Abstract
Hepatitis E virus is the aetiological agent of acute hepatitis E, a self-limiting disease prevalent in developing countries. Molecular analysis of viral genomic RNA from a chronically infected patient confirmed the recent discovery that chronic infection correlated with extensive diversification of the virus quasispecies: the hypervariable region of some virus genomes in this USA patient contained large continuous deletions and a minor proportion of genomes in faeces and serum had acquired a mammalian sequence that encoded 39 aa of S19 ribosomal protein fused to the virus non-structural protein. Genomes with this insert were selected during virus passage in cultured cells to become the predominant species, suggesting that the inserted sequence promoted virus growth. The results demonstrated that hepatitis E virus can mutate dramatically during a prolonged infection and suggests it may be important to prevent or cure chronic infections before new variants with unpredictable properties arise.
Hepatitis E, once believed to be an acute waterborne disease of subtropical regions with inadequate sanitation is now emerging as a zoonotic threat in industrialized countries around the world (Pavio et al., 2010). Previously, hepatitis E was defined as an acute disease (viraemia and faecal shedding lasting 2–7 weeks) that never progressed to chronicity (Ahmad et al., 2011; Meng et al., 2011); however, a steadily increasing number of chronic hepatitis E cases are being reported in immunosuppressed persons such as organ transplant patients or human immunodeficiency virus (HIV)-infected individuals (Kamar et al., 2010a, b, c, 2011a, b; Haagsma et al., 2010).
Since hepatitis E virus (HEV) has been exceedingly difficult to grow in cultured cells, the virus has not been adequately characterized and the epidemiology of infections and pathology of the disease are poorly understood. Okamoto and associates in Japan recently succeeded in adapting both a genotype 3 (JE03-1760F strain) and a genotype 4 isolate to grow relatively efficiently in cell culture (Tanaka et al., 2007, 2009). In each case, the virus was isolated from a patient with acute hepatitis E and successful propagation depended on starting with a high-titrated virus inoculum (Takahashi et al., 2010). The overall sequence and structure of the two Japanese cell culture-adapted virus genomes were similar to others of their respective genotype and point mutations scattered throughout the genome were postulated to be adaptive (Okamoto, 2011). In contrast, the sequence of the cell culture-adapted Kernow C1 genotype 3 strain, the only virus isolate from a chronically infected patient that thus far has been adapted to grow in cell culture, differs significantly from that of other genotype 3 viruses (Shukla et al., 2011). The Kernow strain was isolated from the faeces of an English patient who had been chronically co-infected with HEV and HIV for at least 2 years. The unexpected outcome of culturing the virus on HepG2/C3A hepatoma cells was the discovery that serial passage in these cells selected for a very rare, pre-existing virus–human recombinant genome that incorporated a proportion of the S17 human ribosomal protein gene, in-frame, within the hypervariable (HVR) region of the virus ORF1 non-structural gene region. This result raised the question of whether the insertion of a foreign sequence and cell culture selection of the resulting recombinant was an anomaly and unique to this isolate or whether the lengthy period of chronic infection had permitted the virus to accumulate mutations and other alterations to an unusual extent, in which case similar recombinants might be present in other HEV infections undergoing abnormally long courses of replication in vivo.
In an attempt to answer this question, HEV from an American liver-transplant patient chronically infected with genotype 3 strain (LBPR-0379) was characterized. The first serum collected from the patient was anti-HEV negative in an in-house ELISA (Engle et al., 2002; Yu et al., 2003), whereas one collected 2 days later demonstrated seroconversion with a sample/cut-off (S/C) ratio of 4.10 and 2.10 for anti-IgM and IgG, respectively (S/C >1 = reactive), and an RNA titre (Johne et al., 2010) of 6.08 log10 genome equivalents (GE) ml−1. A third serum collected 10 months later, 1 day prior to the faeces collection, had anti-HEV IgM and IgG S/C ratios of 6.50 and 4.08, respectively, and an RNA titre of 7.35 log10 GE ml−1. Therefore, the patient had been chronically infected for at least 10 months when the third serum was collected. The consensus sequence of virus in the serum was determined for all but the 5′ non-coding region and 30–40 nt of ORF1. Excluding the HVR region (nt 2089–2460 of the sequence with the GenBank accession no. JN564006), this virus shared 99.46 and 98.87 % amino acid identity with the three ORFs of Kernow and JE03-1760F genotype 3 strains, respectively. Faeces and serum from this later date were individually inoculated onto HepG2/C3A cells in a T25 flask and cells were cultured in 5 ml medium at 37 °C in a CO2 incubator as described previously for the Kernow strain (Shukla et al., 2011). Infectious viruses released into the medium after 46–51 days of continuous culture were quantified by a focus-forming assay to determine if the virus from the faeces or serum was replicating.
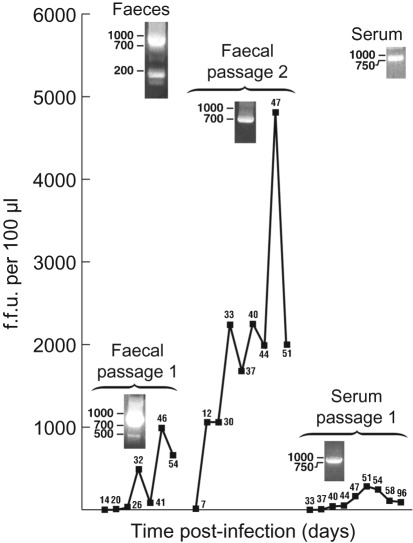
Twenty-six days after inoculation with a 10 % faecal suspension in PBS, cell cultures contained 3–5 % of HEV-infected cells as determined by immunofluorescence microscopy for virus capsid protein (Shukla et al., 2011); quantitative (q)RT-PCR and titration of infectious virus released into the medium indicated that only a low number of infectious viruses were released into the medium at this time (340 f.f.u. and 8×1010 genomes ml−1) (Fig. 1). At day 46, the infectious virus titre had increased almost 30-fold to 9900 f.f.u. and the RNA to 8.75×1010 genomes ml−1 medium; 1 ml of this medium was inoculated onto a monolayer of fresh HepG2/C3A cells in a T25 flask to initiate passage 2. At day 47 of passage 2, almost 50 000 f.f.u. (Supplementary Fig. S1, available in JGV Online) and 9.35×1010 genomes ml−1 medium were present. A similar attempt to culture virus from the serum collected 1 day before faeces were collected was only partially successful: 51 days after inoculation of 1 ml serum onto HepG2/C3A cells in a T25 flask, a peak titre of 2830 f.f.u. and 9.71×1010 genomes ml−1 was detected; thereafter, the titre gradually declined to 910 f.f.u. and 9.24×1010 genomes ml−1 at day 96 when the experiment was terminated.
Fig. 1.
Growth of LBPR-0379 in cultures of HepG2/C3A cells. Cell cultures were inoculated with a 10 % faecal suspension or with serum from a chronically infected patient and incubated at 37 °C. Total medium was harvested and replaced with fresh medium on the indicated days post-infection. Harvested medium was filtered through a 0.45 µm filter and stored at −80 °C. Thawed medium (100 µl) was plated on a monolayer of HepG2/C3A cells in eight-well chamber slides and the number of f.f.u. was determined by immunofluorescence microscopy as described previously (Shukla et al., 2011). Day of harvest is given next to each point. Agarose gels of RT-PCR products from the faeces, serum and cell culture medium at the time of peak infectious virus release are shown next to the molecular masses of DNA markers. In the medium, viral genome titres were ~106-fold higher than infectious virus titres (see text).
The HVR and surrounding regions (nt 1930–2819) of viruses in the faeces and serum and those in the medium at peak shedding times (days 46 and 47 for faecal viruses passage 1 and 2, respectively, and day 51 for serum viruses) were amplified by RT-PCR for sequence comparisons in order to determine if this region of the virus genome more closely resembled that of the acute JE03 virus propagated in Japan or the chronic Kernow isolate from England. The HVR region of LBPR-0379 encompasses 255 nt, encodes 85 aa and is located near the middle of the ~5200 nt long ORF1, which encodes non-structural proteins (Meng et al., 2010) (Fig. 2). Putative ORF1 protein domains, in order from the amino terminus, are methyltransferase, Y domain, papain-like cysteine protease (Karpe & Lole, 2011), HVR, X or macro domain (Ahmad et al., 2011), helicase and RNA-dependent RNA polymerase. It is unresolved whether ORF1 protein functions as an uncleaved polyprotein or whether it is proteolytically cleaved into active subunits. The HVR, Y and X domains have not been assigned functions. Agarose gel electrophoresis of PCR products amplified directly from the faeces revealed two broad bands at ~800 and ~200 bp, whereas PCR products from faecal passage 1 migrated as broad bands of ~800 and ~500 bp (Fig. 1). RT-PCR amplification of serum, serum passage 1 or faecal passage 2 yielded only one band at ~800 bp. Direct sequencing of the ~200 bp product provided a clear consensus sequence that suggested a fairly homogeneous population of genomes that contained a large deletion spanning most of the HVR region: analysis of 55 cDNA clones confirmed this (Table 1). A direct consensus sequence was also obtained for the two RT-PCR products from cell culture-passage 1 faecal virus so they were not cloned. All RT-PCR products of ~800 kb (except that from passage 1 faecal virus) were transformed into Escherichia coli and individual clones were sequenced (Table 1).
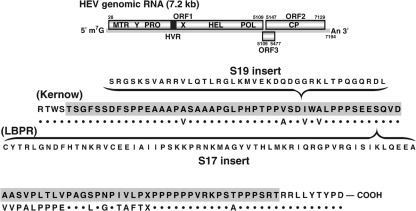
Fig. 2.
HVR sequence comparison of Kernow and LBPR viruses. A diagram of the full-length genome is shown at the top. In the expanded region, the HVR sequence of the Kernow strain is highlighted and that of the LBPR strain is presented underneath as dots for identical amino acids or as letters identifying different ones. X denotes a mixture. The sequence and placement of the S19 and S17 inserts are shown in parentheses.
Table 1. Nucleotide sequence comparisons of LBPR-0379 HVR populations.
| Source | No. nucleotides inserted (position)*/deleted (position) | No. clones | Clones (%) |
| Faeces (~800 bp) | 0/0 | 22/37 | 59.5 |
| 117 (2189)/0 | 13/37 | 35.0 | |
| 0/770 (2031) | 2/37 | 5.4 | |
| (~200 bp) | 0/770 (2031) | 54/55 | 98.2 |
| 0/801 (2018) | 1/55 | 1.8 | |
| Passage 1, faecal (~800 bp) | 117 (2189)/0 | Consensus† | |
| (~500 bp) | 117 (2189)/452 (2343) | Consensus | |
| Passage 2, faecal (~800 bp) | 0/463 (2356) | 2/15 | 13.3 |
| 117 (2189)/0 | 10/15 | 66.7 | |
| 0/760 (1952) | 2/15 | 13.3 | |
| 0/828 (1991) | 1/15 | 6.7 | |
| Serum (~800 bp) | 117(2189)/0 | 44/44 | 100 |
| Passage 1, serum (~800 bp) | 117 (2189)/42 (2381) | 92/92 | 100 |
Numbering according to the sequence with the GenBank accession no. JN564006 which contains the 117 nt insert at position 2189.
PCR products were purified on agarose gels, sequenced directly and the sequence was deposited in GenBank as JQ036303–JQ036304. The consensus sequence for underlined clones was deposited in GenBank as JQ036300–JQ036302, JQ036305–JQ036307. Clones not underlined were shorter than the 200 nt minimum required for deposit in GenBank.
Sequence comparisons demonstrated that the LBPR chronic strain genome structurally resembled the Kernow chronic strain genome rather than the JE03 acute strain genome. Whereas the JE03 strain lacked an insert, both chronic strains not only contained a large nucleotide insertion within the same region of the HVR (117 nt for the LBPR only 31 nt upstream from the 174 nt insertion location in the Kernow strain) (Fig. 2), but also, amazingly, in both cases, the insert was part of a ribosomal protein gene and had been acquired prior to cell culture since it was detected in viruses present in the faeces. The Kernow strain contained host ribosomal protein S17 sequence, while the LBPR strain contained host ribosomal protein S19 sequence. Also, in both cases the viral genome with the insert constituted only a minor species in the faeces, but became the major species during passage in cell culture (Table 1). However, the LBPR virus quasispecies differed from those of the Kernow strain and from known acute strains in containing a subset of viruses with large continuous deletions ranging from 42 to over 700 nt (Table 1).
It will require an infectious cDNA clone to determine why viruses carrying a large insertion in the HVR were preferentially selected during growth in cell culture. There is no evidence that the inserted sequences normally share a common or related function. The acquired S19 and S17 regions are each highly conserved across many species but differ considerably in size (39 vs 58 aa, respectively) and do not share obvious sequence similarities between them. The S19 inserted sequence had identity of 35/39 aa to genes of humans, pigs, mice, dogs and others but its identity of 114/117 nt with the human gene (GenBank accession no. NM_001022.3) is most consistent with a human origin as was true of the S17 insert (GenBank accession no. DQ896701.2). Natural deletions of the magnitude described here have not been reported previously. However, mutants with experimental deletions of up to 79 of the 86 aa within the HVR of genotype 3 swine virus were viable, although attenuated, in pigs (Pudupakam et al., 2009).
Most of the large deletions listed in Table 1 began just before the HVR region, eliminated it entirely, and terminated well within the X region, a macro domain of unknown function. The fact that passage 2 faecal viruses with large deletions were recovered in the medium of cultured cells 47 days post-inoculation, suggests that they may have been viable although the small number of clones identified suggests that they were not robust. The 452 nt deletion present in faecal passage 1 appears to have been selected against by passage 2 since the ~500 bp species was no longer detected (Table 1). In contrast, in serum passage 1, a smaller deletion of 42 nt was selected in conjunction with the 117 nt insert and all clones contained both. It is tempting to conclude that the larger deletion was detrimental and that the smaller deletion was selected because it aided genome packaging by compensating for the increase in genome size due to the 117 nt insertion; however, the fact that the serum virus did not replicate to high titres during cell culture suggests the deletion may also have had an adverse effect.
Finally, it is worth noting that the complexity of the virus population in the serum appeared to be significantly less than that in the faeces (Table 1). It will be informative to discover if this difference in the populations of the two compartments reflects different replication sites, different exit pathways from cells, differential stability or some other mechanism. Quasispecies compartmentalization of HEV between the serum and cerebrospinal fluid has been suggested as a factor that could be related to the development of neurological symptoms (Kamar et al., 2010b). It will also be important to determine if differential compartmentalization is linked to different pathologies.
In conclusion, these results support the idea that chronic infection with HEV can lead to a wide and unpredictable diversification of the viral quasispecies with consequences that need to be further explored.
Acknowledgements
This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases. This work was carried out under an approved protocol of the Carolinas Medical Center, IRB 10-0709B, with the informed consent of the patient.
Footnotes
A supplementary figure is available with the online version of this paper.
References
- Ahmad I., Holla R. P., Jameel S. (2011). Molecular virology of hepatitis E virus. Virus Res 161, 47–58 10.1016/j.virusres.2011.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engle R. E., Yu C., Emerson S. U., Meng X. J., Purcell R. H. (2002). Hepatitis E virus (HEV) capsid antigens derived from viruses of human and swine origin are equally efficient for detecting anti-HEV by enzyme immunoassay. J Clin Microbiol 40, 4576–4580 10.1128/JCM.40.12.4576-4580.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagsma E. B., Riezebos-Brilman A., van den Berg A. P., Porte R. J., Niesters H. G. (2010). Treatment of chronic hepatitis E in liver transplant recipients with pegylated interferon alpha-2b. Liver Transpl 16, 474–477 [DOI] [PubMed] [Google Scholar]
- Johne R., Plenge-Bönig A., Hess M., Ulrich R. G., Reetz J., Schielke A. (2010). Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. J Gen Virol 91, 750–758 10.1099/vir.0.016584-0 [DOI] [PubMed] [Google Scholar]
- Kamar N., Abravanel F., Garrouste C., Cardeau-Desangles I., Mansuy J. M., Weclawiak H., Izopet J., Rostaing L. (2010a). Three-month pegylated interferon-alpha-2a therapy for chronic hepatitis E virus infection in a haemodialysis patient. Nephrol Dial Transplant 25, 2792–2795 10.1093/ndt/gfq282 [DOI] [PubMed] [Google Scholar]
- Kamar N., Izopet J., Cintas P., Garrouste C., Uro-Coste E., Cointault O., Rostaing L. (2010b). Hepatitis E virus-induced neurological symptoms in a kidney-transplant patient with chronic hepatitis. Am J Transplant 10, 1321–1324 10.1111/j.1600-6143.2010.03068.x [DOI] [PubMed] [Google Scholar]
- Kamar N., Rostaing L., Abravanel F., Garrouste C., L’homme S., Esposito L., Basse G., Cointault O., Ribes D. & other authors (2010c). Ribavirin therapy inhibits viral replication on patients with chronic hepatitis E virus infection. Gastroenterology 139, 1612–1618 10.1053/j.gastro.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Kamar N., Bendall R. P., Peron J. M., Cintas P., Prudhomme L., Mansuy J. M., Rostaing L., Keane F., Ijaz S. & other authors (2011a). Hepatitis E virus and neurologic disorders. Emerg Infect Dis 17, 173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamar N., Garrouste C., Haagsma E. B., Garrigue V., Pischke S., Chauvet C., Dumortier J., Cannesson A., Cassuto-Viguier E. & other authors (2011b). Factors associated with chronic hepatitis in patients with hepatitis E virus infection who have received solid organ transplants. Gastroenterology 140, 1481–1489 10.1053/j.gastro.2011.02.050 [DOI] [PubMed] [Google Scholar]
- Karpe Y. A., Lole K. S. (2011). Deubiquitination activity associated with hepatitis E virus putative papain-like cysteine protease. J Gen Virol 92, 2088–2092 10.1099/vir.0.033738-0 [DOI] [PubMed] [Google Scholar]
- Meng X. J. (2010). Recent advances in Hepatitis E virus. J Viral Hepat 17, 153–161 10.1111/j.1365-2893.2009.01257.x [DOI] [PubMed] [Google Scholar]
- Meng X. J., Anderson D. A., Arankalle V. A., Emerson S. U., Harrison T. J., Jameel S., Okamoto H. (2011). Hepeviridae. In Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses, pp 991-998 Edited by King A. M. Q., Adams M. J., Carstens E. B., Lefkowitz E. J. London: Elsevier Academic Press [Google Scholar]
- Okamoto H. (2011). Efficient cell culture systems for hepatitis E virus strains in feces and circulating blood. Rev Med Virol 21, 18–31 10.1002/rmv.678 [DOI] [PubMed] [Google Scholar]
- Pavio N., Meng X. J., Renou C. (2010). Zoonotic hepatitis E: animal reservoirs and emerging risks. Vet Res 41, 46 10.1051/vetres/2010018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudupakam R. S., Huang Y. W., Opriessnig T., Halbur P. G., Pierson F. W., Meng X. J. (2009). Deletions of the hypervariable region (HVR) in open reading frame 1 of hepatitis E virus do not abolish virus infectivity: evidence for attenuation of HVR deletion mutants in vivo. J Virol 83, 384–395 10.1128/JVI.01854-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla P., Nguyen H. T., Torian U., Engle R. E., Faulk K., Dalton H. R., Bendall R. P., Keane F. E., Purcell R. H., Emerson S. U. (2011). Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc Natl Acad Sci U S A 108, 2438–2443 10.1073/pnas.1018878108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Tanaka T., Takahashi H., Hoshino Y., Nagashima S., Jirintai S., Mizuo H., Yazaki Y., Takagi T. & other authors (2010). Hepatitis E virus (HEV) strains in serum samples can replicate efficiently in cultured cells despite the coexistence of HEV antibodies: characterization of HEV virions in blood circulation. J Clin Microbiol 48, 1112–1125 10.1128/JCM.02002-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Takahashi M., Kusano E., Okamoto H. (2007). Development and evaluation of an efficient cell-culture system for hepatitis E virus. J Gen Virol 88, 903–911 10.1099/vir.0.82535-0 [DOI] [PubMed] [Google Scholar]
- Tanaka T., Takahashi M., Takahashi H., Ichiyama K., Hoshino Y., Nagashima S., Mizuo H., Okamoto H. (2009). Development and characterization of a genotype 4 hepatitis E virus cell culture system using a HE-JF5/15F strain recovered from a fulminant hepatitis patient. J Clin Microbiol 47, 1906–1910 10.1128/JCM.00629-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Engle R. E., Bryan J. P., Emerson S. U., Purcell R. H. (2003). Detection of immunoglobulin M antibodies to hepatitis E virus by class capture enzyme immunoassay. Clin Diagn Lab Immunol 10, 579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]