Abstract
JC virus (JCV) is the aetiological agent of the demyelinating disease progressive multifocal leukoencephalopathy, an AIDS defining illness and serious complication of mAb therapies. Initial infection probably occurs in childhood. In the working model of dissemination, virus persists in the kidney and lymphoid tissues until immune suppression/modulation causes reactivation and trafficking to the brain where JCV replicates in oligodendrocytes. JCV infection is regulated through binding of host factors such as Spi-B to, and sequence variation in the non-coding control region (NCCR). Although NCCR sequences differ between sites of persistence and pathogenesis, evidence suggests that the virus that initiates infection in the brain disseminates via B-cells derived from latently infected haematopoietic precursors in the bone marrow. Spi-B binds adjacent to TATA boxes in the promoter/enhancer of the PML-associated JCV Mad-1 and Mad-4 viruses but not the non-pathogenic, kidney-associated archetype. The Spi-B-binding site of Mad-1/Mad-4 differs from that of archetype by a single nucleotide, AAAAGGGAAGGGA to AAAAGGGAAGGTA. Point mutation of the Mad-1 Spi-B site reduced early viral protein large T-antigen expression by up to fourfold. Strikingly, the reverse mutation in the archetype NCCR increased large T-antigen expression by 10-fold. Interestingly, Spi-B protein binds the NCCR sequence flanking the viral promoter/enhancer, but these sites are not essential for early viral gene expression. The effect of mutating Spi-B-binding sites within the JCV promoter/enhancer on early viral gene expression strongly suggests a role for Spi-B binding to the viral promoter/enhancer in the activation of early viral gene expression.
Introduction
JC virus (JCV) is the aetiological agent of the often fatal demyelinating disease progressive multifocal leukoencephalopathy (PML). PML is an AIDS defining illness in 1–3 % of human immunodeficiency virus-positive patients and is a complication for patients with autoimmune diseases treated with immune modulatory therapies including natalizumab, rituximab and CellCept (Berger, 2010; Major, 2010). PML is caused by replication of JCV in and destruction of oligodendrocytes, which leads to cognitive deficits and motor dysfunction in patients. Although PML is a rare disease that afflicts the immune suppressed, the majority of the world population has been exposed to, and is seropositive for, JCV (Boothpur & Brennan, 2010). JCV can persist in a variety of compartments including the kidney, CD34+ precursor cells in bone marrow, and B-cells in lymphoid tissues and the brain (Brew et al., 2010). JCV activity in sites of persistence, and ultimately development of PML, is restricted by viral non-coding control region (NCCR) variation and activation of gene expression from the NCCR by essential host cell factors (Marshall & Major, 2010). Maintenance of JCV NCCR sequences in cells of the lymphoid lineage similar to those present in PML tissue prior to PML diagnosis suggests that these cells may act as reservoirs of latent virus (Brew et al., 2010; Major, 2010; Marshall & Major, 2010; Tan et al., 2009).
JCV gene expression and viral DNA replication are directed from specific sequences within the NCCR, which physically separates the early and late gene coding regions (Frisque, 1983; Frisque et al., 1984). The NCCR contains the highly conserved origin of DNA replication (ORI) followed by a highly variable segment that contains promoter/enhancer elements. Promoter/enhancer sequences can differ between sites of persistence and pathogenesis in a single patient and in the ability to direct gene expression (Gosert et al., 2010; Martin et al., 1985; Martin & Li, 1991; Marzocchetti et al., 2008; Tan et al., 2009, 2010). The original isolates of JCV, Mad-1 and Mad-4, were obtained from PML tissue and are considered representative for use in laboratory studies. Another JCV sequence used in the laboratory, archetype, was isolated from the urine of a persistently infected healthy individual and is replication incompetent in tissue culture (Daniel et al., 1996; Hara et al., 1998). Importantly, the promoter/enhancer sequences from JCV present in PML tissues differ between patients (Ault & Stoner, 1993; Gosert et al., 2010; Jensen & Major, 2001; Martin et al., 1985; Marzocchetti et al., 2008; Tan et al., 2010; Vaz et al., 2000) and are associated with increased viral activity over the representative laboratory viruses described above (Gosert et al., 2010; Martin & Li, 1991; Reid et al., 2011). Promoter/enhancer sequences isolated from patient tissues are composed of variations in the combination of five blocks of sequence, designated a–e, that make up the minimal archetype promoter/enhancer associated with persistence in the kidney shown in Fig. 1(c). The PML-associated Mad-1 virus shown in Fig. 1(a) encodes a promoter/enhancer that contains direct tandem repeats of the a–c–e blocks of sequence (Frisque, 1983; Frisque et al., 1984). Additional promoter/enhancer sequences that contain tandem repeats, including Mad-4 shown in Fig. 1(b) and Mad-8, are consistently associated with JCV present in the lymphoid compartment and the brain, where sequences similar to the kidney-associated archetype are rarely detected (Gosert et al., 2010; Jensen & Major, 2001; Tan et al., 2009).
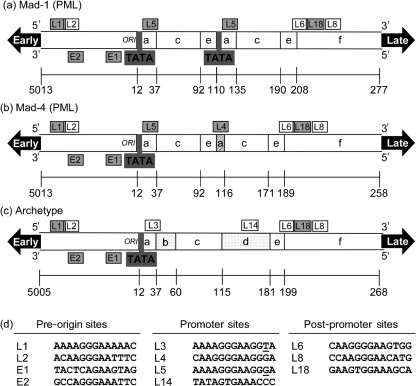
Fig. 1.
Diagram of potential Spi-B-binding sites outside the promoter/enhancer region within the JCV NCCR. The NCCRs from the PML-associated Mad-1 (a), Mad-4 (b) and the non-pathogenic archetype (c) JCVs are represented. The PML-associated promoter/enhancers contain 98 bp tandem repeats. A 19 bp deletion in the second repeat of Mad-4 (diagonal stripes) results in the loss of the second TATA box. The promoter/enhancer of the archetype variant contains two inserts (spotted) in a single 98 bp unit. Sites that bound Spi-B protein in EMSA assays are grey and sites that did not bind protein are white. Sites read on the late strand are named with an L and the sites read on the early strand are named with an E. (d) The Spi-B-binding site sequences are listed according to location in the pre-origin sequence, the promoter or the post-promoter sequence.
Tandem repeat promoter/enhancers contain duplications of elements that are essential to transcriptional activity, such as TATA boxes and transcription factor-binding sites, which contribute to their potency. For example, tandem repeat promoter/enhancers contain multiple binding sites for Oct-6/tst-1/SCIP (Krebs et al., 1995; Sock et al., 1999), purα (Chen & Khalili, 1995) and YB-1 (Chen & Khalili, 1995; Kerr et al., 1994) that promote viral activity in glial cells, as well as NF-1 (Kumar et al., 1996; Monaco et al., 2001) and Spi-B (Marshall et al., 2010) that promote viral activity in lymphoid cells and glial cells. Because JCV persists in cells undergoing lymphoid maturation and may traffic to the brain in B-cells, host factors specific to these cells types, such as Spi-B, could be important for viral dissemination and pathogenesis. In accordance with this hypothesis, Spi-B has been shown to be upregulated in peripheral blood mononuclear cells, a subset of which are capable of harbouring latent JCV, in response to treatment with natalizumab, a known risk factor for the development of PML (Lindberg et al., 2008).
Spi-B is an ETS transcription factor that activates promoters for genes involved in B-cell maturation (Bartel et al., 2000; Gallant & Gilkeson, 2006) as well as some viral promoters (Dekoninck et al., 2003; Erselius et al., 1990; Laux et al., 1994; Petterson & Schaffner, 1987). Spi-B binds to 5′-RGAA-3′ core sequences (Rao et al., 1999) and can cooperate with retinoblastoma protein and TATA-binding protein (TBP) to affect expression of proteins involved in B-cell maturation (Hagemeier et al., 1993; Mao et al., 1996; Rao et al., 1999; Weintraub et al., 1995). Spi-B is expressed at high levels in developing and mature B-cells (Ray et al., 1992; Su et al., 1996), and glial cells (Marshall et al., 2010). Spi-B binding to sites present in the promoter/enhancer of PML-associated JCV variants was recently described (Marshall et al., 2010). Interruption of Spi-B-binding sites by site-directed mutagenesis demonstrated that a unique site present in the second repeat of Mad-4 was essential for viral activity. In this study, we demonstrate that although Spi-B sites outside the promoter/enhancer in the JCV NCCR are not essential, TATA-box-associated Spi-B-binding sites within the promoter/enhancer are essential for efficient early viral gene expression in glial cells. Strikingly, a single nucleotide change between the sequences present in the binding sites located in the Mad-1/Mad-4 and the archetype promoter/enhancers was sufficient to abrogate Spi-B binding and had significant effect on early viral gene expression.
Results
Spi-B binds sites present outside the promoter/enhancer within the JCV NCCR
Potential Spi-B-binding sites were identified in the conserved NCCR sequence upstream of the origin (L1, L2, E1 and E2) and downstream of the promoter/enhancers (L6, L18 and L8) of the Mad-1, Mad-4 and archetype JCVs [shown in Fig. 1(a), (b) and (c), respectively] using the criteria of a RGAA core based on the Spi-B consensus site that was described previously (Laux et al., 1994). Although promoter/enhancer sequences are highly variable between isolates of JCV from PML patients, the sequences flanking either side of the promoter/enhancers are conserved (Jensen & Major, 2001). Binding sites identified on the late strand (named with an L) and sites identified on the early strand (named with an E) are listed in Fig. 1(d).
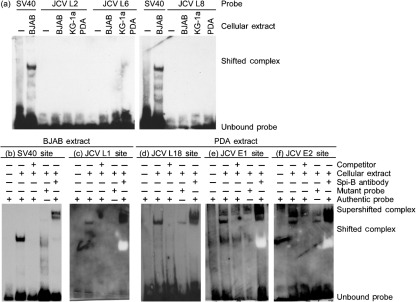
The association of Spi-B protein with potential binding sites was measured by electromobility shift assay (EMSA) using whole-cell extracts from cells susceptible to JCV infection including BJAB (B-cell line), KG-1a (CD34+ surrogate cell line) and progenitor-derived-astrocytes (PDA). Cell extracts were incubated with double-stranded oligonucleotide probes containing the Spi-B-binding sites listed in Table 1, or the SV40 Spi-B site as a positive control (Petterson & Schaffner, 1987). Fig. 2(a) illustrates that a subset of potential binding sites located upstream of the origin (L2) or downstream of the promoter/enhancers (L6 and L8) did not bind protein present in any cell type tested, suggesting the inactivity of these sites in the cells tested. However, the probe for the L1-binding site, located upstream of the origin, bound protein expressed in BJAB as shown in Fig. 2(c). Incubation of the L1 probe with cellular extract resulted in the formation of a single protein–DNA complex consistent among cell types (Table 1). This complex was competed by 200 molar excess of unlabelled oligonucleotide probe. Alteration of the L1 Spi-B site core from GGAA to CCAA abrogated complex formation as demonstrated previously for other Spi-B-binding sites (Dekoninck et al., 2003; Laux et al., 1994; Marshall et al., 2010). Addition of Spi-B antiserum caused a supershift of the original complex, similar to that observed with the SV40 Spi-B-binding site shown in Fig. 2(b), indicating that the protein bound to the L1 probe in the shifted complex is Spi-B. A white band appears in the supershift lanes requiring lengthy exposures (>3 h) and is due to excessive serum proteins present in the Spi-B antiserum.
Table 1. Spi-B-binding sites present outside the promoter/enhancer in the JCV NCCR.
| Binding site | Sequence (5′–3′) | Location (DNA strand) | EMSA binding (BJAB/KG-1a/PDA) |
| L1 | AAAAGGGAAAAAC | Pre-origin (late strand) | +/+/+ |
| L2 | ACAAGGGAATTTC | Pre-origin (late strand) | –/–/– |
| L6 | CAAGGGGAAGTGG | e–f (late strand) | –/–/– |
| L8 | CCAAGGGAACATG | f (late strand) | –/–/– |
| L18 | GAAGTGGAAAGCA | f (late strand) | –/–/+ |
| E1 | TACTCAGAAGTAG | Pre-origin (early strand) | –/–/+ |
| E2 | GCCAGGGAAATTC | Pre-origin (early strand) | –/–/+ |
Fig. 2.
Spi-B binds sites present outside the promoter/enhancer within the JCV NCCR. EMSAs were performed using BJAB, KG-1a or PDA cell extract and biotin-labelled oligonucleotide probes for the JCV L2, L6, L8 Spi-B-binding sites (a) and the SV40 Spi-B-binding site as a positive control for Spi-B protein–DNA complex formation. Representative EMSAs using: BJAB cell extract and biotin labelled oligonucleotide probes for the SV40 (b) and JCV L1 (c) Spi-B-binding sites; or PDA cell extract for the L18 (d), E1 (e) and E2 (f) Spi-B-binding sites are shown. Biotinylated authentic binding site probe was incubated with cell extract alone or in combination with 200-fold excess unlabelled oligonucleotide competitor or Spi-B antiserum (b–f). Biotinylated mutant probe was incubated with cell extract to demonstrate specificity for the Spi-B-binding site core (b–f). The results presented are representative of three independent experiments.
Identical EMSAs were carried out for the potential binding sites E1, E2 and L18; however, these probes only bound protein expressed in PDAs as shown in Fig. 2(d–f). Complex formation was competed by 200 molar excess of unlabelled probe and mutation of the L18 and L1 Spi-B-binding site cores to CCAA resulted in abrogation of complex formation. Formation of the complex with the E2 probe was not significantly affected by the alteration of the Spi-B-binding site core as shown in Fig. 2(f). However, addition of Spi-B antiserum did cause a supershift of the complexes for each binding site, indicating that Spi-B protein was bound to probe even in the case of the E2 site. These results suggest that binding of Spi-B protein to the E2 probe is not completely dependent on the presence of the RGAA core sequence. Several reports indicate that Spi-B binding is also heavily dependent upon the sequence flanking the RGAA core as well as interactions with other protein complex cofactors on the target DNA (Carvalho & Derse, 1993; Pongubala et al., 1992; Wasylyk et al., 1993). For example, the single G to T change in nucleotide sequence between the 3′ end of the JCV L5 and L3 Spi-B-binding sites is sufficient to completely abrogate complex formation (Marshall et al., 2010).
Mutation of Spi-B sites outside the promoter/enhancer region does not affect early viral gene expression in PDAs
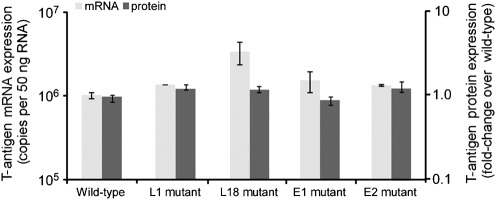
To determine if the Spi-B sites outside the promoter/enhancer that bind Spi-B protein in EMSAs are important for viral activity, site-directed mutagenesis was used to generate plasmids with a GG to CC mutation in the Spi-B site core of the L1-, L18-, E1- and E2-binding sites in Mad-4 JCV. Plasmids containing wild-type Mad-4, L1 mutant Mad-4, L18 mutant Mad-4, E1 mutant Mad-4 and E2 mutant Mad-4 were introduced into cells that permit efficient JCV infection, PDAs, via nucleofection. It is important to note that in this and all subsequent experiments, both the nucleofection and quantification was performed double-blinded in three or more independent experiments. Four days after nucleofection total RNA was isolated and the JCV early gene T-antigen mRNA expression was measured by one-step quantitative reverse transcription-PCR (qRT-PCR). All qRT-PCRs performed in this study include samples tested without reverse transcriptase to determine viral genomic DNA contamination. Eight days after nucleofection, T-antigen protein expression was quantified by blindly counting T-antigen protein-positive cells using immunofluorescence and is expressed as a fold-change over wild-type Mad-4 in Fig. 3. Although T-antigen protein can be detected as early as 5 days post-nucleofection, quantification was performed at 8 days post-nucleofection to allow for sufficient accumulation for consistent measurement by fluorescent microscopy. Fig. 3 demonstrates that mutation of the L1, L18, E1 and E2 Spi-B site cores had no significant affect on T-antigen mRNA or protein expression. These results suggest that although Spi-B can bind to sequences present outside the promoter/enhancer within the JCV NCCR, these sites do not affect early viral gene expression in cells derived from the highly susceptible human astrocytes (progenitor-derived-astrocytes or PDA).
Fig. 3.
Mutation of Spi-B sites outside the promoter/enhancer region does not affect early viral activity in PDAs. JCV T-antigen mRNA is represented as total copies per 50 ng of RNA along with sd (light grey). The number of T-antigen protein-positive cells was quantified on a per cell basis and is represented as a fold-change over the wild-type Mad-4 plasmid along with sd (dark grey).
Point mutation of the L5-binding sites in the Mad-1 promoter/enhancer significantly impairs early viral activity in PDAs
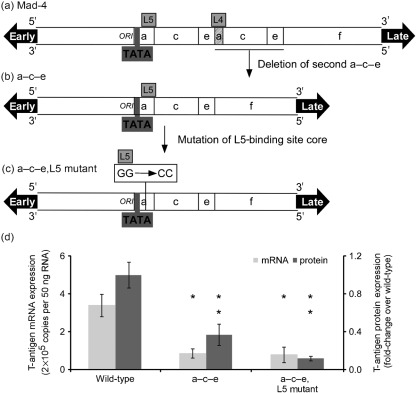
Spi-B-binding sites were previously identified in each of the 98 bp units of the tandem repeat promoter/enhancers of Mad-1 and Mad-4 JCV as shown in Fig. 1. Mutational analysis of these sites revealed that the L4 Spi-B site core present in the second repeat of Mad-4 was required for viral activity in PDAs, while mutation of the L5 Spi-B-binding site core present in the first repeat did not affect viral activity (Marshall et al., 2010). Given the potency of the L4-binding site on viral activity, the intact L4 site in the second repeat of the L5 Spi-B-binding site mutant may have been able to compensate for the loss of the L5-binding site. Because mutation of the L4 site abrogates viral infection it was important to determine the impact of mutation of the L5-binding site in the absence of a second repeat. Site-directed mutagenesis was used to generate a plasmid with the GG to CC mutation of the L5-binding site core in a single a–c–e promoter/enhancer. Plasmids containing Mad-4, a–c–e, and L5 mutant a–c–e shown in Fig. 4(a–c) were introduced into PDAs via nucleofection. Four days post-nucleofection total RNA was isolated and early T-antigen mRNA expression was measured by one-step qRT-PCR. Eight days post-nucleofection, T-antigen protein expression was quantified by blindly counting T protein-positive immunostained cells. Fig. 4(d) demonstrates that deletion of the second a–c–e significantly reduced both T-antigen mRNA and protein expression. Mutation of the remaining L5-binding site core in the single a–c–e did not significantly affect T-antigen mRNA or protein expression.
Fig. 4.
Deletion of the second repeat in the Mad-4 promoter/enhancer reduces early viral activity in PDAs. The NCCR sequences for Mad-4 (a), single a–c–e Mad-4 (b) or L5 mutant a–c–e Mad-4 (c) are represented in a diagram form. JCV T-antigen mRNA is represented as 2×105 copies per 50 ng RNA along with sd (d, light grey). The number of T-antigen protein-positive cells was quantified on a per cell basis and is represented as a fold-change over the wild-type Mad-4 plasmid along with sd (d, dark grey). An asterisk denotes the following statistically significant changes in value for mRNA: a–c–e, P = 0.0005; L5 mutant a–c–e, P = 0.0009. Two asterisks denote the following statistically significant changes in value for protein: a–c–e, P = 0.0012; L5 mutant a–c–e, P = 0.0008.
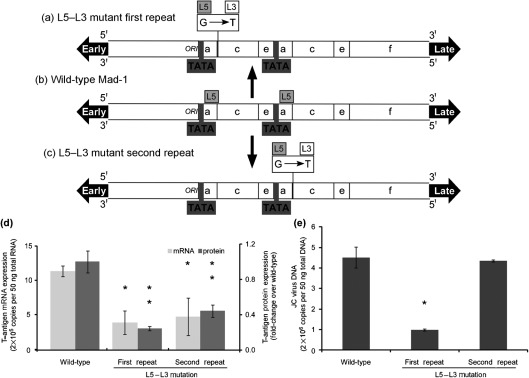
Spi-B protein binding to target sequences is also heavily dependent upon the regions flanking the RGAA core (Carvalho & Derse, 1993; Laux et al., 1994). In the case of TATA box-associated JCV Spi-B-binding sites, a single G to T nucleotide change between the 3′ end of the L5 (Mad-1/Mad-4) and L3 sites (archetype) is sufficient to abrogate complex formation (Marshall et al., 2010). Site-directed mutagenesis was used to generate plasmids with G to T point mutations in the L5 Spi-B-binding sites of the first or second repeat of the Mad-1 promoter/enhancer, in effect converting active L5-binding sites into inactive L3-binding sites. Plasmids containing wild-type Mad-1, and L5 to L3 mutants in the first or second repeat of Mad-1 shown in Fig. 5(a–c) were introduced into PDAs via nucleofection. Four days post-nucleofection, total RNA was isolated and early T-antigen mRNA expression was measured by one-step qRT-PCR. Eight days post-nucleofection, T-antigen protein expression was quantified by blindly counting T-antigen protein-positive immunostained cells. Fig. 5(d) demonstrates that converting an active L5 Spi-B-binding site to an inactive L3-binding site via a single nucleotide change was sufficient to significantly impair the ability of the virus to express the early gene T-antigen. Mutation of the first L5 site reduced T-antigen mRNA expression by 2.9-fold and T protein expression by fourfold. Mutation of the second L5 site reduced T-antigen mRNA expression by 2.4-fold and T-antigen protein expression by 2.3-fold. Because the sequence including the first L5-binding site lies near the JCV origin of DNA replication and has been implicated in the control of late transcription, mutation of this site may also play a role in late events. Ten days post-nucleofection, total DNA was isolated and JCV DNA was detected using quantitative (q)PCR. Fourteen days post-nucleofection, total RNA was isolated and late gene VP-1 mRNA expression was measured by qRT-PCR. Fig. 5(e) shows that point mutation of the L5 site in the first repeat, but not the second repeat, reduced viral DNA replication by fivefold. A similar trend was observed for VP-1 mRNA expression; however, the results were not statistically significant (data not shown). Taken together these results suggest that the L5 Spi-B-binding sites adjacent to TATA boxes in JCV promoter/enhancers contribute to early viral gene expression and that the 3′ sequence flanking the core binding site is important for this function. In addition, the L5 Spi-B site in the first repeat is important to viral DNA replication, and to a lesser extent late gene expression, which is most probably due to the proximity to the origin of DNA replication and late gene start sites.
Fig. 5.
Point mutations converting an L5 Spi-B-binding site to an L3 Spi-B-binding site in the Mad-1 promoter/enhancer reduces early viral activity in PDAs. The NCCR for wild-type Mad-1 (b) and L5–L3 point mutants for the first (a) or second (c) repeats within the Mad-1 promoter/enhancer are represented in a diagram form. JCV T-antigen mRNA is represented as 2×105 copies per 50 ng RNA along with sd (d, light grey). The number of T-antigen protein-positive cells was quantified on a per cell basis and is represented as a fold-change over the wild-type Mad-1 plasmid along with sd (d, dark grey). JC virus DNA is represented as 2×106 copies per 50 ng total DNA along with sd (e). An asterisk denotes the following statistically significant changes in value of mRNA: L5–L3 mutant first repeat, P = 0.0006; L5–L3 mutant second repeat, P = 0.0009. Two asterisks denote the following statistically significant change in value of protein: L5–L3 mutant first repeat, P = 0.0076; L5–L3 mutant second repeat, P = 0.0059. An asterisk denotes a statistically significant change in JC virus DNA for the L5–L3 mutant first repeat, P = 0.01.
A point mutation converting the L3 Spi-B-binding site to an L5 Spi-B-binding site is sufficient to activate early viral gene expression from the archetype promoter in PDAs
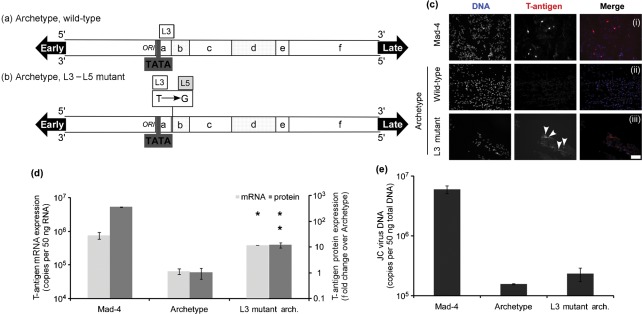
Archetype JCV is replication incompetent in tissue culture (Daniel et al., 1996; Hara et al., 1998) and archetype-like variants of JCV are rarely associated with PML (Gosert et al., 2010; Jensen & Major, 2001). As described previously, the active Spi-B-binding sites present in the promoter/enhancer of Mad-1 and Mad-4 (L5) differ from the inactive site present in archetype (L3) by a single nucleotide on the 3′ end of the binding site (Marshall et al., 2010). Site-directed mutagenesis was used to generate a plasmid with a G to T point mutation in the Spi-B-binding site of the archetype promoter/enhancer, in effect converting the inactive L3-binding site into an active L5-binding site. Plasmids containing Mad-4, wild-type archetype and the L3–L5 mutant archetype shown in Fig. 6(a–b) were introduced into PDAs via nucleofection. Four days post-nucleofection, total RNA was isolated and early gene T-antigen mRNA expression was measured by one-step qRT-PCR. Eight days post-nucleofection, T-antigen protein expression was quantified by blindly counting T-antigen-positive immunostained cells. T-antigen protein expression is represented as a fold-change over archetype. T-antigen protein expression was rarely observed from the wild-type archetype plasmid as shown in Fig. 6(c) panel (ii). T-antigen protein expression was observed from the L3–L5 mutant archetype plasmid as shown in Fig. 6(c) panel (iii) similar to levels observed from the Mad-4 plasmid as shown in Fig. 6(c) panel (i). Strikingly, Fig. 6(d) illustrates that T-antigen protein expression from the mutant promoter was increased 9.3-fold over the wild-type archetype promoter, while T-antigen mRNA expression was increased 6-fold over the wild-type archetype promoter. Ten days post-nucleofection, total DNA was isolated and JCV DNA was detected using qPCR. Fourteen days post-nucleofection, total RNA was isolated and late gene VP-1 mRNA expression was measured by qRT-PCR. Fig. 6(e) shows that creation of an L5 site in the archetype promoter slightly enhances viral DNA replication, while VP-1 RNA and protein expression was not observed up to 21 days post-nucleofection (data not shown). Taken together these results clearly demonstrate that creation of an active Spi-B site is sufficient to activate gene expression from the archetype promoter in PDAs, but is not enough to activate viral DNA replication and late gene expression. This observation may predict changes that occur, which increase a virus’ pathogenic potential in that these viruses have a selective growth advantage in susceptible cells.
Fig. 6.
A point mutation converting the L3 Spi-B-binding site to an L5 Spi-B-binding site is sufficient to activate early viral gene expression from the archetype promoter in PDAs. The NCCR sequences for wild-type archetype (a) and the L3–L5 point mutant archetype (b) are represented in a diagram form. JCV T-antigen mRNA is represented as total copies per 50 ng RNA along with sd (d, light grey). Eight days post-nucleofection, indirect immunofluorescence was performed to visualize T protein-positive cells. In the merged images, DNA is shown in blue and T-antigen is shown in red (c). Mad-4, panel (i); archetype, panel (ii); and L3–L5 mutant archetype, panel (iii). Bar, 500 µm at ×20 magnification. The number of T-antigen protein-positive cells was quantified on a per cell basis and is represented as a fold-change over the archetype plasmid along with sd (d, dark grey). An asterisk denotes the following statistically significant changes in value of mRNA. L3–L5 mutant, P = 5.40×10−5. Two asterisks denote the following statistically significant change in value of protein, 2.08×10−6. JCV DNA is represented as total copies per 50 ng DNA along with sd.
Discussion
NCCR sequences flanking the hypervariable promoter/enhancer of JCV are conserved between viral isolates (Jensen & Major, 2001) and contain Spi-B-binding sites. In this study, sites that actively bound Spi-B protein expressed in JCV susceptible cell types are present both in the early and late sides of the origin of DNA replication (Figs 1 and 2). However, mutation of these sites resulted in no effect on the ability of the virus to initiate early gene expression in PDAs (Fig. 3). The NCCR sequence on the early side of the origin contains a site that binds NF-κB, C/EBPβ and NFAT4, and has been implicated in modulation of JCV gene expression in response to cytokines (Manley et al., 2006; Mayreddy et al., 1996; Ranganathan & Khalili, 1993; Romagnoli et al., 2009; Safak et al., 1999). The binding of NF-1 protein to sites present in NCCR sequence on the late side of the promoter/enhancer has been described previously (Amemiya et al., 1989, 1992); however, binding of NF-1 to this region has been shown to be dispensable for viral gene expression (Kumar et al., 1996). Natural alterations in these flanking sequences are rarely observed and in most cases these sequences are completely conserved between pathogenic and non-pathogenic JCV isolates. Because non-pathogenic JCV isolates, such as archetype, are replication incompetent in tissue culture, it is probable that while host cell factor binding in the flanking regions contributes to viral activity, it is not essential. The hypervariable promoter/enhancer of JCV contains the majority of elements necessary for initiating viral gene expression, including TATA boxes and transcription factor-binding sites (Marshall & Major, 2010). In addition, the promoter/enhancer region varies between each patient, and isolates with changes in this sequence from the archetype and or prototype viruses have been associated with increased viral infection (Gosert et al., 2010; Martin & Li, 1991). For this reason the promoter/enhancer is considered to be the portion of the NCCR that confers tissue tropism and supports pathogenesis.
Mutational analysis of the Spi-B-binding sites present in the promoter/enhancer of Mad-4, Mad-1 and archetype clearly demonstrates that these mutated Spi-B sites present are important for early viral gene expression in PDAs. Spi-B-binding sites in the promoter/enhancer of JCV variants are located directly adjacent to TATA boxes, or in place of the TATA box for the L4 site, that are essential for transcription of early and late viral genes. Spi-B is an ETS transcription factor that can cooperate with pRB and TATA-binding protein (TBP) to alter expression of proteins involved in B-cell maturation (Gallant & Gilkeson, 2006; Rao et al., 1999). TBP binds TATA box elements in promoters and is a subunit of the basal transcription complex TFIID, which increases RNA polymerase II activity (Hochheimer & Tjian, 2003). Recruitment of the TFIID complex to JC viral promoters by Spi-B and TBP is an attractive model for activation of JCV gene expression. Importantly, the binding of SV40 T-antigen to TBP-TAFII (TBP-associated factors) complexes (Zhai et al., 1997) and TFIID (Damania & Alwine, 1996) is critical for transcriptional activation, and evidence suggests a similar role for JCV T-antigen (Rekvig, 1997). The proximity of T-antigen-binding sites, TATA boxes and Spi-B-binding sites on the JCV promoter/enhancer suggests that T-antigen and Spi-B may cooperate in recruitment of transcriptional apparatus components in immune cells, similar to the cooperation of T protein and Oct-6/tst-1/SCIP in glial cells (Krebs et al., 1995; Sock et al., 1999).
As shown in Fig. 6, archetype JCV expresses early T-antigen mRNA at reduced levels compared with Mad-4, and rarely supports expression of observable T-antigen protein in PDAs. Archetype JCV is replication incompetent in tissue culture and is associated with asymptomatic persistence in the kidney of normal healthy individuals (Daniel et al., 1996; Hara et al., 1998). Although archetype-like JCV sequences have been isolated from PML tissue, these viral sequences consistently contain mutations and deletions (Jensen & Major, 2001). The sequence encoding the inactive archetype Spi-B site, AAAAGGGAAGGTA, is conserved among viruses isolated from the urine of healthy individuals (Agostini et al., 1998; Delbue et al., 2005; Elsner & Dörries, 1998; Gosert et al., 2010; O’Connor et al., 2005; Pagani et al., 2003; Reid et al., 2011; Vaz et al., 2000). Fig. 6(c–d) shows the novel observation of one such point mutation that creates an active Spi-B-binding site directly adjacent to the archetype TATA box (L3–L5 mutant archetype), which converts archetype to a biologically active virus capable of expressing both T-antigen mRNA and T-antigen protein at significantly increased levels over wild-type. Importantly, similar mutations that create active Spi-B-binding sites adjacent to the TATA boxes of archetype-like viruses have been described in the brain of a patient with cerebellar atrophy (Roux et al., 2011) and Crohn’s disease patients treated with infliximab (Bellizzi et al., 2011), suggesting that this type of mutation is supported during dissemination in the host. JCV DNA isolated from a patient’s tissue containing similar archetype sequences with an active Spi-B site downstream of the TATA box increases T-antigen gene expression (data not shown, unpublished results). Therefore, the changes observed in the sequences obtained from PML patients may represent alterations that make the virus more successful and ultimately contribute to pathogenesis. Future analyses of alterations and acquisitions of unique transcription factor-binding sites naturally occurring in patients will probably offer more insight into the role of these factors in viral pathogenesis.
In this study, we have identified Spi-B-binding sites that bound Spi-B protein expressed in cells derived from human fetal brain (PDAs), but not from the immune system (KG-1a and BJAB cells) (Fig. 2). These results indicate a difference between Spi-B expressed in the brain and B-cells. Spi-B protein expression appears identical when proteins are examined by protein blotting with Spi-B antibody (Marshall et al., 2010); therefore, the difference is unlikely due to the presence of the second Spi-B isoform, which results from alternative splicing and contains an observable change in molecular mass (Ray-Gallet et al., 1996). In addition, the second Spi-B isoform is missing the ETS domain that binds DNA and has been shown to be incapable of binding Spi-B probes by EMSA. Spi-B can be phosphorylated (Mao et al., 1996), which would not cause a significant shift by protein blotting. Investigation into Spi-B expression in the brain will be essential to any further understanding of this difference. It will be important to determine the physiological role of Spi-B in the brain. Differential binding of Spi-B protein to a viral promoter/enhancer in different cell types may be another way that the NCCR confers tissue tropism and supports development of PML. This work has demonstrated that Spi-B binding to the viral promoter contributes to early viral gene expression in astrocytes; however, Spi-B is naturally expressed at high levels in developing B-cells. B-cells have been shown to support the low levels of JCV infection, and are likely carriers of infectious virus to the brain during reactivation and dissemination leading to the development of PML. Activation of Spi-B gene expression in transitional B-cells that contain latent virus may be an important step in viral reactivation. Further investigation into the molecular interactions that occur between Spi-B, protein co-factors and the JCV NCCR in cells that support latent (haematopoietic progenitors and B-cells) and productive (astrocytes and oligodendrocytes) infection will offer additional insight into molecular pathogenesis, reactivation from latency in lymphocytes and the development of PML.
Methods
Cells and plasmids.
KG-1a cells were maintained in RPMI 1640 medium (Cellgro) supplemented with 20 % FBS (Atlanta Biologicals) and 2 mM l-glutamine (Quality biologics). BJAB cells were maintained in RPMI 1640 medium supplemented with 10 % FBS and 2 mM l-glutamine. Human central nervous system progenitor cells were isolated and differentiated into astrocytes as described previously (Messam et al., 2003). PDAs were maintained in minimal essential medium (Cellgro) supplemented with 10 % FBS, 2 mM l-glutamine and 50 µg gentamicin ml−1.
Plasmids containing the full-length JCV Mad-4 genome [pMad4 (586)], Mad-1 genome (pM1TC), and archetype genome (pCY) were described previously (Frisque et al., 1984; Major et al., 1987; Yogo et al., 1990). Promoter/enhancer repeat deletion mutants of pMad4 (586) were generated as by-products of site-directed mutagenesis studies (unpublished results).
Antibodies.
A mouse mAb for SV40 T-protein (EMD Calbiochem), which cross-reacts with JCV T-protein, was used at 5 µg ml−1. The rabbit pAb for GFAP (Covance) was used at a 1 : 1000 dilution. Fluorescent-labelled antibodies qualified for multiple labelling experiments were obtained from Invitrogen and used at 1–2 µg ml−1. Spi-B antiserum was generously provided by Dr Moreau-Gachelin of Institut Curie (Paris, France) (Laux et al., 1994).
Preparation of whole-cell extracts.
Whole-cell extracts were prepared by a modification of the method of Andrews and Faller (Andrews & Faller, 1991) as described previously (Marshall et al., 2010).
Isolation of total RNA.
Total RNA was isolated from PDAs nucleofected with wild-type and mutant JCV DNA using the RNeasy plus mini kit (Qiagen) according to the manufacturer’s instructions. RNA was eluted from the column in nuclease-free water and quantified on a Nanodrop 8000 (Thermo Scientific).
Isolation of total DNA.
Total DNA was isolated from PDAs nucleofected with wild-type and mutant JCV DNA using the DNeasy blood and tissue kit (Qiagen) according to the manufacturer’s instructions. DNA was eluted from the column in nuclease-free water and quantified on a Nanodrop 8000.
Preparation of Spi-B-binding site mutants.
Site-directed mutagenesis was performed on the pMad4 (586) plasmid, the pM1TC plasmid and the pCY plasmid to alter Spi-B-binding site sequences using the QuikChange II XL site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions. Integrated DNA Technologies (IDT) synthesized sense and antisense oligonucleotide primers, listed in the Supplementary Table S1 (available in JGV Online). DNA sequencing was performed to confirm the presence of each desired mutation as well as the fidelity of the viral sequence by the NINDS DNA sequencing facility.
Nucleofection.
Archetype, Mad-1, Mad-4 and Spi-B-binding site mutant plasmids were introduced into PDAs by nucleofection using the Amaxa basic kit for primary neurons (Lonza) according to the manufacturer’s instructions as described previously (Marshall et al., 2010).
Quantitative immunofluorescence.
Immunofluorescence labelling was conducted on Lipofectamine 2000-transfected and/or nucleofected cell cultures. Cells were fixed with 4 % paraformaldehyde and permeabilized with 0.2 % Triton X-100 before indirect antibody labelling. Samples were mounted with a glycerol-based mounting medium containing the DNA dye DAPI and analysed by fluorescence microscopy by using a Zeiss Axiovert 200M microscope fitted with filters appropriate for DAPI, Alexa-Fluor-488 and Alexa-Fluor-647 excitation. In three independent experiments, two biological replicates were counted per condition. Four representative micrographs per replicate were counted and each micrograph contained between 1000 and 3000 total cells.
qPCR.
JCV DNA present in nucleofected cells was measured using the Taqman gene expression master mix reagents (Applied Biosystems) and the following forward primer (JCT1 5′-AGAGTGTTGGGATCCTGTGTTTT-3′), reverse primer (JCT2 5′-GAGAAGTGGGGATGAAGACCTGTTT-3′) and FAM/TAMRA-labelled probe (JCT1.1 5′-6FAMTCATCACTGGCAAACATTTCTTCATGGCTAMRA-3′). Eight 1 : 10 serial dilutions of 1 ng Mad-1 plasmid (pM1TC) µl−1 were prepared to generate a standard curve to determine JCV copy number.
qRT-PCR.
JCV T-antigen mRNA present in nucleofected cells was measured using the Taqman one-step RT-PCR master mix reagents (Applied Biosystems) and the primer/probes described for the qPCR method. Duplicate reactions were prepared without reverse transcriptase to determine the contribution of signal from contaminating viral genome and mRNA values were adjusted based on contaminating DNA values.
EMSA.
Sense and antisense oligonucleotides containing the authentic and mutant sequences of the SV40 Spi-B-binding site, or the JCV Spi-B-binding sites L1, L2, L6, L8, L18, E1 and E2 listed in the Supplementary Table S2 (available in JGV Online) were synthesized with and without 5′ biotinylation by IDT. The sense and antisense oligonucleotide pairs were annealed to form double-stranded probes at a concentration of 100 ng ml−1. The biotin labelled probes were diluted 1 : 200 in water. Biotin-labelled authentic probe or mutant probe was incubated with 25 µg whole-cell extract from KG-1a cells, BJAB cells or PDA in the presence or absence of 200-fold excess of unlabelled authentic probe (competitor). Supershifts were carried out by incubation of the cellular extracts with 2 µl Spi-B antiserum for 30 min on ice before the addition of probe as described previously (Laux et al., 1994). The reactions were incubated at room temperature for 20 min and resolved by electrophoresis in a 6 % polyacrylamide-TBE DNA retardation gel (Invitrogen). The complexes were transferred to a positively charged nylon membrane and detected using the LightShift chemiluminescent EMSA kit (Thermo Scientific/Pierce).
Acknowledgements
We thank Françoise Moreau-Gachelin of the Institut Curie (Paris, France) for generously providing polyclonal rabbit Spi-B antiserum and human Spi-B plasmid. We thank the DNA sequencing facility at the National Institute of Neurological Disorders and Stroke (NINDS) for aid in sequencing of mutant viral plasmids. We thank all of the members of the Laboratory of Molecular Medicine and Neuroscience at the NINDS for their hard work, support, and valuable input. The intramural program at the NINDS provided support for this work.
Footnotes
Supplementary tables are available with the online version of this paper.
References
- Agostini H. T., Ryschkewitsch C. F., Stoner G. L. (1998). Rearrangements of archetypal regulatory regions in JC virus genomes from urine. Res Virol 149, 163–170 10.1016/S0923-2516(98)80034-4 [DOI] [PubMed] [Google Scholar]
- Amemiya K., Traub R., Durham L., Major E. O. (1989). Interaction of a nuclear factor-1-like protein with the regulatory region of the human polyomavirus JC virus. J Biol Chem 264, 7025–7032 [PubMed] [Google Scholar]
- Amemiya K., Traub R., Durham L., Major E. O. (1992). Adjacent nuclear factor-1 and activator protein binding sites in the enhancer of the neurotropic JC virus. A common characteristic of many brain-specific genes. J Biol Chem 267, 14204–14211 [PubMed] [Google Scholar]
- Andrews N. C., Faller D. V. (1991). A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res 19, 2499 10.1093/nar/19.9.2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ault G. S., Stoner G. L. (1993). Human polyomavirus JC promoter/enhancer rearrangement patterns from progressive multifocal leukoencephalopathy brain are unique derivatives of a single archetypal structure. J Gen Virol 74, 1499–1507 10.1099/0022-1317-74-8-1499 [DOI] [PubMed] [Google Scholar]
- Bartel F. O., Higuchi T., Spyropoulos D. D. (2000). Mouse models in the study of the Ets family of transcription factors. Oncogene 19, 6443–6454 10.1038/sj.onc.1204038 [DOI] [PubMed] [Google Scholar]
- Bellizzi A., Anzivino E., Ferrari F., Di Nardo G., Colosimo M. T., Fioriti D., Mischitelli M., Chiarini F., Cucchiara S., Pietropaolo V. (2011). Polyomavirus JC reactivation and noncoding control region sequence analysis in pediatric Crohn’s disease patients treated with infliximab. J Neurovirol 17, 303–313 10.1007/s13365-011-0036-3 [DOI] [PubMed] [Google Scholar]
- Berger J. R. (2010). Progressive multifocal leukoencephalopathy and newer biological agents. Drug Saf 33, 969–983 10.2165/11537510-000000000-00000 [DOI] [PubMed] [Google Scholar]
- Boothpur R., Brennan D. C. (2010). Human polyoma viruses and disease with emphasis on clinical BK and JC. J Clin Virol 47, 306–312 10.1016/j.jcv.2009.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brew B. J., Davies N. W., Cinque P., Clifford D. B., Nath A. (2010). Progressive multifocal leukoencephalopathy and other forms of JC virus disease. Nat Rev Neurol 6, 667–679 10.1038/nrneurol.2010.164 [DOI] [PubMed] [Google Scholar]
- Carvalho M., Derse D. (1993). The PU.1/Spi-1 proto-oncogene is a transcriptional regulator of a lentivirus promoter. J Virol 67, 3885–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N. N., Khalili K. (1995). Transcriptional regulation of human JC polyomavirus promoters by cellular proteins YB-1 and Pur alpha in glial cells. J Virol 69, 5843–5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damania B., Alwine J. C. (1996). TAF-like function of SV40 large T antigen. Genes Dev 10, 1369–1381 10.1101/gad.10.11.1369 [DOI] [PubMed] [Google Scholar]
- Daniel A. M., Swenson J. J., Mayreddy R. P., Khalili K., Frisque R. J. (1996). Sequences within the early and late promoters of archetype JC virus restrict viral DNA replication and infectivity. Virology 216, 90–101 10.1006/viro.1996.0037 [DOI] [PubMed] [Google Scholar]
- Dekoninck A., Calomme C., Nizet S., de Launoit Y., Burny A., Ghysdael J., Van Lint C. (2003). Identification and characterization of a PU.1/Spi-B binding site in the bovine leukemia virus long terminal repeat. Oncogene 22, 2882–2896 10.1038/sj.onc.1206392 [DOI] [PubMed] [Google Scholar]
- Delbue S., Sotgiu G., Fumagalli D., Valli M., Borghi E., Mancuso R., Marchioni E., Maserati R., Ferrante P. (2005). A case of a progressive multifocal leukoencephalopathy patient with four different JC virus transcriptional control region rearrangements in cerebrospinal fluid, blood, serum, and urine. J Neurovirol 11, 51–57 10.1080/13550280590900382 [DOI] [PubMed] [Google Scholar]
- Elsner C., Dörries K. (1998). Human polyomavirus JC control region variants in persistently infected CNS and kidney tissue. J Gen Virol 79, 789–799 [DOI] [PubMed] [Google Scholar]
- Erselius J. R., Jostes B., Hatzopoulos A. K., Mosthaf L., Gruss P. (1990). Cell-type-specific control elements of the lymphotropic papovavirus enhancer. J Virol 64, 1657–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisque R. J. (1983). Regulatory sequences and virus-cell interactions of JC virus. Prog Clin Biol Res 105, 41–59 [PubMed] [Google Scholar]
- Frisque R. J., Bream G. L., Cannella M. T. (1984). Human polyomavirus JC virus genome. J Virol 51, 458–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant S., Gilkeson G. (2006). ETS transcription factors and regulation of immunity. Arch Immunol Ther Exp (Warsz) 54, 149–163 10.1007/s00005-006-0017-z [DOI] [PubMed] [Google Scholar]
- Gosert R., Kardas P., Major E. O., Hirsch H. H. (2010). Rearranged JC virus non-coding control regions found in progressive multifocal leukoencephalopathy increase virus early gene expression and replication rate. J Virol 84, 10448–10456 10.1128/JVI.00614-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeier C., Bannister A. J., Cook A., Kouzarides T. (1993). The activation domain of transcription factor PU.1 binds the retinoblastoma (RB) protein and the transcription factor TFIID in vitro: RB shows sequence similarity to TFIID and TFIIB. Proc Natl Acad Sci U S A 90, 1580–1584 10.1073/pnas.90.4.1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Sugimoto C., Kitamura T., Aoki N., Taguchi F., Yogo Y. (1998). Archetype JC virus efficiently replicates in COS-7 cells, simian cells constitutively expressing simian virus 40 T antigen. J Virol 72, 5335–5342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheimer A., Tjian R. (2003). Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev 17, 1309–1320 10.1101/gad.1099903 [DOI] [PubMed] [Google Scholar]
- Jensen P. N., Major E. O. (2001). A classification scheme for human polyomavirus JCV variants based on the nucleotide sequence of the noncoding regulatory region. J Neurovirol 7, 280–287 10.1080/13550280152537102 [DOI] [PubMed] [Google Scholar]
- Kerr D., Chang C. F., Chen N., Gallia G., Raj G., Schwartz B., Khalili K. (1994). Transcription of a human neurotropic virus promoter in glial cells: effect of YB-1 on expression of the JC virus late gene. J Virol 68, 7637–7643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs C. J., McAvoy M. T., Kumar G. (1995). The JC virus minimal core promoter is glial cell specific in vivo. J Virol 69, 2434–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K. U., Devireddy L. R., Tang S. C., Pater A., Pater M. M. (1996). Human JC virus nuclear factor 1 binding motifs and large tumor antigen region required for transactivation of late promoter. J Neurochem 67, 473–481 10.1046/j.1471-4159.1996.67020473.x [DOI] [PubMed] [Google Scholar]
- Laux G., Dugrillon F., Eckert C., Adam B., Zimber-Strobl U., Bornkamm G. W. (1994). Identification and characterization of an Epstein–Barr virus nuclear antigen 2-responsive cis element in the bidirectional promoter region of latent membrane protein and terminal protein 2 genes. J Virol 68, 6947–6958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg R. L., Achtnichts L., Hoffmann F., Kuhle J., Kappos L. (2008). Natalizumab alters transcriptional expression profiles of blood cell subpopulations of multiple sclerosis patients. J Neuroimmunol 194, 153–164 10.1016/j.jneuroim.2007.11.007 [DOI] [PubMed] [Google Scholar]
- Major E. O. (2010). Progressive multifocal leukoencephalopathy in patients on immunomodulatory therapies. Annu Rev Med 61, 35–47 10.1146/annurev.med.080708.082655 [DOI] [PubMed] [Google Scholar]
- Major E. O., Vacante D. A., Traub R. G., London W. T., Sever J. L. (1987). Owl monkey astrocytoma cells in culture spontaneously produce infectious JC virus which demonstrates altered biological properties. J Virol 61, 1435–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley K., O’Hara B. A., Gee G. V., Simkevich C. P., Sedivy J. M., Atwood W. J. (2006). NFAT4 is required for JC virus infection of glial cells. J Virol 80, 12079–12085 10.1128/JVI.01456-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C., Ray-Gallet D., Tavitian A., Moreau-Gachelin F. (1996). Differential phosphorylations of Spi-B and Spi-1 transcription factors. Oncogene 12, 863–873 [PubMed] [Google Scholar]
- Marshall L. J., Major E. O. (2010). Molecular regulation of JC virus tropism: insights into potential therapeutic targets for progressive multifocal leukoencephalopathy. J Neuroimmune Pharmacol 5, 404–417 10.1007/s11481-010-9203-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall L. J., Dunham L., Major E. O. (2010). Transcription factor Spi-B binds unique sequences present in the tandem repeat promoter/enhancer of JC virus and supports viral activity. J Gen Virol 91, 3042–3052 10.1099/vir.0.023184-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J. D., Li P. (1991). Enhancer/promoter activities of regulatory regions of representative JC virus isolates. Arch Virol 120, 305–311 10.1007/BF01310486 [DOI] [PubMed] [Google Scholar]
- Martin J. D., King D. M., Slauch J. M., Frisque R. J. (1985). Differences in regulatory sequences of naturally occurring JC virus variants. J Virol 53, 306–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzocchetti A., Wuthrich C., Tan C. S., Tompkins T., Bernal-Cano F., Bhargava P., Ropper A. H., Koralnik I. J. (2008). Rearrangement of the JC virus regulatory region sequence in the bone marrow of a patient with rheumatoid arthritis and progressive multifocal leukoencephalopathy. J Neurovirol 14, 455–458 10.1080/13550280802356837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayreddy R. P., Safak M., Razmara M., Zoltick P., Khalili K. (1996). Transcription of the JC virus archetype late genome: importance of the kappa B and the 23-base-pair motifs in late promoter activity in glial cells. J Virol 70, 2387–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messam C. A., Hou J., Gronostajski R. M., Major E. O. (2003). Lineage pathway of human brain progenitor cells identified by JC virus susceptibility. Ann Neurol 53, 636–646 10.1002/ana.10523 [DOI] [PubMed] [Google Scholar]
- Monaco M. C., Sabath B. F., Durham L. C., Major E. O. (2001). JC virus multiplication in human hematopoietic progenitor cells requires the NF-1 class D transcription factor. J Virol 75, 9687–9695 10.1128/JVI.75.20.9687-9695.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor P., Miller D., Riester K., Yang M., Panzara M., Dalton C., Miszkiel K., Khan O., Rice G., Sheremata W., International Natalizumab Trial Group (2005). Relapse rates and enhancing lesions in a phase II trial of natalizumab in multiple sclerosis. Mult Scler 11, 568–572 10.1191/1352458505ms1205oa [DOI] [PubMed] [Google Scholar]
- Pagani E., Delbue S., Mancuso R., Borghi E., Tarantini L., Ferrante P. (2003). Molecular analysis of JC virus genotypes circulating among the Italian healthy population. J Neurovirol 9, 559–566 [DOI] [PubMed] [Google Scholar]
- Petterson M., Schaffner W. (1987). A purine-rich DNA sequence motif present in SV40 and lymphotropic papovavirus binds a lymphoid-specific factor and contributes to enhancer activity in lymphoid cells. Genes Dev 1, 962–972 10.1101/gad.1.9.962 [DOI] [PubMed] [Google Scholar]
- Pongubala J. M., Nagulapalli S., Klemsz M. J., McKercher S. R., Maki R. A., Atchison M. L. (1992). PU.1 recruits a second nuclear factor to a site important for immunoglobulin kappa 3′ enhancer activity. Mol Cell Biol 12, 368–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan P. N., Khalili K. (1993). The transcriptional enhancer element, kappa B, regulates promoter activity of the human neurotropic virus, JCV, in cells derived from the CNS. Nucleic Acids Res 21, 1959–1964 10.1093/nar/21.8.1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S., Matsumura A., Yoon J., Simon M. C. (1999). SPI-B activates transcription via a unique proline, serine, and threonine domain and exhibits DNA binding affinity differences from PU.1. J Biol Chem 274, 11115–11124 10.1074/jbc.274.16.11115 [DOI] [PubMed] [Google Scholar]
- Ray D., Bosselut R., Ghysdael J., Mattei M. G., Tavitian A., Moreau-Gachelin F. (1992). Characterization of Spi-B, a transcription factor related to the putative oncoprotein Spi-1/PU.1. Mol Cell Biol 12, 4297–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Gallet D., Tavitian A., Moreau-Gachelin F. (1996). An alternatively spliced isoform of the Spi-B transcription factor. Biochem Biophys Res Commun 223, 257–263 10.1006/bbrc.1996.0881 [DOI] [PubMed] [Google Scholar]
- Reid C. E., Li H., Sur G., Carmillo P., Bushnell S., Tizard R., McAuliffe M., Tonkin C., Simon K. & other authors (2011). Sequencing and analysis of JC virus DNA from natalizumab-treated PML patients. J Infect Dis 204, 237–244 10.1093/infdis/jir256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekvig O. (1997). Polyoma induced autoimmunity to DNA; experimental systems and clinical observations in human SLE. Lupus 6, 325–326 10.1177/096120339700600324 [DOI] [PubMed] [Google Scholar]
- Romagnoli L., Wollebo H. S., Deshmane S. L., Mukerjee R., Del Valle L., Safak M., Khalili K., White M. K. (2009). Modulation of JC virus transcription by C/EBPβ. Virus Res 146, 97–106 10.1016/j.virusres.2009.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux D., Bouldouyre M. A., Mercier-Delarue S., Seilhean D., Zagdanski A. M., Delaugerre C., Simon F., Molina J. M., Legoff J. (2011). JC virus variant associated with cerebellar atrophy in a patient with AIDS. J Clin Microbiol 49, 2196–2199 10.1128/JCM.02057-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safak M., Gallia G. L., Ansari S. A., Khalili K. (1999). Physical and functional interaction between the Y-box binding protein YB-1 and human polyomavirus JC virus large T antigen. J Virol 73, 10146–10157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sock E., Enderich J., Wegner M. (1999). The J domain of papovaviral large tumor antigen is required for synergistic interaction with the POU-domain protein Tst-1/Oct6/SCIP. Mol Cell Biol 19, 2455–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su G. H., Ip H. S., Cobb B. S., Lu M. M., Chen H. M., Simon M. C. (1996). The Ets protein Spi-B is expressed exclusively in B cells and T cells during development. J Exp Med 184, 203–214 10.1084/jem.184.1.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C. S., Dezube B. J., Bhargava P., Autissier P., Wüthrich C., Miller J., Koralnik I. J. (2009). Detection of JC virus DNA and proteins in the bone marrow of HIV-positive and HIV-negative patients: implications for viral latency and neurotropic transformation. J Infect Dis 199, 881–888 10.1086/597117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C. S., Ellis L. C., Wüthrich C., Ngo L., Broge T. A., Jr, Saint-Aubyn J., Miller J. S., Koralnik I. J. (2010). JC virus latency in the brain and extraneural organs of patients with and without progressive multifocal leukoencephalopathy. J Virol 84, 9200–9209 10.1128/JVI.00609-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz B., Cinque P., Pickhardt M., Weber T. (2000). Analysis of the transcriptional control region in progressive multifocal leukoencephalopathy. J Neurovirol 6, 398–409 10.3109/13550280009018304 [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Hahn S. L., Giovane A. (1993). The Ets family of transcription factors [erratum appears in Eur J Biochem 1993 Aug 1;215(3):907]. Eur J Biochem 211, 7–18 10.1111/j.1432-1033.1993.tb19864.x [DOI] [PubMed] [Google Scholar]
- Weintraub S. J., Chow K. N., Luo R. X., Zhang S. H., He S., Dean D. C. (1995). Mechanism of active transcriptional repression by the retinoblastoma protein. Nature 375, 812–816 10.1038/375812a0 [DOI] [PubMed] [Google Scholar]
- Yogo Y., Kitamura T., Sugimoto C., Ueki T., Aso Y., Hara K., Taguchi F. (1990). Isolation of a possible archetypal JC virus DNA sequence from nonimmunocompromised individuals. J Virol 64, 3139–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai W., Tuan J. A., Comai L. (1997). SV40 large T antigen binds to the TBP-TAF(I) complex SL1 and coactivates ribosomal RNA transcription. Genes Dev 11, 1605–1617 10.1101/gad.11.12.1605 [DOI] [PubMed] [Google Scholar]