Abstract
We report the first identification, genetic characterization and disease association studies of several novel species of canine bocaviruses (CBoV). Evolutionary analysis confirmed that CBoV are genetically distinct from the only other known canine bocavirus, minute virus of canines, with which they share less than 63, 62 and 64 % protein identity in NS, NP and VP genes, respectively. Comparative genetic analysis of 37 VP gene variants found in diseased and healthy animals showed that these novel viruses are genetically highly diverse and are common in canine respiratory infections that have remained undetected until now. Interestingly, we observed that a CBoV genotype with a unique deletion in the VP2 gene was significantly more prevalent in animals with respiratory diseases compared with healthy animals.
Parvoviruses, which frequently infect animals through the faecal–oral route, are small, non-enveloped icosahedral viruses with linear ssDNA genomes (Fauquet et al., 2004). They are members of the family Parvoviridae, which comprises two subfamilies, Densovirinae and Parvovirinae, members of which infect non-vertebrate and vertebrate hosts, respectively (Brown, 2010; Fauquet et al., 2004). The International Committee on Taxonomy of Viruses (ICTV) has further classified the subfamily Parvovirinae into six genera: Dependovirus, Bocavirus, Erythrovirus, Parvovirus, Amdovirus and Partetravirus. Bocaviruses are unique among parvoviruses as they contain a third ORF between the non-structural- and structural-coding regions (Kapoor et al., 2010b; Manteufel & Truyen, 2008; Qiu et al., 2007). The genus Bocavirus currently includes the bovine parvoviruses (BPV), minute virus of canines (MVC) (Fauquet et al., 2004), porcine bocaviruses (Cheng et al., 2010), gorilla bocavirus (GBoV) (Kapoor et al., 2010a) and four species of human bocaviruses (HBoV 1–4) (Allander et al., 2005; Arthur et al., 2009; Chieochansin et al., 2007; Kapoor et al., 2009, 2010b). Bocaviruses commonly infect the respiratory and gastrointestinal tract of young animals and humans, and except for BPV, the pathological manifestations of these infections remain largely unknown (Don et al., 2011; Kapoor et al., 2011; Manteufel & Truyen, 2008; Martin et al., 2009, 2010).
First discovered in 1967 in faeces of healthy dogs, MVC is the only known bocavirus that infects dogs. It can cause abortions in bitches and severe respiratory infections in newborn puppies, but infections are mostly subclinical in adult animals (Carmichael et al., 1991). MVC replicates to high titres in Walter Reed/3873D (WRD) canine cells, making it a useful model system to dissect the replication kinetics of genetically similar, but uncultivable, HBoV (Sun et al., 2009). During a metagenomic study conducted to better characterize the respiratory viral flora of domestic animals, we observed several sequences with distant amino acid sequence similarity to animal parvoviruses in respiratory samples from diseased dogs. Extension of these novel sequences using a primer walking approach revealed the presence of a novel bocavirus tentatively named canine bocavirus (CBoV). Thereafter, a consensus PCR assay using primers targeting conserved structural protein motifs was used to determine the prevalence and genetic diversity of CBoV variants in a cohort of respiratory samples obtained from diseased and healthy dogs (Supplementary Table S1, available in JGV Online). Briefly, extracted nucleic acids from each sample were used for two rounds of nested PCR (first round: forward-CBoV-QFX1-f1, 5′-CARTGGTAYGCTCCMATYTTTAA-3′ and reverse-CBoV-QFX1-r1, 5′-TGGCTCCCGTCACAAAAKATRTG-3′; and second round: forward-CBoV-QFX1-f2, 5′-TGGTAYGCTCCMATYTTTAAYGG-3′, reverse-CBoV-QFX1-r2 5′-GCTCCCGTCACAAAAKATRTGAAC-3′). The amplification products (~400nt long) representing the partial CBoV VP1 gene were sequenced for confirmation and to determine viral genetic diversity. Of the 158 animals tested, 36 (23 %) were infected with CBoV variants (Supplementary Table S1).
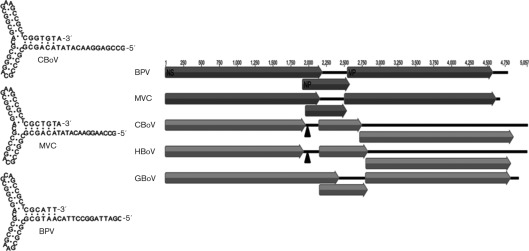
The nearly complete genome of CBoV variant con-161 is 5413 nt (GenBank accession no. JN648103) and bears a high degree of similarity to other known bocaviruses predicted to contain non-coding terminal sequences flanking the three large ORFs (Fig. 1). ORF1 encodes a 648 aa non-structural (NS) protein. ORF2 encodes 712 aa overlapping the VP1/VP2 capsid ORFs. ORF3 encodes a 195 aa NP1 protein. The non-coding region on left-hand side (LHS) terminus, located at the 5′ end of positive-sense ssDNA genomes or at the amino terminus of NS protein, is 306 nt. Its secondary structure folds into an imperfect palindrome and contains a rabbit-ear structure similar to MVC and BPV (Fig. 1). The right-hand side (RHS) non-coding region, found at the 3′ end of positive-sense ssDNA genomes, is not identical to the LHS terminus and forms an imperfect palindrome. The LHS terminus showed highest sequence similarity to the MVC terminus and also contained a NS-binding site (Sun et al., 2009). While the MVC and BPV NS protein-coding regions encode a single long NS protein, recent studies have shown that the homologous region of all four HBoV species encodes two NS proteins of variable length (Chen et al., 2010; Kapoor et al., 2010a). Remarkably, although CBoV is genetically similar to HBoV in NS gene splicing, it is most similar to MVC. The CBoV NS-coding region encodes a shorter NS protein, as well as conserved RNA splicing signals essential to generate a longer NS protein (Fig. 1). Like other bioinformatics analyses and predictions, these observations require experimental validation in subsequent studies.
Fig. 1.
Comparative genomic organization and LHS inverted terminal repeat structure of different bocavirus species. All genomes were aligned starting from first N-terminal amino acid codon of the NS gene (nucleotide positions) to comparatively show the location of the putative RNA splicing region in the NS exon (shown as filled triangle).
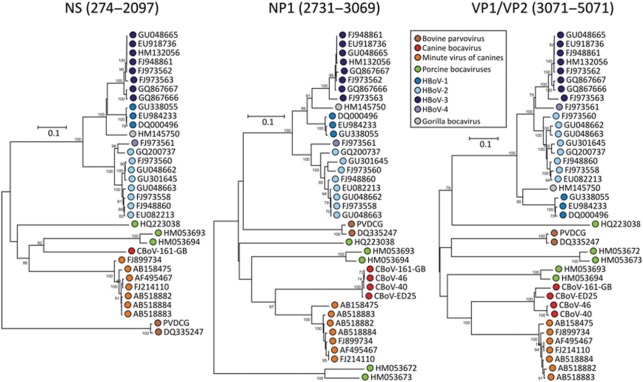
To determine CBoV’s appropriate phylogenetic classification and genetic relatedness to other known parvovirus species, at least one representative virus, as well as the reference genome from each human and animal bocavirus species and their translated protein sequences, was used for generating sequence alignments. The most appropriate protein or nucleotide substitution model was determined using mega, and the method with lowest scores was used to calculate pair-wise distances and to construct phylogenetic trees (Fig. 2). All three CBoV proteins (NS, NP and VP) were genetically most related to corresponding MVC proteins; however, there was more genetic diversity/variability between CBoV and MCV proteins than among different HBoV species or different species of genus Parvovirus (Fig. 2). The ICTV criteria for species classification within the genus Bocavirus specify that members of each species are probably antigenically distinct and that natural infection is confined to a single host species. Species are defined as having <95 % homologous NS gene DNA sequences (http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/). While the antigenic properties of CBoV were not studied here, we found >35 % genetic divergence in the NS protein compared with other known bocaviruses, suggesting that CBoV and its variants represent one or more novel species within the genus Bocavirus.
Fig. 2.
Phylogenetic analyses of inferred amino acid sequences of the three principal ORFs (NS, NP and VP proteins) of human and animal bocaviruses; bootstrap values of >70 % are shown. Bars, represent 0.1 substitutions per amino acid site.
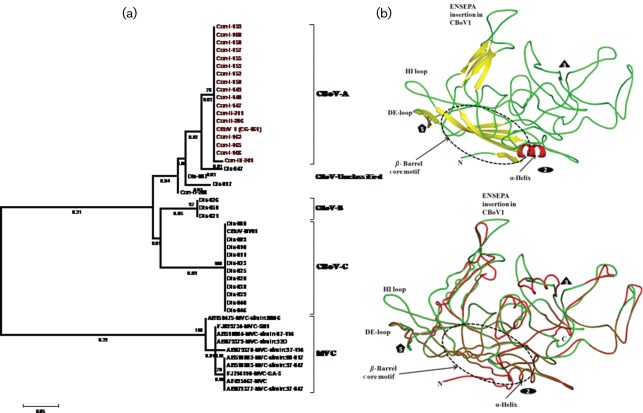
Parvoviral capsid proteins contain determinants of immunogenicity and host cell tropism. Minor genetic changes in these proteins are known to alter the host range and pathogenic potential of parvoviruses (Hoelzer et al., 2008a, b; Parrish & Kawaoka, 2005). Moreover, evolutionary studies confirmed that, unlike other DNA viruses, parvoviruses can evolve rapidly displaying frequent recombination and mutation rates that approach the high mutation rates observed in RNA viruses (Shackelton & Holmes, 2006; Shackelton et al., 2005, 2007). Expecting that genetic diversity in CBoV capsid proteins should influence their pathogenic potential, we classified CBoV variants according to the genetic relatedness in their capsid protein sequences (Fig. 3a). Calculation of pair-wise distances using the 261 nt long VP1/2 region of 35 CBoV variants resulted in up to 37 % (mean diversity of 15 %) nucleotide and 17 % (mean diversity of 7.3 %) protein sequence divergence (GenBank accession nos JN648104–JN648139). Phylogenetic analysis suggested that most CBoV variants can be divided into three major genetic groups, provisionally named CBoV-A to -C, while some were outliers (Fig. 3a). Our results imply that CBoV represents a highly diverse group of novel canine bocaviruses. The extent of genetic diversity observed among CBoV variants characterized in this single study exceeds the maximum genetic diversity known to exist among all MVC variants reported worldwide to date (Fig. 3a). Combined analyses of genetic diversity and PCR prevalence data suggest that CBoV variants of group A were significantly more prevalent in healthy dogs raised in controlled environments than in animals from other groups; and CBoV variants of group C were substantially more prevalent in dogs with respiratory diseases than in healthy animals (Supplementary Table S1).
Fig. 3.
(a) Genetic diversity of structural proteins among CBoV variants found in diseased (Dis-disease) and healthy (Con-control) dogs. For comparison, genetic diversity among MVC variants was included in the analysis (lower branch of the tree); and (b) comparative secondary structure of the capsid protein of CBoV variants was used to decipher the structural changes caused by insertion of six amino acid residues. In the ribbon diagram of CBoV1, the secondary structure elements (β-strand in yellow, helix in red and loop in green) are coloured differently. The icosahedral symmetry axes are represented as oval, triangle and pentagon. In the coil representation of CBoV capsid structure the CBoV-A and -C are shown as red and green, respectively. Bars, represent 0.05 substitutions per amino acid site.
To further investigate genetic diversity between CBoV-A and -C viruses, we acquired the complete capsid gene sequence of its representative variants. The CBoV-A capsid protein showed 89 % protein identity to CBoV-C (GenBank accession no. JN648135) over its entire length (714 aa, data not shown). To determine the location of amino acid position changes on viral capsid structure, we modelled the secondary structure of CBoV-A and -C for comparison. A homology model was made by giving a human B19 crystal structure coordinates (PDB ID:1S58) as a template model with knowledge based protein modelling program, swiss-model (Arnold et al., 2006). The final model was structurally and geometrically consistent and did not reveal either structure or sequence discrepancies. The model geometries were cross-validated with the procheck program (Laskowski et al., 1996) to check the accuracy of the CBoV homology model. Both B19 and CBoV structures were superimposed and resulted in a good r.m.s.d value of 0.7 Å using the coot program (Emsley & Cowtan, 2004). The graphical images were generated with the pymol program (www.pymol.org). The CBoV model contains a high structurally conserved β-barrel motif, a single α-helix and several stretched loops adopt a different conformational position (Fig. 3b). The high structurally similar β-barrel motif region has been implicated for genome packaging and protecting the virus capsid from the environmental damage. The fivefold pore region of the capsid is critical for VP1 externalization, packing of genome and capsid assembly functions (Gurda et al., 2010). The intertwining or flexible loops in the CBoV capsid decorate in the exterior surface region of the capsid and are highly distinguished from all other parvoviruses. The variable loops are important to control various biological properties of the capsid and that includes tissue tropism, transduction and receptor binding (Gurda et al., 2010). We noticed that the six amino acid insertion unique to CBoV-A was located in the variable exposed loop (Fig. 3b). Moreover the structural folding of all four outer surface exposed loops was very different between CBoV-A and -C variants possibly reflecting difference in their biological properties.
To conclude, we report several previously uncharacterized species of canine bocaviruses, their sequences, genomic characteristics and genetic diversity. We also compared the genetic diversity and the differences in prevalence of these novel viruses in sick and healthy animals. Animals infected with CBoV-B1 variants and suffering from respiratory infections were housed in a shelter facility. Animal shelters often house animals likely to have a wide variety of infections (Steneroden et al., 2011). These crowded conditions probably facilitate a higher prevalence of viruses in shelter animals compared with the healthy pet population (Helps et al., 2005). Moreover, we could not rule out the possibility that respiratory diseases in these animals were caused by other pathogenic respiratory viruses, but even then the higher prevalence of CBoV-B1 variants in this population alone suggests that these viruses are more likely to infect diseased animals (opportunistic infections) or that they can cause or enhance the pathology of other infections (co-infections). We note that the HBoV species include several highly divergent viruses (Kapoor et al., 2010b) and therefore more comprehensive disease-association studies should consider the genotype-specific prevalence pattern of these viruses. Comparable and high genetic diversity among CBoV variants makes it a more appropriate model to study HBoV disease-association as well as the evolution and pathogenicity of parvoviruses. Despite distant phylogenetic relatedness, we noticed many similarities between CBoV and HBoV species. Both groups of recently identified viruses are genetically diverse and contain many species and genotypes whose pathogenic potential remains unknown (Kapoor et al., 2010b). Unlike other animal bocaviruses, CBoV and HBoV have splicing signals in the NS gene and are likely to encode more than one NS protein (Kapoor et al., 2010a). Unfortunately, the studies for complete biological characterization of HBoV pathogenesis are hampered by the lack of a successful cell culture or animal model (Kapoor et al., 2011). Elucidation of the nearly complete CBoV genome, its genetic diversity and prevalence will help to establish a successful cell culture system for these viruses.
Acknowledgements
We thank Natasha Qaisar for excellent technical assistance. We are grateful for high throughput sequencing support provided by Roche 454 Life Sciences. This work was supported by awards from the National Institutes of Health (AI090196, AI081132, AI079231, AI57158, AI070411, AI090055, AI072613 and EY017404), the Defense Threat Reduction Agency and USDA 58-1275-7-370.
Footnotes
A supplementary table is available with the online version of this paper.
References
- Allander T., Tammi M. T., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. (2005). Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A 102, 12891–12896 10.1073/pnas.0504666102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J., Schwede T. (2006). The swiss-model workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- Arthur J. L., Higgins G. D., Davidson G. P., Givney R. C., Ratcliff R. M. (2009). A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathog 5, e1000391 10.1371/journal.ppat.1000391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. E. (2010). The expanding range of parvoviruses which infect humans. Rev Med Virol 20, 231–244 10.1002/rmv.648 [DOI] [PubMed] [Google Scholar]
- Carmichael L. E., Schlafer D. H., Hashimoto A. (1991). Pathogenicity of minute virus of canines (MVC) for the canine fetus. Cornell Vet 81, 151–171 [PubMed] [Google Scholar]
- Chen A. Y., Cheng F., Lou S., Luo Y., Liu Z., Delwart E., Pintel D., Qiu J. (2010). Characterization of the gene expression profile of human bocavirus. Virology 403, 145–154 10.1016/j.virol.2010.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W. X., Li J. S., Huang C. P., Yao D. P., Liu N., Cui S. X., Jin Y., Duan Z. J. (2010). Identification and nearly full-length genome characterization of novel porcine bocaviruses. PLoS ONE 5, e13583 10.1371/journal.pone.0013583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chieochansin T., Chutinimitkul S., Payungporn S., Hiranras T., Samransamruajkit R., Theamboolers A., Poovorawan Y. (2007). Complete coding sequences and phylogenetic analysis of human bocavirus (HBoV). Virus Res 129, 54–57 10.1016/j.virusres.2007.04.022 [DOI] [PubMed] [Google Scholar]
- Don M., Söderlund-Venermo M., Hedman K., Ruuskanen O., Allander T., Korppi M. (2011). Don’t forget serum in the diagnosis of human bocavirus infection. J Infect Dis 203, 1031–1032, author reply 1032–1033 10.1093/infdis/jiq157 [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. (2004). coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (editors) (2004). Virus Taxonomy: The Eighth Report of the International Committee on Taxonomy of Viruses. Oxford, UK: Academic Press [Google Scholar]
- Gurda B. L., Parent K. N., Bladek H., Sinkovits R. S., DiMattia M. A., Rence C., Castro A., McKenna R., Olson N. & other authors (2010). Human bocavirus capsid structure: insights into the structural repertoire of the Parvoviridae. J Virol 84, 5880–5889 10.1128/JVI.02719-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helps C. R., Lait P., Damhuis A., Björnehammar U., Bolta D., Brovida C., Chabanne L., Egberink H., Ferrand G. & other authors (2005). Factors associated with upper respiratory tract disease caused by feline herpesvirus, feline calicivirus, Chlamydophila felis and Bordetella bronchiseptica in cats: experience from 218 European catteries. Vet Rec 156, 669–673 [DOI] [PubMed] [Google Scholar]
- Hoelzer K., Shackelton L. A., Holmes E. C., Parrish C. R. (2008a). Within-host genetic diversity of endemic and emerging parvoviruses of dogs and cats. J Virol 82, 11096–11105 10.1128/JVI.01003-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzer K., Shackelton L. A., Parrish C. R., Holmes E. C. (2008b). Phylogenetic analysis reveals the emergence, evolution and dispersal of carnivore parvoviruses. J Gen Virol 89, 2280–2289 10.1099/vir.0.2008/002055-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Slikas E., Simmonds P., Chieochansin T., Naeem A., Shaukat S., Alam M. M., Sharif S., Angez M. & other authors (2009). A newly identified bocavirus species in human stool. J Infect Dis 199, 196–200 10.1086/595831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Mehta N., Esper F., Poljsak-Prijatelj M., Quan P. L., Qaisar N., Delwart E., Lipkin W. I. (2010a). Identification and characterization of a new bocavirus species in gorillas. PLoS ONE 5, e11948 10.1371/journal.pone.0011948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Simmonds P., Slikas E., Li L., Bodhidatta L., Sethabutr O., Triki H., Bahri O., Oderinde B. S. & other authors (2010b). Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J Infect Dis 201, 1633–1643 10.1086/652416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Hornig M., Asokan A., Williams B., Henriquez J. A., Lipkin W. I. (2011). Bocavirus episome in infected human tissue contains non-identical termini. PLoS ONE 6, e21362 10.1371/journal.pone.0021362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996). AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8, 477–486 10.1007/BF00228148 [DOI] [PubMed] [Google Scholar]
- Manteufel J., Truyen U. (2008). Animal bocaviruses: a brief review. Intervirology 51, 328–334 10.1159/000173734 [DOI] [PubMed] [Google Scholar]
- Martin E. T., Taylor J., Kuypers J., Magaret A., Wald A., Zerr D., Englund J. A. (2009). Detection of bocavirus in saliva of children with and without respiratory illness. J Clin Microbiol 47, 4131–4132 10.1128/JCM.01508-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E. T., Fairchok M. P., Kuypers J., Magaret A., Zerr D. M., Wald A., Englund J. A. (2010). Frequent and prolonged shedding of bocavirus in young children attending daycare. J Infect Dis 201, 1625–1632 10.1086/652405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish C. R., Kawaoka Y. (2005). The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu Rev Microbiol 59, 553–586 10.1146/annurev.micro.59.030804.121059 [DOI] [PubMed] [Google Scholar]
- Qiu J., Cheng F., Johnson F. B., Pintel D. (2007). The transcription profile of the bocavirus bovine parvovirus is unlike those of previously characterized parvoviruses. J Virol 81, 12080–12085 10.1128/JVI.00815-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelton L. A., Holmes E. C. (2006). Phylogenetic evidence for the rapid evolution of human B19 erythrovirus. J Virol 80, 3666–3669 10.1128/JVI.80.7.3666-3669.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelton L. A., Parrish C. R., Truyen U., Holmes E. C. (2005). High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc Natl Acad Sci U S A 102, 379–384 10.1073/pnas.0406765102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelton L. A., Hoelzer K., Parrish C. R., Holmes E. C. (2007). Comparative analysis reveals frequent recombination in the parvoviruses. J Gen Virol 88, 3294–3301 10.1099/vir.0.83255-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steneroden K. K., Hill A. E., Salman M. D. (2011). A needs-assessment and demographic survey of infection-control and disease awareness in western US animal shelters. Prev Vet Med 98, 52–57 10.1016/j.prevetmed.2010.11.001 [DOI] [PubMed] [Google Scholar]
- Sun Y., Chen A. Y., Cheng F., Guan W., Johnson F. B., Qiu J. (2009). Molecular characterization of infectious clones of the minute virus of canines reveals unique features of bocaviruses. J Virol 83, 3956–3967 10.1128/JVI.02569-08 [DOI] [PMC free article] [PubMed] [Google Scholar]