Abstract
Accumulation of baculovirus defective interfering particle (DIP) and few polyhedra (FP) mutants is a major limitation to continuous large-scale baculovirus production in insect-cell culture. Although overcoming these mutations would result in a cheaper platform for producing baculovirus biopesticides, little is known regarding the mechanism of FP and DIP formation. This issue was addressed by comparing DIP production of wild-type (WT) Autographa californica multiple nucleopolyhedrovirus (AcMNPV) with that of a recombinant AcMNPV (denoted Ac-FPm) containing a modified fp25k gene with altered transposon insertion sites that prevented transposon-mediated production of the FP phenotype. In addition to a reduction in the incidence of the FP phenotype, DIP formation was delayed on passaging of Ac-FPm compared with WT AcMNPV. Specifically, the yield of DIP DNA in Ac-FPm was significantly lower than in WT AcMNPV up to passage 16, thereby demonstrating that modifying the transposon insertion sites increases the genomic stability of AcMNPV. A critical component of this investigation was the optimization of a systematic method based on the use of pulsed-field gel electrophoresis (PFGE) to characterize extracellular virus DNA. Specifically, PFGE was used to detect defective genomes, determine defective genome sizes and quantify the amount of defective genome within a heterogeneous genome population of passaged virus.
Introduction
Establishment of low-cost, large-scale baculovirus production in a continuous insect-cell culture system is highly desirable for biopesticide production (Black et al., 1997; DuPont, 1996; Gard, 1997; Moscardi, 1999; Murhammer, 1996). The major obstacle to developing such a continuous system is that baculoviruses undergo genetic alterations during passaging in a bioreactor to produce few polyhedra (FP) (Fraser et al., 1983; Harrison & Summers, 1995) and defective interfering particle (DIP) (Kool et al., 1991; Pijlman et al., 2001; Wickham et al., 1991) mutants. Accumulation of these mutants results in decreased polyhedra productivity and reduced virulence, i.e. the baculovirus becomes an ineffective biopesticide (Kool et al., 1991; Kompier et al., 1988; van Lier et al., 1990). It has been estimated that baculoviruses produced in a continuous bioreactor system would cost approximately 50 % less (lower capital and operating costs) than baculoviruses produced in a batch bioreactor system if FP and DIP accumulation could be eliminated (Tramper & Vlak, 1986). Moreover, this ‘passage effect’ has also been demonstrated to be a problem in the large-scale batch production of recombinant proteins/vaccines with the baculovirus expression vector system (BEVS), as the mutants can accumulate during the few passages required to produce the necessary baculovirus stock (Parekh, 2008; Pijlman et al., 2003; Slansky, 2008).
Genetic alterations in passaged baculoviruses include deletions of genome fragments, insertions of transposon-mediated host-cell or viral genome sequences, point mutations and frame-shift mutations (Krell, 1996; Kumar & Miller, 1987). Among these, the DIP mutants (Kool et al., 1991; Pijlman et al., 2001) are replication-defective deletion mutant viruses that arise during passaging and compete with the growth of the normal wild-type (WT) virus. A sharp decrease in recombinant protein production with the BEVS is associated with DIP mutants that often lack the foreign gene of interest or other genes involved in very late gene expression (van Lier et al., 1992, 1994). Specifically, Autographa californica multiple nucleopolyhedrovirus (AcMNPV) DIPs are rapidly generated in cell culture (within two passages) and are heterogeneous in size at late passages (Lee & Krell, 1992; Pijlman et al., 2001). Wickham et al. (1991) showed that 60 % of the AcMNPV genomes were defective at higher passages, but it is not known if this was an equilibrium value (i.e. a constant ratio of DIP mutants to standard AcMNPV) or whether an equilibrium value is even reached upon passaging AcMNPV.
Genome deletions in DIPs most commonly occur between distant homologous repeat (hr) regions in AcMNPV (see Supplementary Table S1, available in JGV Online), Spodoptera exigua multiple nucleopolyhedrovirus (Pijlman et al., 2003) and the Thailand isolate of white spot syndrome virus (WSSV; family Whispoviridae) (van Hulten et al., 2001). These observations suggest that a possible mechanism for genome deletion could be recombination between any two of the eight hr regions present in the AcMNPV genome. hr regions are 500–800 bp AT-rich segments containing two to eight highly conserved direct, inverted repeated sequences of about 72 bp. hr sequences share 65–87 % identity and function as replication origins and transcription enhancers (Cochran & Faulkner, 1983; Guarino et al., 1986). Additionally, evidence of DNA insertions in AcMNPV mutants has been found near hr sites (i.e. hr2, hr3, hr4 and hr5), suggesting that these sites may have a role in genomic alterations/deletions (Burand & Summers, 1982; Potter & Miller, 1980). However, many fundamental questions regarding the mechanism of DIP formation, the role of hr regions in genome deletion, the specific sequences involved in deletion, the size of defective genomes and the ratio of defective to standard particles during passaging remain unanswered.
As a first step towards understanding these processes, a method to study DIP accumulation in passaged baculoviruses was designed and optimized by modifying previously described methods (Lee & Krell, 1992; Pfaller et al., 1994; Vanarsdall et al., 2007). This involved extracting extracellular virus DNA in situ, restriction enzyme digestion at a single site to linearize the circular genome (using AvrII, which cuts only at nt 11750) and pulsed-field gel electrophoresis (PFGE) of the linearized DNA. This method was used to compare DIP formation by passaged WT AcMNPV and by Ac-FPm, an AcMNPV variant that was designed to overcome FP mutant accumulation (Giri et al., 2010). Moreover, the mechanism of AcMNPV DIP formation was investigated by determining the size of the defective genomes obtained from restriction enzyme analysis using AvrII, EcoNI and a combination of AvrII and EcoNI.
Results
To study DIP formation, AcMNPV and Ac-FPm were serially passaged at a high m.o.i. and the defective virion particle sizes, defective genome sizes and their relative quantities were determined. FP mutant formation during serial passage of the viruses has been examined previously (Giri et al., 2010).
Analysis of AcMNPV budded virus (BV) DIPs by transmission electron microscopy (TEM)
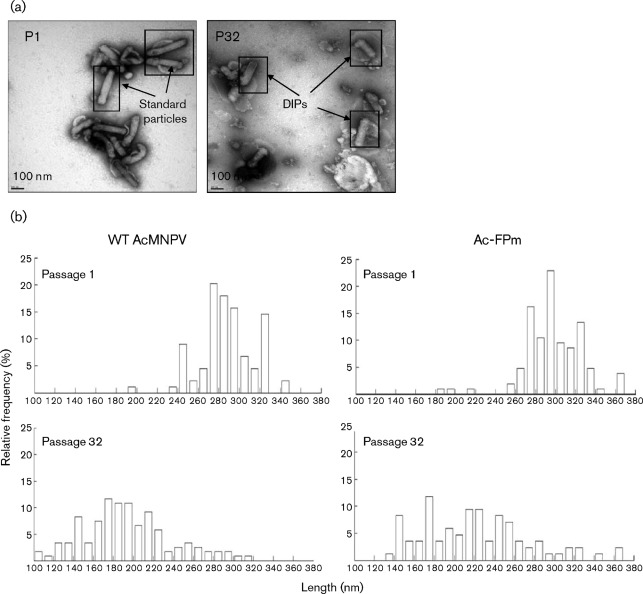
The size (length) distribution of standard particles and DIPs in the AcMNPV BV populations from passages 1 and 32 were examined by TEM (Fig. 1a) and the size range, mean±sd, median and mode are given in Table 1. The WT BV length decreased from 288±26 nm (mean±sd) in passage 1 to 193±44 in passage 32 (Fig. 1b; P<0.01). The percentage of the WT BV population of length >270 nm decreased from 87 % at passage 1 to 7 % at passage 32 (Fig. 1b). Similarly, the mean Ac-FPm BV length decreased from 298±29 nm in passage 1 to 222±53 in passage 32 (Fig. 1b; P<0.01) and the percentage of the Ac-FPm BV population of length >270 nm decreased from 90 % at passage 1 to 16 % at passage 32. Clearly, both BV populations became more heterogeneous, with a wider size range upon passaging (Fig. 1b; Table 1). Although there was no significant difference between the mean lengths of WT AcMNPV and Ac-FPm BV at passage 1 (P>0.01), the mean Ac-FPm BV length was significantly greater than the mean WT BV length at passage 32 (P<0.01). Similarly, the median length of WT AcMNPV and Ac-FPm BV at passage 1 was similar but the median length for Ac-FPm (218 nm) was longer than that of WT AcMNPV (190 nm) at passage 32. The mode (175 nm) for both BV populations, however, was the same, indicating that the most frequent DIP length was the same for the two viruses.
Fig. 1.
TEM analysis of AcMNPV DIPs. The size range of BVs increased and the mean BV size decreased upon passaging. (a) Electron micrographs of WT AcMNPV BV particles at initial and late passages. P1, Passage 1, with standard size particles; P32, passage 32 with smaller DIPs. (b) Size distribution of AcMNPV and Ac-FPm BV particles at passages 1 and 32 obtained by TEM analysis (size interval, 10 nm). Relative frequency is the percentage of BV particles within a given size interval. WT BV length at passage 1 was 288±26 nm (mean±sd) and passage 32 was 193±44 nm, whilst Ac-FPm BV length at passage 1 was 298±29 nm and at passage 32 was 222±53 nm. Statistical analysis was performed by analysis of variance (ANOVA) in matlab; a total of 85 BVs were measured per sample.
Table 1. Statistical parameters for the size distribution of WT AcMNPV and Ac-FPm at passages 1 and 32.
| Parameter | WT AcMNPV | Ac-FPm | ||
| Passage 1 | Passage 32 | Passage 1 | Passage 32 | |
| Virion size range (width) (nm) | 190–342 (152) | 107–314 (207) | 189–368 (179) | 139–361 (222) |
| Mean±sd (nm) | 288±26 | 193±44 | 298±29 | 222±53 |
| Median (nm) | 286 | 190 | 296 | 218 |
| Mode (nm) | 275 | 175 | 295 | 175 |
Comparison of defective genomes in passaged WT and Ac-FPm by PFGE analysis: defective genome formation is delayed in Ac-FPm
Detection of defective genomes.
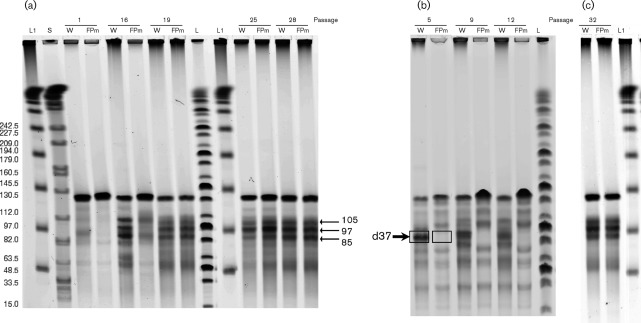
PFGE analysis of linearized baculovirus DNA demonstrated that both WT AcMNPV and Ac-FPm at passage 1 contained the standard genome size of 134 kb (Fig. 2a). At passage 5, a significant amount of defective WT BV with a genome size less than that of the standard genome, i.e. with a defective genome, was detected (Figs 2b and 3a). Although a trend of increased defective genome frequency with increasing passage number was observed for both WT and Ac-FPm viruses, analysis of the defective genome in early passages (passages 1–12) indicated that DIP DNA was less prominent for Ac-FPm than for the WT virus (P<0.05; Fig. 3a). There was no significant difference (P<0.05) in defective genome formation between the viruses at late passages (passages 19–32; Figs 2a, c, and 3a).
Fig. 2.
Detection and size distribution of AcMNPV and Ac-FPm BV DNA linearized with AvrII at different passages by PFGE. DNA was obtained from purified virions. (a) Comparison of DIP DNA formation in AcMNPV and Ac-FPm at passages 1, 16, 19, 25 and 28. (b) Reduced DIP DNA production in Ac-FPm compared with WT AcMNPV at early passages (passage 5, 9 and 12). The arrow indicates the defective genome d37 with 37 % of the genome deleted. (c) Size distribution of the standard and defective genomes of WT AcMNPV and Ac-FPm at passage 32. Lanes: L, MidRange I PFG ladder; L1, λ DNA ladder, with sizes shown in kb in (a); S, whole Salmonella DNA digested with XbaI (used as a control for electrophoresis and size marker); W, WT AcMNPV DNA; FPm, Ac-FPm DNA. Arrows indicate sizes (kb) of DIP DNAs.
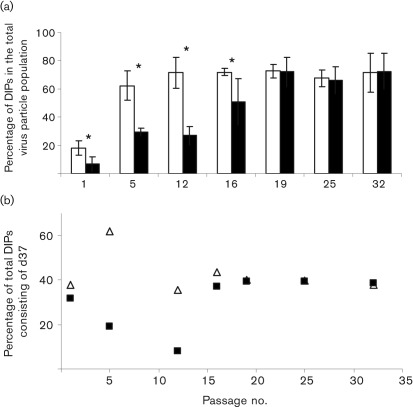
Fig. 3.
Quantification of DIP production from PFGE analysis of WT AcMNPV and Ac-FPm BV DNA. (a) Comparison of the percentage of DIPs in virus stocks at different passages showing that DIP accumulation was reduced in Ac-FPm (filled bars) compared with WT AcMNPV (empty bars) during early passages, but reached the same equilibrium values at later passages. Error bars represent the 95 % confidence limit. *, P<0.05 (Student’s t-test, n = 3, DNA samples were run three times in agarose gels). (b) d37 accumulation was delayed in Ac-FPm (▪) compared with the WT AcMNPV (▵). At some higher passages, the value for WT AcMNPV was the same as for Ac-FPm and is therefore not visible.
Quantification of defective genomes.
The defective genome sizes and relative proportions of each defective genome for WT AcMNPV and Ac-FPm were determined by quantifying the band intensities on the gel of linearized viral genomes (Fig. 2). The percentage of total defective genomes (85, 97 and 105 kb) in WT AcMNPV BV stock increased with increasing passage number, and by passage 12 and thereafter ~70 % of the virus particles were deletion mutants (i.e. DIPs) (Fig. 3a). In contrast, only 33 % of the Ac-FPm virus particles were DIPs at passage 12. By passage 19 and thereafter, ~70 % of the Ac-FPm virus particles were DIPs (Fig. 3a). The distribution of WT AcMNPV reached an equilibrium of ~29 % 85 kb DIPs, ~25 % 97 kb DIPs, ~17 % 105 kb DIPs and ~29 % 134 kb normal-sized virus particles by passage 16 (Table 2). The distribution of Ac-FPm reached comparable equilibrium values by passage 19. These results demonstrated that removal of the transposon insertion sites in Ac-FPm delayed the accumulation of DIPs, but that eventually the proportion and distribution of DIPs in the WT and modified virus populations reached the same values.
Table 2. Range of defective virion sizes and corresponding percentage deletion in the context of defective genome size.
| Standard particle | DIP 1 | DIP 2 | DIP 3 | |
| AcMNPV genome size (kb) | 134 | 105 | 97 | 85 |
| Deletion (%) | – | 22 | 28 | 37 |
| Relative amount of genome (%) | 29 | 17 | 25 | 29 |
| Abundant virion size range and (mean) (nm) | 260–340 (300) | 220–230 (225) | 210–220 (215) | 170–180 (175) |
| Predicted defective virion size corresponding to DIP genome (nm) | 300 | 225 | 215 | 175 |
| Deletion (%) | – | 25 | 28 | 42 |
| Relative amount of virion particles (%) | 30 | 18 | 23 | 30 |
Size distribution of the defective genome population.
PFGE analysis was used to investigate genome alterations by monitoring the size distribution of WT and Ac-FPm baculovirus genomes during serial passaging. At passage 5, almost 62 % of WT AcMNPV DIPs consisted of the 85 kb mutant (denoted as d37 to indicate that 37 % of the genome was deleted) (Figs 2b and 3b). Conversely, only about 19 % of passage 5 Ac-FPm DIPs consisted of d37 mutants (P<0.05, Student’s t-test, Fig. 3b). Additional defective genomes of 97 and 105 kb corresponded to deletions of 28 and 22 %, respectively (Table 2). The relative frequencies of the three WT AcMNPV DIPs reached equilibrium by passage 16 with the 85, 97 and 105 kb DIPs comprising approximately 44, 32 and 25 % of the total DIPs, respectively (Fig. 2a, c). Comparable equilibrium values of the relative frequencies of the three Ac-FPm DIPs were reached by passage 19 (Fig. 2a, c).
PFGE of BV DNA digested by AvrII and EcoNI.
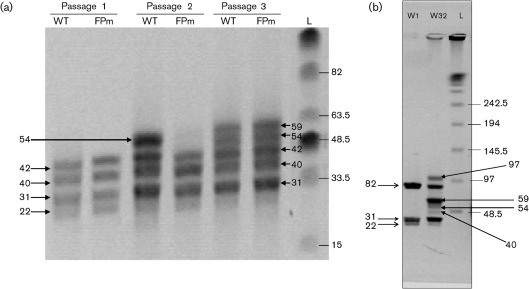
Additional verification of the delayed DIP accumulation in passaged Ac-FPm was achieved by a double digestion of WT and Ac-FPm BV DNA with AvrII and EcoNI. These enzymes cut the WT AcMNPV genome into four segments of 42, 40, 31 and 22 kb (Fig. 4a). At passage 16, an additional band of 54 kb appeared in the digested WT AcMNPV genome but not in the Ac-FPm genome (Fig. 4a). At passage 19, however, the 54 kb band and an additional 59 kb band were observed for both WT and Ac-FPm viruses. This result further demonstrated that DIP formation was delayed in Ac-FPm compared with WT AcMNPV.
Fig. 4.
(a) PFGE of WT AcMNPV and Ac-FPm BV DNA digested with AvrII and EcoNI at different passages. DIP formation was delayed in Ac-FPm compared with AcMNPV. The left arrow at 54 kb indicates the defective genome formation at passage 16 and the two right arrows at 54 and 59 kb indicate the defective genome formation at passage 19. (b) PFGE of WT AcMNPV BV DNA digested with EcoNI. The arrows on the left indicate the DNA fragment sizes (kb) corresponding to standard DNA of WT AcMNPV and the arrows on the right indicate the sizes of DNA bands appearing due to the formation of DIPs at passage 32. Lanes: L, λ DNA ladder (kb); WT, WT AcMNPV DNA; FPm, Ac-FPm DNA; W1, WT AcMNPV DNA at passage 1; W32, WT AcMNPV DNA at passage 32.
PFGE of BV DNA digested by EcoNI.
As the AvrII restriction site may be lost during DIP production, EcoNI digestion was also performed with WT AcMNPV BV DNA from passages 1 and 32 to obtain all possible sizes of WT AcMNPV DIP DNAs. WT AcMNPV at passage 1 (134 kb) yielded three fragments of 82, 31 and 22 kb when digested with EcoNI (Fig. 4b). In contrast, the passage 32 virus containing defective genomes yielded four additional bands of 97, 59, 54 and 40 kb (Fig. 4b). Southern blots were also performed to verify that all the bands obtained from AvrII and EcoNI digestion consist of AcMNPV DNA (data not shown).
Discussion
Delayed DIP formation in Ac-FPm: link between FP and DIP mutation
DIP accumulation in Ac-FPm was delayed compared with WT AcMNPV as a result of modifying the transposon insertion sites (i.e. TTAA sites) in the baculovirus fp25k gene. However, by passage 19, both the frequency (Fig. 3a) and distribution (i.e. among the 85, 97 and 105 kb defective genomes; Fig. 2a) of the WT AcMNPV and Ac-FPm defective genomes were essentially the same. In this context, it is natural to ask why DIP formation is delayed in Ac-FPm. Previous investigations predicted that an FP mutant could be an intermediate step in DIP mutant generation during serial passaging (Kool et al., 1991). Pijlman et al. (2001) demonstrated that the deletion junction sites of the AcMNPV defective genomes contained TTAA insertion sequences, and Carstens (1987) described an AcMNPV spontaneous deletion mutant resulting from transposon insertions at 2.6 and 46 map units (m.u.), followed by deletion of the intervening sequence. These previous results suggest that DIPs are formed by a process that involves transposon insertions (including insertions within the fp25k gene), followed by deletion of the intervening sequence; in other words, transposon insertions (which result in FP mutant formation) are a critical step in DIP formation. The delayed Ac-FPm DIP formation in the current study is consistent with this mechanism, as modification of the TTAA sites (to TTCA, TCAA, TTGA, TGAA and TAAA) in the fp25k gene delayed transposon insertion, thereby delaying FP mutant formation. Further sequencing analysis across the putative recombination sites, however, is required to confirm this DIP formation mechanism.
Like all other biological mutations, DIP formation is a chance-driven process, and it is possible that DIPs may be generated and may accumulate at different rates, even when the same virus is passaged. In this context, the question can be raised as to whether the difference between the rate of WT and Ac-FPm accumulation is probabilistic or is due to the specific mutation in fp25k locus. It is well known from previous literature that significant DIP accumulation occurs during early passages (passages 3–4) for WT AcMNPV (Lee & Krell, 1992; Pijlman et al., 2001). In contrast, Ac-FPm viruses do not reach comparable levels of DIP accumulation until passage 16. Additional passaging experiments using a number of individually picked virus plaques of WT and FPm viruses would enable us to answer the question posed above.
Size of defective genomes obtained from PFGE analysis
PFGE analysis of WT and Ac-FPm virus from passage 1 showed a major band at 134 kb, and the WT AcMNPV also contained small amounts of DIP DNA (Fig. 2a). Although WT AcMNPV and Ac-FPm were plaque purified by the same method (Kelly et al., 2007), it is likely that the small amounts of DIPs were formed during amplification of the plaque-purified WT AcMNPV isolate (passage 0) to obtain a sufficient amount of DNA for the PFGE experiment. This result indicates that the WT AcMNPV DIPs are generated rapidly during virus passaging in suspension cell culture and is consistent with earlier WT AcMNPV DIP analysis by Pijlman et al. (2001) (DIPs found by passage 4) and Lee & Krell (1992) (small amount of DIPs present at passage 1). The Ac-FPm isolate from passage 0 was amplified by a similar method but contained fewer defective genomes when analysed by PFGE (Fig. 2a). Note that, whilst defective genomes were not detectable by eye for Ac-PFm at passage 1 (Fig. 2a), small amounts of defective genomes were detected with the scanning software (see Methods), as indicated in Fig. 3. The AcMNPV defective genome sizes obtained from the present PFGE study (85, 96 and 105 kb) are in the size range (70–100 kb) found by Lee & Krell (1992) in serially passaged virus (passage 15).
Some of the most prevalent defective genome sizes in the literature (Supplementary Table S1) were not found in the present study, and the difference in size could be due to differences in the method used, i.e. based on the PFGE of linearized AcMNPV DNA in this study compared with previous methods based on the PFGE of circular DNA or on electron microscopy. At later passages, smaller genomes ranging from 50 to 65 kb (corresponding to low-intensity bands) could result from multiple deletions (Fig. 2). Continuous passaging resulted in a more heterogeneous population, leading to equilibrium with ~70 % DIPs (Fig. 3a).
PFGE analysis of the AcMNPV defective genome
Previous electron micrographs of AcMNPV BV containing DIP mutants demonstrated that DIPs are smaller (~190 nm) than the standard virus (~330 nm) (Kool et al., 1991; Wickham et al., 1991) but did not indicate the presence of DIPs of different sizes. The current study involved a TEM analysis of BVs that described the physical length and size distribution of the virus particles during late passage. However, this analysis was labour intensive and did not allow the detection of a specific particle size, only a size range. Furthermore, traditional restriction enzyme analysis of baculovirus DNA with enzymes such as HindIII, EcoRI, XhoI, KpnI, SstII, PstI, BamHI and XbaI (Burand & Summers, 1982; Burand et al., 1983; Kool et al., 1991; Lee & Krell, 1992; Pijlman et al., 2002, 2003, 2004) and other approaches for DIP detection, such as a sensitive nested PCR-based assay and real-time PCR (Pijlman et al., 2001; Zwart et al., 2008), are not able to detect the defective genome size distribution or quantify their relative amount within a heterogeneous virus population. Lee & Krell (1992) used PFGE analysis of AcMNPV DNA followed by Southern blotting to detect the size of defective genomes for the first time. However, the individual defective genome sizes were not well defined due to low resolution and it was difficult to quantify their relative abundance. The PFGE analysis of extracellular virus DNA described in this report is useful in (i) detecting defective genome in a virus stock, (ii) determining defective genome sizes and (iii) quantifying defective genomes of specific sizes in a virus stock. Other important advantages of this method include (i) a shorter assay time resulting from the elimination of DNA isolation and Southern blotting and (ii) no handling of radioactive isotopes. PFGE analysis of AcMNPV BV DNA can provide valuable insights into the production of DIPs, which correlates with loss of infectivity. This method can be improved to detect all the defective genomes and corresponding genome lengths by digestion with AvrII and FseI, both of which cut once within the genome, although either site may be lost during DIP formation.
Correlation between the DIP genome length and virion length
Electron microscopic analysis of DIPs showed a broad range of virion sizes from 100 to 230 nm, whereas PFGE analysis of the DIP genome shows three major bands at 105, 97 and 85 kb and some in the 50–60 kb range. Statistical analysis of the virion sizes indicated that the most abundant DIP sizes were 225, 215, 175 and 145 nm, corresponding to a 25, 28, 42 and 52 % reduction, respectively, of the WT mean virion size (300 nm). In contrast, the defective genome of sizes 105, 97 and 85 kb corresponded to 22, 28 and 37 % deletion, respectively. Hence, there is a rough correlation between the most common virion sizes (or small size ranges centred around these most common sizes) and the specific DIP genome sizes (Table 2). Furthermore, a correlation exists between the percentage of DIPs within specific size ranges and genome sizes (Table 2).
Prediction of putative deletion regions for AcMNPV DIPs: is homologous recombination a mechanism of deletion?
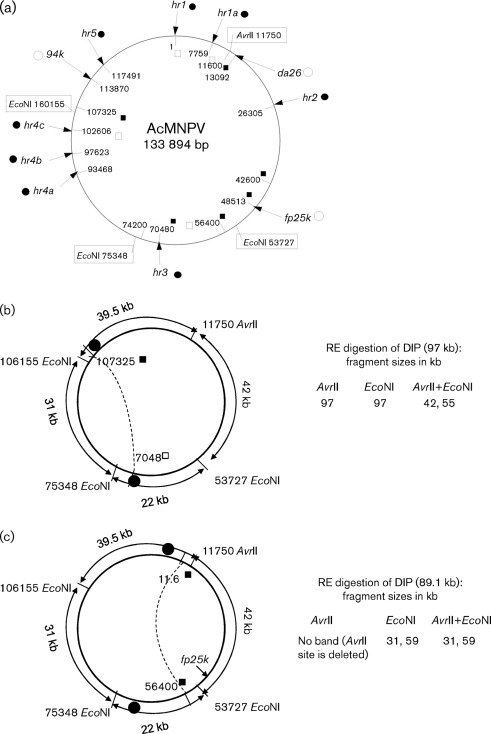
Although the exact mechanism of DIP formation is not clear, the current analysis suggests that DIP generation during late passages follows the same course in WT and Ac-FPm, as DNA fragments of similar sizes were generated (85, 97 and 105 kb when digested with AvrII and 54, 59, 85 and 96 kb when digested with EcoNI). This indicates that there could be a defined pattern of subsequent deletion events, possibly through homologous recombination of hr regions. Note that PCR amplification of the fp25k gene region and sequencing thereof was checked at many passages of WT AcMNPV and Ac-FPm up to passage 32 to confirm that the virus stocks were not cross-contaminated (Giri et al., 2010). It is known from previous studies that spontaneous genomic deletions are likely to occur by excision of the non-hr region (Pijlman et al., 2001, 2002, 2004). For example, a major deletion of 12 kb found in the WSSV genome was located in the largest inter-hr region (van Hulten et al., 2001). Supplementary Table S1 summarizes relevant information about passaged AcMNPV (e.g. deleted fragment size, deletion location) obtained during the current and previous investigations from different laboratories. Based on deletion regions found in AcMNPV DIPs from previous studies and the deletion size analysis from the current PFGE profile (digested with AvrII, EcoNI and EcoNI+AvrII), a computational analysis was performed to find some of the putative deletion regions in passaged AcMNPV. Furthermore, based on the pattern of predicted deletion regions, a hypothetical model of DIP formation is proposed that involves recombination between hr sites (Fig. 5a). For example, Fig. 5(b) shows a DIP model with a 37 kb genomic deletion from nt 70480 kb (hr3) to 107325 within the hr3–hr5 region to yield a 97 kb (134−37 = 97 kb) DIP that coincides with the 97 kb band obtained by AvrII digestion (Fig. 2a). Additionally, the 42 and 55 kb bands (Fig. 5b) coincide with the 42 and 54 kb bands in Fig. 4(b) obtained by digestion with AvrII and EcoNI. Whilst the predicted sizes are expected to match reasonably well with the experimental data, there are considerations that can explain differences: (i) the length of the hr sites is ~800 bp and recombination may occur in any region within this site and (ii) short insertions or deletions within the hr region during passaging will alter the defective genome size.
Fig. 5.
Predicted deletions during AcMNPV defective genome formation. (a) Schematic of AcMNPV genome with the nucleotide positions of the hr regions (hr1, hr1a, hr2, hr3, hr4a, hr4b, hr4c and hr5; •), genes that may have insertions (○), putative sites of recombination during DIP formation (▪), sites that are both hr and putative recombination sites (□) and restriction sites for AvrII and EcoNI (boxes). (b, c) Predicted deletion regions corresponding to the AcMNPV defective genome obtained from the PFGE analysis by AvrII, EcoNI and AvrII+EcoNI digestion. The putative site of recombination (▪), the hr regions (•) and the homologous recombination and consequent deletion (dotted lines) are indicated.
Fig. 5(c) depicts another putative defective genome of size 89 kb that corresponds to a total deletion of 45 kb from nt 11600 to 56400 (134−45 = 89 kb). AvrII digestion does not yield an 89 kb band (as the AvrII site is deleted), but 59 and 31 kb (59+31 = 90 kb) bands are produced from EcoNI and AvrII+EcoNI digestions (Fig. 4a, b). Similarly, many other deletion combinations are consistent with the fragment sizes obtained from the PFGE profile of passaged WT AcMNPV. The predicted recombination sites and the corresponding deletion regions of the AcMNPV DIPs model described here could be confirmed through (i) a detailed restriction mapping of the AcMNPV DIP DNA obtained from PFGE of the passaged virus DNA and (ii) PCR amplification and sequencing across the putative recombination sites/deletion junctions in the defective genomes.
Conclusion
This study indicates that DIP formation can be reduced by inhibiting transposon insertion from the host-cell genome and the subsequent deletion. The next logical step to reduce DIP formation would be to modify other insertion sites in the AcMNPV genome with high frequencies of insertion, e.g. the 13–13.7 m.u. region of the da26 gene (Kumar & Miller, 1987; O’Reilly et al., 1990) and the 86.4–86.6 m.u. region of the 94k gene (Miller & Miller, 1982). Such a strategy may be useful in improving the genetic stability of baculoviruses during the continuous production of biopesticides. Furthermore, this approach may improve recombinant protein production using the BEVS where passaging is required to amplify a small virus stock to obtain a sufficient amount to infect insect cells in a large-scale batch bioreactor.
Methods
Cells and virus.
The Spodoptera frugiperda cell line Sf-21 (Vaughn et al., 1977) was grown at 27 °C in Sf-900II growth medium (Gibco) (without serum and antibiotics) in 125 ml Erlenmeyer flasks (20 ml working volume) on an orbital shaker (Thermo Scientific) at 180 r.p.m. AcMNPV strain E2, which has a genome size of 134 kb (Ayres et al., 1994), was propagated in Sf-21 cells in a 125 ml shaker flask. A recombinant baculovirus denoted Ac-FPm was constructed by modifying the 13 TTAA transposon target sites in the AcMNPV fp25k gene such that the amino acid sequence of the FP25K protein remained unchanged (Giri et al., 2010). Initial stocks of WT AcMNPV and Ac-FPm (passage 0) were obtained following two rounds of plaque purification (Kelly et al., 2007) and used as the starting material for the passaging experiments. Additionally, ~200 ml WT AcMNPV and Ac-FPm BV stocks were prepared for conducting PFGE analysis of linearized virus DNA by amplifying the plaque-purified isolates from passage 0. Sf-900II growth medium supplemented with 10 % FBS (Gibco) was used for virus infections with AcMNPV and Ac-FPm during the passaging experiments (Jarvis & Garcia, 1994). The FBS was added to increase virus stability during storage at 4 °C. Continuous passaging of the two viruses was simulated as described by Giri et al. (2010). At the end of each passage, the cell supernatant containing BV was obtained after centrifuging at 800 g for 10 min.
Electron microscopy.
BV was pelleted by centrifugation from the culture medium through a 20 % (w/w) sucrose cushion at 10 000 g for 1 h. The pellet was resuspended overnight in 1 mM Tris/HCl (pH 7). The resuspended virus was then applied to a carbon-coated, Formvar copper grid for 3 min and stained using uranyl molybdate for 30 s. For each passage, the length of 85 BV particles was measured from electron micrographs taken on a JEOL JEM-1230 transmission electron microscope. The BV lengths from passages 1 and 32 (number of virus particles = 85) were analysed using ANOVA in matlab (Mathworks) to determine whether the mean lengths of the two heterogeneous populations were statistically significantly different at P<0.05. The size distribution for each passage, i.e. the percentage of virus particles corresponding to a given BV particle size range (nm), was examined.
PFGE analysis of extracellular virus.
PFGE analysis of AcMNPV DNA was performed according to the method described by Pfaller et al. (1994) with some modifications as briefly described below.
In situ extraction of viral DNA.
The BV stock (15 ml) was pelleted through a 30 % (w/v) sucrose cushion of 8 ml. Briefly, 8 ml 30 % sucrose solution was pushed underneath the BV solution by a syringe in a tube and centrifuged at 100 000 g for 2 h at 4 °C. The resulting BV pellet was resuspended in 1 % low-melting-point agarose in PBS solution and transferred into a Bio-Rad plug mould (~100 µl BV solution per plug). The agarose in the moulds was solidified by cooling at 4 °C for 15 min. The DNA was isolated in situ by incubating the agarose plugs with 2 ml 10 mM Tris/HCl (pH 8.0), 100 mM EDTA, 1 % N-lauroyl sarcosine and 200 µg proteinase K ml−1 at 50 °C overnight in a six-well plate (Vanarsdall et al., 2007). The agarose plugs were then washed four times (3 ml per wash with gentle shaking for 30 min) with 0.1 M Tris/HCl (pH 7.5), 0.1 M EDTA at room temperature.
Restriction enzyme digestion.
Before restriction enzyme digestion, the agarose plugs were washed another three times (200 µl per wash for 30 min) with a 100 mM Tris/HCl (pH 8.0), 5 mM MgCl2 solution to remove substances that could inhibit restriction enzyme activity in a 96-well plate. The washing buffer was then replaced by the restriction enzyme buffer containing no restriction enzyme and incubated for 1 h at 4 °C. For DNA digestion, the plugs were incubated in a 5 µl AvrII reaction buffer for 1 h followed by overnight incubation with 20 U of AvrII (New England Biolabs) at 37 °C. The reaction was terminated by adding 250 µl 0.6× TBE.
PFGE and quantification of DIP DNA.
The DNA plugs were inserted into the wells of a 1 % agarose gel accommodating up to 24 samples. The DNA was then separated by PFGE in a CHEF-DR III variable angle system (Bio-Rad) using a programmable pulse-inverter. Gels were run at an initial switch time of 5 s and a final switch time of 20 s at 200 V for 23 h. A MidRange PFG I ladder (New England Biolabs) and a λ ladder were used as DNA size markers. The WT and defective genomes were quantified by measuring the DNA band intensities using ImageJ version 1.3 software (National Institutes of Health, Bethesda, MA, USA) after scanning the stained gel (stained with ethidium bromide) in a Typhoon 9400 scanner (GE Healthcare). As the size of the DNA has an impact on DNA band intensity, the defective genome frequency was calculated by taking the DNA fragment size into consideration using the formula: DNA band intensity = (constant)×(size of DNA fragment)×(number of copies of the fragment). This equation was used to determine the relative number of DNA fragments, which corresponded to the relative number of virus particles.
Acknowledgements
We are grateful for the technical assistance provided by Richard Hollis and Jennifer Kroeger from the University of Iowa Hospitals and Clinics, Special Microbiology Laboratory, and Kathy Walters from the Central Microscopy Facility at the University of Iowa, IA, USA. We would also like to thank Professor Richard Roller and Jean Sippy for their advice regarding deletion mutant analysis. Finally, we thank Sharon Braunagel and Max Summers for providing the WT AcMNPV used in this research. This research work was supported by the following grants: Environmental Protection Agency RD-83142101 (D. W. M.), National Science Foundation 0717620 (M. G. F.) and National Institutes of Health GM-51611 (M. G. F.). Scholarship support for L. G. was provided by the University of Iowa, Center for Biocatalysis and Bioprocessing.
Footnotes
A supplementary table is available with the online version of this paper.
References
- Ayres M. D., Howard S. C., Kuzio J., Lopez-Ferber M., Possee R. D. (1994). The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202, 586–605 10.1006/viro.1994.1380 [DOI] [PubMed] [Google Scholar]
- Black B. C., Brennan L. A., Dierks P. M., Gard I. E. (1997). Commercialization of baculoviral insecticides. In The Baculoviruses, pp. 341–388 Edited by Miller L. K. New York: Plenum Press [Google Scholar]
- Burand J. P., Summers M. D. (1982). Alteration of Autographa californica nuclear polyhedrosis virus DNA upon serial passage in cell culture. Virology 119, 223–229 10.1016/0042-6822(82)90082-4 [DOI] [PubMed] [Google Scholar]
- Burand J. P., Wood H. A., Summers M. D. (1983). Defective particles from a persistent baculovirus infection in Trichoplusia ni tissue culture cells. J Gen Virol 64, 391–398 10.1099/0022-1317-64-2-391 [DOI] [Google Scholar]
- Carstens E. B. (1987). Identification and nucleotide sequence of the regions of Autographa californica nuclear polyhedrosis virus genome carrying insertion elements derived from Spodoptera frugiperda. Virology 161, 8–17 10.1016/0042-6822(87)90165-6 [DOI] [PubMed] [Google Scholar]
- Cochran M. A., Faulkner P. (1983). Location of homologous DNA sequences intercepted at five regions in the baculovirus AcMNPV genome. J Gen Virol 45, 961–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPont (1996). Notification to conduct small-scale field testing of a genetically altered baculovirus. EPA No. 352-NMP-4.
- Fraser M. J., Smith G. E., Summers M. D. (1983). Acquisition of host cell DNA sequences by baculoviruses: relationship between host DNA insertions and FP mutants of Autographa californica and Galleria mellonella nuclear polyhedrosis viruses. J Virol 47, 287–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard I. E. (1997). Field testing a genetically modified baculovirus. In Brighton Crop Protection Conference Symposium Proceedings No. 68: Microbial Insecticides: Novelty or Necessity?, pp. 101–114. [Google Scholar]
- Giri L., Li H., Sandgren D., Feiss M. G., Roller R., Bonning B. C., Murhammer D. W. (2010). Removal of transposon target sites from the Autographa californica multiple nucleopolyhedrovirus fp25k gene delays, but does not prevent, accumulation of the few polyhedra phenotype. J Gen Virol 91, 3053–3064 10.1099/vir.0.024430-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino L. A., Gonzalez M. A., Summers M. D. (1986). Complete sequence and enhancer function of the homologous DNA regions of Autographa californica nuclear polyhedrosis virus. J Virol 60, 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R. L., Summers M. D. (1995). Mutations in the Autographa californica multinucleocapsid nuclear polyhedrosis virus 25 kDa protein gene result in reduced virion occlusion, altered intranuclear envelopment and enhanced virus production. J Gen Virol 76, 1451–1459 10.1099/0022-1317-76-6-1451 [DOI] [PubMed] [Google Scholar]
- Jarvis D. L., Garcia A., Jr (1994). Long-term stability of baculoviruses stored under various conditions. Biotechniques 16, 508–513 [PubMed] [Google Scholar]
- Kelly B. J., King L. A., Possee R. D. (2007). Introduction to baculovirus molecular biology. In Baculovirus and Insect Cell Expression Protocols, pp. 25–53 Edited by Murhammer D. W. Totowa, New Jersey: Humana Press [Google Scholar]
- Kompier R., Tramper J., Vlak J. M. (1988). A continuous process for the production of baculovirus using insect-cell cultures. Biotechnol Lett 10, 849–854 10.1007/BF01026994 [DOI] [Google Scholar]
- Kool M., Voncken J. W., van Lier F. L. J., Tramper J., Vlak J. M. (1991). Detection and analysis of Autographa californica nuclear polyhedrosis virus mutants with defective interfering properties. Virology 183, 739–746 10.1016/0042-6822(91)91003-Y [DOI] [PubMed] [Google Scholar]
- Krell P. J. (1996). Passage effect of virus infection in insect cells. Cytotechnology 20, 125–137 10.1007/BF00350393 [DOI] [PubMed] [Google Scholar]
- Kumar S., Miller L. K. (1987). Effects of serial passage of Autographa californica nuclear polyhedrosis virus in cell culture. Virus Res 7, 335–349 10.1016/0168-1702(87)90047-5 [DOI] [PubMed] [Google Scholar]
- Lee H. Y., Krell P. J. (1992). Generation and analysis of defective genomes of Autographa californica nuclear polyhedrosis virus. J Virol 66, 4339–4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. W., Miller L. K. (1982). A virus mutant with an insertion of a copia-like transposable element. Nature 299, 562–564 10.1038/299562a0 [DOI] [PubMed] [Google Scholar]
- Moscardi F. (1999). Assessment of the application of baculoviruses for control of Lepidoptera. Annu Rev Entomol 44, 257–289 10.1146/annurev.ento.44.1.257 [DOI] [PubMed] [Google Scholar]
- Murhammer D. W. (1996). Use of viral insecticides for pest control and production in cell culture. Appl Biochem Biotechnol 59, 199–220 10.1007/BF02787821 [DOI] [Google Scholar]
- O’Reilly D. R., Passarelli A. L., Goldman I. F., Miller L. K. (1990). Characterization of the DA26 gene in a hypervariable region of the Autographa californica nuclear polyhedrosis virus genome. J Gen Virol 71, 1029–1037 10.1099/0022-1317-71-5-1029 [DOI] [PubMed] [Google Scholar]
- Parekh D. (2008). Development and Scale-up of Protein Production Using the Baculovirus Expression System. Boston, MA: Process Sciences, Diosynth Biotechnology, Baculovirus Technology [Google Scholar]
- Pfaller M. A., Hollis R. J., Sader H. S. (1994). Chromosomal restriction fragment analysis by pulsed field gel electrophoresis. In Clinical Microbiology Procedures Handbook, Suppl. 1pp. 10.5c.1–10.5c12 Edited by Isenberg H. D. Washington, DC: American Society for Microbiology [Google Scholar]
- Pijlman G. P., van den Born E., Martens D. E., Vlak J. M. (2001). Autographa californica baculoviruses with large genomic deletions are rapidly generated in infected insect cells. Virology 283, 132–138 10.1006/viro.2001.0854 [DOI] [PubMed] [Google Scholar]
- Pijlman G. P., Dortmans J. C., Vermeesch A. M. G., Yang K., Martens D. E., Goldbach R. W., Vlak J. M. (2002). Pivotal role of the non-hr origin of DNA replication in the genesis of defective interfering baculoviruses. J Virol 76, 5605–5611 10.1128/JVI.76.11.5605-5611.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijlman G. P., van Schijndel J. E., Vlak J. M. (2003). Spontaneous excision of BAC vector sequences from bacmid-derived baculovirus expression vectors upon passage in insect cells. J Gen Virol 84, 2669–2678 10.1099/vir.0.19438-0 [DOI] [PubMed] [Google Scholar]
- Pijlman G. P., de Vrij J., van den End F. J., Vlak J. M., Martens D. E. (2004). Evaluation of baculovirus expression vectors with enhanced stability in continuous cascaded insect-cell bioreactors. Biotechnol Bioeng 87, 743–753 10.1002/bit.20178 [DOI] [PubMed] [Google Scholar]
- Potter K. N., Miller L. K. (1980). Correlating genetic mutation of a baculovirus with the physical map of the DNA genome. ICN-UCLA Symp Mol Cell Biol 18, 71–80 [Google Scholar]
- Slansky J. (2008). Generation of Antigen-specific Antitumor Immunity Using Insect Cells Infected with Recombinant Baculoviruses. Boston, MA: Health Sciences Center, University of Colorado, Baculovirus Technology [Google Scholar]
- Tramper J., Vlak J. M. (1986). Some engineering and economic aspects of continuous cultivation of insect cells for the production of baculoviruses. Ann N Y Acad Sci 469, 279–288 10.1111/j.1749-6632.1986.tb26505.x [DOI] [Google Scholar]
- Vanarsdall A. L., Mikhailov V. S., Rohrmann G. F. (2007). Characterization of a baculovirus lacking the DBP (DNA-binding protein) gene. Virology 364, 475–485 10.1016/j.virol.2007.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hulten M. C. W., Witteveldt J., Peters S., Kloosterboer N., Tarchini R., Fiers M., Sandbrink H., Lankhorst R. K., Vlak J. M. (2001). The white spot syndrome virus DNA genome sequence. Virology 286, 7–22 10.1006/viro.2001.1002 [DOI] [PubMed] [Google Scholar]
- van Lier F. L. J., van den End E. J., de Gooijer C. D., Vlak J. M., Tramper J. (1990). Continuous production of baculovirus in a cascade of insect-cell reactors. Appl Microbiol Biotechnol 33, 43–47 [DOI] [PubMed] [Google Scholar]
- van Lier F. L. J., van der Meijs W. C., Grobben N. G., Olie R. A., Vlak J. M., Tramper J. (1992). Continuous β-galactosidase production with a recombinant baculovirus insect-cell system in bioreactors. J Biotechnol 22, 291–298 10.1016/0168-1656(92)90147-2 [DOI] [PubMed] [Google Scholar]
- van Lier F. L. J., van Duijnhoven G. C., de Vaan M. M., Vlak J. M., Tramper J. (1994). Continuous β-galactosidase production in insect cells with a p10 gene based baculovirus vector in a two-stage bioreactor system. Biotechnol Prog 10, 60–64 10.1021/bp00025a007 [DOI] [PubMed] [Google Scholar]
- Vaughn J. L., Goodwin R. H., Tompkins G. J., McCawley P. (1977). The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera; Noctuidae). In Vitro 13, 213–217 10.1007/BF02615077 [DOI] [PubMed] [Google Scholar]
- Vlak J. M., Smith G. E. (1982). Orientation of the genome of Autographa californica nuclear polyhedrosis virus: a proposal. J Virol 41, 1118–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham T. J., Davis T., Granados R. R., Hammer D. A., Shuler M. L., Wood H. A. (1991). Baculovirus defective interfering particles are responsible for variations in recombinant protein production as a function of multiplicity of infection. Biotechnol Lett 13, 483–488 10.1007/BF01049204 [DOI] [Google Scholar]
- Zwart M. P., Erro E., van Oers M. M., de Visser J. A. G. M., Vlak J. M. (2008). Low multiplicity of infection in vivo results in purifying selection against baculovirus deletion mutants. J Gen Virol 89, 1220–1224 10.1099/vir.0.83645-0 [DOI] [PubMed] [Google Scholar]