Abstract
Jos virus (JOSV), originally isolated in Jos, Nigeria in 1967, has remained unclassified despite cultivation in tissue culture, development of animal models of infection and implementation of seroprevalence surveys for infection. Here, we report genetic, ultrastructural and serological evidence that JOSV is an orthomyxovirus distinct from but phylogenetically related to viruses of the genus Thogotovirus.
The family Orthomyxoviridae comprises viruses with six to eight segments of linear, negative-sense, ssRNA genomes. These viruses are currently assigned to six genera: Influenza A virus, Influenza B virus, Influenza C virus, Isavirus, Thogotovirus (Kawaoka et al., 2005) and the recently proposed Quarjavirus (Presti et al., 2009). The influenza viruses and isavirus (infectious salmon anemia virus) are transmitted to their vertebrate hosts by aerosol or through water (Kawaoka et al., 2005) and the thogotoviruses and quarjaviruses are transmitted by ticks (Da Silva et al., 2005; Kawaoka et al., 2005; Presti et al., 2009).
The genus Thogotovirus currently consists of three named viruses: Thogoto (THOV), Dhori (DHOV) and Araguari (ARAV) (Da Silva et al., 2005; Kawaoka et al., 2005). THOV and DHOV are widely distributed in Africa, southern Europe and Central Asia and are associated with ticks and mammals (Hubalek & Halouzka, 1996; Institut Pasteur da Dakar, 2001; Karabatsos, 1985). Two natural human infections have been reported for THOV with one fatality (Moore et al., 1975), and five accidental infections with DHOV have been reported (Butenko et al., 1987). All seven cases presented fever and encephalitis or meningoencephalitis. ARAV was isolated from a marsupial (opossum) collected in northern Brazil (Da Silva et al., 2005); its pathogenic potential is unknown.
Jos virus (JOSV) was originally isolated from cow serum (Bos indicus) in Jos, Nigeria in 1967 (Lee et al., 1974). Thereafter, the virus was repeatedly isolated from Amblyomma and Rhipicephalus (Boophilus) ticks collected in Ethiopia, Guinea, Central African Republic, Nigeria, Ivory Coast and Senegal (Institut Pasteur da Dakar, 2001; Wood et al., 1978). Studies of the field infection rate in ticks in the Central African Republic and Ethiopia found the prevalence to be 3 and 1 %, respectively (Sureau et al., 1976; Wood et al., 1978). Initial efforts to characterize the pathology of JOSV in infected suckling mice showed acute cell necrosis in the liver, lymph nodes, bone marrow and spleen (Fagbami & Ikede, 1978; Lee et al., 1974). Given the lack of genetic information or a serological relationship with other arboviruses, JOSV has remained unclassified.
Here, we report genetic, ultrastructural and serological evidence that JOSV is an orthomyxovirus distinct from but phylogenetically related to viruses of the genus Thogotovirus.
The virus strains used were THOV strain ITAL Ar 126, DHOV strain IAr-611313 and ARAV strain BeAn-174214. The JOSV prototype strain (IBAn-17854) was isolated from newborn mice inoculated with the original infected bovine serum (Lee et al., 1974). The virus killed newborn and 10-day-old mice within 4–5 days when inoculated intracerebrally or intraperitoneally (Fagbami & Ikede, 1978). All virus stocks were obtained from the World Reference Center for Emerging Viruses and Arboviruses at the University of Texas Medical Branch. Methods used to prepare antigens for the complement-fixation (CF) tests and for making immune ascitic fluids have been described previously (Beaty et al., 1989; Travassos da Rosa et al., 1983; Xu et al., 2007). Antigens and antibodies were both prepared in mice. CF titres were recorded as the highest dilutions giving 3+ or 4+ fixation of complement. Titres of 1 : 8 were considered positive. Haemagglutination inhibition (HI) tests were done in microtitre plates as described previously (Travassos da Rosa et al., 1983). HI tests were performed with four haemagglutination units of virus at the optimal pH (5.75) against serial twofold antiserum dilutions starting at 1 : 20. HI titres of 1 : 20 were considered positive. Results obtained in CF tests with ARAV, DHOV, THOV and JOSV are summarized in Supplementary Table S1 (available in JGV Online). All antisera were robustly reactive with their cognate antigens. Antisera to JOSV were modestly cross-reactive with the ARAV, DHOV and THOV; however, JOSV was recognized by the antisera to THOV but not the antisera to ARAV or DHOV.
Results obtained in HI tests are summarized in Supplementary Table S2 (available in JGV Online). JOSV, ARAV, DHOV and THOV had high titre HI activity against their cognate antigens. THOV antisera had HI activity against both JOSV and DHOV (1 : 640 vs 1 : 80, respectively). Similarly, JOSV antisera had HI activity against both DHOV and THOV antigens (1 : 320 vs 1 : 40, respectively). Although DHOV antigen was recognized by both JOSV and THOV antisera, DHOV serum only recognized its cognate antigen.
Transmission electron microscopy was performed as described previously by Popov et al. (1995). Briefly, Vero cells were infected with 0.01 TCID50 per cell, harvested 5 days post-infection, fixed with formaldehyde/glutaraldehyde, post-fixed with 1 % OsO4 and stained with uranyl acetate. Ultrathin sections were stained with lead citrate and examined in a transmission electron microscope at 60 kV. Pleiomorphic ovoid virions 85–120 nm in diameter were observed in the cytoplasm of infected cells. In some instances virions could also be seen budding from the cell surface (Fig. 1).
Fig. 1.
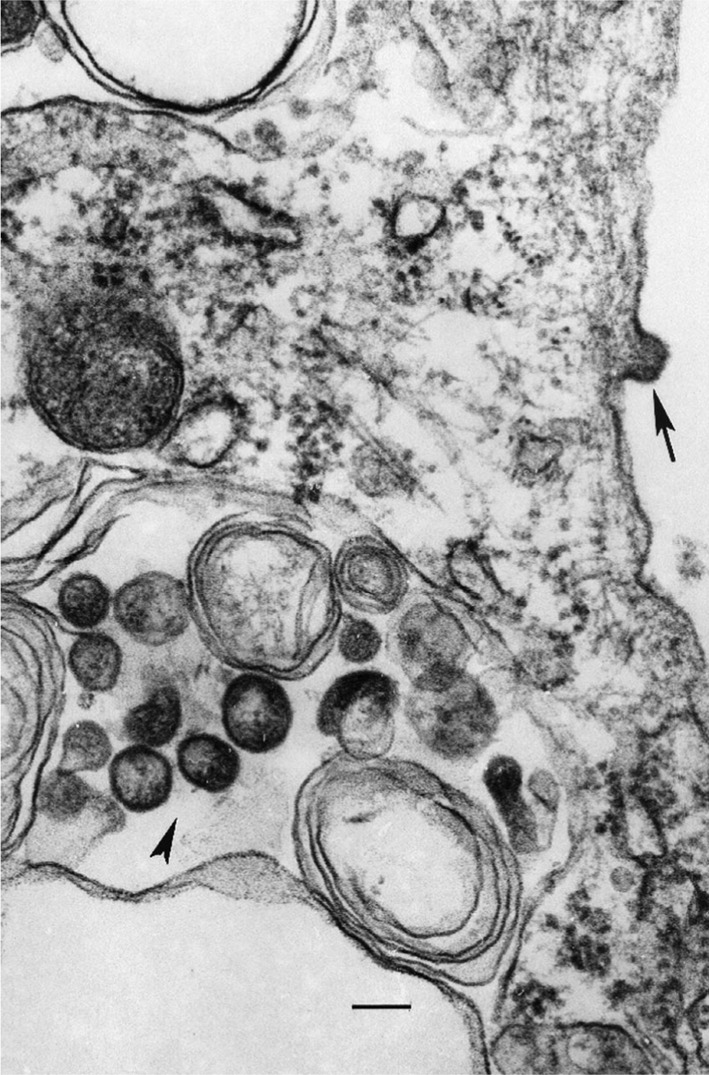
JOSV as observed in the cytoplasm of an infected Vero cell in ultrathin section. Arrow above indicates a virion budding from the cell surface. Bar, 100 nm. Arrowhead indicates virions 100–120 nm in diameter in the cytoplsam.
For genome sequencing, JOS viral stocks were pyrosequenced as described previously (Cox-Foster et al., 2007; Margulies et al., 2005; Palacios et al., 2008). Sequence gaps were completed by PCR and posterior Sanger sequencing, using primers based on pyrosequencing data. For 3′ termini of each segment, two primers (one for segments 1–5; a second for segment 6) with the 13 nt conserved THOV sequence were used for a specific reverse transcription with an additional arbitrary nucleotide on the 5′ end. This primer was designed to bind to the 3′ end of the genomic RNA. For the termini of each segment, we used the Clontech SMART RACE kit (Clontech) for the 5′ termini and 3′ RACE kit (Clontech) for 3′ termini. The sequence of the different segments was verified by Sanger sequencing using primers designed to create products of 1000 bp with 500 bp overlap from the draft sequence. For sequence assembly and analysis Geneious 4.8.3 (Biomatters Inc.) was used.
The assembled data resembled a classical Thogotovirus genus-like genome (GenBank accession nos HM627170–HM627175). Sequence analysis of JOSV indicates the presence of at least six RNA segments coding for seven ORFs corresponding to the polymerase basic protein 2 (PB2, segment 1), polymerase basic protein 1 (PB1, segment 2), acidic polypeptide (PA, segment 3), glycoprotein (GP, segment 4), nucleoprotein (NP, segment 5) and matrix (M) and its long isoform (ML) (segment 6). All remaining contigs and singletons in the pyrosequenced data were properly identified. No additional non-matched data were observed. This was interpreted as an indication that JOSV was composed of at least six segments.
The conserved terminal sequences of the viral RNA (vRNA) are partially complementary like those of THOV and influenza viruses. Indeed, the conserved terminal sequences of JOSV vRNA are identical to those of THOV: 5′-AGAGAUAUCAAGGC-3′ and 3′-UCGUUUUUGUCCG-5′ (segments 1–5) or 3′-UCACCUUUGUCCG-5′ (segment 6). Priming of viral mRNA synthesis in influenza viruses occurs by stealing capped fragments of 10–13 nt from the host (Lamb & Krug, 2001). Although THOV virus mRNA is capped, 5′ RACE analysis indicates that THOV mRNAs do not contain heterogeneous sequences (Weber et al., 1996, 1997). Similarly JOSV mRNAs do not contain heterogeneous sequences (data not shown). In contrast, 5′ RACE of mRNA from the novel Quaranfil virus identified 9–11 nt that are heterogeneous among the different products, a finding consistent with cap stealing (Presti et al., 2009; Weber et al., 1996, 1997).
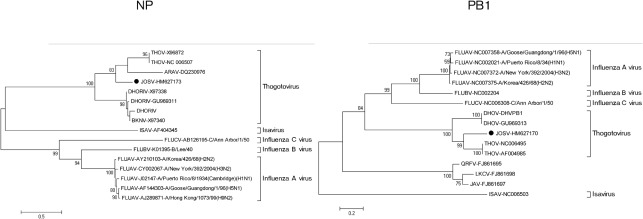
Phylogenetic analysis was performed using a set of orthomyxovirus sequences (16 for the NP segment and 15 for the PB1 segment) comprising all sequences available from GenBank (January 2011). Additionally, the phylogeny of each of the six segments of the members of the genera Thogotovirus and Quarjavirus was analysed with the purpose of clarifying the origin of the segments and for identifying recombination events. All sequences were aligned using the clustal algorithm (as implemented in the mega package version 3) at the nucleotide and amino acid level with additional manual editing to ensure the highest possible quality of alignment. Neighbour-joining analysis at the amino acid level was performed due to the observed high variability of the underlying nucleotide sequences of members of the family Orthomyxoviridae. The statistical significance of tree topology was evaluated by bootstrap resampling of the sequences 1000 times. Phylogenetic analysis was performed using mega software (Kumar et al., 2004). Neighbour-joining analysis at the nucleotide level was performed using the Kimura two-parameter and was evaluated by bootstrap resampling of the sequences 1000 times.
In the phylogenetic analysis of the more conserved ORFs at the family level (NP and PB1), major nodes that represent viruses belonging to the same genus were clearly distinct and confirmed previously reported topologies (Presti et al., 2009). JOSV was clearly associated with the genus Thogotovirus and the proposed genus Quarjavirus (Fig. 2).
Fig. 2.
Phylogenetic analysis of the NP and PB1 ORFs from all orthomyxoviruses. Bar represents the number of amino acid substitutions per site.
Analysis of the six segments at the nucleotide level confirmed the clustering of JOSV with thogotoviruses. Distance similarities of JOSV with other thogotoviruses, quarjaviruses and other members of the family are shown in Supplementary Table S3 (available in JGV Online). Branching inconsistencies were detected when ARAV was compared with JOSV and THOV (Supplementary Fig. S1, available in JGV Online). This may reflect the paucity of sequences used for analysis; only partial sequences of segments 4 and 5 of Araguari are available (575 nt for HA and 526 nt for NP). No evidence of reassortment was found using the Recombination Detection Program (RDP; Darren Martin) (Martin & Rybicki, 2000) and the algorithms Bootscan (Salminen et al., 1995), MaxChi (Smith, 1992), Chimaera (Posada & Crandall, 2001), LARD (Holmes, 1998) and phylip Plot (Felsenstein, 1989) (data not shown).
Finally, topology and targeting predictions were generated by employing SignalP, NetNGlyc, TMHMM (http://www.cbs.dtu.dk/services), TopPred2 (http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html), and integrated predictions in Geneious (Bendtsen et al., 2004; Claros & von Heijne, 1994; Kahsay et al., 2005; Käll et al., 2004; Krogh et al., 2001). The phyre program was used to predict structural similarity of the predicted ORF against known protein structures (Kelley & Sternberg, 2009). The algorithm WWIHS was used to predict areas of likely interaction between viral proteins and cell membrane proteins (Wimley & White, 1996).
ORF analysis showed that the JOSV viral RNA-dependent RNA-polymerase (PB1) contains the pre-A, A, B, C, D and E motifs found in the catalytic domain of negative-strand RNA viruses (Delarue et al., 1990; Müller et al., 1994; Poch et al., 1989; Vieth et al., 2004) (Supplementary Fig. S2, available in JGV Online). The influenza virus PB1 has two nuclear localization domains, not found in thogotoviruses and JOSV; however, a nuclear localization signal was predicted in JOSV by using PredictNLS (Nair & Rost, 2005) (K754RREAEEAIEEMTKRRK) (Supplementary Fig. S2).
Comparison of JOSV with THOV PB2 showed regions of high similarity at the 5′ end, suggesting that their conservation is under selection pressure (data not shown). This region is implicated in the interaction of the PB1 and PB2 subunits of influenza A virus (Perales et al., 1996).
The NP of the family Orthomyxoviridae is the major structural protein that associates with the genomic RNA segments to form the ribonucleoprotein particles. JOSV NP has many protein domains in common with the NPs of influenza viruses, although the amino acid sequence similarity is only 14.6, 16.4 and 17.3 % with FLUCV, FLUBV and FLUAV, respectively. Interestingly, four separate highly conserved short regions (14–30 aa long), initially identified for DHOV by Fuller et al. (1987), were detected (Supplementary Fig. S3, available in JGV Online). They may represent critical domains for conserved functions of this protein family; in fact, one of them includes the nuclear accumulation sequence as defined by Davey et al. (1985) (Supplementary Fig. S4, available in JGV Online). A bipartite nuclear localization signal similar to the one demonstrated in THOV (Weber et al., 1998) was detected in JOSV NP (positions 174–175 and 185–188). Moreover, a second putative bipartite nuclear localization signal was found at position 367–381. This sequence contains an upstream (KR) and a downstream (KGKR) cluster of basic amino acids that are separated by a stretch of 8 aa. It is predicted to have surface exposure. Using a similar approach, a similar motif can also be predicted in THOV. This putative signal overlaps partially with the fourth highly conserved region mentioned above and corresponds with the tail loop of the FLUAV NP. No sequence conservation was found when comparing the JOSV sequence with the regions responsible for influenza virus RNA binding (Albo et al., 1995; Kobayashi et al., 1994). Nonetheless, the predicted secondary structure (consisting of two alpha helices connected by a loop-β-sheet-loop domain) of the RNA-binding domain described by Albo et al. (1995) is conserved and the C-terminal region of the NP has structural similarity to the influenza virus NP as predicted by the phyre program (Kelley & Sternberg, 2009) (Supplementary Fig. S4). Taken together, these data suggest that while the NP gene is derived from a common ancestor among orthomyxoviruses, it followed a separate evolutionary path for the tick-borne viruses.
As previously predicted for THOV (Garry & Garry, 2008), the fourth largest RNA segment of JOSV encodes a putative GP that is similar to the corresponding proteins of ARAV, THOV and baculovirus GP64 with respect to the N-terminal signal sequence, pre-transmembrane and transmembrane domains, cysteine links, sequences with propensity to interface with a lipid bilayer [as identified with by Wimley–White interfacial hydrophobicity scale (WWIHS; Wimley & White, 1996)] and areas of N-glycosylation (Supplementary Fig. S5, available in JGV Online). Furthermore, the alignment shows five areas of high conservation, three of them corresponding to the predicted fusion domain for AcMNPV and THOV (Garry & Garry, 2008) (Supplementary Fig. S5). Thus, based on this structural similarity, the GP of JOSV should be classified as a class III penetrene.
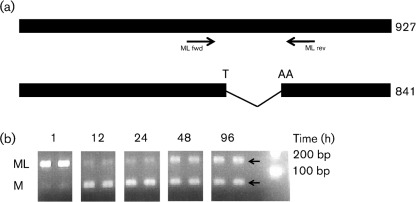
JOSV segment 6 is homologous to THOV segment 6 at the amino acid and nucleotide levels. THOV segment 6 encodes two transcripts: a spliced RNA that encodes the M protein and an unspliced RNA that encodes a C-terminally extended M protein termed ML (matrix protein long) (Hagmaier et al., 2003). RT-PCR analysis of Vero cells infected with JOSV using primers spanning the putative intron revealed the presence of the two expected RNA isoforms. The splicing of JOSV segment 6 transcript results in the formation of an UAA stop codon that terminates the ORF at nucleotide position 813, where the UA originates from the 5′ splice site and the A from the 3′ splice site (Fig. 3a). Moreover, we observed a time-dependent expression of these two isoforms. Whereas ML is expressed as early as 1 h after cell infection, M is expressed only after 12 h post-infection (Fig. 3b). The THOV ML protein has been described as an interferon-antagonist (Jennings et al., 2005) and the early expression of the JOSV protein ML is consistent with it serving a similar role. Moreover, the expression of the M isoform late during the infection is in agreement with its putative role as the major component of the virus particle, like the THOV M protein is (Kochs et al., 2000).
Fig. 3.
Transcripts of JOSV segment 6 modified by splicing. (a) Bars represent unspliced and spliced transcripts of segment 6. The black arrows show the position and orientation of PCR primers spanning the putative intron. (b) Time-course analysis of the expression of mRNA coding for JOSV ML and M (two biological replicates for each time point). Expected product size for ML and M transcript isoforms were 161 and 86 bp (arrows), respectively.
The molecular characterization of JOSV in addition with its similarity with DHOV and THOV, supports the possibility that JOSV should be considered a potential human pathogen. Fagbami and colleagues reported that intracerebral, intraperitoneal or subcutaneous inoculation of newborn mice with JOSV caused a fatal illness within 5 or 6 days with acute hepatocellular necrosis (Fagbami & Ikede, 1978; Mateo et al., 2007). Similar findings have been reported in mice experimentally infected with DHOV (Mateo et al., 2007) and with highly pathogenic influenza viruses (Kawaoka, 1991; Lu et al., 1999), suggesting a common pathogenesis for all of these orthomyxoviruses. Besides being genetically related to THOV, JOSV shares similar temporal and geographical distribution to the pathogenic THOV (Causey et al., 1969). Since JOSV is a tick-borne virus, seroprevalence studies on domestic animals could provide information on the level of circulation. Because of their structural and biochemical similarities to the influenza viruses, their abundance and wide geographical distribution, the ability of orthomyxoviruses to undergo reassortment and the emergence of new virus strains, JOSV and other thogotoviruses may deserve more attention. Their disease potential for humans, livestock and poultry may be overlooked.
Acknowledgements
This work was supported by Google.org, National Institutes of Health award AI57158 (Northeast Biodefense Center - Lipkin), and USAID Predict funding source code 07-301-7119-52258 (Center for Infection and Immunity), and the Department of Defense. A. T. da R., H. G., V. L. P. and R. B. T. were supported by NIH contracts NO1-AI25489 and HHSN272201000040I.
Footnotes
Supplementary material is available with the online version of this paper.
References
- Albo C., Valencia A., Portela A. (1995). Identification of an RNA binding region within the N-terminal third of the influenza A virus nucleoprotein. J Virol 69, 3799–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty B. J., Calisher C. H., Shope R. E. (1989). Arboviruses. In Diagnostic Procedures for Viral, Rickettsial and Chlamydial Infections, pp. 797–855 Edited by Schmidt N. J., Emmons R. W. Washington, DC: American Public Health Association [Google Scholar]
- Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004). Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340, 783–795 10.1016/j.jmb.2004.05.028 [DOI] [PubMed] [Google Scholar]
- Butenko A. M., Leshchinskaia E. V., Semashko I. V., Donets M. A., Mart’ianova L. I. (1987). [Dhori virus – a causative agent of human disease. 5 cases of laboratory infection]. Vopr Virusol 32, 724–729 (in Russian). [PubMed] [Google Scholar]
- Causey O. R., Kemp G. E., Madbouly M. H., Lee V. H. (1969). Arbovirus surveillance in Nigeria, 1964–1967. Bull Soc Pathol Exot 62, 249–253 [PubMed] [Google Scholar]
- Claros M. G., von Heijne G. (1994). TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci 10, 685–686 [DOI] [PubMed] [Google Scholar]
- Cox-Foster D. L., Conlan S., Holmes E. C., Palacios G., Evans J. D., Moran N. A., Quan P. L., Briese T., Hornig M. & other authors (2007). A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318, 283–287 10.1126/science.1146498 [DOI] [PubMed] [Google Scholar]
- Da Silva E. V., Da Rosa A. P., Nunes M. R., Diniz J. A., Tesh R. B., Cruz A. C., Vieira C. M., Vasconcelos P. F. (2005). Araguari virus, a new member of the family Orthomyxoviridae: serologic, ultrastructural, and molecular characterization. Am J Trop Med Hyg 73, 1050–1058 [PubMed] [Google Scholar]
- Davey J., Dimmock N. J., Colman A. (1985). Identification of the sequence responsible for the nuclear accumulation of the influenza virus nucleoprotein in Xenopus oocytes. Cell 40, 667–675 10.1016/0092-8674(85)90215-6 [DOI] [PubMed] [Google Scholar]
- Delarue M., Poch O., Tordo N., Moras D., Argos P. (1990). An attempt to unify the structure of polymerases. Protein Eng 3, 461–467 10.1093/protein/3.6.461 [DOI] [PubMed] [Google Scholar]
- Fagbami A. H., Ikede B. O. (1978). Pathogenicity and pathology of Jos virus infection in mice and tissue culture. Microbios 21, 81–88 [PubMed] [Google Scholar]
- Felsenstein J. (1989). phylip – phylogeny inference package (version 3.2). Cladistics 5, 164–166 [Google Scholar]
- Fuller F. J., Freed Man-Faulstich E. Z., Barnes J. A. (1987). Complete nucleotide sequence of the tick-borne, orthomyxo-like Dhori/Indian/1313/61 virus nucleoprotein gene. Virology 160, 81–87 10.1016/0042-6822(87)90047-X [DOI] [PubMed] [Google Scholar]
- Garry C. E., Garry R. F. (2008). Proteomics computational analyses suggest that baculovirus GP64 superfamily proteins are class III penetrenes. Virol J 5, 28 10.1186/1743-422X-5-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmaier K., Jennings S., Buse J., Weber F., Kochs G. (2003). Novel gene product of Thogoto virus segment 6 codes for an interferon antagonist. J Virol 77, 2747–2752 10.1128/JVI.77.4.2747-2752.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E. C. (1998). Molecular epidemiology of dengue virus – the time for big science. Trop Med Int Health 3, 855–856 10.1046/j.1365-3156.1998.00332.x [DOI] [PubMed] [Google Scholar]
- Hubalek Z., Halouzka J. (1996). Arthropod-borne viruses of vertebrates in Europe. Acta Sc Nat Brno 30, 1–95 [Google Scholar]
- Institut Pasteur da Dakar (2001). Centre Collaboreur OMS de Reference et de Recherche pour les Arbovirus et Virus de Fievres Hemorragiques, Rapport Annuel 2001. Dakar: Institut Pasteur de Dakar [Google Scholar]
- Jennings S., Martínez-Sobrido L., García-Sastre A., Weber F., Kochs G. (2005). Thogoto virus ML protein suppresses IRF3 function. Virology 331, 63–72 10.1016/j.virol.2004.10.015 [DOI] [PubMed] [Google Scholar]
- Kahsay R. Y., Gao G., Liao L. (2005). An improved hidden Markov model for transmembrane protein detection and topology prediction and its applications to complete genomes. Bioinformatics 21, 1853–1858 10.1093/bioinformatics/bti303 [DOI] [PubMed] [Google Scholar]
- Käll L., Krogh A., Sonnhammer E. L. (2004). A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338, 1027–1036 10.1016/j.jmb.2004.03.016 [DOI] [PubMed] [Google Scholar]
- Karabatsos N. (1985). International Catalogue of Arbovirus, including Certain Other Viruses of Vertebrates. San Antonio, Texas: American Society of Tropical Medicine and Hygiene [Google Scholar]
- Kawaoka Y. (1991). Equine H7N7 influenza A viruses are highly pathogenic in mice without adaptation: potential use as an animal model. J Virol 65, 3891–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka Y., Cox N. J., Haller O., Hongo S., Kaverin N., Klenk H. D., Lamb R. A., McCauley J., Palese P. & other authors (2005). Family Orthomyxoviridae. In Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses, pp. 681–693 Edited by Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. San Diego: Elsevier Academic Press [Google Scholar]
- Kelley L. A., Sternberg M. J. (2009). Protein structure prediction on the Web: a case study using the phyre server. Nat Protoc 4, 363–371 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Toyoda T., Adyshev D. M., Azuma Y., Ishihama A. (1994). Molecular dissection of influenza virus nucleoprotein: deletion mapping of the RNA binding domain. J Virol 68, 8433–8436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochs G., Weber F., Gruber S., Delvendahl A., Leitz C., Haller O. (2000). Thogoto virus matrix protein is encoded by a spliced mRNA. J Virol 74, 10785–10789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305, 567–580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. (2004). mega3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5, 150–163 10.1093/bib/5.2.150 [DOI] [PubMed] [Google Scholar]
- Lamb R. A., Krug R. M. (2001). Orthomyoviridae. In Fields Virology, 4th edn, pp. 1487–1532 Edited by Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E. Philadelphia: Lippincott Williams & Wilkins [Google Scholar]
- Lee V. H., Kemp G. E., Madbouly M. H., Moore D. L., Causey O. R., Casals J. (1974). Jos, a new tick-borne virus from Nigeria. Am J Vet Res 35, 1165–1167 [PubMed] [Google Scholar]
- Lu X., Tumpey T. M., Morken T., Zaki S. R., Cox N. J., Katz J. M. (1999). A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J Virol 73, 5903–5911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W. E., Attiya S., Bader J. S., Bemben L. A., Berka J., Braverman M. S., Chen Y. J. & other authors (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D., Rybicki E. (2000). RDP: detection of recombination amongst aligned sequences. Bioinformatics 16, 562–563 10.1093/bioinformatics/16.6.562 [DOI] [PubMed] [Google Scholar]
- Mateo R. I., Xiao S. Y., Lei H., Da Rosa A. P., Tesh R. B. (2007). Dhori virus (Orthomyxoviridae: Thogotovirus) infection in mice: a model of the pathogenesis of severe orthomyxovirus infection. Am J Trop Med Hyg 76, 785–790 [PubMed] [Google Scholar]
- Moore D. L., Causey O. R., Carey D. E., Reddy S., Cooke A. R., Akinkugbe F. M., David-West T. S., Kemp G. E. (1975). Arthropod-borne viral infections of man in Nigeria, 1964–1970. Ann Trop Med Parasitol 69, 49–64 [DOI] [PubMed] [Google Scholar]
- Müller R., Poch O., Delarue M., Bishop D. H., Bouloy M. (1994). Rift Valley fever virus L segment: correction of the sequence and possible functional role of newly identified regions conserved in RNA-dependent polymerases. J Gen Virol 75, 1345–1352 10.1099/0022-1317-75-6-1345 [DOI] [PubMed] [Google Scholar]
- Nair R., Rost B. (2005). Mimicking cellular sorting improves prediction of subcellular localization. J Mol Biol 348, 85–100 10.1016/j.jmb.2005.02.025 [DOI] [PubMed] [Google Scholar]
- Palacios G., Druce J., Du L., Tran T., Birch C., Briese T., Conlan S., Quan P. L., Hui J. & other authors (2008). A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med 358, 991–998 10.1056/NEJMoa073785 [DOI] [PubMed] [Google Scholar]
- Perales B., de la Luna S., Palacios I., Ortín J. (1996). Mutational analysis identifies functional domains in the influenza A virus PB2 polymerase subunit. J Virol 70, 1678–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poch O., Sauvaget I., Delarue M., Tordo N. (1989). Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J 8, 3867–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov V. L., Chen S. M., Feng H. M., Walker D. H. (1995). Ultrastructural variation of cultured Ehrlichia chaffeensis. J Med Microbiol 43, 411–421 10.1099/00222615-43-6-411 [DOI] [PubMed] [Google Scholar]
- Posada D., Crandall K. A. (2001). Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci U S A 98, 13757–13762 10.1073/pnas.241370698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presti R. M., Zhao G., Beatty W. L., Mihindukulasuriya K. A., Travassos da Rosa A. P., Popov V. L., Tesh R. B., Virgin H. W., Wang D. (2009). Quaranfil, Johnston Atoll, and Lake Chad viruses are novel members of the family Orthomyxoviridae. J Virol 83, 11599–11606 10.1128/JVI.00677-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen M. O., Carr J. K., Burke D. S., McCutchan F. E. (1995). Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retroviruses 11, 1423–1425 10.1089/aid.1995.11.1423 [DOI] [PubMed] [Google Scholar]
- Smith J. M. (1992). Analyzing the mosaic structure of genes. J Mol Evol 34, 126–129 10.1007/BF00182389 [DOI] [PubMed] [Google Scholar]
- Sureau P., Cornet J. P., Germain M., Camicas J. L., Robin Y. (1976). [Survey of tick-borne arboviruses in the Central African Republic (1973–1974). Isolation of Dugbe, CHF/Congo, Jos and Bhanja viruses]. Bull Soc Pathol Exot 69, 28–33 (in French). [PubMed] [Google Scholar]
- Travassos da Rosa A. P., Tesh R. B., Pinheiro F. P., Travassos da Rosa J. F., Peterson N. E. (1983). Characterization of eight new phlebotomus fever serogroup arboviruses (Bunyaviridae: Phlebovirus) from the Amazon region of Brazil. Am J Trop Med Hyg 32, 1164–1171 [DOI] [PubMed] [Google Scholar]
- Vieth S., Torda A. E., Asper M., Schmitz H., Günther S. (2004). Sequence analysis of L RNA of Lassa virus. Virology 318, 153–168 10.1016/j.virol.2003.09.009 [DOI] [PubMed] [Google Scholar]
- Weber F., Haller O., Kochs G. (1996). Nucleoprotein viral RNA and mRNA of Thogoto virus: a novel ‘cap-stealing’ mechanism in tick-borne orthomyxoviruses? J Virol 70, 8361–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., Haller O., Kochs G. (1997). Conserved vRNA end sequences of Thogoto-orthomyxovirus suggest a new panhandle structure. Arch Virol 142, 1029–1033 10.1007/s007050050138 [DOI] [PubMed] [Google Scholar]
- Weber F., Kochs G., Gruber S., Haller O. (1998). A classical bipartite nuclear localization signal on Thogoto and influenza A virus nucleoproteins. Virology 250, 9–18 10.1006/viro.1998.9329 [DOI] [PubMed] [Google Scholar]
- Wimley W. C., White S. H. (1996). Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol 3, 842–848 10.1038/nsb1096-842 [DOI] [PubMed] [Google Scholar]
- Wood O. L., Lee V. H., Ash J. S., Casals J. (1978). Crimean-Congo hemorrhagic fever, Thogoto, dugbe, and Jos viruses isolated from ixodid ticks in Ethiopia. Am J Trop Med Hyg 27, 600–604 [DOI] [PubMed] [Google Scholar]
- Xu F., Liu D., Nunes M. R., Travassos da Rosa A. P., Tesh R. B., Xiao S. Y. (2007). Antigenic and genetic relationships among Rift Valley fever virus and other selected members of the genus Phlebovirus (Bunyaviridae). Am J Trop Med Hyg 76, 1194–1200 [PubMed] [Google Scholar]