Abstract
Real-time quaking-induced conversion (RT-QuIC) is an assay in which disease-associated prion protein (PrP) initiates a rapid conformational transition in recombinant PrP (recPrP), resulting in the formation of amyloid that can be monitored in real time using the dye thioflavin T. It therefore has potential advantages over analogous cell-free PrP conversion assays such as protein misfolding cyclic amplification (PMCA). The QuIC assay and the related amyloid seeding assay have been developed largely using rodent-passaged sheep scrapie strains. Given the potential RT-QuIC has for Creutzfeldt–Jakob disease (CJD) research and human prion test development, this study characterized the behaviour of a range of CJD brain specimens with hamster and human recPrP in the RT-QuIC assay. The results showed that RT-QuIC is a rapid, sensitive and specific test for the form of abnormal PrP found in the most commonly occurring forms of sporadic CJD. The assay appeared to be largely independent of species-related sequence differences between human and hamster recPrP and of the methionine/valine polymorphism at codon 129 of the human PrP gene. However, with the same conditions and substrate, the assay was less efficient in detecting the abnormal PrP that characterizes variant CJD brain. Comparison of these QuIC results with those previously obtained using PMCA suggested that these two seemingly similar assays differ in important respects.
Introduction
Creutzfeldt–Jakob disease (CJD) is a fatal neurological condition characterized by neuronal loss, reactive gliosis and spongiform changes in the brain. The condition is unusual in that it can be genetic (associated with mutations in the prion protein gene, PRNP), it can be acquired in the forms of iatrogenic or variant CJD (vCJD) or it can occur in an idiopathic form known as sporadic CJD (sCJD). sCJD occurs worldwide with a relatively uniform incidence of 1–2 cases per million of the population per year. There is marked heterogeneity in the clinico-pathological phenotype in sCJD. In contrast, vCJD has a relatively stereotyped phenotype. All forms of CJD are transmissible under appropriate experimental settings (reviewed by Ironside et al., 2008).
The existence of acquired forms and transmissibility both imply the existence of an infectious agent in CJD. According to the prion hypothesis, the agent is a conformationally altered form of the host-encoded prion protein (PrP) that is converted from a normal cellular isoform (PrPC) to a disease-associated isoform (PrPSc) by a post-translational conformational transition. This renders it higher in β-sheet structures, relatively poorly soluble, prone to aggregation and partially protease resistant (reviewed by Prusiner, 1998; Caughey et al., 2009). This last property has been exploited diagnostically, and protease-resistant prion protein (PrPres) has become a convenient operational definition of PrPSc. Western blot analysis of the N-terminally truncated protease-resistant core fragment size and the relative ratio of the three possible glycoforms have shown that different forms of CJD are associated with different PrPres types. vCJD is characterized by a 19 kDa (type 2) PrPres, with the diglycosylated glycoform predominating (termed type 2B). In contrast, mono- or non-glycosylated PrPres predominates in sCJD and the PrPres can be either type 1 (21 kDa) or type 2, or a mixture of the two isoforms. The PrPres type and the naturally occurring polymorphism at codon 129 of PRNP [which can encode either methionine (M) or valine (V)] are thought to partially underlie the phenotypic variation in sCJD, which is now classified according to these parameters as MM1/MV1, VV1, MM2C (cortical variant), MM2T (thalamic variant or sporadic fatal insomnia), MV2K (kuru plaque variant) or VV2 using a system introduced in 1999 and recently updated to take into account the existence of mixed PrPres types (Parchi et al., 2010).
Transmissibility also adds a public-health dimension to managing these diseases: CJD (most probably sCJD) has been transmitted from person to person via human growth hormone therapy, dura mater grafting, contaminated surgical instruments and corneal transplantation (reviewed by Brown et al., 2006), but current concerns have focused on vCJD, which has recently been shown to be transmissible from asymptomatic blood donors to recipients of red blood cells and blood products (Llewelyn et al., 2004; Peden et al., 2004, 2010; Hewitt et al., 2006; Wroe et al., 2006). Although PrPSc accumulates to readily detectable levels in the brain during the clinical phase, PrPSc levels in the blood are likely to be considerably lower and those present in blood during the asymptomatic pre-clinical phase lower still.
Nevertheless, the need for a blood-screening assay, particularly in the UK where the underlying incidence of vCJD infection may be higher than the level of clinical vCJD cases suggests, has provided an incentive for the development of tests for PrPSc as a surrogate for infectivity in blood (reviewed by Hilton, 2006; Peden et al., 2008; Turner & Ludlam, 2009; Will, 2010). One possible strategy is to use the conversion process to amplify the amount of PrPSc in vitro until a point is reached where PrPSc is readily detectable by conventional means. The efficiency of early methods for PrPSc template-dependent conversion of PrPC was improved dramatically by the introduction of cycles of sonication and incubation in a process that came to be known as protein misfolding cyclic amplification (PMCA) (reviewed by Jones et al., 2011). PMCA and detection of the resultant PrPres by Western blotting has proved to be a sensitive method for the detection of low levels of PrPSc. It has also provided a rapid and flexible in vitro system in which to model aspects of prion replication relevant to human prion disease, such as the effect of PrPSc type, PRNP polymorphisms and transmission barriers (Jones et al., 2007, 2009a, b).
More recently, a third-generation assay has been developed in which recombinant prion protein (recPrP) replaces the source of naturally occurring PrPC in the PMCA assay (usually a brain homogenate) and controlled periodic shaking replaces sonication. The resulting assay, termed quaking-induced conversion (QuIC), has been developed using hamster-adapted sheep scrapie brain tissue and a Western blot readout for PrPres (Atarashi et al., 2007, 2008). It has shown to be sufficiently sensitive to detect PrPSc in cerebrospinal fluid (CSF) from hamsters with scrapie (Atarashi et al., 2008) and has also been shown to detect PrPSc in vCJD brain and CSF from scrapie-infected sheep (Orrú et al., 2009). More recently, the QuIC assay has incorporated aspects of the amyloid seeding assay developed by Colby et al. (2007), employing thioflavin T (ThT) as a reporter of recPrP conversion in real time, producing a real-time QuIC (RT-QuIC) assay for hamster scrapie prions that is specific and has a sensitivity that rivals that of bioassays (Wilham et al., 2010; Atarashi et al., 2011).
Given the potential importance of such in vitro assays for understanding the mechanisms by which abnormal PrP is produced in CJD and the potential public-health applications of a rapid and sensitive test for human prions, we undertook a characterization of RT-QuIC using recPrP ‘substrates’ differing in sequence (including human) and brain PrPSc ‘seeds’ from the major relevant forms of CJD. The results showed that RT-QuIC can be applied to CJD specimens, but comparison of the results with those obtained using PMCA indicated mechanistic differences between these two ostensibly similar assays.
Results
sCJD brain homogenate seeds the conversion of hamster recPrP (recHaPrP)
sCJD brain homogenates seeded heterologous RT-QuIC reactions using recHaPrP, both full-length and N-terminally truncated. The results shown in Figs 1–3 were typical of experiments performed at least three times using brain homogenate seeds prepared on two separate occasions from patients of each subtype of sCJD. The reactions were seeded with 2 µl of a dilution of CJD brain containing approximately 100 fg PrPres. Non-CJD brain tissue controls were added at a dilution of 0.5×10−5 of 100 % brain tissue; this dilution was within the 9×10−7–1×10−5 range of brain frontal cortex tissue dilutions for each sCJD and vCJD patient that gave approximately 100 fg PrPres in 2 µl (see Methods).
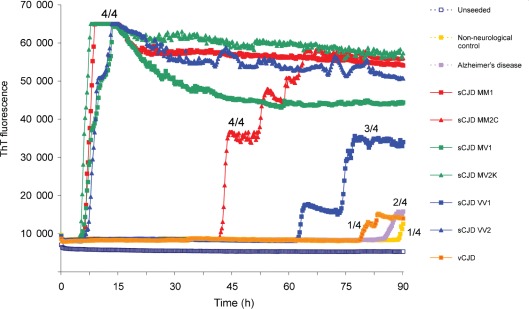
Fig. 1.
sCJD brain homogenate efficiently seeds the conversion of N-terminally truncated recHaPrP. RT-QuIC was performed using N-terminally truncated recHaPrP (aa 90–231) seeded with brain homogenates from sCJD, vCJD (patient 1), Alzheimer’s disease or a non-neurological negative control. Reactions were seeded with the equivalent of 100 fg PrPres from CJD brain homogenate (or an equivalent dilution for negative-control brain homogenate). The results of an unseeded reaction are also shown. The mean ThT fluorescence values per well were plotted against time. Fluorescence counts increased to a maximum of 65 000 per well. The number of samples that became positive out of the four replicates for each seed during the course of the experiment is written as a fraction next to the traces.
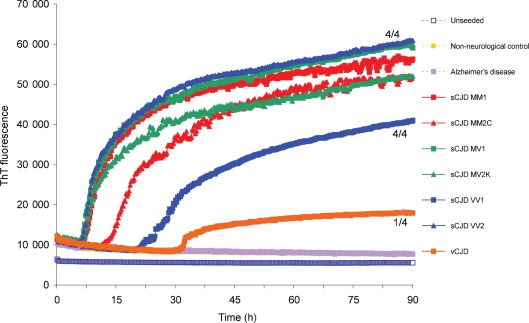
Fig. 3.
sCJD brain homogenate efficiently seeds the conversion of full-length recHaPrP. RT-QuIC was performed using full-length recHaPrP (aa 23–231) seeded with brain homogenates from sCJD, vCJD or Alzheimer’s disease patients at 100 fg PrPres (or an equivalent dilution for the non-neurological negative-control brain homogenate). An unseeded reaction is also shown. The mean ThT fluorescence values per well were plotted against time. The number of samples that became positive out of the four replicates for each seed during the course of the experiment is written as a fraction beside the traces.
A rapid early and near-synchronous seeding of truncated recHaPrP (aa 90–231) was seen with the sCJD MM1, MV1, MV2K and VV2 brain samples that initiated within 10 h and reached a plateau within 15 h (Fig. 1). In contrast, the sCJD MM2C and VV1 samples had considerably and consistently longer lag times. This was particularly true for the sCJD VV1 brain sample, which additionally failed to reach maximal values within the 90 h reaction time and with only three of the four replicates rising above background readings in the example shown in Fig. 1. The unseeded reactions gave baseline values throughout the assays. The vCJD, Alzheimer’s disease, Lewy body dementia (data not shown) and non-neurological disease brain samples gave either no discernible response or gave responses that were inconsistent between replicate wells and were much delayed. Examples of these phenomena can be seen in Fig. 1 in which each trace is marked with the number of replicate wells out of four that gave above-background values during the 90 h reaction time.
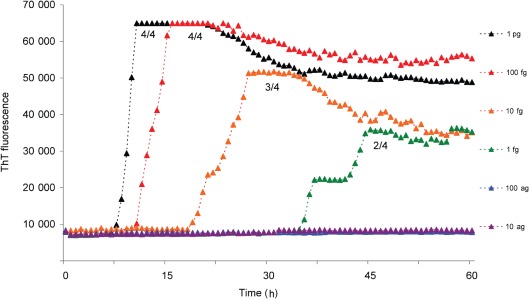
A dose–response analysis using truncated recHaPrP (aa 90–231) substrate conducted with MV2K sCJD seed indicated that both the duration of the lag and the reaction rate (once initiated) were related to dose (Fig. 2). The kinetics of fibrilization of the truncated recHaPrP substrate conformed to equation (1) given in Methods, which has been used previously to describe amyloid formation by other proteins. The rate of conversion correlated with the amount of sCJD seed introduced into the RT-QuIC reaction, i.e. a tenfold reduction in seed resulted in an approximate twofold decrease in k (rate constant). Under these conditions, RT-QuIC could detect MV2K sCJD brain seed down to a dilution of 1 fg PrPres (Fig. 2), although at this concentration of seed only two of the four replicates became positive. This amount of seed corresponded to 2 µl of a 9×10−8 dilution of 100 % brain tissue or 1.8×10−10 g brain tissue, and indicated a high analytical sensitivity (i.e. a low limit of detection).
Fig. 2.
Dose–response curves for RT-QuIC using sCJD (MV2K subtype). RT-QuIC reactions using recHaPrP (aa 90–231) substrate were seeded with a range of sCJD MV2K brain seed amounts ranging from the equivalent of 1 pg to 10 ag PrPres. The mean ThT fluorescence values per well were plotted against time. The number of samples that became positive out of the four replicates for each dilution of seed during the course of the experiment is written as a fraction beside each trace.
The kinetics of the RT-QuIC reaction was found to change when the truncated recHaPrP (aa 90–231) was replaced with full-length recHaPrP (aa 23–231) (Fig. 3) and conformed better to equation (2) shown in Methods. Each of the sCJD brain samples (MM1, MV1, VV1, MM2C, MV2K and VV2) produced responses rising above baseline within 25 h and gave similar, sigmoidal curves. The order of appearance was the same as with the truncated recHaPrP substrate, with sCJD MM1, MV1, MV2K and VV2 appearing early, followed by MM2C and then VV1. Inconsistent and low responses from the vCJD brain samples was also a feature of RT-QuIC with full-length recHaPrP, whereas signals from unseeded and non-CJD samples remained at baseline values throughout the 90 h reaction times (Fig. 3).
The conversion of recPrP substrate by sCJD seeds in the RT-QuIC reactions showed a reasonable degree of consistency, particularly for samples that gave rapid and early responses. For example 100 fg sCJD MM1 sample resulted in the conversion of truncated recHaPrP (aa 90–231) with a lag time of 6.0±1.4 h (mean±sd, n = 4) and the conversion of full-length recHaPrP (aa 23–231) with a lag time of 5.4±2.0 (n = 4), where n = 4 denotes samples run in quadruplicate on four separate occasions.
Full-length human recPrP (recHuPrP) is converted with similar kinetics to full-length recHaPrP
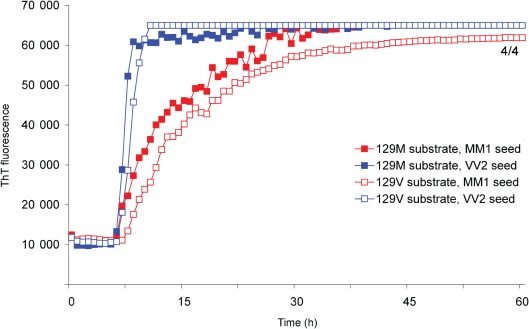
Next we replaced the full-length hamster sequence with full-length recHuPrP (aa 23–231) to produce a homologous RT-QuIC reaction. Both MM1 and VV2 sCJD brain homogenate seeds promoted the rapid and early conversion of full-length recHuPrP (Fig. 4) at dilutions similar to those used to stimulate the conversion of full-length recHaPrP (Fig. 3). The reactions were again specific in that no response was seen using equivalent dilutions of Alzheimer’s disease or Lewy body dementia samples or a brain homogenate from an individual without neurological disease (data not shown). We found that recHuPrP 129M and 129V substrates were both able to detect sCJD seeds of subtypes MM1 and VV2 (Fig. 4) without an apparent preference for matching of the codon 129 genotype between seed and substrate. The kinetics of fibrilization appeared to be more rapid for RT-QuIC reactions seeded with sCJD VV2 brain homogenate, irrespective of whether the recHuPrP substrate was 129M or 129V (Fig. 4).
Fig. 4.
sCJD MM1 and VV2 subtypes both efficiently seed recHuPrP irrespective of whether M or V is present at position 129. sCJD brain homogenate of subtype MM1 or VV2, diluted to 100 fg PrPres, was used to seed RT-QuIC reactions using either recHuPrP-129M or recHuPrP-129V substrate. The mean ThT fluorescence values per well were plotted against time. The number of samples that became positive out of the four replicates for each seed during the course of the experiment is written as a fraction beside the traces.
vCJD brain is less efficient at seeding RT-QuIC reactions
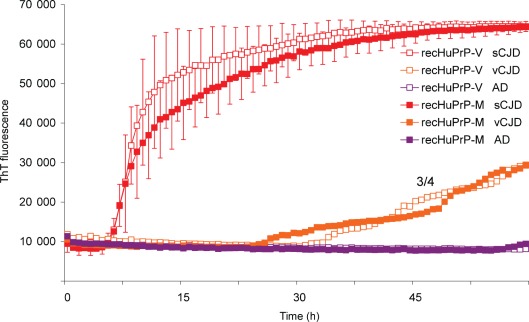
vCJD brain was less efficient at seeding homologous RT-QuIC reactions than sCJD, even though the assays were primed with equivalent amounts of seed PrPres. Fig. 5 shows a direct comparison of seeding RT-QuIC reactions using full-length recHuPrP with sCJD or vCJD brain seed containing 100 fg PrPres (corresponding to 2 µl 1×10−5 dilution of a 10 % brain homogenate). The recHuPrP-129M and -129V substrates produced similar results with vCJD seeds, giving a delayed and slower conversion than sCJD. The response, as judged by lag periods between sCJD (MM1 and VV2) and vCJD brain, were significantly different, as shown in Supplementary Table S1 (available in JGV Online).
Fig. 5.
vCJD frontal cortex brain homogenate seeds the conversion of recHuPrP much less efficiently than sCJD homogenate. recHuPrP-129M or recHuPrP-129V was seeded with dilutions of sCJD or vCJD (patient 2) brain homogenate containing the equivalent of 100 fg PrPres per reaction. As a control, reactions were seeded with an equivalent dilution of Alzheimer’s disease (AD) brain homogenate (purple filled and opened squares). The mean ThT fluorescence values per well were plotted against time. The variability of sCJD-seeded reactions where recHuPrP-129M (filled red squares) and recHuPrP-129V (open red squares) were used as a substrate is shown. The recHuPrP-129M curve represents the mean of nine sCJD-seeded reactions (seven seeded with MM1 and two with VV2). The recHuPrP-129V curve represents the mean of four MM1- and two VV2-seeded reactions. Vertical bars represent sd. These mean curves were compared with representative traces from vCJD-seeded reactions (orange filled and open squares). The number of vCJD samples that became positive out of the four replicates is shown as a fraction beside the trace. A similar result was obtained using brain seeds from vCJD patient 3 (data not shown).
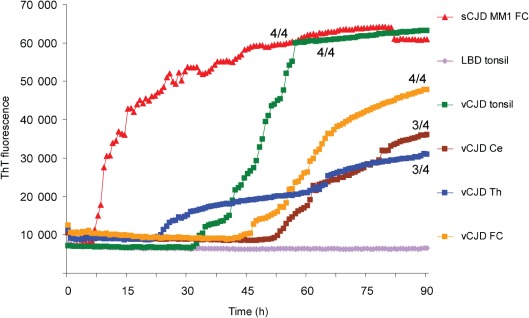
Samples of thalamus and cerebellum homogenate from two different cases of vCJD were also assayed by RT-QuIC using recHuPrP substrate. These homogenates were titrated for PrPres and diluted to the same levels of PrPres used to analyse the sCJD and vCJD frontal cortex homogenates. Similar results were obtained for vCJD thalamus and cerebellum compared with vCJD frontal cortex, suggesting that, compared with sCJD and regardless of the brain region of origin, vCJD PrPSc was less able to stimulate the conversion of recPrP substrates using the conditions used in this RT-QuIC assay (Fig. 6).
Fig. 6.
vCJD thalamus, cerebellum and tonsil seed the conversion of recHuPrP less efficiently than sCJD brain homogenate. recHuPrP-129M was seeded with diluted samples containing the equivalent of 100 fg PrPres from sCJD MM1 brain frontal cortex (FC), vCJD brain frontal cortex, vCJD cerebellum (Ce) and vCJD medial thalamus (Th), all from vCJD patient 3. Similar reactions, again using recHuPrP-129M as a substrate, were seeded with diluted samples of vCJD tonsil from a fourth vCJD patient containing the equivalent of 100 fg PrPres or an equivalent dilution of tonsil from a neurological control patient with Lewy body dementia (LBD). The number of vCJD samples that became positive out of the four replicates during the course of the experiment is shown as a fraction beside the traces.
In vCJD, PrPSc is readily detectable in tissues outside the central nervous system. In particular, lymphoreticular tissues such as spleen, lymph nodes and tonsil are consistently affected. We therefore investigated whether PrPSc extracted from vCJD tonsil was more efficient at stimulating the conversion of recPrP than PrPSc from vCJD brain. Homogenates of vCJD tonsil (10 %, w/v) were prepared and the amount of PrPSc per unit of volume was estimated by titrating for PrPres by Western blotting. Dilutions of vCJD tonsil homogenate containing femtogram amounts of PrPres produced a positive response, in contrast to equivalent dilutions of non-CJD tonsil. As an example, Fig. 6 shows the result for a RT-QuIC reaction using recHuPrP-129M substrate seeded with 100 fg vCJD tonsil PrPres. However, the lag times (>30 h) for these responses were again considerably longer than that of sCJD brain.
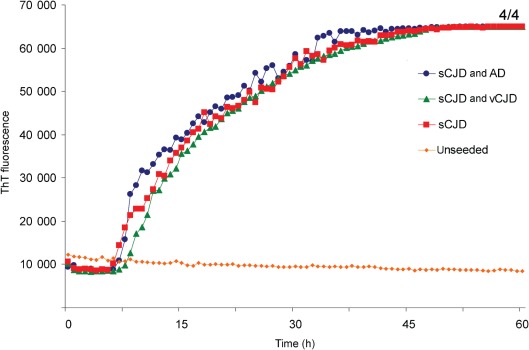
The presence of inhibitors of the RT-QuIC reaction in the vCJD brain was discounted by competition experiments involving the mixing of vCJD and sCJD brain homogenates. In the experiment shown in Fig. 7, a 10 % sCJD MM1 brain homogenate was diluted to 100 fg PrPres µl−1, which was a 2×10−4 dilution. This dilution was mixed 1 : 1 with PBS or with vCJD brain homogenate diluted to 10 pg PrPres µl−1 (or a 2×10−2 dilution) or the same dilution of an Alzheimer’s disease brain homogenate. Two microlitres of these mixtures was added to the RT-QuIC reactions using recHuPrP-129 M as substrate. The presence of a 100-fold excess of either vCJD or Alzheimer’s disease brain homogenate did not alter the conversion of substrate caused by sCJD MM1 seed (Fig. 7). A range of vCJD or Alzheimer’s disease brain homogenate dilutions (2×10−2–2×10−6) were pre-mixed 1 : 1 with the 2×10−4 dilution of sCJD seed but none altered the apparent kinetics of sCJD-seeded conversion (data not shown). We concluded that the poor seeding ability of vCJD brain homogenate was not due to the presence of an endogenous inhibitory factor that also interfered with sCJD-seeded conversion.
Fig. 7.
The presence of vCJD brain homogenate does not affect the ability of sCJD brain homogenate to seed the conversion of recHuPrP. Homogenates (10 %, w/v) of vCJD (patient 2) and Alzheimer’s disease (AD) brain frontal cortex (from a patient who was MM at PRNP codon 129) were both diluted to 2×10−2 with N2 buffer. A 10 % homogenate of sCJD MM1 brain was diluted 2×10−4 and mixed 1 : 1 with N2 buffer or the 2×10−2 dilutions of either Alzheimer’s disease or vCJD brain homogenates. Two microlitres of these mixtures was added to RT-QuIC reactions using recHuPrP-129M as a substrate. The mean ThT fluorescence values per well were plotted against time. The number of samples that became positive out of the four replicates during the course of the experiment is shown as a fraction beside the traces.
There is evidence to suggest that the most infectious PrPSc oligomers are relatively small (Silveira et al., 2005). Given that vCJD brain neuropathology is characterized by the presence of numerous large plaques containing PrPSc in the form of amyloid, we investigated the effect of sonicating vCJD brain homogenate in the hope of reducing the mean size of PrP aggregates prior to the serial dilution step. The sonication conditions chosen were those used in a single round of our PMCA protocol (Jones et al., 2007). No obvious increase or decrease was observed in the efficiency of vCJD brain homogenate to seed the conversion of recHuPrP-129M following sonication (data not shown).
Discussion
This is the first systematic investigation of the potential for human brain PrPSc from different forms of CJD to seed the RT-QuIC assay and is also the first to investigate whether the sequence of the recPrP used has a direct bearing on whether a given CJD brain homogenate can seed the RT-QuIC reaction. Our data showed that sCJD brain homogenates can seed heterologous RT-QuIC reactions using recHaPrP, whether full-length or N-terminally truncated, and that replacing the hamster sequence with full-length recHuPrP produced similar results. Likewise, full-length human PrP with M or V at position 129 appeared to be interchangeable in RT-QuIC seeded with CJD brain homogenates. When equivalent amounts of sCJD brain PrPSc (as judged by titration of PrPres against recPrP) were used, rapid and early seeding was seen with the most commonly occurring forms of sCJD (MM1/MV1, VV2 and MV2K). Interestingly, Atarashi et al. (2011) also showed that recHuPrP converted equally well in the presence of sCJD MM1 and MM2 brain seeds, in addition to the majority of CSF samples from sCJD patients, irrespective of the PrPres type or the PRNP codon 129 genotype. We did, however, find increased lag times and less reproducible responses when using MM2C and VV1 brain samples. Whether this is a general feature of these different forms of CJD, perhaps reflecting something intrinsic to PrPSc, or something particular to these specific cases or samples will require further investigation of a larger series of sCJD cases.
A dose–response experiment conducted with MV2K sCJD seed and truncated recHaPrP substrate indicated that both the duration of the lag and the reaction rate (once initiated) were related to dose, and it is interesting to note that the RT-QuIC reaction kinetics, when recHaPrP (aa 90–231) was used as a substrate, conformed to equations used previously to describe amyloid formation by other proteins. The biochemical sensitivity of sCJD PrPSc detection (as determined for sCJD MV2K) compared favourably with other sensitive assays, detecting PrPres in the femtogram range or as little as 2 µl of a ~10−7 dilution of 100 % brain tissue. Although the exact PrPSc molecular species involved in the initiation of the seeding of recPrP in RT-QuIC is not currently known, cross-seeding between amyloidogenic molecules did not appear to occur to any great extent under the conditions used, given that the results from Alzheimer’s disease and Lewy body dementia brain homogenates were very similar to those obtained with normal human brain homogenates.
It was therefore all the more surprising that vCJD brain was considerably slower at seeding RT-QuIC reactions than sCJD, even though the assays were primed with equivalent amounts of seed PrPres. We were able to discount certain explanations for this phenomenon. Case-to-case variation could be discounted by the analysis of samples from three different vCJD cases. Matrix effects could be discounted by the testing of three different regions of the vCJD brain and by testing of vCJD tonsil, which also accumulates PrPSc. The presence of inhibitors of the RT-QuIC reaction in the variant CJD brain was discounted by competition experiments involving the mixing of vCJD and sCJD brain homogenates. In addition, seeding with a wide range of vCJD brain homogenate volumes or pre-sonication to reduce aggregate size did not result in efficient seeding with vCJD brain homgenates. We are therefore forced to conclude that competence to seed RT-QuIC reactions depends on molecular properties of PrPSc that are at least partially independent of PrPres type and codon 129 genotype. One possibility is that this relates to the glycosylation of vCJD PrPSc.
The amounts of the various brain homogenates used to prime the RT-QuIC assays were normalized according to the amounts of PrPres, the protease-resistant component of PrPSc. In principle the seeding activity could be a protease-resistant and/or protease-sensitive component of PrPSc (Safar et al., 1998, 2005). This raised the possibility that the discrepancy between the seeding efficiencies of vCJD and sCJD brain homogenate might be accounted for by differences in the proportion of protease-sensitive and -resistant PrPSc. In this present study, we noted that, for sCJD MV2K, a tenfold change in the level of seed only produced an approximately twofold increase in lag time (Fig. 2). Therefore, the small differences (~20–40 %) in the relative levels of protease-sensitive PrPSc observed between vCJD and sCJD brain homogenates (Choi et al., 2011) are unlikely to account for the dramatic differences in the lag time for conversion in RT-QuIC reactions.
QuIC and PMCA assays have strong formal similarities. Both are essentially seeded prion protein conversion assays. However, there are several potentially significant differences between the two tests. First, PMCA reaction conditions are biochemically complex. Although cell-free, the conversion occurs in a crude homogenate containing proteins, lipids and nucleic acids. Secondly, in PMCA, the substrate (PrPC) is naturally produced and glycosylated and contains a glycosylphosphatidylinsitol (GPI) anchor by which its interaction with membranes is effected. In contrast, QuIC employs purified, bacterially expressed PrP that is devoid of glycans and has no GPI anchor, and that has been through a cycle of denaturation and refolding during purification. The physical forces used to accelerate the reactions also differ. QuIC employs periodic shaking, whereas PMCA depends on periodic sonication. A further difference in the real-time variant of QuIC is that ThT fluorescence is monitored rather than the production of PrPres in PMCA and QuIC. The results we have shown here using RT-QuIC contrast sharply with our previous experience of PMCA. Unlike RT-QuIC, non-CJD human brain, human platelets and humanized transgenic mouse brain all support the amplification of vCJD PrPSc in the PMCA assay but do so in a codon 129-compatible manner (Jones et al. 2007, 2009b). Humanized transgenic mouse brain also supports PMCA of sporadic CJD brain seeds, but again this occurs in a codon 129-dependent manner (Jones et al., 2009b), partially replicating the transmission properties of sCJD agents transmitted to humanized transgenic mouse lines (Bishop et al., 2010). Issues of species-specific PrP sequence and prion agent strain can also affect amplification in the PMCA assay (Jones et al., 2011). Throughout these studies, PMCA has shown considerable fidelity in maintaining the PrPres type of the input seed, in terms of both fragment size and glycosylation ratio (Jones et al., 2007, 2008, 2009b).
It seems likely that the outcomes of RT-QuIC and PMCA reactions depend on the exact conditions employed (biochemical, chemical and physical) in addition to the nature of the seeds and substrates, and it is therefore likely that conditions could be found in which vCJD PrPSc is recognized in the RT-QuIC assay. Indeed, a preparative PrPSc-specific immunoprecipitation method that renders vCJD tissue homogenates compatible with a modified version of RT-QuIC (termed eQuIC) has been developed that allows highly sensitive detection of vCJD brain material diluted in human plasma (Orrú et al., 2011). Nevertheless, the different results obtained using human materials in QuIC and PMCA could be taken to indicate that they differ mechanistically and therefore that they have different relationships to PrP accumulation in situ. It may be that RT-QuIC is ideally suited to applications in which a sensitive and specific test for PrPSc is required, whereas PMCA may provide a more physiological model, better suited to understanding the role of PrP, auxiliary molecules and microenvironments in the conversion of PrPC to PrPSc.
Methods
CJD and control brain tissues.
Brain frontal cortex tissue was taken from three cases of vCJD and from examples of six sCJD cases, defined by the combination of their PRNP codon 129 genotype (MM, MV or VV) and their PrPres type (type 1 or 2) according to the nomenclature of Parchi et al. (1999) and Gambetti et al. (2003) as MM1/MV1, MM2C, MV1, MV2K, VV1 and VV2. All cases used had been thoroughly characterized and the diagnosis reached using internationally accepted criteria. In each case, samples of ~100 mg were taken from the frontal cortex for preparation as seed material for RT-QuIC. Samples of cerebellum and medial thalamus were taken from two of the three vCJD cases mentioned above. In addition, tonsil tissue was taken from a fourth case of vCJD. Frontal cortex tissue was also taken from non-CJD cases, which were used as negative controls and expected to contain PrPC in the absence of PrPSc. Some of these were cases were referred to the National CJD Research & Surveillance Unit (NCJDRSU) as being suspected of having CJD but received an alternative diagnosis. The neurological control cases used were three cases of Alzheimer’s disease (two MM and one VV at PRNP codon 129) and one case of Lewy body dementia (VV at PRNP codon 129). Ethical approval for the use of brain tissue from the Medical Research Council (MRC) NCJDRSU Brain and Tissue Bank was covered by LREC 2000/4/157 (Professor J. W. Ironside). In addition, two further negative controls from individuals without neurodegenerative diseases (one MV and the other VV at PRNP codon 129) were obtained from the MRC Sudden Death Brain and Tissue Bank. These are referred to in the text as non-neurological controls.
Frozen tissue samples were weighed and homogenized in PBS (Sigma-Aldrich) containing 1 mM EDTA, 150 mM NaCl, 0.5 % Triton X-100 and Complete Mini EDTA-free Protease Inhibitor Cocktail (Roche), using a buffer volume that gave a final tissue concentration of 10 % (w/v). The homogenization was performed in a 2 ml centrifuge tube using an Eppendorf micropestle. The homogenate was cleared by centrifugation at 420 g for 5 min at 4 °C. The supernatants were vortexed and separated into 20 µl aliquots and stored at −80 °C.
Sonication of brain homogenate.
To investigate the effect of prior sonication on the ability of brain homogenate to promote the conversion of recPrP in RT-QuIC, samples (100 µl) were each placed in an individual well of a thin-walled 96-well PCR plate and the wells were sealed. The plate was transferred to the plate holder of a microplate horn sonicator (model 3000; Misonix) in a 37 °C incubator and subjected to a 40 s burst of sonication (85 % potency, 294–297 W). This was equivalent to one round of our PMCA protocol (Jones et al., 2007).
Determination of the titre of PrPres in CJD brain homogenates.
The amounts of brain homogenate introduced into the RT-QuIC assays were normalized according to the titre of PrPres. Samples of vCJD or sCJD brain homogenate cleared by low-speed centrifugation were digested for 1 h at 37 °C with 50 µg proteinase K ml−1 and digestion was terminated with 100 mM PefaBloc (Roche). Diluted samples of these proteinase K-treated brain homogenates were separated on Novex 10 % Bis-Tris gels (Invitrogen) alongside dilutions of purified recHuPrP. The separated proteins were then transferred to PVDF membrane and immunoblotted with anti-PrP mAb 3F4, as described previously (Yull et al., 2006). The blots were developed using ECL Plus (GE Biosciences). The samples of brain homogenate ranged from 300 to 150 µg equivalent wet weight of brain tissue, whereas the mass of recHuPrP analysed ranged from 10 to 0.3 ng. The blots were exposed to ECL HyperFilm (GE Biosciences) for 30 s, 3 min or 30 min.
The levels of PrPres per unit of brain extract were estimated by comparing the densities of the single Western blot bands corresponding to purified recHuPrP and the combined densities of the three bands corresponding to PrPres in the brain extracts. The densities of the bands were determined by scanning the exposures using a Bio-Rad GS-800 Densitometer and analysing the scans using Bio-Rad Quantity One software. For the vCJD and sCJD frontal cortex samples used in this study, estimates of the levels of PrPres varied from 4.6 to 59 µg PrPres (g brain tissue)−1 (wet mass).
Dilution of brain homogenates for RT-QuIC.
Aliquots of 10 % brain homogenate were thawed at room temperature. Brain homogenates were diluted by serial 1 : 10 dilutions using N2 buffer [PBS containing 0.1 % SDS and 1× N2 supplement (Invitrogen)]. Usually, for each dilution, a 10 µl sample was mixed with 90 µl N2, with ~8 s of vortex mixing between each stage of a serial dilution. Brain homogenate was diluted so that a 2 µl volume of the final dilution would contain a defined amount of PrPres. For vCJD and sCJD frontal cortex tissue samples, the dilution of (100 %) brain tissue required for 100 fg PrPres in 2 µl varied from 9×10−7 to 1×10−5.
Expression and purification of RT-QuIC substrates.
recHaPrP (aa 23–231 and 90–231; GenBank accession no. K02234) and recHuPrP (aa 23–231, with either M or V at codon 129; GenBank accession no. M13899) were prepared according to the method of Wilham et al. (2010) with minor modifications. Briefly, the DNA sequences encoding hamster or human PrP proteins as they are expressed in cells (aa 23–231) were amplified to include a terminal stop codon and ligated into the pRSET A vector (Invitrogen) at the NdeI and EcoRI sites. Plasmids were transformed into Escherichia coli Rosetta cells (Invitrogen), and PrP sequences were expressed using the Overnight Express Autoinduction system (Merck). Cell pellets from 0.5 l cultures were lysed with BugBuster master mix (Merck) to isolate the inclusion bodies. The inclusion bodies were washed twice with 0.1× BugBuster, pelleted by centrifugation and frozen at −20 °C. Pellets were later resuspended and denatured in 8 M guanidine/HCl (pH 8.0), centrifuged at 16 000 g for 5 min and the supernatant bound to 20 ml Ni-NTA Superflow resin (Qiagen) equilibrated in denaturing buffer [100 mM sodium phosphate (pH 8.0), 10 mM Tris/HCl, 6 M guanidine/HCl]. The resin was loaded onto a column and the denatured protein was refolded using a linear gradient of refolding buffer [100 mM sodium phosphate (pH 8.0), 10 mM Tris/HCl] over 5 h at a flow rate of 1 ml min−1. Bound protein was eluted with a linear gradient of elution buffer [100 mM sodium phosphate (pH 5.8), 10 mM Tris/HCl, 500 mM imidazole] at 1 ml min−1 over 60 min. Eluted protein was diluted twofold into dialysis buffer [10 mM sodium phosphate (pH 5.8)] filtered with a 0.2 µm syringe filter and dialysed. The concentration of recPrP was determined using bicinchoninic acid reagent (Thermo Scientific) and adjusted to 0.2–0.5 mg ml−1. Aliquots were stored at −80 °C. The purity of the recPrP was estimated as >99 % by SDS-PAGE and immunoblotting (data not shown).
RT-QuIC.
This was performed as described by Wilham et al. (2010) but with a few alterations. Frozen aliquots of recPrP (full-length or truncated) were allowed to thaw at room temperature (~20 °C). The protein solution was then filtered through a 100 kDa Nanosep centrifugal concentrator (VWR). To determine the concentration of the filtered recPrP, samples were taken and diluted 1 : 10 in PBS containing 0.1 % SDS and the absorbance was measured at 280 nm in an Ultrospec 3000 spectrophotometer (GE Life Sciences). The protein concentration was calculated by multiplying by the dilution factor and using an extinction coefficient of 2.6 l g−1 cm−1 for full-length recPrP (aa 23–232) and 1.4 l g−1 cm−1 for truncated recPrP (aa 90–231).
The 100 µl RT-QuIC reactions were set up in quadruplicate in the wells of a clear-bottomed black 96-well microplate (Fisher Scientific). Stock solutions containing the reaction components were filtered through a 0.22 µM Millex PES filter prior to making a master mix. recPrP was the final addition to the master mix, which was mixed by gently inverting the tube and then dispensed into the wells in 98 µl volumes. After adding 2 µl of the appropriately diluted brain homogenate (see above) to the wells, the final reactions contained 1× PBS, 0.002 % SDS, 0.02 % N2, 324 mM NaCl (including the NaCl present in PBS), 1 mM EDTA, 10 µM ThT and 0.1 mg recPrP ml−1. The plates were sealed with sealing film and inserted into a FLUOstar OPTIMA microplate reader (BMG Labtech). The plates were incubated at 42 °C and shaken intermittently (1 min shaking, 1 min at rest) at 600 r.p.m. in a double orbital configuration and fluorescence readings were taken at 480 nm every 15 min from the bottom of the wells after excitation with 20 flashes per well at 450 nm. ThT emission fluorescence counts increased to a maximum limit of 65 000 per well when recPrP conversion occurred in all four wells of the quadruplicate reactions.
Graphs were constructed by importation of the data into Microsoft Excel. The traces represent the mean of the four replicate tests included in each run and traces on individual graphs show samples analysed together on one plate during a single run. The graphs also show the number of wells with above-background values in the form of n/4, where n is the number of positive wells. A summary of the brain homogenate seeds and recPrP substrates used in this study is shown in Table 1.
Table 1. Summary of brain homogenate seeds and recombinant PrP substrates used in this study.
A positive result for RT-QuIC within a 60 h run time is denoted ‘+’, whereas no reaction is denoted ‘−’. References are given for the relevant figure. Where no reference is given to a figure, the analysis was performed, with the given result, but the data are not shown. nt, Not tested.
| Seed (brain homogenates) | Substrate (recPrP) | |||||
| Diagnosis | PRNP codon 129/PrPres type | No. cases tested | Full-length recHaPrP (aa 23–231) | Truncated recHaPrP (aa 90–231) | Full-length recHuPrP-129M (aa 23–231) | Full-length recHuPrP-129V (aa 23–231) |
| sCJD | MM1 | 1 | + (Fig. 3) | + (Fig. 1) | + (Figs 4, 5 and 6) | + (Figs 4 and 5) |
| MV1 | 1 | + (Fig. 3) | + (Fig. 1) | + | + | |
| VV1 | 1 | + (Fig. 3) | + (Fig. 1) | + | + | |
| MM2 | 1 | + (Fig. 3) | + (Fig. 1) | + | + | |
| MV2 | 1 | + (Fig. 3) | + (Figs 1 and 2) | + | + | |
| VV2 | 1 | + (Fig. 3) | + (Fig. 1) | + (Figs 4 and 5) | + (Figs 4 and 5) | |
| vCJD | MM2 | 3* | + (Fig. 3) | − (Fig. 1) | + (Fig. 5) | + (Fig. 5) |
| Alzheimer’s disease | MM | 2 | − (Fig. 3) | − (Fig. 1) | − (Fig. 5) | − |
| VV | 1 | − | nt | − | nt | |
| Lewy body dementia | VV | 1† | − | − (Fig. 1) | − | nt |
| Non-neurological control | MM | 1 | − (Fig. 3) | − (Fig. 1) | − | nt |
| MV | 1 | − | − | − | nt | |
| VV | 1 | − | − | − | nt | |
A fourth vCJD case provided tonsil tissue.
A second Lewy body dementia case provided tonsil tissue.
Kinetics of fibrilization.
In most cases where truncated recHaPrP (aa 90–231) was used as a substrate, the time-dependent changes in the fluorescence of ThT (F) could be approximately described using the following equation:
| Equation 1 |
where t is the time from the start of the assay, A is the base level of ThT fluorescence during the lag phase, B is the difference between the maximum ThT fluorescence and the basal level, k is the rate constant of fibril growth (h−1) and tm is the observed time at the midpoint of transition. When truncated recPrP was used as a substrate, the lag time tl of fibril formation was calculated as: tl = tm−2/k. This equation has been used previously by Nielsen et al. (2001) and Chia et al. (2010) to model fibrilization. However, in cases where full-length recPrP (recHaPrP or recHuPrP, aa 23–231) was used a substrate, the kinetics of fibrilization appeared to be different. Once fibrilization had been initiated, it appeared to be better approximated using the following equation in which t is the time after the initiation of fibrilization:
| Equation 2. |
In assays where full-length recPrP was used as a RT-QuIC substrate, the lag time was determined simply as the last time point before the mean ThT fluorescence value rapidly rose above the baseline.
Acknowledgements
This work was funded by grants from the Scottish Government Health Directorates Chief Scientist Office (CZB/4/688) to M. W. H. and from Alliance BioSecure to A. J. E. G. This work was also supported in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. The National CJD Surveillance Unit Brain and Tissue Bank is supported by the MRC (UK), and the NCJDRSU as a whole is funded by the Department of Health, UK, and the Scottish Government. The views expressed in the publication are those of the authors and not necessarily those of the Department of Health, UK. The authors would like to thank the patients and their relatives for the opportunity to conduct research on tissue specimens.
Footnotes
A supplementary table is available with the online version of this paper.
References
- Atarashi R., Moore R. A., Sim V. L., Hughson A. G., Dorward D. W., Onwubiko H. A., Priola S. A., Caughey B. (2007). Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nat Methods 4, 645–650 10.1038/nmeth1066 [DOI] [PubMed] [Google Scholar]
- Atarashi R., Wilham J. M., Christensen L., Hughson A. G., Moore R. A., Johnson L. M., Onwubiko H. A., Priola S. A., Caughey B. (2008). Simplified ultrasensitive prion detection by recombinant PrP conversion with shaking. Nat Methods 5, 211–212 10.1038/nmeth0308-211 [DOI] [PubMed] [Google Scholar]
- Atarashi R., Satoh K., Sano K., Fuse T., Yamaguchi N., Ishibashi D., Matsubara T., Nakagaki T., Yamanaka H., et al. (2011). Ultrasensitive human prion detection in cerebrospinal fluid by real-time quaking-induced conversion. Nat Med 17, 175–178 10.1038/nm.2294 [DOI] [PubMed] [Google Scholar]
- Bishop M. T., Will R. G., Manson J. C. (2010). Defining sporadic Creutzfeldt–Jakob disease strains and their transmission properties. Proc Natl Acad Sci U S A 107, 12005–12010 10.1073/pnas.1004688107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P., Brandel J. P., Preece M., Sato T. (2006). Iatrogenic Creutzfeldt–Jakob disease: the waning of an era. Neurology 67, 389–393 10.1212/01.wnl.0000231528.65069.3f [DOI] [PubMed] [Google Scholar]
- Caughey B., Baron G. S., Chesebro B., Jeffrey M. (2009). Getting a grip on prions: oligomers, amyloids, and pathological membrane interactions. Annu Rev Biochem 78, 177–204 10.1146/annurev.biochem.78.082907.145410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia R., Tattum M. H., Jones S., Collinge J., Fisher E. M., Jackson G. S. (2010). Superoxide dismutase 1 and tgSOD1 mouse spinal cord seed fibrils, suggesting a propagative cell death mechanism in amyotrophic lateral sclerosis. PLoS ONE 5, e10627 10.1371/journal.pone.0010627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. P., Gröner A., Ironside J. W., Head M. W. (2011). Comparison of the level, distribution and form of disease-associated prion protein in variant and sporadic Creutzfeldt–Jakob diseased brain using conformation-dependent immunoassay and Western blot. J Gen Virol 92, 727–732 10.1099/vir.0.026948-0 [DOI] [PubMed] [Google Scholar]
- Colby D. W., Zhang Q., Wang S., Groth D., Legname G., Riesner D., Prusiner S. B. (2007). Prion detection by an amyloid seeding assay. Proc Natl Acad Sci U S A 104, 20914–20919 10.1073/pnas.0710152105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetti P., Kong Q., Zou W., Parchi P., Chen S. G. (2003). Sporadic and familial CJD: classification and characterisation. Br Med Bull 66, 213–239 10.1093/bmb/66.1.213 [DOI] [PubMed] [Google Scholar]
- Hewitt P. E., Llewelyn C. A., Mackenzie J., Will R. G. (2006). Creutzfeldt–Jakob disease and blood transfusion: results of the UK Transfusion Medicine Epidemiological Review study. Vox Sang 91, 221–230 10.1111/j.1423-0410.2006.00833.x [DOI] [PubMed] [Google Scholar]
- Hilton D. A. (2006). Pathogenesis and prevalence of variant Creutzfeldt–Jakob disease. J Pathol 208, 134–141 10.1002/path.1880 [DOI] [PubMed] [Google Scholar]
- Ironside J. W., Ghetti B., Head M. W., Piccardo P., Will R. G. (2008). Prion diseases. In Greenfield's Neuropathology, Vol. 2. 8th edn, pp. 1197–1273 Edited by Love S., Louis D. N., Ellison D. W. London: Hodder and Arnold [Google Scholar]
- Jones M., Peden A. H., Prowse C. V., Gröner A., Manson J. C., Turner M. L., Ironside J. W., MacGregor I. R., Head M. W. (2007). In vitro amplification and detection of variant Creutzfeldt–Jakob disease PrPSc. J Pathol 213, 21–26 10.1002/path.2204 [DOI] [PubMed] [Google Scholar]
- Jones M., Peden A. H., Wight D., Prowse C., Macgregor I., Manson J., Turner M., Ironside J. W., Head M. W. (2008). Effects of human PrPSc type and PRNP genotype in an in-vitro conversion assay. Neuroreport 19, 1783–1786 10.1097/WNR.0b013e328318edfa [DOI] [PubMed] [Google Scholar]
- Jones M., Peden A. H., Yull H., Wight D., Bishop M. T., Prowse C. V., Turner M. L., Ironside J. W., MacGregor I. R., Head M. W. (2009a). Human platelets as a substrate source for the in vitro amplification of the abnormal prion protein (PrP) associated with variant Creutzfeldt–Jakob disease. Transfusion 49, 376–384 10.1111/j.1537-2995.2008.01954.x [DOI] [PubMed] [Google Scholar]
- Jones M., Wight D., Barron R., Jeffrey M., Manson J., Prowse C., Ironside J. W., Head M. W. (2009b). Molecular model of prion transmission to humans. Emerg Infect Dis 15, 2013–2016 10.3201/eid1512.090194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M., Peden A. H., Head M. W., Ironside J. W. (2011). The application of in vitro cell-free conversion systems to human prion diseases. Acta Neuropathol 121, 135–143 10.1007/s00401-010-0708-8 [DOI] [PubMed] [Google Scholar]
- Llewelyn C. A., Hewitt P. E., Knight R. S., Amar K., Cousens S., Mackenzie J., Will R. G. (2004). Possible transmission of variant Creutzfeldt–Jakob disease by blood transfusion. Lancet 363, 417–421 10.1016/S0140-6736(04)15486-X [DOI] [PubMed] [Google Scholar]
- Nielsen L., Frokjaer S., Brange J., Uversky V. N., Fink A. L. (2001). Probing the mechanism of insulin fibril formation with insulin mutants. Biochemistry 40, 8397–8409 10.1021/bi0105983 [DOI] [PubMed] [Google Scholar]
- Orrú C. D., Wilham J. M., Hughson A. G., Raymond L. D., McNally K. L., Bossers A., Ligios C., Caughey B. (2009). Human variant Creutzfeldt–Jakob disease and sheep scrapie PrPres detection using seeded conversion of recombinant prion protein. Protein Eng Des Sel 22, 515–521 10.1093/protein/gzp031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrú C. D., Wilham J. M., Raymond L. D., Kuhn F., Schroeder B., Raeber A. J., Caughey B. (2011). Prion disease blood test using immunoprecipitation and improved quaking-induced conversion. MBio 2, e00078-11 10.1128/mBio.00078-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchi P., Giese A., Capellari S., Brown P., Schulz-Schaeffer W., Windl O., Zerr I., Budka H., Kopp N., et al. (1999). Classification of sporadic Creutzfeldt–Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann Neurol 46, 224–233 [DOI] [PubMed] [Google Scholar]
- Parchi P., Cescatti M., Notari S., Schulz-Schaeffer W. J., Capellari S., Giese A., Zou W. Q., Kretzschmar H., Ghetti B., Brown P. (2010). Agent strain variation in human prion disease: insights from a molecular and pathological review of the National Institutes of Health series of experimentally transmitted disease. Brain 133, 3030–3042 10.1093/brain/awq234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden A. H., Head M. W., Ritchie D. L., Bell J. E., Ironside J. W. (2004). Preclinical vCJD after blood transfusion in a PRNP codon 129 heterozygous patient. Lancet 364, 527–529 10.1016/S0140-6736(04)16811-6 [DOI] [PubMed] [Google Scholar]
- Peden A. H., Head M. W., Jones M., MacGregor I. R., Turner M. L., Ironside J. W. (2008). Advances in the development of a screening test for variant Creutzfeldt–Jakob disease. Expert Opin Med Diagn 2, 207–219 10.1517/17530059.2.2.207 [DOI] [PubMed] [Google Scholar]
- Peden A., McCardle L., Head M. W., Love S., Ward H. J., Cousens S. N., Keeling D. M., Millar C. M., Hill F. G., Ironside J. W. (2010). Variant CJD infection in the spleen of a neurologically asymptomatic UK adult patient with haemophilia. Haemophilia 16, 296–304 10.1111/j.1365-2516.2009.02181.x [DOI] [PubMed] [Google Scholar]
- Prusiner S. B. (1998). Prions. Proc Natl Acad Sci U S A 95, 13363–13383 10.1073/pnas.95.23.13363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safar J., Wille H., Itri V., Groth D., Serban H., Torchia M., Cohen F. E., Prusiner S. B. (1998). Eight prion strains have PrPSc molecules with different conformations. Nat Med 4, 1157–1165 10.1038/2654 [DOI] [PubMed] [Google Scholar]
- Safar J. G., Geschwind M. D., Deering C., Didorenko S., Sattavat M., Sanchez H., Serban A., Vey M., Baron H., et al. (2005). Diagnosis of human prion disease. Proc Natl Acad Sci U S A 102, 3501–3506 10.1073/pnas.0409651102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira J. R., Raymond G. J., Hughson A. G., Race R. E., Sim V. L., Hayes S. F., Caughey B. (2005). The most infectious prion protein particles. Nature 437, 257–261 10.1038/nature03989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M. L., Ludlam C. A. (2009). An update on the assessment and management of the risk of transmission of variant Creutzfeldt–Jakob disease by blood and plasma products. Br J Haematol 144, 14–23 10.1111/j.1365-2141.2008.07376.x [DOI] [PubMed] [Google Scholar]
- Wilham J. M., Orrú C. D., Bessen R. A., Atarashi R., Sano K., Race B., Meade-White K. D., Taubner L. M., Timmes A., Caughey B. (2010). Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog 6, e1001217 10.1371/journal.ppat.1001217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will B. (2010). Variant CJD: where has it gone, or has it? Pract Neurol 10, 250–251 10.1136/jnnp.2010.223693 [DOI] [PubMed] [Google Scholar]
- Wroe S. J., Pal S., Siddique D., Hyare H., Macfarlane R., Joiner S., Linehan J. M., Brandner S., Wadsworth J. D., et al. (2006). Clinical presentation and pre-mortem diagnosis of variant Creutzfeldt–Jakob disease associated with blood transfusion: a case report. Lancet 368, 2061–2067 10.1016/S0140-6736(06)69835-8 [DOI] [PubMed] [Google Scholar]
- Yull H. M., Ritchie D. L., Langeveld J. P., van Zijderveld F. G., Bruce M. E., Ironside J. W., Head M. W. (2006). Detection of type 1 prion protein in variant Creutzfeldt–Jakob disease. Am J Pathol 168, 151–157 10.2353/ajpath.2006.050766 [DOI] [PMC free article] [PubMed] [Google Scholar]