Abstract
We recently demonstrated that the N-acyl-homoserine lactone [autoinducer (AI)-1] and LuxS (AI-2)-based quorum-sensing (QS) systems exerted positive and negative regulation, respectively, on the virulence of a diarrhoeal isolate SSU of Aeromonas hydrophila. However, the role of a newly identified, two-component-based QseBC QS system in the regulation of bacterial virulence in general is not well understood, with only a limited number of studies showing its function in bacterial pathogenesis. In this report, we identified and characterized the QseBC QS system in A. hydrophila SSU and found that, as was the case with enterohaemorrhagic Escherichia coli, the open reading frames for the qseB (the response regulator) and qseC (the sensor histidine kinase) genes overlapped by 4 bp at the ATGA motif. Our data provide evidence that deletion of the qseB gene from A. hydrophila resulted in attenuation of bacterial virulence in a septicaemic mouse model of infection and diminished swimming and swarming motility, and the mutant bacteria formed denser biofilms compared with those from the parental strain of A. hydrophila. The decrease in the virulence of the A. hydrophila ΔqseB mutant correlated with reduced production of protease and the cytotoxic enterotoxin, which has associated haemolytic activity. The swimming and swarming motility, haemolytic activity, protease production and biofilm formation were restored in the qseBC-complemented strain to a level similar to that of the wild-type A. hydrophila SSU. Our study is the first, to our knowledge, to report a functional QseBC QS system in A. hydrophila which may be linked to AI-1 and AI-2 QS systems in modulating bacterial virulence, possibly through the cyclic diguanosine monophosphate.
Introduction
In bacteria, two-component systems are widely used signal transduction mechanisms that facilitate in eliciting an adaptive response to various environmental stimuli, particularly through changes in gene transcription (Beier & Gross, 2006; Hoch, 2000). The two-component systems are typically composed of a membrane-associated sensor histidine kinase and a cytoplasmic transcriptional regulator (Beier & Gross, 2006; Hoch, 2000). In most cases, the stimuli sensed by these systems are transformed into a cellular signal via autophosphorylation of sensor kinase at the conserved histidine residue. The signal is then transmitted onto the response regulator, following phosphorylation at the aspartate residue, which results in its activation by undergoing conformational changes. The activated response regulator then exerts its regulation on the transcription of various target genes (Beier & Gross, 2006; Hoch, 2000).
One such two-component system that responds to a quorum sensing (QS) signal is QseBC (QseC, a sensor histidine kinase, and QseB, a response regulator), which was first discovered in enterohaemorrhagic Escherichia coli (EHEC) (Sperandio et al., 2002). Later, this system was also found in other pathogens, such as Salmonella enterica serovar Typhimurium (Bearson & Bearson, 2008; Moreira et al., 2010), Edwardsiella tarda (Wang et al., 2011) and uropathogenic E. coli (UPEC) (Kostakioti et al., 2009). In addition to responding to an autoinducer (AI)-3, QseC sensor kinase has been reported to also respond to eukaryotic hormones typified by epinephrine and/or norepinephrine (Clarke et al., 2006; Waldor & Sperandio, 2007; Walters & Sperandio, 2006). Further, by using adrenergic receptor antagonists, it is possible to block the effects of AI-3 and epinephrine/norepinephrine, perhaps indicative of a similar structure for these molecules and a similar signalling pathway (Walters & Sperandio, 2006). The role of the QseBC system in the pathogenesis of EHEC, Francisella tularensis and S. Typhimurium was recently reported (Rasko et al., 2008), as deletion of the QseC histidine kinase-encoding gene attenuated virulence of these bacteria (Rasko et al., 2008). Further, a synthetic compound (LED209) that interferes with the QseC signalling inhibited the in vitro virulence of EHEC, S. Typhimurium and F. tularensis, and it also modulated in vivo virulence of the latter two pathogens (Rasko et al., 2008). In UPEC, a recent study has shown that in addition to kinase activity, QseC has phosphatase activity that is critical in modulating the regulatory activity of QseB (Kostakioti et al., 2009).
Initially, only four Aeromonas species were recognized; however, through developments in the molecular postgenomic era, there are now 30 known species of Aeromonas (http://www.bacterio.cict.fr/). Among these, A. hydrophila, A. caviae and A. veronii biovar sobria are the most common species known to cause the majority of human infections (Janda, 1991). Our case-control study reported the presence of aeromonads in 7.2 % of children suffering from diarrhoea in Bangladesh, with only 3.3 % of the healthy children excreting aeromonds in their stools (Albert et al., 2000). Likewise, in our 27 month prospective study, we showed a similar isolation rate of this organism from the paediatric population suffering from diarrhoea (7.3 %) at the Mercy Hospital in Chicago, with only 2.2 % of the control healthy children excreting aeromonads in the stools (Challapalli et al., 1988). In addition, our recent study has clearly shown transmission of Aeromonas species from water to humans (Khajanchi et al., 2010). A PubMed search using the word ‘Aeromonas’ generated approximately 663 citations between 1980 and 1994 (Janda & Abbott, 2010). However, to date, this number has increased to 5398, which illustrates that over the years there has been an enormous increase in studying the genus Aeromonas by the scientific and medical community.
In humans, Aeromonas species cause diarrhoea and various extra-intestinal infections (Galindo et al., 2006; Janda, 2002; Vila et al., 2003), which include septicaemia, cellulitis, wound infections, urinary tract infections, soft tissue infections and, occasionally, meningitis and peritonitis (Galindo et al., 2006; Horneman et al., 2007; Vila et al., 2003). This pathogen can cause haemolytic uraemic syndrome and necrotizing fasciitis, particularly in children with a compromised immune status (Abuhammour et al., 2006; Figueras et al., 2007). In addition, this waterborne pathogen receives significant attention during natural disasters, as aeromonads were most frequently isolated from wounds of patients following the 2004 tsunami in Thailand (Hiransuthikul et al., 2005). Further, an increased isolation rate of Aeromonas species was noted in floodwater samples after Hurricane Katrina in New Orleans (Presley et al., 2006).
In addition to causing human diseases, aeromonads are also associated with various severe diseases in both cold- and warm-blooded animals, and these include furunculosis in fish (commonly caused by A. salmonicida), ulcerative stomatitis in snakes and lizards, ‘red leg’ disease in frogs, septicaemia in dogs and septic arthritis in calves (Janda & Abbott, 2010).
A. hydrophila produces a number of virulence factors which function together to cause diseases in the host (Chopra & Houston, 1999; Khajanchi et al., 2010; Krovacek et al., 1994; Sha et al., 2002). In our laboratory, we have identified and characterized new virulence factors/mechanisms that contribute to virulence in the diarrhoeal isolate SSU of A. hydrophila. Some of the key virulence factors that have been characterized are: (i) a cytotoxic enterotoxin (Act), a type 2 secretion system (T2SS) toxin that functions as a haemolysin, a cytotoxin or an enterotoxin, depending upon the target cells (Chopra et al., 2000; Galindo et al., 2004; Sha et al., 2002); (ii) a new T3SS effector, AexU, which leads to ADP-ribosylation of the host cell proteins, resulting in their death via apoptosis (Sierra et al., 2007, 2010); and (iii) two T6SS effectors, such as haemolysin-coregulated protein (Hcp) and the valine glycine repeat G (VgrG) family of proteins (Suarez et al., 2008). VgrG1 of A. hydrophila possesses actin ADP-ribosylating activity associated with its carboxyl-terminal vegetative insecticidal protein-2 (VIP-2) domain that induces cell rounding followed by host cell apoptosis (Suarez et al., 2010a). Hcp, on the other hand, modulates innate immunity by inhibiting phagocytosis of A. hydrophila, thus allowing bacterial multiplication and spread to different organs of mice, resulting in their death (Suarez et al., 2010b).
In bacteria, the cell-to-cell signalling system, known as QS, has been identified as a global regulator which controls virulence mechanisms at appropriate times, depending on the physiological conditions in the environment as well as in the host. In Gram-negative bacteria, three QS circuits have been identified, of which AI-3/QseBC is the least studied. Recently, we characterized the role of N-acyl-homoserine lactone-mediated (AI-1) and LuxS-based (AI-2) QS systems in the regulation of virulence factors in A. hydrophila SSU (Khajanchi et al., 2009; Kozlova et al., 2008). We demonstrated that the ahyRI-based AI-1 QS system of A. hydrophila was a positive regulator of bacterial virulence, as disruption of the ahyRI genes reduced metalloprotease production, biofilm formation, secretion of the type T6SS effectors, e.g. Hcp and VgrG family of proteins, and mortality in a septicaemic mouse model of infection (Khajanchi et al., 2009). In contrast, we showed that the LuxS-based QS system (AI-2) negatively regulated the virulence of this pathogen, as the luxS mutant resulted in increased biofilm formation and enhanced mortality in an animal model compared with the wild-type (WT) bacterium (Kozlova et al., 2008). We also provided data showing that the bacterial second messenger cyclic-diguanosine monophosphate (c-di-GMP) (Hengge, 2009; Römling & Simm, 2009) affected the virulence-associated phenotype in WT and an ahyRI mutant of A. hydrophila SSU (Kozlova et al., 2011).
By our sequence annotation of A. hydrophila ATCC 7966 strain (Seshadri et al., 2006), we identified qseB and qseC genes in A. hydrophila SSU, which exhibited a 99 and 96 % homology at the amino acid level with the corresponding genes of strain ATCC 7966. A 4 bp overlap with the ATGA motif was found in the open reading frames (ORFs) for qseB and qseC genes in which the translation stop codon of QseB overlapped with the translation start codon for QseC. A similar genomic organization for qseBC genes was identified in EHEC (Clarke & Sperandio, 2005b); however, QseB exhibited 51 % and QseC showed only 31 % identity with the corresponding proteins of E. coli.
In this study, we identified and characterized the role of the QseBC system in the regulation of virulence in A. hydrophila SSU by generating a ΔqseB isogenic mutant. We demonstrated that the QseBC system in A. hydrophila positively regulated both swimming and swarming motility, and haemolytic activity of Act and protease production, while negatively modulating the biofilm formation. Since QseBC functions as both a positive and negative regulator in controlling in vitro virulence of A. hydrophila, we indeed observed a marginal, but statistically significant, attenuation of virulence in an in vivo septicaemic mouse model when the qseB gene was deleted from the WT strain. As mentioned above, QseBC has been studied recently in other bacterial species (Bearson & Bearson, 2008; Kostakioti et al., 2009; Sperandio et al., 2002; Wang et al., 2011), and this regulatory system controls different sets of virulence genes in various bacteria. Consequently, more information is needed as to how this regulatory system may be modulating bacterial virulence in general. We believe Aeromonas represents a model organism to study the QseBC system, as it has characteristics of E. coli, Vibrio and Pseudomonas (Kozlova et al., 2011). In addition, it possesses functional type 2, 3 and 6 secretion systems through which bacteria secrete a variety of virulence factors. The presence of this network of virulence factors results in very complex regulatory mechanisms that turn virulence genes ‘off’ and ‘on’ based on QS signalling molecules present in a given environment. Therefore, our study is timely and important in better understanding the functional role of QseBC QS in the regulation of virulence in A. hydrophila, as currently there are no data available, to our knowledge, on the QseBC system of A. hydrophila.
Methods
Bacterial strains, plasmids and chemicals.
The bacterial strains and plasmids used in this study are listed in Table 1. LB medium was supplemented with l-arabinose (0.2 %) when the ggdef-domain-containing gene (GGDEF-domain-containing proteins increase c-di-GMP within bacterial cells) was expressed from the pBAD/Myc-HisB : : ggdef plasmid (Table 1) under the control of an arabinose-inducible pBAD promoter (Kozlova et al., 2011). The kinetic growth data showed no difference in the growth rates of WT A. hydrophila SSU versus the qseB mutant strain (data not shown). However, we always normalized the data with the same c.f.u. of bacteria for any minor variations that we might observe in the growth rates among the strains tested.
Table 1. Strains and plasmids used in this study.
Abbreviations: Rif, rifampicin; Km, kanamycin; Sm, streptomycin; Sp, spectinomycin; Ap, ampicillin; Tc, tetracycline.
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
| A. hydrophila SSU | CDC | |
| ATCC 7966 | Environmental isolated strain | ATCC |
| SSU-R | Rifr strain of A. hydrophila SSU | Laboratory stock |
| ΔqseB | qseB gene deletion mutant of A. hydrophila SSU-R strain Rifr Kmr | This study |
| ΔahyRI | ahyRI gene deletion mutant of A. hydrophila SSU-R strain Rifr Smr Spr | Khajanchi et al. (2009) |
| ΔluxS | luxS mutant of A. hydrophila SSU-R strain Rifr Kmr | Kozlova et al. (2008) |
| ΔqseB/pBR322-qseBC | qseB mutant complemented with the qseBC genes (ATCC) via pBR322 Rifr Kmr Apr | This study |
| E. coli | ||
| DH5α | Production of recombinant plasmids. recA gyrA | Life Technologies |
| SM10 | Kmr, λpir | Edwards et al. (1998) |
| Plasmids | ||
| pCR2.1 | TA cloning vector Apr Kmr | Invitrogen |
| pCR2.1/qseB | TA cloning vector carrying qseB gene Apr Kmr | This study |
| pUC-4K | Contains a 1.2 kb kanamycinr gene cassette | Amersham |
| pCR2.1/qseB-Km | TA cloning vector harbouring the qseB gene disrupted by Km cassette, Apr Kmr | This study |
| pDMS197 | Suicide vector; R6K ori, sacB, Tcr | Edwards et al. (1998) |
| pDMS197/qseB-Km | Suicide vector containing qseB gene with Km cassette, Tcr Kmr | This study |
| pBR322 | Apr Tcr | Amersham |
| pBR322-qseBC | Contains the qseBC genes (ATCC), Apr | This study |
| pBAD/Myc-HisB | Vector, araBAD promoter Apr | Invitrogen |
| pBAD : : ggdef | GGDEF-domain-encoded gene of A. hydrophila cloned into pBAD/Myc-HisB ara Kmr Apr | Laboratory stock |
Generation of an isogenic qseB mutant of A. hydrophila SSU.
The qseB single-knockout mutant of A. hydrophila SSU was generated by using a double crossover homologous recombination method. The qseB gene (690 bp) was amplified by PCR employing genomic DNA (gDNA) of A. hydrophila SSU-R (Table 1) as the template, and a pair of primers (qseB-N, 5′-ATGCGGATCCTGTTGGTGGAAGA-3′ and qseB-C, 5′-CATGCCCGGTGGTCCCGGCGCTG-3′). The PCR product was then cloned in the TA cloning vector pCR2.1 (Invitrogen). Subsequently, the pCR2.1-qseB recombinant plasmid (r-plasmid) was transformed into E. coli DH5α (Table 1). Within the qseB coding region, a unique BlpI restriction enzyme site exists; the pCR2.1-qseB r-plasmid was thus linearized by BlpI digestion. A kanamycin resistance (Kmr) gene cassette flanked by the BlpI restriction site from the plasmid pUC4K (GE Healthcare) was inserted at the BlpI site of pCR2.1-qseB r-plasmid to generate a pCR2.1-qseB-Km plasmid (Table 1). After digestion with the KpnI/XbaI restriction enzymes, the DNA fragment from the pCR2.1-qseB-Km r-plasmid was ligated to a pDMS197 suicide vector at the KpnI/XbaI sites, and the resulting plasmid (pDMS197-qseB-Km) was transformed into an E. coli SM10 λpir strain (Edwards et al., 1998) (Table 1). The recombinant E. coli (pDMS197-qseB-Km) was then conjugated with the WT A. hydrophila SSU-R. The transconjugants were selected based on resistance to appropriate antibiotics and sucrose and subjected to further analysis (Sha et al., 2002). The identity of the qseB mutant was confirmed by Southern blot analysis.
Southern blot analysis.
From the WT A. hydrophila SSU and its qseB mutant strain, the gDNA was isolated, digested with KpnI/XbaI restriction enzymes and subjected to 0.8 % agarose gel electrophoresis. Southern blot analysis was performed as described previously (Sha et al., 2002; Xu et al., 1998). The PCR products from the qseB gene, Kmr gene cassette, and the pDMS197 vector digested by using XbaI–KpnI were employed as probes.
Complementation of the A. hydrophila SSU ΔqseB mutant.
The qseBC genes were amplified by PCR using gDNA of A. hydrophila ATCC 7966 as the template and two primers QseBCN-HindIII (5′-GGGAAGCTTGCATCGACCCCAACTTCTTCT-3′) and QseBCC-NheI (5′-GGGGCTAGCTGGAGCACATGGTGACGGT-3′; the restriction endonuclease sites are underlined in both primers). Since qseBC genes are highly homologous between A. hydrophila strains SSU and ATCC 7966, we used qseBC from the latter strain for complementation, as we annotated the genome of this strain and the upstream sequences containing the promoter regions for these genes were available (Seshadri et al., 2006). We included a 249 bp flanking upstream DNA sequence containing the potential promoter region of the qseBC genes for complementation studies. This DNA fragment (2648 bp) was cloned in pBR322 vector (Tcr Apr) at the HindIII–NheI sites and transformed into the E. coli DH5α strain (Table 1). The pBR322/qseBC (Tcs Apr) recombinant plasmid was isolated from the E. coli strain and electroporated into an A. hydrophila SSU ΔqseB mutant (Sha et al., 2002) (Table 1).
We complemented the qseB mutant with both the qseBC genes, as they exist in an operon, and we suspected that deletion of the qseB gene would have impacted the transcript level of the qseC gene.
Swimming and swarming motility assay.
LB medium with 0.3 % Difco Bacto-agar was used to characterize the swimming motility, while Difco nutrient broth with 0.5 % Eiken agar (Eiken Chemical) was employed for measuring the swarming motility of WT A. hydrophila SSU, its ΔqseB mutant, and the complemented strain, as described in our previous studies (Khajanchi et al., 2009; Kozlova et al., 2008). For both the swimming and swarming motility assay, media supplemented with ampicillin (450 µg ml−1) were used for the complemented strain. Briefly, the overnight cultures grown in LB medium in the presence of the respective antibiotics were adjusted to the same optical density, and equal numbers of WT A. hydrophila, its ΔqseB mutant bacteria or the complemented strain (1×108 c.f.u.) were stabbed into agar plates. The swimming and swarming agar plates were incubated at 37 and 30 °C, respectively, for 16–18 h and then motilities were assessed by examining migration of bacteria through the agar from the centre towards the periphery of the plate.
Measurement of the haemolytic activity.
For measuring the haemolytic activity associated with Act of WT A. hydrophila SSU, its ΔqseB mutant or the complemented strain, the culture filtrates from bacteria grown for 18 h in LB medium at 37 °C with shaking (180 r.p.m.) were first treated with trypsin [final concentration 0.05 % to activate Act (Sha et al., 2004)] at 37 °C for 1 h and then subjected to a haemolytic assay by using rabbit red blood cells (RBCs), as described previously (Sha et al., 2002). Briefly, 100 µl Dulbecco’s PBS (DPBS) was added to each well of a 96-well microtitre plate. Aliquots (100 µl) of culture filtrates were added to the first well, followed by a serial twofold dilution, with subsequent addition of 100 µl 2.5 % rabbit erythrocytes (Colorado Serum). The plate was incubated at 37 °C for 1 h and observed for the lysis of RBCs. The supernatant was taken from those wells that showed a partial lysis of rabbit erythrocytes, and the release of haemoglobin was then evaluated by measuring absorbance at 540 nm. The haemolytic activity titres were calculated as the absorbance value of the haemoglobin release multiplied by the dilution of the culture filtrates. The haemolytic units were reported per ml cell filtrate per 1×108 c.f.u.
For the neutralization assays, culture filtrates of WT and ΔqseB mutant strains were mixed with either pre-immune (control) or hyper-immune rabbit sera (laboratory stock, 1 : 10 dilution) containing antibodies to Act (Erova et al., 2007; Khajanchi et al., 2009) before we measured the haemolytic activity.
Measurement of the protease activity.
Protease activity was measured in culture filtrates of overnight-grown WT A. hydrophila, its ΔqseB mutant or the complemented strain, as described earlier (Erova et al., 2006; Khajanchi et al., 2010). The protease activity was calculated per ml cell filtrate per 1×108 c.f.u. The hide azure powder substrate (Calbiochem) was used for measuring protease activity because of the sensitivity and rapidity of the assay. Further, this substrate could detect both metallo and serine proteases, which are the two major classes of this enzyme produced by Aeromonas species (Swift et al., 1999). The substrate incubated with DPBS alone served as a negative control.
Crystal violet (CV) biofilm assay.
The WT A. hydrophila SSU, its ΔqseB mutant or the complemented strain were grown in 3 ml LB broth contained in polystyrene tubes at 37 °C for 24 h with shaking. Biofilm formation was quantified according to the procedure described elsewhere (Khajanchi et al., 2009; O’Toole & Kolter, 1998). The biofilm formation results were normalized to 1×109 c.f.u. to account for any minor differences in the growth rate of various bacterial strains used. The experiment was repeated independently three times.
Scanning electron microscopy (SEM) of biofilms.
SEM of biofilm formation of A. hydrophila SSU and its ΔqseB mutant was performed by using 13 mm diameter thermanox plastic coverslips. After 48 h incubation, unattached cells were removed, and then the coverslips were fixed and stained with ruthenium red, and samples were examined in a Hitachi S4700 field emission scanning electron microscope (Hitachi High Technologies America) by using the procedure described in our previous studies (Khajanchi et al., 2009; Kozlova et al., 2008).
Western blot analysis.
Overnight cultures of WT and ΔqseB mutant strains were diluted 1 : 20 in fresh LB medium and grown for 2 h (OD600~0.8) and/or 4 h (OD600~1.4) at 37 °C with shaking at 180 r.p.m. Western blot analysis was performed to measure production and secretion of T6SS effector Hcp in the LB medium (Khajanchi et al., 2009). We also examined production of T3SS effector AexU in the bacterial whole-cell lysates as well as in the insoluble fraction collected after co-culturing of WT and ΔqseB mutant of A. hydrophila SSU with HeLa cells in the Dulbecco’s modified Eagle medium (DMEM) (Khajanchi et al., 2009; Sha et al., 2007).
Animal experiments.
Groups of 10 Swiss Webster mice (Taconic Farms) were infected by the intraperitoneal route with 5×107 c.f.u. (WT or its ΔqseB mutant) in accordance with the approved animal care protocol. One group of mice was inoculated with DPBS (n = 10) and served as a control. Deaths were recorded for 16 days post-infection. The animal experiments were repeated three times.
Statistical analysis.
All of the experiments were performed in triplicate, and wherever appropriate, the data were analysed by using the Student’s t test, and a P-value of ≤0.05 was considered significant. The data were presented as an arithmetic mean±sd. The animal data were analysed by using the Kaplan–Meier’s survival estimates.
Results
Characterization of the two-component QseBC QS system from A. hydrophila SSU
By analysing the protein sequences of QseB and QseC in the NCBI conserved domains database, a Pfam protein sequence search (http://pfam.sanger.ac.uk/) and SMART analysis (http://smart.embl-heidelberg.de/), it was revealed that QseB possesses two domains: a receiver domain (REC) and a transcriptional regulatory protein/C-terminal helix–turn–helix (HTH) domain (Supplementary Fig. S1a, available with the online version of this paper). QseC, on the other hand, harbours three domains: a HAMP/transmembrane domain, a His-kinase (HisKA) and a ATPase domain (Supplementary Fig. S1b).
Deletion of the qseB gene from A. hydrophila SSU diminishes both swimming and swarming motility
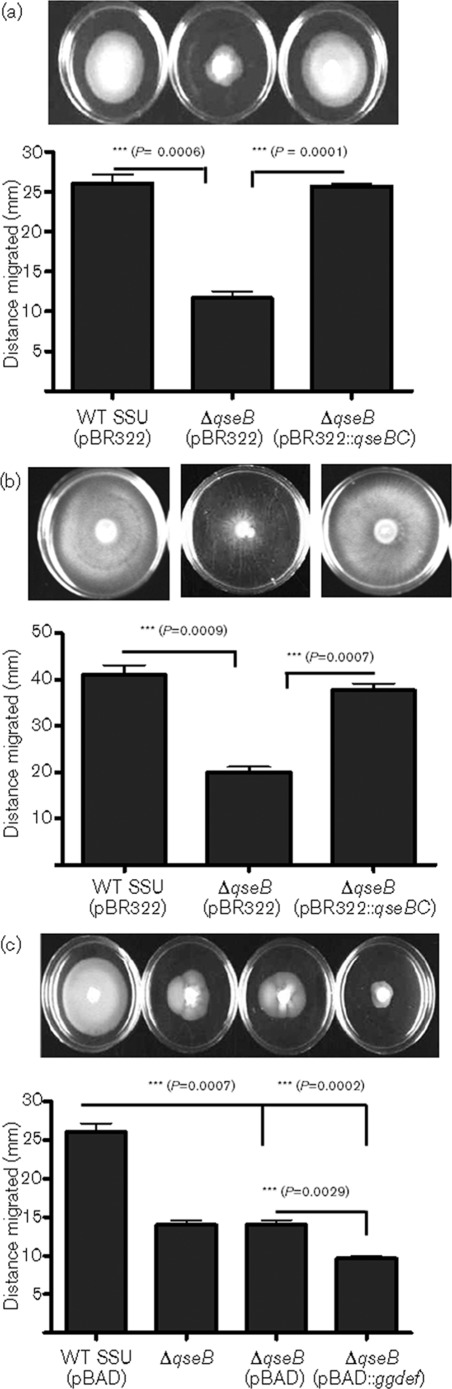
To characterize the role of the QseBC system in the regulation of virulence in A. hydrophila SSU, we deleted the qseB gene by double crossover homologous recombination and subsequently generated a complemented strain with both of the qseBC genes that were supplied in trans. Motility is considered to be an important virulence factor in the pathogenesis of Aeromonas-associated infections, as it facilitates pathogens to adhere and invade the host cells (Kirov et al., 2002; Kirov, 2003; Kirov et al., 2004). Aeromonas species possess polar flagella for swimming motility and lateral flagella for swarming motility (Kirov et al., 2002; Kirov, 2003; Kirov et al., 2004). To cause infection by avoiding the host defence, the motility of the pathogens must be tightly regulated. In this study, we examined whether the two-component QseBC system involved in QS regulates motility in A. hydrophila. The results of the motility assay revealed that the ΔqseB mutant had significantly reduced migration in the swimming and the swarming agar plates, compared with that of the WT A. hydrophila SSU (Fig. 1a, b). The latter finding indicated that both the swimming and swarming motility in A. hydrophila SSU were regulated by the QseBC QS system. Further, the swimming and the swarming motility were restored in the qseBC-complemented strain (Fig. 1a, b). Importantly, when the GGDEF-domain-containing protein (AHA0701h) was overproduced in the ΔqseB mutant (Kozlova et al., 2011), the swimming motility of this strain was further reduced compared with the ΔqseB mutant harbouring the pBAD vector alone (Fig. 1c).
Fig. 1.
Swimming and swarming motility of WT A. hydrophila SSU, the ΔqseB mutant and its qseBC-complemented strain. (a) LB medium with 0.3 % Difco Bacto-agar (supplemented with ampicillin for the complemented strain) was used to characterize the swimming motility. The ΔqseB mutant showed significantly decreased migration compared with WT A. hydrophila SSU. The qseBC-complemented strain migrated in a manner similar to that of the WT strain. Asterisks (***) indicate a significant difference in migration between WT A. hydrophila SSU and its ΔqseB mutant (P = 0.0006), and between the ΔqseB mutant and the qseBC-complemented strain (P = 0.0001). (b) Difco nutrient broth with 0.5 % Eiken agar was used to determine swarming motility. A strong swarming response was observed for the WT A. hydrophila SSU and the qseBC-complemented strain, while reduced swarming motility was noted for the ΔqseB mutant. Asterisks (***) indicate a significant difference in migration between the WT A. hydrophila SSU and the ΔqseB mutant (P = 0.0009), and between the ΔqseB mutant and the qseBC-complemented strain (P = 0.0007). (c) Influence of ggdef encoding gene overexpression in the ΔqseB mutant on swimming motility. The ΔqseB mutant with ggdef overexpression did not show any migration but grew at the inoculation site. Asterisks (***) indicate statistically significant differences between the WT and qseB mutant (P = 0.0007), and the qseB mutant and the qseB mutant with an overexpressed ggdef-encoding gene (P = 0.0029). Three independent experiments were performed, and the arithmetic means±sd are plotted. Statistical analyses between the WT and the mutant and complemented strains were performed by using Student’s t test.
Haemolytic activity of T2SS-associated Act is significantly reduced in the ΔqseB mutant
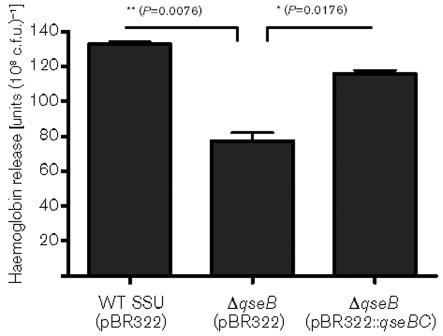
We earlier showed that Act is one of the most potent virulence factors that contributes to the pathogenesis of A. hydrophila SSU (Chopra et al., 2000; Galindo et al., 2004; Sha et al., 2002). To examine whether the QseBC-system regulates Act production, we measured the haemolytic activity associated with Act in the culture supernatant of WT A. hydrophila SSU, the ΔqseB mutant and the qseBC-complemented strain. Interestingly, we found that the haemoglobin release from rabbit RBCs was significantly reduced when culture filtrate from the ΔqseB mutant was used, compared with culture filtrate from the WT strain, and the haemoglobin release was restored in the qseBC complemented strain (Fig. 2).
Fig. 2.
Measurement of the haemolytic activity associated with Act of WT A. hydrophila SSU, ΔqseB mutant and its qseBC-complemented strain by using rabbit RBCs. Haemoglobin release was quantified by measuring absorbance at 540 nm, and the haemolytic titres were calculated as the absorbance value of the haemoglobin release multiplied by the dilution of the culture filtrate. The data were normalized to 1×108 c.f.u. to account for any minor differences in the growth rates between the various bacterial strains. Three independent experiments were performed, and the arithmetic means±sd are plotted. Asterisks indicate statistically significant difference in haemoglobin release between the WT and its ΔqseB mutant (**P = 0.0076) as well as between the ΔqseB mutant and the qseBC-complemented strain (*P = 0.0176) by Student’s t test.
To demonstrate that the majority of this haemolytic activity was associated with Act, we neutralized the toxin by using specific antibodies. Indeed we noted much reduced and similar residual haemolytic activity in the culture supernatants of both WT and ΔqseB mutant. This residual activity is contributed by another haemolysin that we characterized in isolate SSU of A. hydrophila (Erova et al., 2007). These data indicated that the haemolytic activity associated with this haemolysin gene was not altered by deletion of the qseB gene and that the QseBC system specifically controlled the haemolytic activity of Act in A. hydrophila SSU.
The ΔqseB mutant produces lower amounts of protease
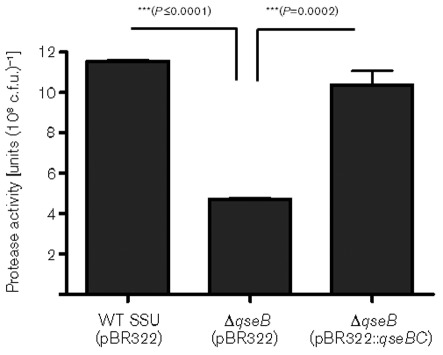
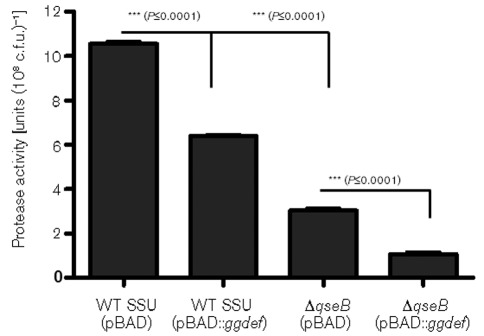
Earlier studies have shown that the pathogenic and virulence characteristics of A. hydrophila were associated in part with the production of proteases (Leung & Stevenson, 1988; Sakai, 1985). Consequently, we measured protease production and found that the ΔqseB mutant exhibited a significantly reduced level of protease production when compared with the WT A. hydrophila SSU, and this enzyme activity was restored in the qseBC-complemented strain (Fig. 3), possibly meaning that the QseBC system also controlled protease production in A. hydrophila. Further, to investigate the influence of overproduction of GGDEF protein on protease level, we examined protease activity in WT A. hydrophila SSU and the ΔqseB mutant with overproduced GGDEF protein. Interestingly, our data showed that the protease level was decreased when GGDEF protein was overproduced in WT A. hydrophila SSU, when compared with the parental strain with the empty pBAD vector. In addition, the ΔqseB mutant with overproduced GGDEF protein further reduced protease activity compared to that in the ΔqseB mutant with the pBAD vector alone (Fig. 4). These data clearly indicate a link between the QseBC QS system with A. hydrophila virulence (motility, haemolytic activity associated with Act and protease production) and c-di-GMP.
Fig. 3.
Protease activity in the culture supernatants of WT A. hydrophila SSU, the ΔqseB mutant and its qseBC-complemented strain. The data were normalized to 1×108 c.f.u. to account for any minor differences in the growth rates between the various bacterial strains. Three independent experiments were performed, and the arithmetic means±sd are plotted. Asterisks (***) indicate statistically significant differences in protease activity between the WT and its ΔqseB mutant (P≤0.0001), as well as between the ΔqseB mutant and the qseBC-complemented strain (P = 0.0002) by Student’s t test.
Fig. 4.
Influence of ggdef-encoding gene overexpression in the WT and the ΔqseB mutant on protease production of A. hydrophila SSU. The data were normalized to 1×108 c.f.u. to account for any minor differences in the growth rates between the various bacterial strains. The results were reproduced in three independent experiments, and the error bars represent sd. Asterisks (***) indicate statistically significant differences in protease activity between the WT and WT overexpressing the ggdef-encoding gene (P≤0.0001), WT and the ΔqseB mutant (P≤0.0001), and the ΔqseB mutant and the ΔqseB mutant overexpressing the ggdef-encoding gene (P≤0.0001) by Student’s t test.
To examine whether the regulation of protease production by QseBC is dependent on cell density, we measured the protease activity of WT A. hydrophila SSU and its ΔqseB mutant at different bacterial growth phases (exponential, mid-exponential, late-exponential, early stationary and late stationary). Although a slight reduction in protease activity was noted at the mid- and late-exponential phases in the ΔqseB mutant compared with the WT strain, protease production by the ΔqseB mutant was significantly reduced in late stationary phase compared with that produced by the WT strain, indicating that QseBC regulates protease production in A. hydrophila SSU at high cell density (data not shown).
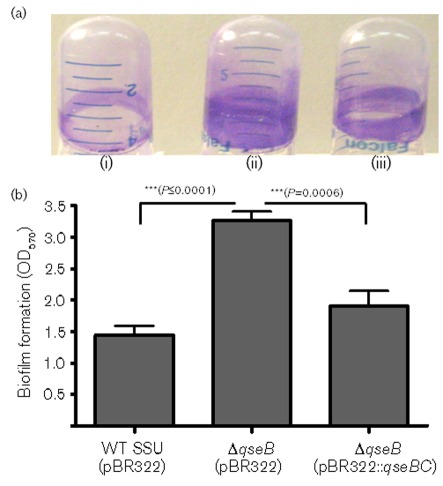
CV staining and light microscopic observations on biofilm formation demonstrate a more efficient attachment of the ΔqseB mutant to the polystyrene tubes and Thermonox coverslips
To measure the solid surface-associated biofilm formation, we performed a CV staining assay and examined biofilm formation on Thermonox coverslips by using light microscopy. The biofilm formation was observed after 24 h growth of WT, ΔqseB mutant and its qseBC complemented strain in LB medium. The ΔqseB mutant formed a significantly increased, solid-surface-associated biofilm in polystyrene tubes, with a more than twofold increase in the CV staining when compared with that of the WT A. hydrophila SSU strain (Fig. 5). Likewise, the ΔqseB mutant also produced more aggregated biofilms which were uniformly distributed all over the coverslip (Supplementary Fig. S2b, available with the online version of this paper) compared with the biofilms of the WT strain which produced less aggregation (Supplementary Fig. S2a). In addition, the qseBC-complemented strain produced biofilms that were similar to those from the WT strain when examined by CV staining on polystyrene tubes (Fig. 5), as well as on the Thermonox coverslips when examined by using light microscopy (Supplementary Fig. S2c). We used ΔahyRI (biofilm-deficient) and ΔluxS (enhances biofilm) mutants as controls to compare biofilm formation by different QS mutant strains (Supplementary Fig. S3). These data suggested to us that, similar to the ΔluxS mutant, the QseBC QS system also negatively controlled biofilm formation in A. hydrophila SSU.
Fig. 5.
Measurement of biofilm mass by CV staining of biofilms formed on polystyrene by the WT A. hydrophila SSU (i), ΔqseB mutant (ii) and its qseBC-complemented strain (iii). Biofilms were quantified after 24 h incubation at 37 °C. (a) Adherent bacteria were stained with 1 % CV and washed with distilled water; the extracted colour (with 95 % ethanol) was measured at A570. (b) The data were normalized to 1×109 c.f.u. to account for any minor differences in the growth rates of various bacterial strains used. The results were reproduced in three independent experiments and the error bars represent sd. Asterisks (***) represent statistically significant differences in biofilm formation between the WT and its ΔqseB mutant (P≤0.0001), and between the ΔqseB mutant and the qseBC-complemented strain (P = 0.0006) by Student’s t test.
A 3D structured and/or denser biofilm is observed in the ΔqseB mutant when examined by SEM
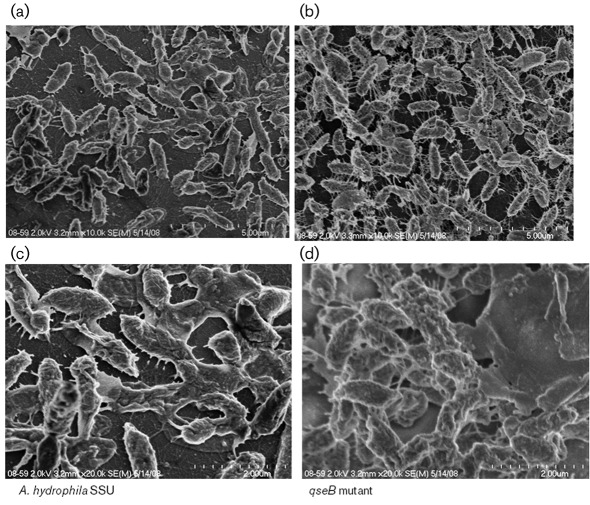
To investigate in detail the surface architecture of bacterial cells aggregated in biofilms formed by the WT and its ΔqseB mutant, we performed SEM. We used ruthenium red, which binds strongly to negatively charged polysaccharides, and represents an excellent method for visualization of the surface properties of bacteria. We observed that the ΔqseB mutant produced very thick intercellular filaments and bundles of aggregated cells that formed a dense biofilm (Fig. 6b). In addition, at a higher magnification, the ΔqseB mutant was found to have formed a 3D structure of biofilm covered with thick exopolysaccharides (Fig. 6d), when we compared these findings with the flattened biofilms formed by the WT bacterium which were less aggregated and connected with fewer filaments (Fig. 6a, c). The SEM observations further confirmed CV staining results that the ΔqseB mutant produced denser biofilms than those produced by the WT, indicating that the QseBC QS system functioned as a negative regulator of biofilm formation in A. hydrophila SSU.
Fig. 6.
Representative SEM images of biofilm formation by WT A. hydrophila SSU and its ΔqseB mutant after 48 h culture at 37 °C on Thermonox coverslips stained with ruthenium red. Compact aggregated cells were well connected with filaments, and denser 3D biofilms were formed by the ΔqseB mutant (b, d) compared with the less aggregated cells connected with fewer filaments and flattened biofims produced by WT bacteria (a, c). SEM images (a) and (b) are at low magnification (×10 000) and (c) and (d) are at higher magnification (×20 000).
Production of AexU, a T3SS effector, and Hcp, a T6SS effector, is not affected in the ΔqseB mutant of A. hydrophila SSU
To demonstrate any regulation of the QseBC QS system on the T3SS, we examined the expression and production of AexU in bacterial cell pellets of WT A. hydrophila SSU and its ΔqseB mutant grown in LB medium. We also infected HeLa cells with WT A. hydrophila and its ΔqseB mutant in DMEM and monitored the production of AexU in bacterial cells by Western blot analysis. We found that the ΔqseB mutant had similar levels of AexU to the WT when grown in LB medium (Supplementary Fig. S4a) and also during co-culture with HeLa cells (Supplementary Fig. S4b). These data indicated that the QseBC QS system had no effect on the production of the T3SS effector, AexU.
Similarly, the QseBC system did not regulate the production and secretion of the T6SS effector, Hcp, as the Western blot analysis data showed that both the WT and the ΔqseB mutant produced and secreted Hcp at similar levels (Supplementary Fig. S5).
The ΔqseB mutant shows a marginally decreased virulence in an animal model
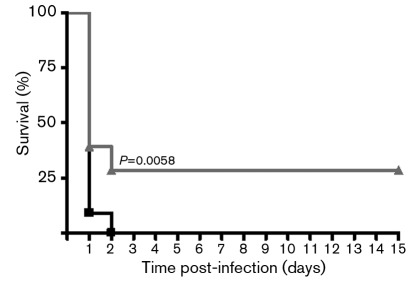
By using in vitro experiments, we demonstrated that the QseBC QS system positively regulated swimming and swarming motility, protease and Act production, and negatively modulated biofilm formation. To further examine whether these changes in virulence factors regulated by the QseBC QS system had any influence on the virulence of A. hydrophila SSU in vivo, we injected mice via the intraperitoneal route with the ΔqseB mutant and the WT strain of A. hydrophila at a lethal dose of 5×107 c.f.u. (Fig. 7). We noted that 100 % of the animals infected with WT A. hydrophila SSU died within 2–3 days. However, mice infected with the ΔqseB mutant strain exhibited 30 % lower mortality over a test period of 16 days, suggesting that a marginal but statistically significant (P = 0.0058) bacterial attenuation occurred when we deleted the qseB gene from A. hydrophila (Fig. 7).
Fig. 7.
Marginal decrease in in vivo virulence of the ΔqseB mutant in a septicaemic mouse model of A. hydrophila infection. Swiss Webster mice were injected via the intraperitoneal route with 5×107 c.f.u. of WT A. hydrophila SSU (n = 22; ▪). The same dose was used to infect mice with the ΔqseB mutant (n = 27; ▴), and both groups were monitored for death over a 16 day period. The percentage of surviving mice over time combined from three independent experiments is shown. The data were analysed by using Kaplan–Meier’s survival estimates showing statistically significant differences in animal survival between the ΔqseB mutant and the WT A. hydrophila SSU (P = 0.0058).
Discussion
In this study, we demonstrated the role of the QseBC two-component system in controlling virulence of A. hydrophila SSU, which possesses all three functional QS circuits and might establish a complex QS network to exert its regulatory role during pathogenesis (Khajanchi et al., 2009; Kozlova et al., 2008). Therefore, A. hydrophila SSU represents an excellent model organism to study the role of different QS networks in bacterial pathogenesis.
Flagella are not only important for bacterial movement but also contribute to pathogenesis by aiding in an organism’s adherence to the target host cells and biofilm formation (Kirov et al., 2002; Kirov, 2003; Kirov et al., 2004). Aeromonas species possess two distinct flagellar systems: a polar flagellum for swimming motility and several lateral flagella for swarming motility over surfaces (Kirov et al., 2004). We observed that both the swimming and swarming motility were diminished in A. hydrophila SSU when the qseB gene was deleted, suggesting to us that qseB is a regulator of both polar and lateral flagella. In accordance with our data, recent studies also showed that QseBC regulated the swimming motility of several pathogens such as E. coli (Sperandio et al., 2002), S. Typhimurium (Bearson & Bearson, 2008; Moreira et al., 2010) and Edwardsiella tarda (Wang et al., 2011). However, the investigators in these reports only examined the swimming and not the swarming motility.
In the present study, we noted that in addition to swimming motility, QseB also regulated the swarming motility of A. hydrophila SSU. Further, Clarke & Sperandio (2005a) reported that QseBC regulated flagella and motility through the flagellar master regulator FlhDC. They also demonstrated that in order to control motility, QseB directly bound to the flhDC promoter at both low and high affinity binding sites (Clarke & Sperandio, 2005a). Our future studies will examine the specific mechanism(s) of how QseBC regulates motility by controlling both the polar and lateral flagella systems in A. hydrophila SSU.
Interestingly, the ahyRI-mediated (AI-1) QS did not regulate either swimming or swarming motilities (Khajanchi et al., 2009), but deletion of the luxS gene (AI-2 QS) reduced the motility of A. hydrophila SSU (Kozlova et al., 2008).
In contrast with our study in A. hydrophila, it was noted that the qseB single mutant and the qseBC double mutant showed motility phenotypes similar to that of the WT S. Typhimurium (Bearson et al., 2010). However, in the latter study, the authors reported a decreased motility in the qseC mutant compared to that of the WT S. Typhimurium strain. Based on these observations, they concluded that the decreased motility in the qseC mutant was due to the negative regulation of QseB in the absence of QseC (Bearson et al., 2010).
Studies in E. coli have shown that QseC is an adrenergic receptor for AI-3 and host hormones epinephrine and/or norepinephrine (Clarke et al., 2006). Indeed, addition of epinephrine and/or AI-3 in the growth medium increased expression of many virulence genes, including that of the flagellar regulon genes in E. coli (Kendall et al., 2007). Likewise, addition of norepinephrine in the soft agar increased the motility of S. Typhimurium (Bearson & Bearson, 2008; Bearson et al., 2010). However, since supplementing the medium with norepinephrine also increased the motility of the QseC mutant, the authors questioned the role of QseBC in the motility of S. Typhimurium in response to norepinephrine (Bearson et al., 2010).
Our initial data indicated that addition of epinephrine and/or norepinephrine to the soft agar did not influence the motility of A. hydrophila. However, further study is necessary to demonstrate the effect of AI-3, epinephrine and norepinephrine on the QseBC-mediated regulation of virulence mechanisms in A. hydrophila.
In this report, we showed that QseB positively regulated protease production in A. hydrophila SSU. Likewise, in our recent study, we observed that AI-1 QS positively modulated protease activity, particularly that of metalloprotease (Khajanchi et al., 2009). It will be very intriguing to delineate the interaction of these three different QS systems to control various virulence mechanisms in A. hydrophila SSU, as our data indeed suggested the possibility of cross-talk between the AI-1 and AI-2 QS systems of A. hydrophila (Kozlova et al., 2011). In this context, we observed that the gene transcript levels for qseB and qseC were increased in the ahyRI mutant, while the ahyR and ahyI genes were downregulated in the qseB mutant (unpublished data). Based on these observations and the reduction of protease activity in both the qseB and the ahyRI mutant, we speculate that QseB regulates protease production indirectly through the AI-1 QS system. Thus, these data also suggested to us that there could be cross-talk between AI-1 and the QseBC QS systems in A. hydrophila, which will be explored further in our future studies.
Act is one of the most potent virulence factors of A. hydrophila SSU, which possesses several biological activities (Chopra et al., 2000; Galindo et al., 2004). Further, the Act mutant of A. hydrophila SSU is significantly attenuated in an animal model, indicating that Act contributes to in vivo virulence of this pathogen (Xu et al., 1998). In this study, we further showed that QseB regulated Act production, which could be very important for A. hydrophila as it is establishing an infection in the host. In a similar fashion in EHEC, QseC also regulated Shiga toxin by controlling the transcription of another two-component system QseEF (Hughes et al., 2009). It is interesting that we also detected a homologue of a QseEF two-component system in A. hydrophila SSU (unpublished data). In the future, we intend to study cross-talk between the QseBC and QseEF systems in the pathogenesis of A. hydrophila and to delineate specific mechanisms that regulate the biological activity of Act. Importantly, we observed that AI-1- and AI-2-mediated QS had no influence on the haemolytic activity of Act, which may mean that different QS systems in A. hydrophila SSU regulate different sets of virulence factors (Khajanchi et al., 2009; Kozlova et al., 2008).
Most bacteria in nature are not present as free-floating, isolated cells; rather they prefer to form surface-associated communities known as biofilms (Costerton et al., 1987; Costerton et al., 1999). Bacteria present in biofilms are more robust in nature in that they have the ability to withstand chemical and physical stresses and are more resistant to host defences than when they are free-living or in a planktonic state (Costerton et al., 1999; Singh et al., 2006). Therefore, biofilm formation is considered one of the most important virulence mechanisms that contribute to human disease transmission and pathogenesis (Huq et al., 2008; Oggioni et al., 2006). Several studies also pointed out that QS plays a crucial role in controlling biofilm development and establishing efficient infections in both Gram-positive (Oggioni et al., 2006; Petersen et al., 2006) and Gram-negative bacteria (de Kievit, 2009; Zhu & Mekalanos, 2003).
Previously, we demonstrated that AhyRI (AI-1) QS positively regulated biofilm formation in A. hydrophila SSU (Khajanchi et al., 2009), while this was negatively regulated by LuxS/AI-2-based QS (Kozlova et al., 2008). In the present study, we further showed that QseBC, in a manner similar to the luxS system, also negatively regulated biofilm formation in A. hydrophila. In agreement with our study, Moreira et al. (2006) showed that deletion of the qseA gene (encoding LysR-type regulator), which is also involved in AI-3 mediated QS circuits (Kaper & Sperandio, 2005) from enteropathogenic E. coli (EPEC), enhanced biofilm formation. However, we observed differences in solid surface biofilm formation by using CV staining and light microscopy at 24 h, as well as noticeable differences in the architecture of biofilm by using SEM at 48 and 72 h between the WT and the qseB mutant of A. hydrophila. On the other hand, Moreira et al. (2006) noticed that the qseA mutant formed denser/more compact biofilms than those seen with WT EPEC at earlier time points, such as at 6 and 12 h, following light microscopy examination and measurement of the biofilm biomass (c.f.u. cm−2).
In contrast with our findings, it was shown that inactivation of the qseC gene in Aggregatibacter actinomycetemcomitans reduced biofilm growth (Novak et al., 2010). These data indicate that regulation of biofilm formation by the QseBC system is distinct in different bacterial pathogens. Further, it was also observed that alterations in the biofilm formation ability of Aggregatibacter actinomycetemcomitans through the QseBC system was dependent on the AI-2/LuxS-based QS system (Novak et al., 2010). The production of AI-3 could be altered by cellular metabolism when the luxS gene is mutated in EHEC (Walters et al., 2006). Therefore, it is worth examining whether these AI-2 and QseBC QS systems in A. hydrophila SSU are linked in controlling biofilm formation.
In addition, we also showed that overproduction of c-di-GMP, the bacterial intracellular second messenger, altered biofilm formation and motility in A. hydrophila SSU in a QS-dependent manner involving both AI-1 and AI-2 systems (Kozlova et al., 2011). The loss in the motility and reduction of protease activity by GGDEF overproduction in the qseB mutant, which we described here, may mean that the QseBC QS system is involved in the c-di-GMP-dependent regulatory network in A. hydrophila. Because we argued that a balance of the transcriptional level of gene expression between luxS and ahyR was regulated by the modulation of c-di-GMP (Kozlova et al., 2011), we are interested in further studying a network connection between all three QS systems and c-di-GMP signalling in A. hydrophila SSU.
We also demonstrated that AI-1- and AI-2-mediated QS systems had opposite effects on the virulence of A. hydrophila in a septicaemic mouse model (Khajanchi et al., 2009; Kozlova et al., 2008). While deletion of the ahyRI QS genes attenuated the bacterium, the luxS mutant of A. hydrophila SSU had increased virulence (Khajanchi et al., 2009; Kozlova et al., 2008). Similar to the luxS mutant, deletion of the qseB gene from A. hydrophila enhanced in vitro biofilm formation; however, the qseB mutant was less virulent compared with the WT A. hydrophila in an in vivo model. In an animal model, we expected a marginal decrease in the virulence of the qseB mutant compared with that of the WT bacterium. This was based on in vitro experiments, where we noted that in addition to serving as a negative regulator of biofilm formation, QseBC also positively regulated some of the important virulence factors, such as Act and protease, and the latter (reduced Act and protease production) could possibly balance overall the pathogenicity of A. hydrophila. In agreement with our study, other investigators also showed that the qseC mutant and/or interference of QseC signalling in different bacteria resulted in attenuation of bacteria in different animal models tested (Bearson & Bearson, 2008; Kostakioti et al., 2009; Novak et al., 2010; Rasko et al., 2008). Although the present study has provided some evidence that the QseBC system might influence the virulence of A. hydrophila in an animal model, future in vivo studies are needed to elucidate the roles of AI-3 and the QseBC system in the virulence of A. hydrophila.
In conclusion, we characterized the role of the QseBC two-component system in order to better understand how different QS systems regulate the virulence of A. hydrophila. We have shown that QseBC both positively and negatively regulates various virulence factors/mechanisms of A. hydrophila SSU that could play an important role in fine-tuning the expression of virulence genes at an appropriate time to facilitate the establishment of infection by the pathogen in a highly efficient manner.
Acknowledgements
B. K. K. is a recipient of the J. W. McLaughlin Predoctoral Fellowship, UTMB. The funding obtained from the Environmental Protection Agency and National Institutes of Health (AI041611) grants are duly acknowledged. We thank Ms Mardelle Susman for editing the manuscript.
Abbreviations:
- AI
autoinducer
- CV
crystal violet
- EHEC
enterohaemorrhagic Escherichia coli
- gDNA
genomic DNA
- QS
quorum sensing
- RBC
red blood cell
- SEM
scanning electron microscopy
- T2SS, T3SS, T6SS
type X secretion system
- WT
wild-type
Footnotes
Five supplementary figures are available with the online version of this paper.
References
- Abuhammour W., Hasan R. A., Rogers D. (2006). Necrotizing fasciitis caused by Aeromonas hydrophilia in an immunocompetent child. Pediatr Emerg Care 22, 48–51. 10.1097/01.pec.0000195755.66705.f8 [DOI] [PubMed] [Google Scholar]
- Albert M. J., Ansaruzzaman M., Talukder K. A., Chopra A. K., Kuhn I., Rahman M., Faruque A. S., Islam M. S., Sack R. B., Mollby R. (2000). Prevalence of enterotoxin genes in Aeromonas spp. isolated from children with diarrhea, healthy controls, and the environment. J Clin Microbiol 38, 3785–3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearson B. L., Bearson S. M. (2008). The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb Pathog 44, 271–278. 10.1016/j.micpath.2007.10.001 [DOI] [PubMed] [Google Scholar]
- Bearson B. L., Bearson S. M., Lee I. S., Brunelle B. W. (2010). The Salmonella enterica serovar Typhimurium QseB response regulator negatively regulates bacterial motility and swine colonization in the absence of the QseC sensor kinase. Microb Pathog 48, 214–219. 10.1016/j.micpath.2010.03.005 [DOI] [PubMed] [Google Scholar]
- Beier D., Gross R. (2006). Regulation of bacterial virulence by two-component systems. Curr Opin Microbiol 9, 143–152. 10.1016/j.mib.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Challapalli M., Tess B. R., Cunningham D. G., Chopra A. K., Houston C. W. (1988). Aeromonas-associated diarrhea in children. Pediatr Infect Dis J 7, 693–698. 10.1097/00006454-198810000-00005 [DOI] [PubMed] [Google Scholar]
- Chopra A. K., Houston C. W. (1999). Enterotoxins in Aeromonas-associated gastroenteritis. Microbes Infect 1, 1129–1137. 10.1016/S1286-4579(99)00202-6 [DOI] [PubMed] [Google Scholar]
- Chopra A. K., Xu X., Ribardo D., Gonzalez M., Kuhl K., Peterson J. W., Houston C. W. (2000). The cytotoxic enterotoxin of Aeromonas hydrophila induces proinflammatory cytokine production and activates arachidonic acid metabolism in macrophages. Infect Immun 68, 2808–2818. 10.1128/IAI.68.5.2808-2818.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M. B., Sperandio V. (2005a). Transcriptional regulation of flhDC by QseBC and sigma (FliA) in enterohaemorrhagic Escherichia coli. Mol Microbiol 57, 1734–1749. 10.1111/j.1365-2958.2005.04792.x [DOI] [PubMed] [Google Scholar]
- Clarke M. B., Sperandio V. (2005b). Transcriptional autoregulation by quorum sensing Escherichia coli regulators B and C (QseBC) in enterohaemorrhagic E. coli (EHEC). Mol Microbiol 58, 441–455. 10.1111/j.1365-2958.2005.04819.x [DOI] [PubMed] [Google Scholar]
- Clarke M. B., Hughes D. T., Zhu C., Boedeker E. C., Sperandio V. (2006). The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci U S A 103, 10420–10425. 10.1073/pnas.0604343103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J. W., Cheng K. J., Geesey G. G., Ladd T. I., Nickel J. C., Dasgupta M., Marrie T. J. (1987). Bacterial biofilms in nature and disease. Annu Rev Microbiol 41, 435–464. 10.1146/annurev.mi.41.100187.002251 [DOI] [PubMed] [Google Scholar]
- Costerton J. W., Stewart P. S., Greenberg E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- de Kievit T. R. (2009). Quorum sensing in Pseudomonas aeruginosa biofilms. Environ Microbiol 11, 279–288. 10.1111/j.1462-2920.2008.01792.x [DOI] [PubMed] [Google Scholar]
- Edwards R. A., Keller L. H., Schifferli D. M. (1998). Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207, 149–157. 10.1016/S0378-1119(97)00619-7 [DOI] [PubMed] [Google Scholar]
- Erova T. E., Pillai L., Fadl A. A., Sha J., Wang S., Galindo C. L., Chopra A. K. (2006). DNA adenine methyltransferase influences the virulence of Aeromonas hydrophila. Infect Immun 74, 410–424. 10.1128/IAI.74.1.410-424.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erova T. E., Sha J., Horneman A. J., Borchardt M. A., Khajanchi B. K., Fadl A. A., Chopra A. K. (2007). Identification of a new hemolysin from diarrheal isolate SSU of Aeromonas hydrophila. FEMS Microbiol Lett 275, 301–311. 10.1111/j.1574-6968.2007.00895.x [DOI] [PubMed] [Google Scholar]
- Figueras M. J., Aldea M. J., Fernández N., Aspíroz C., Alperi A., Guarro J. (2007). Aeromonas hemolytic uremic syndrome. A case and a review of the literature. Diagn Microbiol Infect Dis 58, 231–234. 10.1016/j.diagmicrobio.2006.11.023 [DOI] [PubMed] [Google Scholar]
- Galindo C. L., Fadl A. A., Sha J., Gutierrez C., Jr, Popov V. L., Boldogh I., Aggarwal B. B., Chopra A. K. (2004). Aeromonas hydrophila cytotoxic enterotoxin activates mitogen-activated protein kinases and induces apoptosis in murine macrophages and human intestinal epithelial cells. J Biol Chem 279, 37597–37612. 10.1074/jbc.M404641200 [DOI] [PubMed] [Google Scholar]
- Galindo C. L., Gutierrez C., Jr, Chopra A. K. (2006). Potential involvement of galectin-3 and SNAP23 in Aeromonas hydrophila cytotoxic enterotoxin-induced host cell apoptosis. Microb Pathog 40, 56–68. 10.1016/j.micpath.2005.11.001 [DOI] [PubMed] [Google Scholar]
- Hengge R. (2009). Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7, 263–273. 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- Hiransuthikul N., Tantisiriwat W., Lertutsahakul K., Vibhagool A., Boonma P. (2005). Skin and soft-tissue infections among tsunami survivors in southern Thailand. Clin Infect Dis 41, e93–e96. 10.1086/497372 [DOI] [PubMed] [Google Scholar]
- Hoch J. A. (2000). Two-component and phosphorelay signal transduction. Curr Opin Microbiol 3, 165–170. 10.1016/S1369-5274(00)00070-9 [DOI] [PubMed] [Google Scholar]
- Horneman A. J., Ali A., Abbott S. (2007). Aeromonas. Manual of Clinical Microbiology, 9th edn Edited by Murray P. Washington, DC: American Society for Microbiology. [Google Scholar]
- Hughes D. T., Clarke M. B., Yamamoto K., Rasko D. A., Sperandio V. (2009). The QseC adrenergic signaling cascade in enterohemorrhagic E. coli (EHEC). PLoS Pathog 5, e1000553. 10.1371/journal.ppat.1000553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq A., Whitehouse C. A., Grim C. J., Alam M., Colwell R. R. (2008). Biofilms in water, its role and impact in human disease transmission. Curr Opin Biotechnol 19, 244–247. 10.1016/j.copbio.2008.04.005 [DOI] [PubMed] [Google Scholar]
- Janda J. M. (1991). Recent advances in the study of the taxonomy, pathogenicity, and infectious syndromes associated with the genus Aeromonas. Clin Microbiol Rev 4, 397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda J. M. (2002). Aeromonas and Plesiomonas. In Molecular Medical Microbiology, vol. 2, pp. 1237–1270. Edited by Sussman M. London, UK: Academic Press; 10.1016/B978-012677530-3/50278-6 [DOI] [Google Scholar]
- Janda J. M., Abbott S. L. (2010). The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev 23, 35–73. 10.1128/CMR.00039-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper J. B., Sperandio V. (2005). Bacterial cell-to-cell signaling in the gastrointestinal tract. Infect Immun 73, 3197–3209. 10.1128/IAI.73.6.3197-3209.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall M. M., Rasko D. A., Sperandio V. (2007). Global effects of the cell-to-cell signaling molecules autoinducer-2, autoinducer-3, and epinephrine in a luxS mutant of enterohemorrhagic Escherichia coli. Infect Immun 75, 4875–4884. 10.1128/IAI.00550-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajanchi B. K., Sha J., Kozlova E. V., Erova T. E., Suarez G., Sierra J. C., Popov V. L., Horneman A. J., Chopra A. K. (2009). N-Acylhomoserine lactones involved in quorum sensing control the type VI secretion system, biofilm formation, protease production, and in vivo virulence in a clinical isolate of Aeromonas hydrophila. Microbiology 155, 3518–3531. 10.1099/mic.0.031575-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajanchi B. K., Fadl A. A., Borchardt M. A., Berg R. L., Horneman A. J., Stemper M. E., Joseph S. W., Moyer N. P., Sha J., Chopra A. K. (2010). Distribution of virulence factors and molecular fingerprinting of Aeromonas species isolates from water and clinical samples: suggestive evidence of water-to-human transmission. Appl Environ Microbiol 76, 2313–2325. 10.1128/AEM.02535-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov S. M. (2003). Bacteria that express lateral flagella enable dissection of the multifunctional roles of flagella in pathogenesis. FEMS Microbiol Lett 224, 151–159. 10.1016/S0378-1097(03)00445-2 [DOI] [PubMed] [Google Scholar]
- Kirov S. M., Tassell B. C., Semmler A. B., O’Donovan L. A., Rabaan A. A., Shaw J. G. (2002). Lateral flagella and swarming motility in Aeromonas species. J Bacteriol 184, 547–555. 10.1128/JB.184.2.547-555.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov S. M., Castrisios M., Shaw J. G. (2004). Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect Immun 72, 1939–1945. 10.1128/IAI.72.4.1939-1945.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostakioti M., Hadjifrangiskou M., Pinkner J. S., Hultgren S. J. (2009). QseC-mediated dephosphorylation of QseB is required for expression of genes associated with virulence in uropathogenic Escherichia coli. Mol Microbiol 73, 1020–1031. 10.1111/j.1365-2958.2009.06826.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlova E. V., Popov V. L., Sha J., Foltz S. M., Erova T. E., Agar S. L., Horneman A. J., Chopra A. K. (2008). Mutation in the S-ribosylhomocysteinase (luxS) gene involved in quorum sensing affects biofilm formation and virulence in a clinical isolate of Aeromonas hydrophila. Microb Pathog 45, 343–354. 10.1016/j.micpath.2008.08.007 [DOI] [PubMed] [Google Scholar]
- Kozlova E. V., Khajanchi B. K., Sha J., Chopra A. K. (2011). Quorum sensing and c-di-GMP-dependent alterations in gene transcripts and virulence-associated phenotypes in a clinical isolate of Aeromonas hydrophila. Microb Pathog 50, 213–223. 10.1016/j.micpath.2011.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krovacek K., Pasquale V., Baloda S. B., Soprano V., Conte M., Dumontet S. (1994). Comparison of putative virulence factors in Aeromonas hydrophila strains isolated from the marine environment and human diarrheal cases in southern Italy. Appl Environ Microbiol 60, 1379–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung K. Y., Stevenson R. M. (1988). Tn5-induced protease-deficient strains of Aeromonas hydrophila with reduced virulence for fish. Infect Immun 56, 2639–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira C. G., Palmer K., Whiteley M., Sircili M. P., Trabulsi L. R., Castro A. F., Sperandio V. (2006). Bundle-forming pili and EspA are involved in biofilm formation by enteropathogenic Escherichia coli. J Bacteriol 188, 3952–3961. 10.1128/JB.00177-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira C. G., Weinshenker D., Sperandio V. (2010). QseC mediates Salmonella enterica serovar typhimurium virulence in vitro and in vivo. Infect Immun 78, 914–926. 10.1128/IAI.01038-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak E. A., Shao H., Daep C. A., Demuth D. R. (2010). Autoinducer-2 and QseC control biofilm formation and in vivo virulence of Aggregatibacter actinomycetemcomitans. Infect Immun 78, 2919–2926. 10.1128/IAI.01376-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole G. A., Kolter R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30, 295–304. 10.1046/j.1365-2958.1998.01062.x [DOI] [PubMed] [Google Scholar]
- Oggioni M. R., Trappetti C., Kadioglu A., Cassone M., Iannelli F., Ricci S., Andrew P. W., Pozzi G. (2006). Switch from planktonic to sessile life: a major event in pneumococcal pathogenesis. Mol Microbiol 61, 1196–1210. 10.1111/j.1365-2958.2006.05310.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen F. C., Ahmed N. A., Naemi A., Scheie A. A. (2006). LuxS-mediated signalling in Streptococcus anginosus and its role in biofilm formation. Antonie van Leeuwenhoek 90, 109–121. 10.1007/s10482-006-9065-y [DOI] [PubMed] [Google Scholar]
- Presley S. M., Rainwater T. R., Austin G. P., Platt S. G., Zak J. C., Cobb G. P., Marsland E. J., Tian K., Zhang B. & other authors (2006). Assessment of pathogens and toxicants in New Orleans, LA, following Hurricane Katrina. Environ Sci Technol 40, 468–474. 10.1021/es052219p [DOI] [PubMed] [Google Scholar]
- Rasko D. A., Moreira C. G., Li R., Reading N. C., Ritchie J. M., Waldor M. K., Williams N., Taussig R., Wei S. & other authors (2008). Targeting QseC signaling and virulence for antibiotic development. Science 321, 1078–1080. 10.1126/science.1160354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U., Simm R. (2009). Prevailing concepts of c-di-GMP signaling. Contrib Microbiol 16, 161–181. 10.1159/000219379 [DOI] [PubMed] [Google Scholar]
- Sakai D. K. (1985). Loss of virulence in a protease-deficient mutant of Aeromonas salmonicida. Infect Immun 48, 146–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri R., Joseph S. W., Chopra A. K., Sha J., Shaw J., Graf J., Haft D., Wu M., Ren Q. & other authors (2006). Genome sequence of Aeromonas hydrophila ATCC 7966T: Jack of all trades. J Bacteriol 188, 8272–8282. 10.1128/JB.00621-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha J., Kozlova E. V., Chopra A. K. (2002). Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infect Immun 70, 1924–1935. 10.1128/IAI.70.4.1924-1935.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha J., Kozlova E. V., Fadl A. A., Olano J. P., Houston C. W., Peterson J. W., Chopra A. K. (2004). Molecular characterization of a glucose-inhibited division gene, gidA, that regulates cytotoxic enterotoxin of Aeromonas hydrophila. Infect Immun 72, 1084–1095. 10.1128/IAI.72.2.1084-1095.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha J., Wang S. F., Suarez G., Sierra J. C., Fadl A. A., Erova T. E., Foltz S. M., Khajanchi B. K., Silver A. & other authors (2007). Further characterization of a type III secretion system (T3SS) and of a new effector protein from a clinical isolate of Aeromonas hydrophila – Part I. Microb Pathog 43, 127–146. 10.1016/j.micpath.2007.05.002 [DOI] [PubMed] [Google Scholar]
- Sierra J. C., Suarez G., Sha J., Foltz S. M., Popov V. L., Galindo C. L., Garner H. R., Chopra A. K. (2007). Biological characterization of a new type III secretion system effector from a clinical isolate of Aeromonas hydrophila – Part II. Microb Pathog 43, 147–160. 10.1016/j.micpath.2007.05.003 [DOI] [PubMed] [Google Scholar]
- Sierra J. C., Suarez G., Sha J., Baze W. B., Foltz S. M., Chopra A. K. (2010). Unraveling the mechanism of action of a new type III secretion system effector AexU from Aeromonas hydrophila. Microb Pathog 49, 122–134. 10.1016/j.micpath.2010.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Paul D., Jain R. K. (2006). Biofilms: implications in bioremediation. Trends Microbiol 14, 389–397. 10.1016/j.tim.2006.07.001 [DOI] [PubMed] [Google Scholar]
- Sperandio V., Torres A. G., Kaper J. B. (2002). Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol Microbiol 43, 809–821. 10.1046/j.1365-2958.2002.02803.x [DOI] [PubMed] [Google Scholar]
- Suarez G., Sierra J. C., Sha J., Wang S., Erova T. E., Fadl A. A., Foltz S. M., Horneman A. J., Chopra A. K. (2008). Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb Pathog 44, 344–361. 10.1016/j.micpath.2007.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez G., Sierra J. C., Erova T. E., Sha J., Horneman A. J., Chopra A. K. (2010a). A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J Bacteriol 192, 155–168. 10.1128/JB.01260-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez G., Sierra J. C., Kirtley M. L., Chopra A. K. (2010b). Role of Hcp, a type 6 secretion system effector, of Aeromonas hydrophila in modulating activation of host immune cells. Microbiology 156, 3678–3688. 10.1099/mic.0.041277-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift S., Lynch M. J., Fish L., Kirke D. F., Tomás J. M., Stewart G. S., Williams P. (1999). Quorum sensing-dependent regulation and blockade of exoprotease production in Aeromonas hydrophila. Infect Immun 67, 5192–5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila J., Ruiz J., Gallardo F., Vargas M., Soler L., Figueras M. J., Gascon J. (2003). Aeromonas spp. and traveler’s diarrhea: clinical features and antimicrobial resistance. Emerg Infect Dis 9, 552–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldor M. K., Sperandio V. (2007). Adrenergic regulation of bacterial virulence. J Infect Dis 195, 1248–1249. 10.1086/513281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters M., Sperandio V. (2006). Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect Immun 74, 5445–5455. 10.1128/IAI.00099-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters M., Sircili M. P., Sperandio V. (2006). AI-3 synthesis is not dependent on luxS in Escherichia coli. J Bacteriol 188, 5668–5681. 10.1128/JB.00648-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang Q., Yang M., Xiao J., Liu Q., Wu H., Zhang Y. (2011). QseBC controls flagellar motility, fimbrial hemagglutination and intracellular virulence in fish pathogen Edwardsiella tarda. Fish Shellfish Immunol 30, 944–953. 10.1016/j.fsi.2011.01.019 [DOI] [PubMed] [Google Scholar]
- Xu X. J., Ferguson M. R., Popov V. L., Houston C. W., Peterson J. W., Chopra A. K. (1998). Role of a cytotoxic enterotoxin in Aeromonas-mediated infections: development of transposon and isogenic mutants. Infect Immun 66, 3501–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Mekalanos J. J. (2003). Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev Cell 5, 647–656. 10.1016/S1534-5807(03)00295-8 [DOI] [PubMed] [Google Scholar]