, WNV has demonstrated adaptation to North American mosquitoes, with increased dissemination capacity being observed with the emergent WN02 genotype (Moudy et al., 2007), demonstrating that, despite high avian viraemogenesis, evidence exists for the presence of selective forces for increased mosquito transmissibility. In contrast, SLEV appears to depend upon a specific association between the virus and a select number of highly susceptible Culex vectors, to which the virus has adapted to maintain infectivity at low oral doses; the long history of SLEV with North American vectors would support the potential for such co-adaptation (Auguste et al., 2009; Baillie et al., 2008). High oral infectivity is probably a by-product of the considerably lower avian viraemias observed following infection with different SLEV strains (Reisen et al., 2004, 2005). Although SLEV viraemias of nestlings have been observed to approach titres commonly observed in adult birds infected with WNV (Mahmood et al., 2004), the evolutionary significance of these temporally and host-restricted viraemias as a selective factor remains unresolved. Fitness constraints imposed by the two-host transmission cycles of these viruses have probably resulted in compromised fitness in both vector and host (Ciota et al., 2009; Coffey et al., 2008). This study was designed to identify specific genetic elements that dictate differential avian- and mosquito-infection phenotypes observed between these viruses; however, future studies are warranted to assess fitness trade-offs and their relative effect on driving the contrasting evolutionary strategies of WNV and SLEV. For instance, assessing the effects of high oral infectivity and avian viraemogenesis of WNV could shed light on vector-infection restrictions that have resulted in adaptations for higher dissemination rates in North American Culex mosquitoes (Moudy et al., 2007) as opposed to higher midgut infectivity, as has been observed for SLEV (Reisen et al., 2008a).
Specific genetic differences between WNV and SLEV have not been implicated in differential cytopathogenicity or the aforementioned dissimilar vector-infectivity and avian-replication/virulence phenotypes. Chimerization of flaviviruses has previously been used for the identification of genetic determinants of virulence and for functional vaccine development (Chambers et al., 1999; Charlier et al., 2010; Huang et al., 2000; Komar et al., 2009; Pletnev & Men, 1998; Pletnev et al., 1992; Pripuzova et al., 2009). Incorporation of the prME genes from tick-borne encephalitis virus into a dengue virus 4 (DENV-4) backbone restricted entry and subsequent RNA replication of the resultant chimera in C6/36 cells (Pletnev et al., 1992), supporting the notion that structural genetic elements play a role in modulating mosquito infectivity. However, chimeras replacing prME genes from DENV-4 with Langat virus prME were viable in C6/36 cells, whereas the parental Langat virus was unable to grow in mosquito cells (Pletnev & Men, 1998). Additionally, a mutant virus generated with the prME genes from WNV inserted into the same DENV backbone failed to infect Cx. tarsalis mosquitoes, indicating that NS genetic determinants also play a role determining vector infectivity (Hanley et al., 2005). Inclusion of NS2A/NS4B genes of the 17D yellow fever vaccine strain into the wild-type Asibi virus has previously demonstrated significant dissemination impairment in A. aegypti (McElroy et al., 2006a). ChimeriVax-WNV (Johnson et al., 2003) and DENV-1–4 tetravalent (Johnson et al., 2004) viruses demonstrated that NS genes mediated in vitro mosquito replication, and specific NS genetic determinants have been identified that restrict in vivo replication of a PDK-53 DENV-2 vaccine candidate (Brault et al., 2011). Replacement of the prME genes of YFV-17D/DENV-2 with the prME genes of Modoc virus, a virus incapable of infecting mosquito cells, failed to reduce growth of the chimera significantly in comparison with YFV-17D and DENV-2 in C6/36 cells, also indicating that genetic elements exclusive of the prME genes dictate replication in mosquito cells (Charlier et al., 2010). Inclusion of the variable region of the 3′-UTR from tick-borne Langat virus in a DENV-4 genetic backbone demonstrated restricted in vitro vertebrate cell growth, but had no distinguishable effect on growth in mosquito cells, indicating the differential effect of specific mutations on host-specificity phenotypes (Tumban et al., 2011). Studies performed with yellow fever virus structural and NS chimeras indicate a potential cumulative role of both gene regions to restrict in vivo mosquito infectivity and dissemination, respectively (McElroy et al., 2006a, b). A similar multigenic scenario could encode enhanced infectivity of SLEV in Culex mosquito vectors, with structural genes governing receptor-mediated virus entry and NS genes modulating intracellular replication through mosquito cell-specific RNA interference (RNAi) antagonism (Keene et al., 2004).
Higher peak growth titres were observed for the parental WNV than for SLEV in C6/36 cells, but no statistical differences in peak titres were observed in CT cells. Although the overall mean titre analyses implicated the prME structural proteins with defining CT-cell growth phenotypes, both structural and NS gene regions were associated with modulation of C6/36 growth. Although differences were observed between WNV and SLEV growth patterns in both mosquito cell types, WNV consistently grew to higher titre in both mosquito cell lines. This finding somewhat contradicts the notion that SLEV would be capable of replicating more efficiently in mosquitoes than WNV, as would be expected based on higher observed SLEV in vivo infection rates (Reisen et al., 2005). The finding that C6/36 cells lack a functional RNAi pathway could dictate such a discordance between mosquito in vitro and in vivo infection models (Brackney et al., 2010), as could differences in virus internalization predicated by altered receptor expression in non-differentiated cell-culture progenitor cultured cell types. For instance, SLEV could have evolved specific structural protein motifs to bind specifically to Culex midgut epithelial-cell receptors that are not present in cultured mosquito cells derived from eggs. Delayed uptake of viral RNA and consequent lag in viral RNA replication in C6/36 cells could have resulted in lower peak titres due to SLEV structural proteins limiting entry into these cells. Once the cells were infected, enhanced intracellular replication could have been afforded by the presence of WNV NS genes in the absence of a functional RNAi response. This could explain the observed enhanced growth of the SLEV-prME/WNV.IC chimeric virus compared with SLEV.IC virus in C6/36 cells. It is intriguing that, in this study, overall growth patterns were determined to be dictated by prME gene regions for CT cells, a cell line derived from a relevant enzootic SLEV vector, although viruses expressing the SLEV prME exhibited lower growth levels than the WNV prME-expressing viruses. Virus entry and replicative capacity in cultured cells may not be reflective of the physiological environment that modulates a virus’s ability to traverse mosquito-midgut and salivary-gland barriers (Ciota et al., 2007). Further studies in vivo will be needed to determine the influence of flavivirus genetic components on oral infection thresholds and tissue tropisms in mosquitoes.
Plaque-morphology and impedance data indicated that NS genes influenced cytopathogenicity profiles for WNV and SLEV; however, because the chimeric viruses maintained homologous capsid proteins, the role that this protein serves in eliciting differential cytopathogenicity cannot be excluded. WNV exhibited CPE more rapidly than SLEV on Vero cells, as measured by the timing of plaque formation and the decreased cellular impedance. Several flaviviral replicon systems have identified mutations in the NS1, NS2A, NS3, NS4B and NS5 genes associated with diminished capacities for eliciting CPE, suggesting that a variety of NS genetic differences between WNV and SLEV could be responsible for the alternative cytopathogenicity phenotypes observed (Liu et al., 2004; Rossi et al., 2007).
WNV grows to high titres in competent avian hosts and causes variable mortality, dependent upon the relative susceptibility of the avian host species (Brault et al., 2004; Langevin et al., 2005); SLEV, in contrast, generates low virus titres in birds and has not been associated with avian mortality (Reisen et al., 2003, 2004, 2005, 2008b). In DEF cells, the WNV-prME/SLEV.IC chimeric virus exhibited a similar growth profile to that of SLEV.IC, whereas SLEV-prME/WNV.IC demonstrated a comparable profile to WNV.IC. These data, in contrast to our finding that structural elements dictate invertebrate-cell growth differences, indicated that genetic elements exclusive of the prME gene region of WNV mediate increased virus replication in DEF cells. Previous studies using infectious cDNA clones of WNV strains with different avian-pathogenicity phenotypes (Brault et al., 2004) implicated a single NS viral helicase mutation in altered viraemogenic potential and mortality in American crows (Brault et al., 2007). These reports, coupled with the findings herein, demonstrate that genetic elements exclusive of the prME genes govern replicative fitness of WNV/SLEV chimeras in an avian cell line. Once again, the role of the capsid protein cannot be excluded, due to the chimerization of only the prME gene regions. The finding that the capsid protein of WNV is capable of serving as an apoptosis agonist (Yang et al., 2008) as well as a cellular serine/threonine phosphatase inhibitor (Hunt et al., 2007) could indicate the importance of the capsid for increased vertebrate pathogenesis of WNV; however, avian-inoculation experiments performed with identical WNV genetic backbones differing by single NS amino acid substitutions have demonstrated dramatic differences in avian virulence, indicating that such virulence variation is not associated with capsid polymorphisms between WNV strains (Brault et al., 2007).
Single point mutations in the NS4B and NS5 regions of WNV have demonstrated altered plaque-morphology, murine-neuroinvasiveness and temperature-sensitivity phenotypes (Wicker et al., 2006). The NS1 protein may also modulate the interferon (IFN) response by inhibiting signal transduction of the TLR3 receptor in addition to inhibiting complement activation by binding to regulatory factors H, C4 and C5 (Avirutnan et al., 2010; Chung et al., 2006; Wilson et al., 2008). Incorporation of an A30P mutation in the NS2A protein of Kunjin virus was shown to disable IFN-α/β induction (Liu et al., 2006), and the NS5 protein of WNV has been determined experimentally to effectively suppress IFN-mediated gene expression and inhibit IFN-α/β induction of the JAK/STAT pathway (Laurent-Rolle et al., 2010).
This study has ascertained that genetic regions exclusive of the prME genes are influential in flavivirus in vitro replication in mammalian and avian cell types, whereas flaviviral structural genetic elements modulate virus growth in mosquito (CT) cells. These data could potentially explain the disparate virological and epidemiological host and vector profiles exhibited by WNV and SLEV. However, the potential that in vitro models do not reflect in vivo physiological factors that might dictate variability in vector infectivity of differentiated midgut epithelial cells or viraemogenesis in an avian host will necessitate further in vivo characterizations of these parental and chimeric viruses in biologically relevant mosquito and avian species. The close genetic identity between WNV and SLEV (71.5 % amino acid identity for the prME genes) makes this an intriguing model for the elucidation of specific genetic determinants associated with the structural or NS genes for modulation of vector infectivity, as well as extra-prME determinants for modulation of vertebrate viraemogenesis. Future studies that utilize these viruses and further refined chimeras to assess specific genetic motifs for altered vertebrate pathogenesis and midgut epithelial infection of reservoir avian species and mosquito vectors will be necessary to assess more completely the hypothesis that WNV and SLEV have evolved differential strategies for enzootic maintenance. Identification of flaviviral genetic determinants of vertebrate host tropism and mosquito infectivity will provide a wealth of information on the evolutionary history of these viruses, allow for potential risk assessments of the emergence of viruses with altered epidemiological patterns and provide insight into novel control methodologies.
Methods
SLEV infectious cDNA.
SLEV strain IMP115, isolated from a pool of Cx. tarsalis mosquitoes from Imperial Valley, CA, USA, in 2003 with one Vero cell passage, was sequenced in its entirety and used for generation of an SLEV infectious cDNA. At least 2 µg total nucleic acids was used for RT-PCR. The genome of the IMP115 SLEV strain was amplified in six overlapping fragments spanning appropriate unique restriction sites. Viral cDNAs were synthesized from RNA by using reverse transcriptase according to the manufacturer’s recommendations (SuperScript II; Invitrogen). Amplicons (primer sequences available as Supplementary Table S1 in JGV Online) were generated by using the high-fidelity Pfu Ultra Hotstart polymerase (Stratagene) and products were cloned into the pCR-XL-TOPO vector (Invitrogen) and sequenced directly. In order to clone viral cDNAs into the final constructions, a specific low-copy-number, kanamycin-resistant vector pMA3, with convenient unique restriction sites, was created (sequence available upon request). Due to bacterial toxicity/stability issues encountered, a two-plasmid cloning strategy was utilized similar to that reported previously (Kinney et al., 2006). The first amplicon (nt 1–2012 of the 5′ end of the SLEV genome) was cloned into a separate 5′ (68M) plasmid, whereas the remaining five amplicons were engineered sequentially into a 3′ (79M) plasmid.
Constructing the SLEV prME/WNV.IC chimera.
Internal BamHI and DraIII restriction sites at genomic nucleotide positions 466 and 2472 within the prME coding region of the 68M plasmid described above were ablated by mutagenesis. The mutated 68M plasmid was used as a template for generation of an SLEV prME region with 5′ and 3′ flanking restriction sites for incorporation into the 5′ WNV plasmid. Primers utilized for the generation of this amplicon were designed to contain BamHI and DraIII restriction sites at the 5′ and 3′ ends of the SLEV prME fragment. Amplicons were generated by using the high-fidelity Pfu Ultra Hotstart polymerase (Stratagene), and the subsequent BamHI–SLEV prME–DraIII fragments were inserted in a pCR-XL-TOPO vector (Invitrogen). A BamHI restriction site was created upstream of the prM gene of the 5′ WNV receptor plasmid [the first plasmid of the bipartite system for strain NY99 (Kinney et al., 2006)]. The (BamHI–prME–DraIII) fragment was cloned directly into the 5′ WNV plasmid with the newly created BamHI and existing DraIII sites in the 5′ WNV plasmid. The BamHI site was removed from the final clone by site-directed mutagenesis to restore the native amino acid identity upstream of the prM gene in the chimeric plasmid. This modified chimeric plasmid containing the SLEV prME genes was subsequently used for in vitro ligation with the WNV 3′ plasmid, for generation of SLEV prME/WNV.IC virus described below.
Constructing the WNV prME/SLEV.IC chimera.
The prME gene region of WNV was amplified from the 5′ WNV plasmid with flanking primers containing the BglII and BssHII restriction sequences and the product was subcloned into a pCR-XL-TOPO vector. The 68M SLEV 5′ plasmid was modified to contain an additional BglII site at nt 462 for direct incorporation of the WNV prME BglII/BssHII amplicon. The presence of the AgeI in vitro ligation site used for the generation of the recombinant parental SLEV at the C terminus of the E protein necessitated the incorporation of 126 nt of SLEV sequence from the 5′ portion of the SLEV plasmid 79M at the 3′ end of plasmid 68M for inclusion of the complete prME gene sequences. To allow for inclusion of this fragment into the chimeric WNV prME 5′ plasmid, a NotI site was incorporated by mutagenesis after the last viral nucleotide. Direct PCR amplification of the 126 nt SLEV fragment from plasmid 79M with corresponding flanking NotI and BssHII sites at nt 10940 and 2152 and subsequent ligation into the modified 68M plasmid were performed. The BglII and NotI sites were removed from the modified 68M plasmid and the corresponding 126 nt sequence was removed from the 79M plasmid by mutagenesis. This cloning process ablated the AgeI restriction site used for in vitro ligation of the SLEV 5′ and 3′ plasmids and, as such, necessitated the use of the introduced BssHII site for in vitro ligation.
Rescue of viruses from infectious cDNAs.
For generation of virus from the SLEV infectious cDNA, plasmids 68M and 79M were digested with SphI and NotI, respectively, treated with calf intestinal alkaline phosphatase (New England Biolabs), purified over spin columns (QIAquick PCR Purification kit; Qiagen) and digested with AgeI. Ligation was performed at the AgeI site for generation of in vitro transcription template. The AgeI restriction site was also used for in vitro ligation of the chimeric SLEV prME 5′ plasmid with the WNV 3′ plasmid. Ligation of the WNV prME 5′ plasmid with the SLEV 3′ plasmid was performed with an engineered BssHII site.
Following in vitro ligation, cDNAs were treated with proteinase K, extracted with phenol : chloroform : isoamyl alcohol (25 : 24 : 1; Amresco) and chloroform, and ethanol-precipitated. Viral genomic RNA was transcribed for 2 h at 37 °C (AmpliScribe T7 Transcription kit; Epicentre). Reactions included 6 mM m7G-(5′)ppp-(5′)A cap analogue (New England Biolabs), 20 % of the manufacturer-recommended concentration of ATP in order to facilitate incorporation of the cap analogue, and in vitro-ligated template cDNA. Transcribed RNA was treated with DNase I (Epicentre), extracted with phenol : chloroform : isoamyl alcohol and chloroform, and precipitated with ethanol. RNA pellets were resuspended in 20 µl RNase-free dH2O and 2 µl aliquots were analysed by non-denaturing agarose gel electrophoresis to visualize genome-length RNA transcripts. Transcribed RNA was transfected by electroporation into C6/36 cells (Kinney et al., 1997) in six-well plates using a Petri pulser (ECM 630; BTX). Supernatants were harvested 7 days post-transfection and RT-PCR and plaque assay on Vero cells were performed to confirm the presence of viral RNA and infectious virus, respectively.
Similarly, chimeric SLEV-prME/WNV.IC and WNV-prME/SLEV.IC viruses were recovered from in vitro ligation of the modified 5′ plasmids containing the reciprocal prME genes with either the 3′ plasmids of WNV or SLEV. All rescued viruses were sequenced fully to ensure complete sequence identity with parental strains and to ensure the exclusion of any spurious mutations that might have occurred during the cloning, assembly or rescuing processes.
IFA.
IFA was performed with the parental and clone-derived chimeric viruses. Briefly, confluent chamber-well slides of Vero cells were inoculated individually with parental or chimeric viruses at an m.o.i. of 10. Medium was removed 24 h p.i. and cells were scraped and added to each well of a 12-well spot slide. The slides were dried for 30 min and fixed with an acetone : DPBS (3 : 1) mixture. Slides were washed and incubated with a 1 : 400 dilution of either WNV-specific (3.67) or SLEV-specific (4A4C-4) mAbs (CDC). The slides were incubated in a humid chamber at 37 °C for 1 h, washed and incubated similarly for 1 h with a goat anti-mouse FITC-conjugated antibody (Fisher Scientific) at a dilution of 1 : 20 with (2 %) Evans’ blue used as a counterstain. Following DPBS washes, 1,4-diazabicyclo-octane (DABCO, anti-fade solution) was added to each well and coverslips were added to each slide and scored by fluorescence microscopy.
Growth-profile experiments.
C6/36, CT (Chao & Ball, 1976), DEF (ATCC CCL-141) and Vero cells were utilized for in vitro growth-profile assays. C6/36 and Vero cells were maintained at 28 and 37 °C, respectively, in Dulbecco’s modified Eagle’s medium (Gibco-BRL) containing 5 % FBS (Hyclone), penicillin (100 U ml–1) and streptomycin (100 µg ml–1). CT and DEF cells were maintained in Schneider’s Drosophila media (Lonza BioWhittaker) and Eagle’s MEM containing 5 % FBS and penicillin (100 U ml–1) and streptomycin (100 µg ml−1) at 28 and 37 °C, respectively. All cells were grown to 90 % confluence in triplicate 25 cm2 flasks prior to inoculation with virus at an m.o.i. of 0.1. Virus was absorbed for 1 h at 37 °C, after which cells were washed twice with DPBS and fresh medium was added. Aliquots (50 µl) of supernatant were collected daily for 7 days p.i., added to 450 µl MEM containing 20 % FBS and stored at −80 °C until titrated by Vero-cell plaque assay for quantification and plaque-morphology (3 and 7 days p.i.) assessment. Each growth-profile experiment for all cell types was replicated to assure the reproducibility of the reported findings.
Cell-viability measurements.
A real-time cell-analysis system using electrical impedance (formerly RT-CES System; ACEA Biosciences) was utilized for quantification of CPE elicited by the respective viruses as described previously (Fang et al., 2011). Impedance was expressed as a dimensionless parameter, the cell index, representing cell status based on the change of impedance of cells relative to cell-negative control wells. Initial cell concentration was measured, and decreasing impedance related to cell death was plotted over time. Vero cells were trypsinized, washed twice with DPBS, added to wells and infected at an m.o.i. of 0.1 (six wells per virus). Real-time impedance values were measured every 15 min until day 5 p.i.
Statistical analysis.
A repeated-measures ANOVA statistically compared plaque sizes, impedance values at selected time points, and growth-profile data over time among viruses, with viral means grouped by an a posteriori Tukey–Kramer multiple comparison test with α = 0.05 (ncss statistical software). Peak titres for specific time points were compared in a mixed-model ANOVA. Both viruses and time points were treated as fixed effects throughout.
Supplementary tables
Acknowledgements
Funding for these studies was provided by the Pacific Southwest Regional Center for Excellence (AI065359) and by grants from NIH (AI061822, AI55607), CDC (CI000235) and the University of California Mosquito Research Program. We would like to thank Jason Velez for providing cell-culture support for these studies.
Footnotes
A supplementary table is available with the online version of this paper.
References
- Auguste A. J., Pybus O. G., Carrington C. V. (2009). Evolution and dispersal of St. Louis encephalitis virus in the Americas. Infect Genet Evol 9, 709–715 10.1016/j.meegid.2008.07.006 [DOI] [PubMed] [Google Scholar]
- Avirutnan P., Fuchs A., Hauhart R. E., Somnuke P., Youn S., Diamond M. S., Atkinson J. P. (2010). Antagonism of the complement component C4 by flavivirus nonstructural protein NS1. J Exp Med 207, 793–806 10.1084/jem.20092545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie G. J., Kolokotronis S. O., Waltari E., Maffei J. G., Kramer L. D., Perkins S. L. (2008). Phylogenetic and evolutionary analyses of St. Louis encephalitis virus genomes. Mol Phylogenet Evol 47, 717–728 10.1016/j.ympev.2008.02.015 [DOI] [PubMed] [Google Scholar]
- Brackney D. E., Scott J. C., Sagawa F., Woodward J. E., Miller N. A., Schilkey F. D., Mudge J., Wilusz J., Olson K. E. & other authors (2010). C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl Trop Dis 4, e856 10.1371/journal.pntd.0000856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault A. C., Langevin S. A., Bowen R. A., Panella N. A., Biggerstaff B. J., Miller B. R., Komar N. (2004). Differential virulence of West Nile strains for American crows. Emerg Infect Dis 10, 2161–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault A. C., Huang C. Y., Langevin S. A., Kinney R. M., Bowen R. A., Ramey W. N., Panella N. A., Holmes E. C., Powers A. M., Miller B. R. (2007). A single positively selected West Nile viral mutation confers increased virogenesis in American crows. Nat Genet 39, 1162–1166 10.1038/ng2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault A. C., Kinney R. M., Maharaj P. D., Green E. N. G., Reisen W. K., Huang C. H. (2011). Replication of the PDK-53 dengue 2 virus vaccine candidate in Aedes aegypti is modulated by a mutation in the 5′ untranslated region and amino acid substitutions in nonstructural proteins 1 and 3. Vector Borne Zoonotic Dis 11, 683–689 10.1089/vbz.2010.0150 [DOI] [PubMed] [Google Scholar]
- Chambers T. J., Nestorowicz A., Mason P. W., Rice C. M. (1999). Yellow fever/Japanese encephalitis chimeric viruses: construction and biological properties. J Virol 73, 3095–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao J., Ball G. H. (1976). A comparison of amino acid utilization by cell lines of Culex tarsalis and Culex pipiens. In Invertebrate Tissue Culture: Applications in Medicine, Biology and Agriculture, pp. 263–266 Edited by Kurstak E., Maramorosch K. Academic Press: New York [Google Scholar]
- Charlier N., Davidson A., Dallmeier K., Molenkamp R., De Clercq E., Neyts J. (2010). Replication of not-known-vector flaviviruses in mosquito cells is restricted by intracellular host factors rather than by the viral envelope proteins. J Gen Virol 91, 1693–1697 10.1099/vir.0.019851-0 [DOI] [PubMed] [Google Scholar]
- Chung K. M., Liszewski M. K., Nybakken G., Davis A. E., Townsend R. R., Fremont D. H., Atkinson J. P., Diamond M. S. (2006). West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory protein factor H. Proc Natl Acad Sci U S A 103, 19111–19116 10.1073/pnas.0605668103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciota A. T., Lovelace A. O., Jones S. A., Payne A., Kramer L. D. (2007). Adaptation of two flaviviruses results in differences in genetic heterogeneity and virus adaptability. J Gen Virol 88, 2398–2406 10.1099/vir.0.83061-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciota A. T., Jia Y., Payne A. F., Jerzak G., Davis L. J., Young D. S., Ehrbar D., Kramer L. D. (2009). Experimental passage of St. Louis encephalitis virus in vivo in mosquitoes and chickens reveals evolutionarily significant virus characteristics. PLoS One 4, e7876 10.1371/journal.pone.0007876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey L. L., Vasilakis N., Brault A. C., Powers A. M., Tripet F., Weaver S. C. (2008). Arbovirus evolution in vivo is constrained by host alternation. Proc Natl Acad Sci U S A 105, 6970–6975 10.1073/pnas.0712130105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Ye P., Wang X., Xu X., Reisen W. (2011). Real-time monitoring of flavivirus induced cytopathogenesis using cell electric impedance technology. J Virol Methods 173, 251–258 10.1016/j.jviromet.2011.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard L. B., Roth A. E., Reisen W. K., Scott T. W. (2002). Vector competence of California mosquitoes for West Nile virus. Emerg Infect Dis 8, 1385–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley K. A., Goddard L. B., Gilmore L. E., Scott T. W., Speicher J., Murphy B. R., Pletnev A. G. (2005). Infectivity of West Nile/dengue chimeric viruses for West Nile and dengue mosquito vectors. Vector Borne Zoonotic Dis 5, 1–10 10.1089/vbz.2005.5.1 [DOI] [PubMed] [Google Scholar]
- Hardy J. L., Reeves W. C. (1990). Experimental studies on infection in vectors. In Epidemiology and Control of Mosquito-Borne Arboviruses in California, 1943–1987, pp. 145–250 Edited by Reeves W. C. Sacramento, CA: California Mosquito and Vector Control Association [Google Scholar]
- Houk E. J., Arcus Y. M., Hardy J. L., Kramer L. D. (1990). Binding of western equine encephalomyelitis virus to brush border fragments isolated from mesenteronal epithelial cells of mosquitoes. Virus Res 17, 105–117 10.1016/0168-1702(90)90072-J [DOI] [PubMed] [Google Scholar]
- Huang C. Y.-H., Butrapet S., Pierro D. J., Chang G.-J. J., Hunt A. R., Bhamarapravati N., Gubler D. J., Kinney R. M. (2000). Chimeric dengue type 2 (vaccine strain PDK-53)/dengue type 1 virus as a potential candidate dengue type 1 virus vaccine. J Virol 74, 3020–3028 10.1128/JVI.74.7.3020-3028.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T. A., Urbanowski M. D., Kakani K., Law L. M., Brinton M. A., Hobman T. C. (2007). Interactions between the West Nile virus capsid protein and the host cell-encoded phosphatase inhibitor, I2PP2A. Cell Microbiol 9, 2756–2766 10.1111/j.1462-5822.2007.01046.x [DOI] [PubMed] [Google Scholar]
- Johnson B. W., Chambers T. V., Crabtree M. B., Arroyo J., Monath T. P., Miller B. R. (2003). Growth characteristics of the veterinary vaccine candidate ChimeriVax-West Nile (WN) virus in Aedes and Culex mosquitoes. Med Vet Entomol 17, 235–243 10.1046/j.1365-2915.2003.00438.x [DOI] [PubMed] [Google Scholar]
- Johnson B. W., Chambers T. V., Crabtree M. B., Guirakhoo F., Monath T. P., Miller B. R. (2004). Analysis of the replication kinetics of the ChimeriVax-DEN 1, 2, 3, 4 tetravalent virus mixture in Aedes aegypti by real-time reverse transcriptase-polymerase chain reaction. Am J Trop Med Hyg 70, 89–97 [PubMed] [Google Scholar]
- Keene K. M., Foy B. D., Sanchez-Vargas I., Beaty B. J., Blair C. D., Olson K. E. (2004). RNA interference acts as a natural antiviral response to O’nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc Natl Acad Sci U S A 101, 17240–17245 10.1073/pnas.0406983101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney R. M., Butrapet S., Chang G.-J. J., Tsuchiya K. R., Roehrig J. T., Bhamarapravati N., Gubler D. J. (1997). Construction of infectious cDNA clones for dengue 2 virus: strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology 230, 300–308 10.1006/viro.1997.8500 [DOI] [PubMed] [Google Scholar]
- Kinney R. M., Huang C. Y., Whiteman M. C., Bowen R. A., Langevin S. A., Miller B. R., Brault A. C. (2006). Avian virulence and thermostable replication of the North American strain of West Nile virus. J Gen Virol 87, 3611–3622 10.1099/vir.0.82299-0 [DOI] [PubMed] [Google Scholar]
- Komar N., Langevin S., Hinten S., Nemeth N., Edwards E., Hettler D., Davis B., Bowen R., Bunning M. (2003). Experimental infection of North American birds with the New York 1999 strain of West Nile virus. Emerg Infect Dis 9, 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar N., Langevin S., Monath T. P. (2009). Use of a surrogate chimeric virus to detect West Nile virus-neutralizing antibodies in avian and equine sera. Clin Vaccine Immunol 16, 134–135 10.1128/CVI.00220-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin S. A., Brault A. C., Panella N. A., Bowen R. A., Komar N. (2005). Variation in virulence of West Nile virus strains for house sparrows (Passer domesticus). Am J Trop Med Hyg 72, 99–102 [PubMed] [Google Scholar]
- Laurent-Rolle M., Boer E. F., Lubick K. J., Wolfinbarger J. B., Carmody A. B., Rockx B., Liu W., Ashour J., Shupert W. L. & other authors (2010). The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J Virol 84, 3503–3515 10.1128/JVI.01161-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. J., Chen H. B., Wang X. J., Huang H., Khromykh A. A. (2004). Analysis of adaptive mutations in Kunjin virus replicon RNA reveals a novel role for the flavivirus nonstructural protein NS2A in inhibition of beta interferon promoter-driven transcription. J Virol 78, 12225–12235 10.1128/JVI.78.22.12225-12235.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. J., Wang X. J., Clark D. C., Lobigs M., Hall R. A., Khromykh A. A. (2006). A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice. J Virol 80, 2396–2404 10.1128/JVI.80.5.2396-2404.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood F., Chiles R. E., Fang Y., Barker C. M., Reisen W. K. (2004). Role of nestling mourning doves and house finches as amplifying hosts of St. Louis encephalitis virus. J Med Entomol 41, 965–972 10.1603/0022-2585-41.5.965 [DOI] [PubMed] [Google Scholar]
- Main O. M., Hardy J. L., Reeves W. C. (1977). Growth of arboviruses and other viruses in a continuous line of Culex tarsalis cells. J Med Entomol 14, 107–112 [DOI] [PubMed] [Google Scholar]
- McElroy K. L., Tsetsarkin K. A., Vanlandingham D. L., Higgs S. (2006a). Manipulation of the yellow fever virus non-structural genes 2A and 4B and the 3′non-coding region to evaluate genetic determinants of viral dissemination from the Aedes aegypti midgut. Am J Trop Med Hyg 75, 1158–1164 [PubMed] [Google Scholar]
- McElroy K. L., Tsetsarkin K. A., Vanlandingham D. L., Higgs S. (2006b). Role of the yellow fever virus structural protein genes in viral dissemination from the Aedes aegypti mosquito midgut. J Gen Virol 87, 2993–3001 10.1099/vir.0.82023-0 [DOI] [PubMed] [Google Scholar]
- Moudy R. M., Meola M. A., Morin L. L., Ebel G. D., Kramer L. D. (2007). A newly emergent genotype of West Nile virus is transmitted earlier and more efficiently by Culex mosquitoes. Am J Trop Med Hyg 77, 365–370 [PubMed] [Google Scholar]
- Pletnev A. G., Men R. (1998). Attenuation of the Langat tick-borne flavivirus by chimerization with mosquito-borne flavivirus dengue type 4. Proc Natl Acad Sci U S A 95, 1746–1751 10.1073/pnas.95.4.1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletnev A. G., Bray M., Huggins J., Lai C. J. (1992). Construction and characterization of chimeric tick-borne encephalitis/dengue type 4 viruses. Proc Natl Acad Sci U S A 89, 10532–10536 10.1073/pnas.89.21.10532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pripuzova N. S., Tereshkina N. V., Gmyl L. V., Dzhivanyan T. I., Rumyantsev A. A., Romanova L. Iu., Mustafina A. N., Lashkevich V. A., Karganova G. G. (2009). Safety evaluation of chimeric Langat/dengue 4 flavivirus, a live vaccine candidate against tick-borne encephalitis. J Med Virol 81, 1777–1785 10.1002/jmv.21587 [DOI] [PubMed] [Google Scholar]
- Reisen W. K., Chiles R. E., Green E. N., Fang Y., Mahmood F. (2003). Previous infection protects house finches from re-infection with St. Louis encephalitis virus. J Med Entomol 40, 300–305 10.1603/0022-2585-40.3.300 [DOI] [PubMed] [Google Scholar]
- Reisen W. K., Chiles R., Martinez V., Fang Y., Green E., Clark S. (2004). Effect of dose on house finch infection with western equine encephalomyelitis and St. Louis encephalitis viruses. J Med Entomol 41, 978–981 10.1603/0022-2585-41.5.978 [DOI] [PubMed] [Google Scholar]
- Reisen W. K., Fang Y., Martinez V. M. (2005). Avian host and mosquito (Diptera: Culicidae) vector competence determine the efficiency of West Nile and St. Louis encephalitis virus transmission. J Med Entomol 42, 367–375 10.1603/0022-2585(2005)042[0367:AHAMDC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Reisen W. K., Fang Y., Martinez V. M. (2006). Effects of temperature on the transmission of West Nile virus by Culex tarsalis (Diptera: Culicidae). J Med Entomol 43, 309–317 10.1603/0022-2585(2006)043[0309:EOTOTT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Reisen W. K., Barker C. M., Fang Y., Martinez V. M. (2008a). Does variation in Culex (Diptera: Culicidae) vector competence enable outbreaks of West Nile virus in California? J Med Entomol 45, 1126–1138 10.1603/0022-2585(2008)45[1126:DVICDC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Reisen W. K., Lothrop H. D., Wheeler S. S., Kennsington M., Gutierrez A., Fang Y., Garcia S., Lothrop B. (2008b). Persistent West Nile virus transmission and the apparent displacement St. Louis encephalitis virus in southeastern California, 2003-2006. J Med Entomol 45, 494–508 10.1603/0022-2585(2008)45[494:PWNVTA]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins C. S. (1973). Introduction, spread and present abundance of the house sparrow in North America. Ornithol Monogr 14, 3–9 [Google Scholar]
- Rossi S. L., Fayzulin R., Dewsbury N., Bourne N., Mason P. W. (2007). Mutations in West Nile virus nonstructural proteins that facilitate replicon persistence in vitro attenuate virus replication in vitro and in vivo. Virology 364, 184–195 10.1016/j.virol.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Tumban E., Mitzel D. N., Maes N. E., Hanson C. T., Whitehead S. S., Hanley K. A. (2011). Replacement of the 3′ untranslated variable region of mosquito-borne dengue virus with that of tick-borne Langat virus does not alter vector specificity. J Gen Virol 92, 841–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker J. A., Whiteman M. C., Beasley D. W. C., Davis C. T., Zhang S., Schneider B. S., Higgs S., Kinney R. M., Barrett A. D. T. (2006). A single amino acid substitution in the central portion of the West Nile virus NS4B protein confers a highly attenuated phenotype in mice. Virology 349, 245–253 10.1016/j.virol.2006.03.007 [DOI] [PubMed] [Google Scholar]
- Wilson J. R., de Sessions P. F., Leon M. A., Scholle F. (2008). West Nile virus nonstructural protein 1 inhibits TLR3 signal transduction. J Virol 82, 8262–8271 10.1128/JVI.00226-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M. R., Lee S. R., Oh W., Lee E. W., Yeh J. Y., Nah J. J., Joo Y. S., Shin J., Lee H. W. & other authors (2008). West Nile virus capsid protein induces p53-mediated apoptosis via the sequestration of HDM2 to the nucleolus. Cell Microbiol 10, 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.