Abstract
Rubella virus (RUBV) replicates slowly and to low titre in vertebrate cultured cells, with minimal cytopathology. To determine whether a cellular stress response is induced during such an infection, the formation of Ras-GAP-SH3 domain-binding protein (G3BP)-containing stress granules (SGs) in RUBV-infected cells was examined. Late in infection, accumulation of G3BP granules was detected, albeit in fewer than half of infected cells. Active virus RNA replication was required for induction of these granules, but they were found to differ from SGs induced by arsenite treatment both in composition (they did not uniformly contain other SG proteins, such as PABP and TIA-1) and in resistance to cycloheximide treatment. Thus, bona fide SGs do not appear to be induced during RUBV infection. The distribution of G3BP, either on its own or in granules, did not overlap with that of dsRNA-containing replication complexes, indicating that it played no role in virus RNA synthesis. However, G3BP did co-localize with viral ssRNAs in perinuclear clusters, suggesting an interaction that could possibly be important in a post-replicative role in virus replication, such as encapsidation.
Introduction
Rubella virus (RUBV) is a positive-strand RNA virus with a genome of approximately 10 kb, and belongs to the family Togaviridae (Frey, 1994). RUBV is the sole member of the genus Rubivirus, but has a genetic coding and replication strategy similar to those of members of the genus Alphavirus, the other togavirus genus, which includes Sindbis virus and Semliki Forest virus (SFV), among others. In RUBV-infected cells, viral RNA synthesis is mediated by two non-structural replicase proteins, P150 and P90. These non-structural proteins are produced from a polyprotein precursor, P200 (NH2-P150-P90-COOH), that is translated directly from the 5′-proximal ORF on the genomic RNA and undergoes self-cleavage by way of an embedded protease at the C terminus of P150. After cleavage, P150 and P90 remain in a complex (Forng & Frey, 1995). In the process of RNA synthesis, the input genomic RNA serves as a template for a genome-length negative-strand RNA, which in turn serves as a template for the synthesis of two positive-strand RNAs, the genomic RNA and a subgenomic RNA. The subgenomic RNA contains the 3′-proximal ORF of the genomic RNA and serves as the mRNA for the translation of this ORF, which encodes the virion structural proteins, capsid protein (CP) and envelope glycoproteins E1 and E2 (NH2-CP-E2-E1-COOH), which are processed by host-cell proteases. As is the case for all positive-strand RNA viruses, RUBV RNA replication occurs in replication complexes (RCs), which are associated with cytoplasmic membranes (Kujala et al., 1999; Lee et al., 1992, 1994; Magliano et al., 1998; Matthews et al., 2009).
During stress, as in the case of heat shock, oxidative stress or infection by some viruses, it is generally accepted that cells stop translating proteins through the cap-dependent route and take precautionary measures to endure or resolve harsh stimuli. Stress granules (SGs), large cytoplasmic mRNA–protein aggregates, are formed after translation stalls and polyribosomes disassemble, and serve to silence the translation of mRNAs until stress is removed or the cell dies through apoptosis (Kedersha et al., 1999, 2005; Tourrière et al., 2003). After binding nucleotide sequence elements within mRNA that are exposed upon polysome disassembly, Ras-GAP-SH3 domain-binding protein (G3BP1, herein referred to as G3BP) and T-cell internal antigen-1 (TIA-1) form SGs through self-aggregation (Anderson & Kedersha, 2002; Kedersha & Anderson, 2002; Kedersha et al., 1999; Tourrière et al., 2003). G3BP is an RNA-binding protein involved in RNA metabolism and signal transduction. It was initially found to take part in regulating the Ras-GTPase-activating protein (RasGAP) through an interaction in the SH3 domain of RasGAP (Parker et al., 1996). Recently, however, much attention has been cast on its role in regulating SGs. G3BP has five conserved domains: nuclear transport factor 2-like (NTF2), an acid-rich region, an RNA-recognition motif (RRM), an arginine–glycine-rich box (RGG) and, depending on splicing of its transcript, several PXXP motifs. The numerous domains in G3BP attest to its role as a multifunctional protein (Tocque et al., 1997; Tourrière et al., 2001, 2003). During the stress process, the poly-A-binding protein (PABP) that is associated with the poly-A tail of mRNAs is also sequestered into SGs (Kedersha et al., 2000). After stress is removed, the mRNAs are released for translation on ribosomes or degradation in processing bodies (P-bodies) (Anderson & Kedersha, 2002; Beckham & Parker, 2008; Kedersha & Anderson, 2002).
SGs are believed to play a defensive role against virus infection by downregulating the translation of viral or host mRNAs that encode proteins required for virus replication (Beckham & Parker, 2008); however, it has been suggested that some viruses may take advantage of SG formation as an aggregating point for RC assembly. Whilst numerous viruses, including members of the families Flaviviridae (Emara & Brinton, 2007), Togaviridae (alphaviruses) (McInerney et al., 2005) and Picornaviridae (Mazroui et al., 2006; White et al., 2007), all of which are positive-strand RNA viruses, have been shown to modulate stress proteins, little is actually known about the role of the cellular stress response during virus infection. Emerging evidence points to the involvement of G3BP in the replication of alphaviruses through interactions with the non-structural proteins (Cristea et al., 2010; Gorchakov et al., 2008), and thus it may also play a similar role in the replication of RUBV.
In contrast to the alphaviruses, which replicate rapidly, cytolytically and to high titre in vertebrate cells, with shutdown of both host transcription and translation, RUBV replicates slowly and to low titre with minimal cytopathology (Frey, 1994). The purpose of this study was to determine whether RUBV induces a stress response in infected cells, as detected by the generation of G3BP-containing SGs. Concomitantly, we were interested in gaining insight into any role that G3BP SGs might play in RUBV replication. This study shows that, during late infection [48 h post-infection (p.i.)], RUBV induces the formation of G3BP granules, which were, however, distinct from bona fide SGs induced by arsenite treatment. Although these granules were formed concomitantly with active viral RNA synthesis and non-structural protein accumulation, the distributions of neither the granules nor G3BP on its own overlapped with the distribution of RCs, indicating that G3BP, in either form, was not involved directly in virus RNA synthesis. However, G3BP did occasionally co-localize with viral ssRNA and P150, suggesting a possible post-synthetic role in virus RNA replication.
Results
RUBV induces the formation of G3BP granules compositionally distinct from SGs
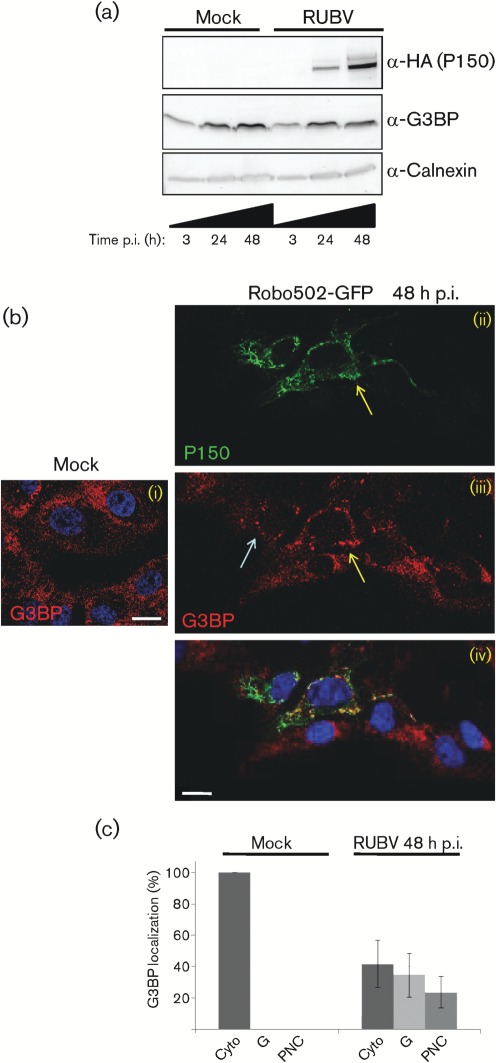
The levels of the cellular stress protein G3BP were measured and compared between mock- and RUBV-infected cells during a time-course of 3–48 h p.i.. Western blotting of cellular lysates revealed no differences in G3BP levels between mock-infected cells and cells infected with Robo502/P150-HA, a virus expressing a haemagglutinin (HA)-tagged P150 (m.o.i. = 3), at 3 or 24 h p.i. (Fig. 1a). However, compared with mock-infected cell lysates, those from Robo502/P150-HA-infected cells showed a small decrease in G3BP levels by 48 h p.i. (Fig. 1a). There were no differences in the subcellular location of G3BP between mock- and Robo502/P150-GFP-infected cells by 24 h p.i. (not shown). However, by 48 h p.i., the subcellular distribution of G3BP had changed from being a diffuse cytoplasmic pattern to accumulating in distinct round granules (blue arrows) or into elongated perinuclear clusters (yellow arrows), which sometimes contained P150 (Fig. 1b). Besides co-localizing with RCs in infected cells, P150 was previously reported to also accumulate in subcellular regions lacking RCs (Matthews et al., 2009), and these perinuclear clusters may be such sites. The subcellular location of G3BP was examined in >100 mock-infected or infected cells from two or more independent mock-infected or infected cultures (Fig. 1c) and this analysis revealed that, among the infected cells, the G3BP pattern was similar to the mock-infected pattern in approximately 40 % of the cells. However, approximately 35 % of the infected cells contained G3BP granules and the remaining approximately 20 % contained G3BP perinuclear clusters (Fig. 1c).
Fig. 1.
Analysis of G3BP during RUBV infection. (a) Mock- or Robo502/P150-HA-infected cells (m.o.i. = 3) were analysed at 3, 24 and 48 h p.i. by Western blotting probed against HA (to detect HA-tagged P150), G3BP and calnexin (as an internal control). This experiment was repeated twice with similar results and thus a representative blot is shown. (b) The subcellular distributions of P150 (ii; green) and G3BP (iii; red) in Robo502/P150-GFP-infected cells (m.o.i. = 0.3) at 48 h p.i. are shown, with the merge in (iv). G3BP was detected by rabbit anti-G3BP and donkey anti-rabbit–Alexa Fluor 595. Blue arrows point to G3BP granules and yellow arrows point to perinuclear clusters. Similarly stained mock-infected cells are shown to the left in (i). Nuclei were stained with Hoechst 33342. Bars, 10 µm. (c) G3BP localization was scored in at least 100 mock- or 100 Robo502/P150-HA-infected cells (m.o.i. = 3) in at least 15 randomly chosen fields of view (infected cells were identified by dsRNA staining). G3BP distribution was categorized as solely cytoplasmic (Cyto), present in granules resembling stress granules (G) or present in perinuclear clusters (PNC). Error bars represent sd. These experiments were conducted at least twice.
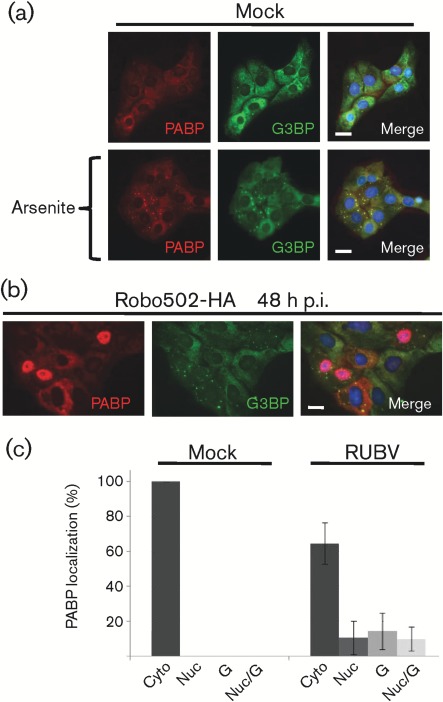
The appearance of G3BP granules in the cytoplasm of infected cells was consistent with the hypothesis that SGs form during RUBV infection. PABP was used as a second marker for SGs. In mock-infected cells, PABP and G3BP were distributed in a diffuse cytoplasmic pattern (Fig. 2a, top panel). As expected, mock-infected cells treated with arsenite showed strict co-localization of PABP with G3BP in SGs (Fig. 2a, bottom panel). Surprisingly, however, in Robo502/P150-HA-infected cells, approximately 75 % of RUBV-infected cells lacked PABP granules (Fig. 2b, c). Interestingly, in about 10 % of infected cells, PABP had localized to the nucleus and, in another about 10 % of infected cells, PABP was localized in both the nucleus and granules (Fig. 2c). Taken together, approximately 35 % of the infected cells had changes in PABP localization, compared with untreated mock-infected cells. The contrast in the percentage of infected cells with G3BP granules (Fig. 1c) against those with PABP granules (Fig. 2c) was the first indication that the granules observed during RUBV infection are not the same as those produced as a result of arsenite treatment.
Fig. 2.
Analysis of PABP during late RUBV infection. (a) Untreated (top panels) or arsenite-treated (bottom panels) cells were co-stained for PABP (red) and G3BP (green). PABP was detected by mouse anti-PABP and anti-mouse–Alexa Fluor 595, while G3BP was detected by rabbit anti-G3BP and chicken anti-rabbit–Alexa Fluor 488. Nuclei were stained with Hoechst 33342. Bars, 10 µm. (b) As (a), except that cells were infected with Robo502/P150-HA (m.o.i. = 3) and analysed at 48 h p.i. (c) At 48 h p.i., PABP localization was scored in at least 100 mock- or 100 Robo502/P150-HA-infected cells (m.o.i. = 3) (identified by dsRNA staining) in at least 15 randomly chosen fields of view (from at least two different experiments). PABP distribution was categorized as solely cytoplasmic (Cyto), nuclear (Nuc), granules resembling stress granules (G) or both nuclear and granules (Nuc/G). Error bars represent sd.
Thus, the composition of the G3BP granules formed during RUBV infection was investigated further by determining the percentage of co-localization of resident SG proteins in the virus-induced structures versus those formed by arsenite treatment. To this end, cells with G3BP granules were scored for the presence or absence of either PABP or TIA-1 in the granules, as determined by immunofluorescence staining (i.e. G3BP/PABP or G3BP/TIA-1 co-staining). In arsenite-treated cells, virtually all of the cells with G3BP-positive SGs exhibited PABP or TIA-1 co-localization in the SGs with G3BP (Fig. 3a, b, respectively). However, at 48 h p.i., in only about 55 % of infected cells with G3BP granules did these granules also contain PABP (Fig. 3a), corresponding to roughly 20 % of the total population of infected cells, and in <40 % of infected cells with G3BP granules did the granules also contain TIA-1 (Fig. 3b), roughly 15 % of total infected cells. Additionally, following treatment with cycloheximide, the arsenite-induced SGs disassembled, but the virus-induced G3BP granules largely remained intact (Fig. 3c). Thus, whilst some granules in RUBV-infected cells appeared to be similar to arsenite-stimulated ones (these granules might be in the initial stages of RUBV-specific disruption), the remainder of the granules did not appear to function like arsenite-induced SGs.
Fig. 3.
Analysis of G3BP granule composition and dynamics. (a) The localization of PABP in cells containing G3BP granules was tabulated in arsenite-treated (Ars+) Vero cells or Robo502/P150-HA-infected cells (m.o.i. = 3, 48 h p.i.) (at least 100 cells total for each sample). Three distinct localization patterns of PABP were observed in RUBV-infected cells positive for G3BP granules: nuclear (Nuc), granules (G) or both (Nuc/G). (b) The presence of TIA-1 in G3BP granules was determined by co-staining [TIA-1 (rabbit) and G3BP (chicken) antibody] of arsenite-treated Vero cells or Robo502/P150-HA-infected Vero cells (m.o.i. = 3) at 48 h p.i. At least 100 different cells that contained granules were counted from 15 different fields of view from each sample and the percentage of cells that contained G3BP granules with co-localizing TIA-1 was determined. (c) Vero cells treated for 35 min with 0.5 mM sodium arsenite were then exposed to 10 µg cycloheximide ml−1 (arsenite remained on cells during cycloheximide treatment) for an additional 35 min (+), while Robo502/P150-HA-infected cells (m.o.i. = 3) were exposed to 10 µg cycloheximide ml−1 for the entire 70 min (+). Vero cells treated with arsenite for 70 min, but not exposed to cycloheximide, and untreated Robo502/P150-HA-infected cells served as controls (−). The cells were then stained with anti-G3BP antibody, and 100 cells from at least 15 different fields of view were scored for the presence of G3BP granules (infected cells in the Robo502/P150HA-infected culture were identified by dsRNA staining). Error bars represent sd from at least two independent experiments.
G3BP granules form concomitantly with virus RNA synthesis
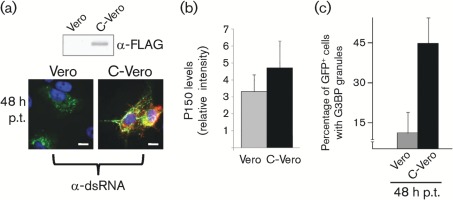
A permissive/non-permissive pair of continuous cell lines was used to determine whether the presence of the virus replicase proteins or the presence of the virus proteins and active RNA replication was required for G3BP granule induction. A RUBV replicon construct that lacks the structural protein ORF and expresses a GFP-tagged P150 that replicates only in the presence of CP was employed. Vero cells are non-permissive for this replicon, whereas C-Vero cells that are stably transfected with the CP [Western blot shows expression of FLAG-tagged CP in Fig. 4(a, top)] are permissive (Tzeng et al., 2006). In both cell lines, following transfection with the replicon, translation of P150 and P90 occurs, but only in C-Vero cells can RNA synthesis be detected. As can be seen in Fig. 4(a, bottom), GFP-tagged P150 was produced in both replicon-transfected Vero and C-Vero cells, but only the replicon-transfected C-Vero cells contained dsRNA (stained in red). Fig. 4(b) shows that the levels of P150 are approximately 1.5-fold higher in C-Vero cells than in similarly transfected Vero cells. Fig. 4(c) shows the percentages of GFP–P150-positive cells that contained G3BP granules at 48 h post-transfection. Roughly 45 % of such cells contained G3BP granules in the C-Vero culture, whilst the figure was approximately 10 % in the Vero cells. Thus, G3BP granules formed concomitantly with viral RNA replication.
Fig. 4.
G3BP granules form concomitantly with viral RNA replication and NSP accumulation. Vero (replication non-permissive) or C-Vero (replication-permissive) cells were transfected with RUBrep/P150-GFP in vitro RNA transcripts and analysed for the percentage of GFP-positive (i.e. successfully transfected) cells that contained G3BP granules by immunofluorescence using antibodies against G3BP. (a) Western blotting of Vero and C-Vero cell lysates for FLAG-tagged CP (approx. 30 kDa) is shown at top, whilst below are micrographs showing Vero or C-Vero cells at 48 h post-transfection with RUBrep/P150-GFP transcripts and stained for dsRNA (red) (GFP-tagged P150 is green in these micrographs and nuclei were counterstained with Hoechst 33342). (b) Quantification by ImageQuant of P150 levels on Western blots from 48 h post-transfection lysates (Vero or C-Vero) using GFP antibodies (mean of two independent experiments). (c) Quantification of GFP-positive cells containing G3BP granules. At least 100 cells from 15 different fields of view were analysed from at least two different experiments. Error bars represent sd.
G3BP granules co-localize with ssRNA, but not dsRNA
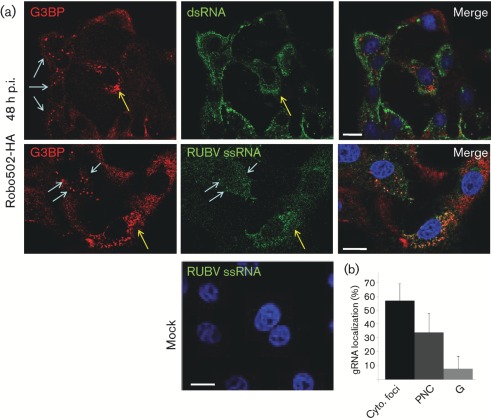
In order to determine whether G3BP granules were involved in viral RNA synthesis, infected Vero cells were co-stained for G3BP and dsRNA, a marker for RCs. In Robo502/P150-HA-infected cells at 48 h p.i., the distribution of G3BP granules did not overlap significantly with that of RCs (Fig. 5a, top panel), indicating no involvement. As G3BP is an RNA-binding protein, we subsequently used fluorescence in situ hybridization (FISH) to analyse whether viral ssRNA accumulated in the G3BP granules. Both nonsense (not shown) and RUBV-specific probes were used for analysis, with only the specific probes producing a signal in infected cells. No signal was detected in mock-infected cells. As shown in Fig. 5(a, middle row) and Fig. 5(b), whilst in a small percentage (approx. 10 %) of infected cells, viral ssRNA was observed in G3BP granules (Fig. 5a, bottom panel, blue arrows), in the majority of the infected cells (approx. 90 %), viral ssRNA did not co-localize with G3BP granules. However, viral ssRNA localized in perinuclear clusters in about 35 % of infected cells and G3BP was found in these structures 40–50 % of the time (corresponding to roughly 15–20 % of total infected cells). Therefore, G3BP granules and perinuclear clusters do not appear to be involved directly in viral RNA synthesis.
Fig. 5.
Analysis of G3BP subcellular localization with viral ss- and dsRNA. (a) In the top panels, Robo502/P150-HA-infected cells (m.o.i. = 3, at 48 h p.i.) were stained red for G3BP with rabbit anti-G3BP/goat anti-rabbit–TRITC conjugate and green for dsRNA using mouse anti-dsRNA/goat anti-mouse–FITC conjugate, with the merged image shown on the right. In the bottom panels, similarly infected cells were probed with nick-translated, dUTP–Alexa Fluor 594-labelled DNA from the Robo502 plasmid to detect viral ssRNA (the FISH probe is pseudo-coloured green) and G3BP (red) as above. Mock-infected cells stained with RUBV-specific probes are shown (bottom). Blue arrows point to SGs and yellow arrows point to perinuclear clusters. Bars, 10 µm. (b) The primary localization of ssRNA [cytoplasmic foci, perinuclear clusters (PNC) or granules (G)] was counted in at least 100 cells from 15 different fields of view (from at least two different experiments). Bars represent sd.
Discussion
This study was initiated to determine whether a stress response is induced during RUBV infection. To this end, G3BP levels and subcellular distribution were analysed in RUBV-infected cells at different times p.i. Whilst RUBV did not dramatically alter the expression levels of G3BP, it did induce the formation of what initially seemed to be SGs in the late stages of infection (i.e. 48 h p.i.), concomitant with peak accumulation of viral macromolecules (Hemphill et al., 1988). However, induction was not uniform and accumulation of G3BP into the apparent SGs was detected in fewer than half of infected cells. In fact, granule formation tended to coincide with cells exhibiting the highest levels of the markers used to detect infection (GFP-tagged P150 and dsRNA) (data not shown). These granules required viral RNA replication for induction, but unexpectedly proved to be compositionally and functionally distinct from arsenite-induced SGs. The RUBV-induced granules often did not contain other known SG proteins such as PABP and TIA-1 and, furthermore, unlike arsenite-induced SGs, were resistant to dispersion by cycloheximide. Taken together, a stress response induced by RUBV infection appears to require a threshold level of virus replication (that is surpassed only in some cells late in infection) and then the formation of SGs is somehow countered.
Whilst RUBV and the alphaviruses share a common replication strategy, the interaction of these viruses with infected cells differs profoundly. Namely, whilst alphaviruses replicate robustly in vertebrate cells, inducing both complete transcriptional and translational shutdown of the host, RUBV replication occurs more slowly and to lower titres with minimal cytopathology (Frey, 1994). The results of the current study also demonstrate distinct differences between RUBV and alphaviruses in the induction and regulation of the stress response during infection. SFV induces the formation of SGs during the early phases of virus infection but, during the later stages, the SGs disappear in the vicinity of ongoing viral RNA synthesis (McInerney et al., 2005). As the composition of the granules formed during SFV infection was determined to be consistent with that found in functional SGs, SFV does not seem to alter the formation or function of these SGs. Instead, SFV has evolved translational enhancer sequences in the subgenomic RNA that allow for efficient translation, despite the host translational shutoff that occurs during infection (McInerney et al., 2005). In contrast, our study shows that a cellular stress response is initiated in a non-uniform manner only during the late stages of RUBV infection and that the G3BP granules formed are not like functional SGs induced by arsenite treatment. This might in part explain why there is only a modest decrease in total protein synthesis during the late stages of RUBV infection (Hemphill et al., 1988).
Recently, a possible role for G3BP in alphavirus RNA replication was suggested, as it was reported that G3BP interacts with the alphavirus replicase proteins nsP3 (Gorchakov et al., 2008) and nsP4 (Cristea et al., 2010). In contrast, data from this study provided no evidence that G3BP was associated with RUBV RCs. However, G3BP appeared to associate with the viral ssRNA in perinuclear clusters in 15–20 % of RUBV-infected cells, which sometimes contained P150. The dynamic localization status of G3BP makes the significance of the G3BP/ssRNA co-localization more difficult to interpret, but our analysis may have only captured a smaller (or possibly larger) percentage of the co-localizing events. Nonetheless, the existence of an interaction between G3BP and RUBV ssRNAs would not be surprising, considering the known RNA-binding capacity of G3BP (Parker et al., 1996; Tourrière et al., 2001, 2003) and its general role in the stress response. It is possible that P150 in these structures is behaving as an intermediary for virus RNA transfer from RCs to the sites of virus RNA encapsidation. We hypothesize that G3BP accumulates around virus positive-strand RNAs that have reached high levels during the late stages of RUBV infection, but G3BP fails to transfer virus RNAs to SGs, as demonstrated by the lack (<10 %) of RUBV genomes in the G3BP granules. Whilst the function of the co-localization between virus ssRNA and G3BP remains to be determined, we and others (Beatch & Hobman, 2000) have found that CP also accumulates in the perinuclear region, suggesting that these virus-specific perinuclear clusters may be where encapsidation is taking place.
Consistent with previous reports on PABP localization during stress (Kedersha et al., 2000), PABP localized in G3BP SGs during arsenite treatment. However, PABP showed a dramatically different redistribution in response to RUBV infection, in particular migration to the nucleus. Nuclear relocalization of PABP has been observed in rotavirus-infected cells following ejection from the small ribosomal initiation complex during the induction of SG formation (Harb et al., 2008). Herpes simplex virus also causes nuclear localization of PABP by activating JNK and p38 mitogen-activated protein kinases that are sensitive to oxidative stress. However, when oxidative stress was applied to uninfected Vero cells via arsenite treatment, we observed no nuclear PABP localization. Other studies have shown that PABP shuttles between the nucleus and cytoplasm and accumulates in the nucleus upon transcription shutoff (Afonina et al., 1998; Brune et al., 2005); however, RUBV is not known to induce transcriptional shutoff (Hemphill et al., 1988). We tried to simulate transcriptional shutoff by treating uninfected cells with various doses and exposures of actinomycin D, but observed no nuclear localization of PABP (data not shown). Finally, under plasmid-directed expression, low-level expression of PABP resulted in a predominantly cytoplasmic distribution, whilst high-level expression led to nuclear localization (Afonina et al., 1998). Ilkow et al. (2008) reported higher levels of PABP expression in RUBV-infected cells. The nuclear localization of PABP that we observed probably reflects the higher protein levels that appeared to reside in RUBV-infected cells. In fact, many of the RUBV-infected cells with nuclear PABP exhibited a more intense signal than was observable in the cytoplasm of mock-infected cells. Collectively, these data support the notion that PABP nuclear localization is the result of higher levels of PABP expression during RUBV infection.
In conclusion, RUBV is capable of inducing a cellular stress response during the late stages of infection, albeit in a non-uniform manner, marked by subcellular redistribution of G3BP, PABP and TIA-1. However, the organization of this response seems to be disrupted in that functional SGs are not formed. Whilst some G3BP was found to co-localize with the virus-specific perinuclear clusters that contained multiple virus components, G3BP does not appear to play a direct role in the synthesis of RUBV RNA. However, G3BP may play a post-replicative role in RUBV infection, possibly as an intermediary in the processing of virus ssRNA from RCs to the sites of encapsidation. The function of the virus-specific perinuclear clusters is currently under further investigation.
Methods
Cells, viruses and replicons.
The Vero African green monkey kidney cell line (obtained from ATCC) was used in this study along with a previously published cell line, C-Vero (Tzeng et al., 2006), that stably expresses the RUBV CP. Cells were maintained at 35 °C and 5 % CO2 in Dulbecco’s modified Eagle’s medium (Mediatech) with 5 % FBS (Atlanta Biologicals) and infections were performed in 1 % FBS/PBS as described previously (Tzeng et al., 2006). The viruses used in this study were produced from the infectious clones Robo502/P150-HA (Tzeng et al., 2006) and Robo502/P150-GFP (Matthews et al., 2010) as described by Pugachev et al. (2000). RUBrep/P150-GFP is a derivative of Robo502/P150-GFP that contains a CAT reporter gene in place of the structural protein ORF. In vitro transcripts of RUBrep/P150-GFP, synthesized as described previously (Tzeng et al., 2006) using linearized plasmid template, were transfected into Vero cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s directions. Accordingly, approximately 5 µg in vitro-transcribed RNA (estimated by gel electrophoresis and ethidium bromide staining) and 5 µl Lipofectamine 2000 per monolayer in 60 mm culture plates were used for transfection. Mock-transfected cells received only Lipofectamine 2000.
Immunofluorescence.
At appropriate time points, Vero cells grown at low density (30–40 % confluent) on glass coverslips and infected or transfected accordingly were simultaneously fixed and permeabilized with ice-cold methanol for 5 min. After washing and equilibration in PBS, the cells were stained with the following antibodies diluted in 2 % BSA/PBS solution: rabbit or chicken anti-G3BP (recognizing G3BP1), Sigma, 1/500; mouse anti-PABP, Sigma, 1/200; mouse anti-dsRNA, Scientific Consultants, 1/1000. Rabbit anti-TIA-1 was obtained from Sigma. In some experiments, primary antibodies were detected by goat anti-mouse or anti-rabbit secondary antibodies with the desired conjugate for red (TRITC) or green (FITC) fluorescence (obtained from Sigma). In other experiments, primary antibodies were detected by donkey anti-chicken, -rabbit, or -mouse secondary antibodies conjugated with Alexa Fluor 488 or 595 (Invitrogen). Nuclei were stained with Hoechst 33342 (Invitrogen).
FISH assay.
This assay was performed essentially as described by Jiménez-García & Spector (1993) with some minor modifications. Briefly, cells harvested at the appropriate time point were fixed in 4 % formaldehyde in PBS before permeabilization by 0.5 % Triton X-100 for 10 min at room temperature. Coverslips were washed in PBS and then 2× SSC. Next, nick-translated probes labelled with Alexa Fluor 594–dUTP (Invitrogen) and purified from free nucleotides using Ambion NucAway columns as described in the manufacturer’s protocol were boiled for 10 min in 50 % formamide, 10 % dextran sulfate, 2× SSC and 1 µg E. coli tRNA ml−1, and stored on ice until being added to each coverslip and incubated overnight at 42 °C. Coverslips were washed in 50 % formamide/2× SSC for 15 min at 42 °C, briefly washed with 2× SSC at 42 °C and once with 1× SSC at room temperature before mounting or proceeding to immunofluorescence.
Microscopy.
Images in Fig. 2 were acquired on a Zeiss Axioplan epifluorescence wide-field microscope with a 40× objective and processed with AxioVision software. The remaining images were acquired on a Zeiss LSM700 confocal microscope using a 63× objective with immersion oil and ZEN software. Images were processed with LSM Image Browser or LSM700 software.
Western blotting.
Lysates from infected or transfected cells were prepared essentially as described previously (Tzeng et al., 2006). Briefly, cells in 60 mm plates were lysed in 500 µl lysis buffer [10 mM Tris (pH 8.0), 150 mM NaCl, 1 % Triton X-100, 0.5 % sodium deoxycholate, 0.1 % SDS and 1× protease inhibitor cocktail (EDTA-free, Roche)] at the appropriate time point. After clearing insoluble debris by high-speed centrifugation (10 min at 16 000 g in an Eppendorf tabletop #5415D centrifuge), lysates were adjusted to 1× with Laemmli sample buffer, heat-denatured by boiling and 5 % of each lysate was loaded onto an SDS-PAGE gel, resolved and then transferred to nitrocellulose membranes for probing with the appropriate antibodies. P150–HA and P150–GFP were detected by mouse anti-HA, Roche, 1/1000, or rabbit anti-GFP, Clontech, 1/40, respectively. The other proteins were detected with the following: rabbit anti-G3BP, Sigma, 1/1000; rabbit anti-calnexin, Sigma, 1/5000. Each of the primary antibodies was visualized on the blot with an appropriate secondary antibody, i.e. anti-rabbit or -mouse that was conjugated to alkaline phosphatase (Promega, 1/5000) and subsequently reacted with NBT/BCIP (Roche) for colour development according to the manufacturer’s suggestions.
Acknowledgements
This work was supported by NIH grant AI21389. We thank Dr Wen-Pin Tzeng for constructing the RUBrep/P150-GFP replicon.
References
- Afonina E., Stauber R., Pavlakis G. N. (1998). The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J Biol Chem 273, 13015–13021 10.1074/jbc.273.21.13015 [DOI] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. (2002). Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones 7, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatch M. D., Hobman T. C. (2000). Rubella virus capsid associates with host cell protein p32 and localizes to mitochondria. J Virol 74, 5569–5576 10.1128/JVI.74.12.5569-5576.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham C. J., Parker R. (2008). P bodies, stress granules, and viral life cycles. Cell Host Microbe 3, 206–212 10.1016/j.chom.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune C., Munchel S. E., Fischer N., Podtelejnikov A. V., Weis K. (2005). Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. RNA 11, 517–531 10.1261/rna.7291205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristea I. M., Rozjabek H., Molloy K. R., Karki S., White L. L., Rice C. M., Rout M. P., Chait B. T., MacDonald M. R. (2010). Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: role for G3BP1 and G3BP2 in virus replication. J Virol 84, 6720–6732 10.1128/JVI.01983-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara M. M., Brinton M. A. (2007). Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc Natl Acad Sci U S A 104, 9041–9046 10.1073/pnas.0703348104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forng R. Y., Frey T. K. (1995). Identification of the rubella virus nonstructural proteins. Virology 206, 843–853 10.1006/viro.1995.1007 [DOI] [PubMed] [Google Scholar]
- Frey T. K. (1994). Molecular biology of rubella virus. Adv Virus Res 44, 69–160 10.1016/S0065-3527(08)60328-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R., Garmashova N., Frolova E., Frolov I. (2008). Different types of nsP3-containing protein complexes in Sindbis virus-infected cells. J Virol 82, 10088–10101 10.1128/JVI.01011-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harb M., Becker M. M., Vitour D., Baron C. H., Vende P., Brown S. C., Bolte S., Arold S. T., Poncet D. (2008). Nuclear localization of cytoplasmic poly(A)-binding protein upon rotavirus infection involves the interaction of NSP3 with eIF4G and RoXaN. J Virol 82, 11283–11293 10.1128/JVI.00872-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemphill M. L., Forng R. Y., Abernathy E. S., Frey T. K. (1988). Time course of virus-specific macromolecular synthesis during rubella virus infection in Vero cells. Virology 162, 65–75 10.1016/0042-6822(88)90395-9 [DOI] [PubMed] [Google Scholar]
- Ilkow C. S., Mancinelli V., Beatch M. D., Hobman T. C. (2008). Rubella virus capsid protein interacts with poly(A)-binding protein and inhibits translation. J Virol 82, 4284–4294 10.1128/JVI.02732-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-García L. F., Spector D. L. (1993). In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell 73, 47–59 10.1016/0092-8674(93)90159-N [DOI] [PubMed] [Google Scholar]
- Kedersha N., Anderson P. (2002). Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans 30, 963–969 10.1042/BST0300963 [DOI] [PubMed] [Google Scholar]
- Kedersha N. L., Gupta M., Li W., Miller I., Anderson P. (1999). RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J Cell Biol 147, 1431–1442 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Cho M. R., Li W., Yacono P. W., Chen S., Gilks N., Golan D. E., Anderson P. (2000). Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 151, 1257–1268 10.1083/jcb.151.6.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 169, 871–884 10.1083/jcb.200502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujala P., Ahola T., Ehsani N., Auvinen P., Vihinen H., Kääriäinen L. (1999). Intracellular distribution of rubella virus nonstructural protein P150. J Virol 73, 7805–7811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y., Marshall J. A., Bowden D. S. (1992). Replication complexes associated with the morphogenesis of rubella virus. Arch Virol 122, 95–106 10.1007/BF01321120 [DOI] [PubMed] [Google Scholar]
- Lee J. Y., Marshall J. A., Bowden D. S. (1994). Characterization of rubella virus replication complexes using antibodies to double-stranded RNA. Virology 200, 307–312 10.1006/viro.1994.1192 [DOI] [PubMed] [Google Scholar]
- Magliano D., Marshall J. A., Bowden D. S., Vardaxis N., Meanger J., Lee J. Y. (1998). Rubella virus replication complexes are virus-modified lysosomes. Virology 240, 57–63 10.1006/viro.1997.8906 [DOI] [PubMed] [Google Scholar]
- Matthews J. D., Tzeng W. P., Frey T. K. (2009). Determinants of subcellular localization of the rubella virus nonstructural replicase proteins. Virology 390, 315–323 10.1016/j.virol.2009.05.019 [DOI] [PubMed] [Google Scholar]
- Matthews J. D., Tzeng W. P., Frey T. K. (2010). Analysis of the function of cytoplasmic fibers formed by the rubella virus nonstructural replicase proteins. Virology 406, 212–227 10.1016/j.virol.2010.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R., Sukarieh R., Bordeleau M. E., Kaufman R. J., Northcote P., Tanaka J., Gallouzi I., Pelletier J. (2006). Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2α phosphorylation. Mol Biol Cell 17, 4212–4219 10.1091/mbc.E06-04-0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney G. M., Kedersha N. L., Kaufman R. J., Anderson P., Liljeström P. (2005). Importance of eIF2α phosphorylation and stress granule assembly in alphavirus translation regulation. Mol Biol Cell 16, 3753–3763 10.1091/mbc.E05-02-0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker F., Maurier F., Delumeau I., Duchesne M., Faucher D., Debussche L., Dugue A., Schweighoffer F., Tocque B. (1996). A Ras-GTPase-activating protein SH3-domain-binding protein. Mol Cell Biol 16, 2561–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugachev K. V., Galinski M. S., Frey T. K. (2000). Infectious cDNA clone of the RA27/3 vaccine strain of rubella virus. Virology 273, 189–197 10.1006/viro.2000.0408 [DOI] [PubMed] [Google Scholar]
- Tocque B., Delumeau I., Parker F., Maurier F., Multon M. C., Schweighoffer F. (1997). Ras-GTPase activating protein (GAP): a putative effector for Ras. Cell Signal 9, 153–158 10.1016/S0898-6568(96)00135-0 [DOI] [PubMed] [Google Scholar]
- Tourrière H., Gallouzi I. E., Chebli K., Capony J. P., Mouaikel J., van der Geer P., Tazi J. (2001). RasGAP-associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation-dependent localization. Mol Cell Biol 21, 7747–7760 10.1128/MCB.21.22.7747-7760.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J. M., Bertrand E., Tazi J. (2003). The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol 160, 823–831 10.1083/jcb.200212128 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tzeng W. P., Matthews J. D., Frey T. K. (2006). Analysis of rubella virus capsid protein-mediated enhancement of replicon replication and mutant rescue. J Virol 80, 3966–3974 10.1128/JVI.80.8.3966-3974.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. P., Cardenas A. M., Marissen W. E., Lloyd R. E. (2007). Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2, 295–305 10.1016/j.chom.2007.08.006 [DOI] [PubMed] [Google Scholar]