Abstract
The close interactions of dogs with humans and surrounding wildlife provide frequent opportunities for cross-species virus transmissions. In order to initiate an unbiased characterization of the eukaryotic viruses in the gut of dogs, this study used deep sequencing of partially purified viral capsid-protected nucleic acids from the faeces of 18 diarrhoeic dogs. Known canine parvoviruses, coronaviruses and rotaviruses were identified, and the genomes of the first reported canine kobuvirus and sapovirus were characterized. Canine kobuvirus, the first sequenced canine picornavirus and the closest genetic relative of the diarrhoea-causing human Aichi virus, was detected at high frequency in the faeces of both healthy and diarrhoeic dogs. Canine sapovirus constituted a novel genogroup within the genus Sapovirus, a group of viruses also associated with human and animal diarrhoea. These results highlight the high frequency of new virus detection possible even in extensively studied animal species using metagenomics approaches, and provide viral genomes for further disease-association studies.
Introduction
Dogs (Canis lupus familiaris) are one of the most popular pets on Earth, with an estimated population of over 400 million worldwide (Coppinger & Coppinger, 2001). As companion animals, dogs have extensive contact with humans, sharing their spaces, playing with children, producing faecal waste, and occasionally biting and scratching. Dogs are also frequently in contact with wildlife through hunting or scavenging. The large population of dogs and their close interactions with humans therefore make them a potential risk factor for zoonotic virus transmission, as exemplified by rabies virus and rotavirus (Cook, 1989; Lackay et al., 2008; Tsugawa & Hoshino, 2008).
Numerous viral infections are known to affect the health of dogs (Buonavoglia & Martella, 2007; Willis, 2000; Wright, 1973). Canine distemper virus (a paramyxovirus), a common viral threat for dogs, is highly contagious and often fatal, causing fever, vomiting and diarrhoea, respiratory symptoms, seizures and paralysis. Canine distemper virus is distributed globally and its host range is broad, including many mammalian species of the families Canidae, Mustelidae, Procyonidae, Ursidae and Viverridae (Griot et al., 2003; Martella et al., 2008b, 2010). Canine parvovirus 2 (CPV2) is found worldwide, is highly contagious and can also be lethal, causing the symptoms of fever, vomiting and haemorrhagic diarrhoea. CPV2 is genetically and antigenically closely related to feline panleukopenia virus (feline distemper virus) (Martella et al., 2005; Parrish, 1999; Parrish & Kawaoka, 2005; Pollock & Coyne, 1993; Truyen, 1999). Canine coronavirus (CCoV) also causes vomiting, diarrhoea and/or respiratory symptoms, and different strains have been identified, including enteric CCoVs, canine respiratory coronavirus and canine pantrophic coronavirus (Decaro & Buonavoglia, 2008; Erles & Brownlie, 2008; Pratelli, 2006). Other viruses such as canine rotavirus, adenovirus, herpesvirus, influenza virus and parainfluenza virus have also been reported as potential dog pathogens (Decaro et al., 2008; Dubovi & Njaa, 2008; Nicholls & Stanley, 1999; Renshaw et al., 2010).
In this study, we used next-generation sequencing to characterize the faecal viruses in diarrhoea specimens from sheltered and privately owned dogs. We report here on the detection of viral pathogens known to cause canine diarrhoea, as well as the first canine kobuvirus and sapovirus.
Results
Viruses in faeces of diarrhoeic dogs
After enrichment of viral particles in the faeces of 18 diarrhoeic dogs using filtration and nuclease treatment, the viral RNA and DNA were extracted, amplified by random RT-PCR and pyrosequenced. Approximately 276 000 high-quality nucleotide sequence reads with a mean length of 200 bases (after removal of the primer and adjacent 8 nt) were generated and binned according to their faecal source of origin. Contigs were then formed within each bin and, together with singletons >100 bases, were classified based on best blast scores (E-value <10−3) to taxonomically assigned sequences in the GenBank non-redundant database. Approximately 10 % of the total reads were assigned to eukaryotic viral sequences, including sequences from DNA and RNA virus families including Parvoviridae, Picornaviridae, Caliciviridae and Coronaviridae (Table 1).
Table 1. Summary of clinical information and eukaryotic viruses detected by the pyrosequencing run.
| ID | Age | Total no. reads | Eukaryotic virus detected (no. reads) | Dog source | Diagnosis |
| AN210b | Adult | 28 626 | Rotavirus (1) | Shelter O | Bloody diarrhoea |
| AN210c | Adult | 22 930 | None | ||
| AN210d | Adult | 11 751 | Sapovirus (396) | ||
| AN210e | Adult | 26 067 | None | ||
| AN211a | 36 months | 13 981 | Densovirus (3) | Shelter I | Bloody diarrhoea |
| AN211b | 12 months | 1 924 | CPV2 (6) | ||
| AN211c | Adult | 2 258 | CPV2 (5) | ||
| AN211d | 24 months | 25 298 | Kobuvirus (22) | ||
| AN195 | 18 months | 20 744 | CCoV (9978) | Shelter G | Conjunctivitis, diarrhoea |
| AN196 | 12 months | 5 132 | CPV2 (1), Sapovirus (8), Brome mosaic virus (1) | ||
| AN201 | 24 months | 16 463 | CCoV (2298) | ||
| AN203 | 120 months | 8 694 | CCoV (1) | ||
| AN213 | 5 months | 26 099 | Kobuvirus (14) | Sheltered | Bloody diarrhoea |
| AN98 | 4 months | 11 320 | CPV2 (2232) | Sheltered | Bloody diarrhoea |
| AN155 | 36 months | 14 545 | CPV2 (3) | Owned | Myocardial necrosis, diarrhoea |
| AN109 | 5 months | 5 323 | Rotavirus (3), Kobuvirus (3), CPV2 (702), Kashmir bee virus (1) | Sheltered | Bloody diarrhoea |
| AN214 | Adult | 16 060 | CPV2 (2), CCoV (13) | Sheltered | Bloody diarrhoea |
| AN126 | Adult | 19 412 | CPV2 (11787) | Owned | Bloody diarrhoea |
Both known and previously uncharacterized viruses were detected. Diarrhoea-associated dog viruses included CPV2, CCoV and rotavirus. CPV2 was found in eight samples and was abundant in terms of total number of viral sequence reads in dogs AN98, AN109 and AN126 (Table 1). The long viral contigs from these samples were highly similar (99 % nucleotide similarity) to reported CPV2 strains. CCoV was detected in four samples including three dogs from the same shelter G outbreak. In this shelter, dogs AN195 and AN201 showed a high number of CCoV sequence reads (Table 1) and for both samples >90 % of the viral genome was covered. CCoV AN195 and AN201 were highly similar to each other, with ~99 % nucleotide identity. Phylogenetic analysis based on the spike glycoprotein region showed that CCoV AN195 and AN201 clustered closely with porcine transmissible gastroenteritis virus-like CCoVs (CCoV subtype IIb strains; data not shown), which originated from a double recombination event with transmissible gastroenteritis virus (Decaro et al., 2009, 2010).
Sequence reads of CPV2 and CCoV accounted for >98 % of all mammalian virus reads identified in these samples. Rotavirus sequences were also found in dogs AN210b (n = 1) and AN109 (n = 3). One sequence (212 bp) from AN109 showed 99 % nucleotide identity to human rotavirus G4 capsid protein VP7 gene; the other three sequences aligned with conserved, phylogenetically uninformative regions.
The presence of viral sequences related to insect viruses including densovirus, Kashmir bee virus and plant viruses such as brome mosaic virus probably reflected the omnivorous diet of dogs and/or contamination of their food.
Canine kobuvirus
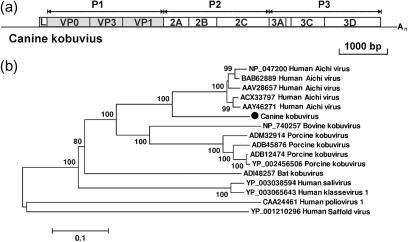
In three dog stool samples from different shelters (AN211d, AN213 and AN109), we found sequences closely related to the human Aichi virus. By connecting gaps between sequenced viral fragments and using 5′- and 3′RACE, the complete genome of the kobuvirus in AN211d was obtained. The resulting genome was 8134 bp with a G+C content of 58 mol%, excluding the poly(A) tail. The AN211d genome contained a large ORF of 7329 bp, encoding a putative polyprotein precursor of 2442 aa, a partial 5′ untranslated region (UTR) of 561 bp and a complete 3′UTR of 244 bp (Fig. 1a). AN211d was most closely related to human Aichi virus, with 78 % amino acid and 77 % nucleotide similarity and a non-synonymous : synonymous substitution rate ratio of 0.2, indicating strong selection pressure to maintain the protein sequences.
Fig. 1.
(a) Genome organization of canine kobuvirus (CaKoV). The P1 region (shaded) was used to construct the phylogenetic tree. (b) Phylogenetic analysis of CaKoV, known kobuviruses and kobu-like viruses based on the complete amino acid sequence of the P1 region using the neighbour-joining method with p-distances and 1000 bootstrap replicates. GenBank accession numbers are shown. Bar, 10 % estimated phylogenetic divergence.
We provisionally named the virus canine kobuvirus (CaKoV). The partial genome sequences of CaKoV sequences from AN213 and AN109 shared 93–98 % nucleotide similarity with the CaKoV in AN211d.
The CaKoV-AN211d polyprotein sequence was analysed for potential proteolytic cleavage sites based on alignment with the Aichi virus sequence. The polyprotein was deduced to consist of the expected proteolytic cleavage protein comprising leader (L), VP0, VP3, VP1, 2A–2C and 3A–3D proteins. The L protein was 172 aa, and was of similar length and had 54 % amino acid similarity to those of Aichi viruses. The conserved RNA-dependent RNA polymerase (RdRp) motifs KDELR, YGDD and FLKR were detected (Yamashita et al., 1998).
According to the International Committee on Taxonomy of Viruses (ICTV) (http://www.picornastudygroup.com/definitions/genus_definition.htm), members of the same picornavirus genus share >40, >40 and >50 % amino acid similarity in their P1, P2 and P3 regions, respectively. The P1, P2 and P3 regions of CaKoV-AN211d shared 75, 83 and 86 % amino acid similarity with Aichi virus, respectively. Based on these criteria, AN211d belongs to the genus Kobuvirus. Intra-species comparison of Aichi virus strains showed that their complete P1, P2 and P3 regions shared more than 93, 96 and 96 % amino acid similarity, respectively. Porcine kobuvirus strains shared more than 91, 95 and 93 % amino acid similarity in the P1, P2 and P3 regions, respectively. CaKoV is therefore tentatively classified as a novel species, closely related to human Aichi virus, in the genus Kobuvirus. Phylogenetic analysis based on the P1 region confirmed that CaKoV-AN211d segregated from the tightly clustered Aichi viruses as a distinct genetic lineage (Fig. 1b). A phylogenetic tree of the 3D (RdRp) region showed a similar tree topology (data not shown).
Canine sapovirus
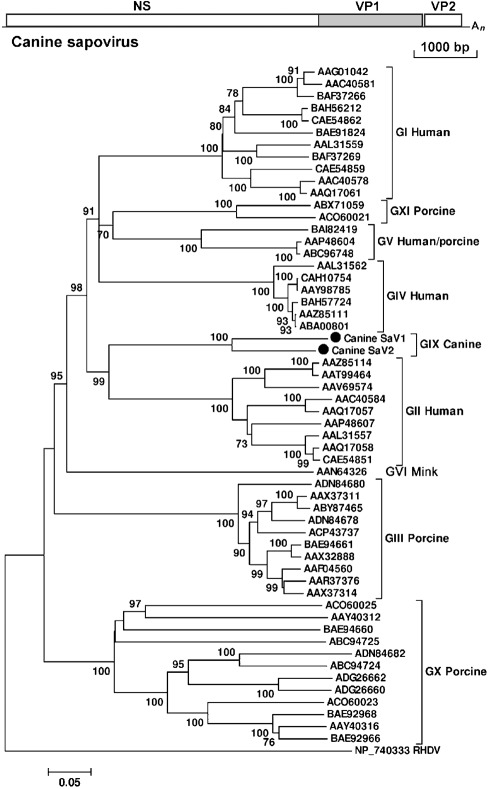
Sequences related to sapoviruses, previously known as sapporo-like viruses, were also detected in two stool samples from different shelters (AN210d and AN196). AN210d had 396 reads, which assembled into 25 contigs covering approximately 90 % of the viral genome. AN196 had eight reads that covered approximately 15 % of the viral genome. The sequence gaps between identified viral fragments were filled by RT-PCR, and 5′- and 3′RACE and genome walking were used to acquire the rest of the genome. The nearly complete viral genome sequence generated from AN210d was 7141 bp excluding its 3′ poly(A) tail. Compared with other sapoviruses, we estimate that approximately 300 bp may be missing from the 5′ end of the viral genome. A partial genome of 2578 bp was acquired from AN196, sharing 76 % nucleotide similarity and 82 % amino acid similarity with the canine sapovirus from AN210d. The two viruses were temporarily named canine sapovirus 1 and 2 (CaSaV1 and -2).
The CaSaV1-AN210d strain was predicted to contain two major ORFs. The partial ORF1 was 6495 bp (2164 aa), encoding the expected non-structural proteins and VP1 capsid protein (556 aa). ORF2, consisting of 501 bp, was predicted to encode a VP2 protein of 166 aa (Fig. 2a). Four nucleotides separated ORF1 and ORF2. ORF3, encoding a small basic protein and only present in some sapovirus genogroups, was not detected. The 3′UTR was 141 bp, longer than that of other sapoviruses. The predicted polyprotein encoded by ORF1 contained highly conserved or slightly modified calicivirus motifs including those of the 2C helicase (GAPGIGK/GPPGIGKT), 3C protease (GDCG) and RdRp (GLPSG and YGDD). The PPG motif was also present in the predicted VP1 (Yang et al., 2009).
Fig. 2.
(a) Genome organization of canine sapovirus. The VP1 region (shaded) was used to construct the phylogenetic tree. (b) Phylogenetic analysis of canine sapovirus and representative sapovirus strains with a complete VP1 region selected from GenBank, based on the complete amino acid sequence of the VP1 region using the neighbour-joining method with p-distances and 1000 bootstrap replicates. GenBank accession numbers are shown. Bar, 5 % estimated phylogenetic divergence. Rabbit haemorrhagic disease virus (RHDV) from the genus Lagovirus in the family Caliciviridae was used as the outgroup. CaSaV1 and -2 were grouped as a distinct clade named genogroup IX (GIX); the traditional genogroups I–V are indicated, and mink sapovirus was named as GVI; two new porcine sapovirus genogroups have been named as GX and GXI (pending an ICTV review; GVII and GVIII are already taken based on RdRp-based classification).
CaSaV1-AN210d shared up to 52 % amino acid similarity in the polyprotein region with the closest sapoviruses, whilst the VP2 protein shared 48 % amino acid similarity. The VP1 region of AN210d ORF1 shared 31–52 % amino acid similarity with known sapovirus strains, with the highest similarities to the GII human sapovirus strains and the lowest similarities to porcine enteric sapoviruses. By comparing all available complete VP1 amino acid sequences available in GenBank, we found that different VP1s within GI shared >75 % amino acid similarity, whilst strains within GII shared >77 % amino acid similarity and strains within GIII shared >83 % amino acid similarity. The VP1 of CaSaV1-AN210d shared less than 46, 52 and 42 % with GI, GII and GIII strains, respectively, falling outside the outer range of intra-genogroup genetic distances. The intergenus amino acid similarities of the full-length capsid protein of the four genera of caliciviruses (Sapovirus, Norovirus, Vesivirus and Lagovirus) have been reported as 15–27 % (Martella et al., 2008b; Smiley et al., 2002). Based on the above criteria, CaSaV1-AN210d was classified as a novel genogroup (GIX) within the genus Sapovirus. We did not use GVII or GVIII, as these genotype names are already used for other sapovirus whose VP1 sequences are not available (Barry et al., 2008; Nakamura et al., 2010). GX and GXI are also similarly used. The RdRp of GVII or GVIII members did not cluster with those of CaSaV1-AN210d or CaSaV2-AN196, confirming that they belong to different genotypes.
Phylogenetic analysis of all available complete sapovirus VP1 sequences from GenBank indicated that CaSaV1-AN210d and CaSaV2-AN196 belonged to the same genogroup (Fig. 2b). Phylogenetic analysis of the RdRp region showed a similar tree topology (data not shown).
Prevalence of CaSaV1 and CaKoV
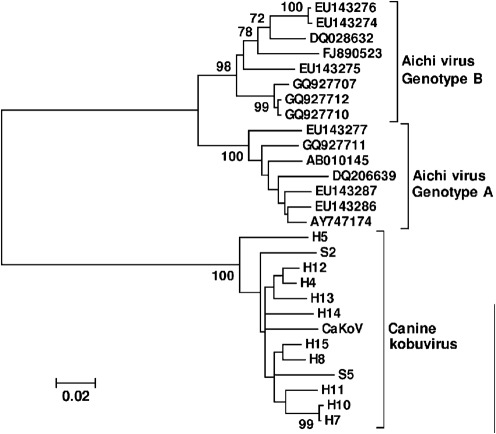
We next tested for the presence of CaSaV1 and CaKoV in the faeces of 200 healthy and 200 diarrhoeic dogs using real-time PCR. CaSaV1 was detected in one healthy and one diarrhoeic dog, whilst CaKoV was found in 14 healthy and six diarrhoeic dogs. No significant difference of CaKoV prevalence was found in healthy versus diarrhoeic dog samples (χ2 test, P>0.05). The detection of CaKoV was therefore not associated with diarrhoea in this dog population. The prevalence of CaSaV1 was too low to be tested for disease association. A partial VP1 region (~330 bp) of CaKoV was amplified and sequenced. Of the 20 real-time CaKoV PCR-positive samples, VP1 could be amplified from 12 samples (ten from healthy and two from diarrhoeic samples), showing >91 % nucleotide similarity. Phylogenetic analyses based on this partial VP1 region revealed limited genetic diversity. No genetic clustering of VP1 from the three symptomatic cases (including CaKoV-AN211d) was observed (Fig. 3).
Fig. 3.
Phylogenetic analyses of CaKoV strains and Aichi virus strains based on the nucleotide sequence of the partial VP1 region using the neighbour-joining method with p-distances and 1000 bootstrap replicates. CaKoV was isolated from diarrhoeic dog AN211d. H, Virus from a healthy dog faecal sample; S, virus from a diarrhoeic dog faecal sample. Bar, 2 % estimated phylogenetic divergence.
Discussion
Our study examined the viral nucleic acids in the faeces of 18 dogs with diarrhoea. Mammalian viruses were found in 15 samples. In three samples, CPV2 plus one or two other mammalian viruses were identified. Whether co-infections result in more severe symptoms will require further studies.
Viruses reported to cause canine diarrhoea detected here were CPV2, CCoV and rotavirus. Canine rotavirus may also infect humans, as rotavirus strains in children with diarrhoea can share very close genetic relationships with canine rotavirus strains with G3 specificities, indicating probable zoonotic transmission (De Grazia et al., 2007; Tsugawa & Hoshino, 2008). One of the rotavirus sequences found in this study was highly similar to the human rotavirus G4.
The genus Kobuvirus in the family Picornaviridae currently contains three species recognized by the ICTV: the human Aichi virus, bovine kobuvirus and porcine kobuvirus. Other kobuviruses were recently characterized, including salivirus/klassevirus from human faeces (Greninger et al., 2009; Holtz et al., 2009; Li et al., 2009) and as a partial genome from bat guano (Li et al., 2010). Aichi virus and salivirus/klassevirus have been associated with human gastroenteritis (Greninger et al., 2009; Holtz et al., 2009; Li et al., 2009), whilst bovine and porcine kobuviruses have been detected in both healthy and diarrhoeic cattle and pigs (Reuter et al., 2009; Yamashita et al., 1998, 2003; Yu et al., 2009). Bovine kobuvirus has also been detected in the faeces of healthy sheep (Reuter et al., 2010). The kobuvirus genome has a typical picornavirus polyprotein organization including an N-termini L protein (Reuter et al., 2009; Yamashita et al., 1998, 2003). Phylogenetically, CaKoV is the closest reported genetic relative of the human Aichi virus, reflecting a relatively recent common origin and possible cross-species transmission. The detection of CaKoV1 in an animal (AN213) just recovering from haemorrhagic diarrhoea may reflect a pathogenic role in this sporadic case or may be the coincidental finding of a usually commensal virus. CaKoV was also identified in an unrelated animal (AN109) co-infected with CPV2 and rotavirus, providing alternative explanations for this animal’s diarrhoea. CaKoV was also found in AN211d, a member of a shelter outbreak in which CPV2 was detected in two out of four diarrhoeic animals. Real-time RT-PCR showed that the rate of detection of CaKoV was relatively high, with no difference in prevalence between healthy and diarrhoeic dogs. Based on the dog sampling performed here, the pathogenicity of these novel viruses, if any, is therefore likely to be low, at least in the mostly adult canine population tested here.
Sapoviruses are positive-sense ssRNA viruses of approximately 7.4 kb with two or three ORFs, in the genus Sapovirus of the family Caliciviridae (Koopmans et al., 2005) and reported to cause diarrhoea in humans, pigs and mink (Chiba et al., 1979; Martella et al., 2008c; Usuku et al., 2008; Guo et al., 2001). Sapovirus classification has been based mostly on capsid sequences, but other genomic regions such as the RdRp have produced similar phylogenies (Farkas et al., 2004; L’Homme et al., 2009; Martella et al., 2008b; Yang et al., 2009). Based on capsid VP1 sequences, sapoviruses have been classified into five distinct genogroups (GI–GV), with human sapoviruses belonging to genogroups GI, GII, GIV and GV; porcine sapoviruses to genogroup GIII and unclassified sapoviruses; and mink sapovirus to other unclassified sapoviruses (Farkas et al., 2004; Hansman et al., 2007; Okada et al., 2006). We have described here a new putative sapovirus genogroup (GIX) consisting of the first reported canine sapoviruses (CaSaV1 and -2). CaSaV1 was detected in one (AN210d) of four animals in a dog shelter diarrhoea outbreak. In the dog shedding CaSaV2 (AN196), the co-detection of CPV2 provides a credible aetiological agent for its diarrhoea. When CaSaV1 was tested in the faeces of 200 diarrhoeic and 200 healthy dogs using real-time PCR, only one infection was detected in each group, indicating that CaSaV1 infection was rare in this particular dog population. Further prevalence measurements in diarrhoeic and healthy control dogs will be required to determine the pathogenic potential of CaSaV. The wide genetic diversity between CaSaV1 and -2 also indicates that they have the potential to differ in pathogenicity.
Only 10 % of the sequences generated here could confidently be related to eukaryotic viruses, whilst the rest were related to canine host sequences, phages or bacteria, or were taxonomically unclassifiable. Showing a high percentage of sequence read hits to eukaryotic virus were three samples with CPV2 (61 % for AN126, 20 % for AN98 and 13 % for AN109) and two CCoV infections (48 % for AN195 and 14 % for AN201). The low percentage of reads for the other viral infections presumably reflects lower viral loads at the time of sample collection.
The characterization of two novel canine viruses from only 18 diarrhoeic faeces samples from a single geographical region (CA, USA) indicates that a significant number of dog viruses probably remain uncharacterized, as exemplified by the recent characterization of a close relative of human hepatitis C virus in canine respiratory secretions (Kapoor et al., 2011). Determination of their viral genome sequence followed by epidemiological studies of their pathogenicity or direct inoculations will further our understanding of the aetiology of infectious canine diseases. Expansion of the known diversity of mammalian viruses will also facilitate ongoing sequence similarity-based virus discovery.
Methods
Animal specimen collection.
A total of 18 faecal specimens of dogs with diarrhoea were collected by the Shelter Medicine Service at the Center for Companion Animal Health, School of Veterinary Medicine at University of California (Davis, CA, USA) from January 2009 to August 2010 from diarrhoea outbreaks in three animal shelters in California (shelters O, I and G) and from individual cases of sheltered and privately owned dogs (Table 1). All dog faecal samples were collected during a diarrhoea episode and tested negative by a commercial canine parvovirus antigen test kit. AN213 was collected 2 days after clinical resolution and did not undergo a CPV2 antigen test. Faecal specimens were stored in small plastic bags and frozen at −80 °C.
Sample preparation and viral nucleic acid extraction.
Faecal samples were processed as described previously (Victoria et al., 2009). Briefly, faecal samples were resuspended by vigorous vortexing in Hanks’ buffered saline solution (Gibco-BRL) at a concentration of ~0.5 g ml−1. The stool suspensions were centrifuged at 10 000 g for 3 min and the supernatants filtered through a 0.45 µm filter (Millipore) to remove intact bacteria and other large cellular debris. The partially purified viral particle-containing filtrates were digested with a mixture of DNases and RNases to remove unprotected nucleic acids (not in viral capsids or otherwise protected from digestion) (Allander et al., 2001). DNA and RNA were then extracted using a QIAamp Viral RNA Mini kit (Qiagen).
Library construction and pyrosequencing.
Viral nucleic acid libraries were constructed by random PCR amplification using RT and PCR primers with degenerate (8 nt) 3′ ends, as described previously (Victoria et al., 2009). Random PCR products were checked by 1 % agarose gel and DNA concentrations determined by measuring the absorbance at 260 nm. The PCR products were pooled and separated by 2 % agarose gel, and fragments of 500–1000 bp were excised and extracted. The resulting purified product was prepared using a GS FLX General Library Preparation kit (454 Life Sciences) and the library of ssDNA fragments was sequenced on a single pyrosequencing gasket using a Genome Sequencer FLX Instrument (454 Life Sciences).
Bioinformatics.
The pyrosequencing reads were sorted into corresponding samples according to their unique sequence tag (20 fixed bases of the random PCR primer). Primer sequences plus the adjacent 8 nt were then trimmed from each read. Trimmed reads from each sample were assembled de novo using the mira assembly program (Chevreux, 2005), with a criterion of ≥95 % identity over 35 bp for the resulting singlets and contigs. Assembled sequences >100 bp were compared with the GenBank non-redundant nucleotide and protein databases using blastn and blastx, respectively. Sequences were classified into eukaryotic viruses, phages, bacteria and eukaryotes based on the taxonomic origin of the best-hit sequence. An E-value of 0.001 was used as the cut-off level for significant hits. The high-quality sequences and contigs of veterinary sample metagenomes have been deposited in the GenBank short-read archive under accession number SRA044889.
Genome sequencing.
In order to acquire the complete genome of target viruses, multiple primer sets were designed based on the sequences obtained by pyrosequencing as above to fill gaps between fragments. The extremities of the RNA genomes were acquired using 3′- and 5′RACE (Invitrogen) and genome walking.
Phylogenetic analysis.
Reference viral sequences from different virus families were obtained from GenBank. Amino acid sequence alignments were generated using clustal w, implemented in mega4.1 with default settings (Kumar et al., 2008). Aligned sequences were trimmed to match the genomic regions of the viral sequences obtained in this study, and phylogenetic trees were generated by mega4.1 using the neighbour-joining method with amino acid p-distances and 1000 bootstrap replicates. The GenBank accession numbers of the viral sequences used in the phylogenetic analyses are shown in the trees.
Prevalence study of CaKoV and CaSaV1.
Real-time PCR was used for the detection of canine kobuvirus and sapovirus 1 (TaqMan Universal PCR Master Mix; Applied Biosystems) in RNA extracts of 200 diarrhoeal and 200 healthy dog faecal samples. cDNA was produced with random primers and Superscript III (Invitrogen) according to standard reverse transcription protocols. The oligonucleotide primer pairs and TaqMan probe for CaKoV were as follows: Dogkobu forward primer (5′- CGGATTCCATATTGGCCCTTG-3′), Dogkobu reverse primer (5′- ACAGCGTGAAATTGCGGTGG-3′) and Dogkobu probe (5′-CCGAGAAGCCCCCGGTGGAAC-3′) for the 2C gene region; and for CaSaV1 were Dogsap1 forward primer (5′- ACACACGATCCAAATTCACCAA-3′), Dogsap1 reverse primer (5′- TGCCAGACAGACCTCCAATTG-3′) and Dogsap1 probe (5′- TAATTGACGTGCAAGCCCGCTCG-3′) for the RdRp gene region. The real-time PCR conditions were 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min using a real-time PCR instrument (LightCycler 480; Roche Applied Science). Synthetic DNA fragments (~150 bp) of the corresponding regions were used to produce a standard curve and determine analytical performance. CaKoV-positive samples were then amplified for the partial VP1 region with the nested primers Dogkobu-F1 (5′-TCTCCTGCTTCACSTACATCG-3′), Dogkobu-F2 (5′-TTCGCATCTCYAACCCMAATG-3′), Dogkobu-R1 (5′-ACGGGAGAGGGAGATYTGTTT-3′) and Dogkobu-R2 (5′-ATCTGCACGMACGATGTAGAC-3′) and sequenced. Primers were designed based on the VP1 region of CaKoV and Aichi virus.
Acknowledgements
The work was supported by the Blood Systems Research Institute and NIH R01 HL083254 to E. D. The UC Davis Center for Companion Animal Health supports the research of P. A. P.
References
- Allander T., Emerson S. U., Engle R. E., Purcell R. H., Bukh J. (2001). A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc Natl Acad Sci U S A 98, 11609–11614 10.1073/pnas.211424698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry A. F., Alfieri A. F., Alfieri A. A. (2008). High genetic diversity in RdRp gene of Brazilian porcine sapovirus strains. Vet Microbiol 131, 185–191 10.1016/j.vetmic.2008.02.021 [DOI] [PubMed] [Google Scholar]
- Buonavoglia C., Martella V. (2007). Canine respiratory viruses. Vet Res 38, 355–373 10.1051/vetres:2006058 [DOI] [PubMed] [Google Scholar]
- Chevreux, B. (2005). mira: an automated genome and EST assembler. PhD thesis, Ruprecht-Karls University, Heidelberg, Germany. [Google Scholar]
- Chiba S., Sakuma Y., Kogasaka R., Akihara M., Horino K., Nakao T., Fukui S. (1979). An outbreak of gastroenteritis associated with calicivirus in an infant home. J Med Virol 4, 249–254 10.1002/jmv.1890040402 [DOI] [PubMed] [Google Scholar]
- Cook G. C. (1989). Canine-associated zoonoses: an unacceptable hazard to human health. Q J Med 70, 5–26 [PubMed] [Google Scholar]
- Coppinger R., Coppinger L. (2001). Dogs: a Startling New Understanding of Canine Origin, Behavior & Evolution. New York: Scribner [Google Scholar]
- Decaro N., Buonavoglia C. (2008). An update on canine coronaviruses: viral evolution and pathobiology. Vet Microbiol 132, 221–234 10.1016/j.vetmic.2008.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Martella V., Buonavoglia C. (2008). Canine adenoviruses and herpesvirus. Vet Clin North Am Small Anim Pract 38, 799–814, viii 10.1016/j.cvsm.2008.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Mari V., Campolo M., Lorusso A., Camero M., Elia G., Martella V., Cordioli P., Enjuanes L., Buonavoglia C. (2009). Recombinant canine coronaviruses related to transmissible gastroenteritis virus of swine are circulating in dogs. J Virol 83, 1532–1537 10.1128/JVI.01937-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Mari V., Elia G., Addie D. D., Camero M., Lucente M. S., Martella V., Buonavoglia C. (2010). Recombinant canine coronaviruses in dogs, Europe. Emerg Infect Dis 16, 41–47 10.3201/eid1601.090726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Grazia S., Martella V., Giammanco G. M., Gòmara M. I., Ramirez S., Cascio A., Colomba C., Arista S. (2007). Canine-origin G3P[3] rotavirus strain in child with acute gastroenteritis. Emerg Infect Dis 13, 1091–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubovi E. J., Njaa B. L. (2008). Canine influenza. Vet Clin North Am Small Anim Pract 38, 827–835, viii 10.1016/j.cvsm.2008.03.004 [DOI] [PubMed] [Google Scholar]
- Erles K., Brownlie J. (2008). Canine respiratory coronavirus: an emerging pathogen in the canine infectious respiratory disease complex. Vet Clin North Am Small Anim Pract 38, 815–825, viii 10.1016/j.cvsm.2008.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas T., Zhong W. M., Jing Y., Huang P. W., Espinosa S. M., Martinez N., Morrow A. L., Ruiz-Palacios G. M., Pickering L. K., Jiang X. (2004). Genetic diversity among sapoviruses. Arch Virol 149, 1309–1323 10.1007/s00705-004-0296-9 [DOI] [PubMed] [Google Scholar]
- Greninger A. L., Runckel C., Chiu C. Y., Haggerty T., Parsonnet J., Ganem D., DeRisi J. L. (2009). The complete genome of klassevirus - a novel picornavirus in pediatric stool. Virol J 6, 82 10.1186/1743-422X-6-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griot C., Vandevelde M., Schobesberger M., Zurbriggen A. (2003). Canine distemper, a re-emerging morbillivirus with complex neuropathogenic mechanisms. Anim Health Res Rev 4, 1–10 [DOI] [PubMed] [Google Scholar]
- Guo M., Evermann J. F., Saif L. J. (2001). Detection and molecular characterization of cultivable caliciviruses from clinically normal mink and enteric caliciviruses associated with diarrhea in mink. Arch Virol 146, 479–493 10.1007/s007050170157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansman G. S., Oka T., Katayama K., Takeda N. (2007). Human sapoviruses: genetic diversity, recombination, and classification. Rev Med Virol 17, 133–141 10.1002/rmv.533 [DOI] [PubMed] [Google Scholar]
- Holtz L. R., Finkbeiner S. R., Zhao G., Kirkwood C. D., Girones R., Pipas J. M., Wang D. (2009). Klassevirus 1, a previously undescribed member of the family Picornaviridae, is globally widespread. Virol J 6, 86 10.1186/1743-422X-6-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Simmonds P., Gerold G., Qaisar N., Jain K., Henriquez J. A., Firth C., Hirschberg D. L., Rice C. M., et al. (2011). Characterization of a canine homolog of hepatitis C virus. Proc Natl Acad Sci U S A 108, 11608–11613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans M. K., Green K. Y., Ando T., Clarke I. N., Estes M. K., Matson D. O., Nakata S., Neill J. D., Thiel H.-J. (2005). Caliciviridae. In Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses, pp. 843–851 Edited by Fauquet C., Mayo M., Maniloff J., Desselberger U., Ball L. San Diego: Academic Press [Google Scholar]
- Kumar S., Nei M., Dudley J., Tamura K. (2008). mega: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9, 299–306 10.1093/bib/bbn017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackay S. N., Kuang Y., Fu Z. F. (2008). Rabies in small animals. Vet Clin North Am Small Anim Pract 38, 851–861, ix 10.1016/j.cvsm.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Homme Y., Sansregret R., Plante-Fortier E., Lamontagne A. M., Lacroix G., Ouardani M., Deschamps J., Simard G., Simard C. (2009). Genetic diversity of porcine Norovirus and Sapovirus: Canada, 2005–2007. Arch Virol 154, 581–593 10.1007/s00705-009-0344-6 [DOI] [PubMed] [Google Scholar]
- Li L., Victoria J., Kapoor A., Blinkova O., Wang C., Babrzadeh F., Mason C. J., Pandey P., Triki H., et al. (2009). A novel picornavirus associated with gastroenteritis. J Virol 83, 12002–12006 10.1128/JVI.01241-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Victoria J. G., Wang C., Jones M., Fellers G. M., Kunz T. H., Delwart E. (2010). Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol 84, 6955–6965 10.1128/JVI.00501-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V., Decaro N., Elia G., Buonavoglia C. (2005). Surveillance activity for canine parvovirus in Italy. J Vet Med B Infect Dis Vet Public Health 52, 312–315 [DOI] [PubMed] [Google Scholar]
- Martella V., Elia G., Buonavoglia C. (2008a). Canine distemper virus. Vet Clin North Am Small Anim Pract 38, 787–797, vii–viii 10.1016/j.cvsm.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Martella V., Lorusso E., Banyai K., Decaro N., Corrente M., Elia G., Cavalli A., Radogna A., Costantini V., et al. (2008b). Identification of a porcine calicivirus related genetically to human sapoviruses. J Clin Microbiol 46, 1907–1913 10.1128/JCM.00341-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V., Bányai K., Lorusso E., Bellacicco A. L., Decaro N., Mari V., Saif L., Costantini V., De Grazia S., et al. (2008c). Genetic heterogeneity of porcine enteric caliciviruses identified from diarrhoeic piglets. Virus Genes 36, 365–373 10.1007/s11262-008-0198-0 [DOI] [PubMed] [Google Scholar]
- Martella V., Bianchi A., Bertoletti I., Pedrotti L., Gugiatti A., Catella A., Cordioli P., Lucente M. S., Elia G., Buonavoglia C. (2010). Canine distemper epizootic among red foxes, Italy, 2009. Emerg Infect Dis 16, 2007–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Saga Y., Iwai M., Obara M., Horimoto E., Hasegawa S., Kurata T., Okumura H., Nagoshi M., Takizawa T. (2010). Frequent detection of noroviruses and sapoviruses in swine and high genetic diversity of porcine sapovirus in Japan during Fiscal Year 2008. J Clin Microbiol 48, 1215–1222 10.1128/JCM.02130-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls P. K., Stanley M. A. (1999). Canine papillomavirus – a centenary review. J Comp Pathol 120, 219–233 10.1053/jcpa.1998.0278 [DOI] [PubMed] [Google Scholar]
- Okada M., Yamashita Y., Oseto M., Ogawa T., Kaiho I., Shinozaki K. (2006). Genetic variability in the sapovirus capsid protein. Virus Genes 33, 157–161 10.1007/s11262-005-0051-7 [DOI] [PubMed] [Google Scholar]
- Parrish C. R. (1999). Host range relationships and the evolution of canine parvovirus. Vet Microbiol 69, 29–40 10.1016/S0378-1135(99)00084-X [DOI] [PubMed] [Google Scholar]
- Parrish C. R., Kawaoka Y. (2005). The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu Rev Microbiol 59, 553–586 10.1146/annurev.micro.59.030804.121059 [DOI] [PubMed] [Google Scholar]
- Pollock R. V., Coyne M. J. (1993). Canine parvovirus. Vet Clin North Am Small Anim Pract 23, 555–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A. (2006). Genetic evolution of canine coronavirus and recent advances in prophylaxis. Vet Res 37, 191–200 10.1051/vetres:2005053 [DOI] [PubMed] [Google Scholar]
- Renshaw R. W., Zylich N. C., Laverack M. A., Glaser A. L., Dubovi E. J. (2010). Pneumovirus in dogs with acute respiratory disease. Emerg Infect Dis 16, 993–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G., Boldizsár A., Pankovics P. (2009). Complete nucleotide and amino acid sequences and genetic organization of porcine kobuvirus, a member of a new species in the genus Kobuvirus, family Picornaviridae. Arch Virol 154, 101–108 10.1007/s00705-008-0288-2 [DOI] [PubMed] [Google Scholar]
- Reuter G., Boros A., Pankovics P., Egyed L. (2010). Kobuvirus in domestic sheep, Hungary. Emerg Infect Dis 16, 869–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley J. R., Chang K. O., Hayes J., Vinjé J., Saif L. J. (2002). Characterization of an enteropathogenic bovine calicivirus representing a potentially new calicivirus genus. J Virol 76, 10089–10098 10.1128/JVI.76.20.10089-10098.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truyen U. (1999). Emergence and recent evolution of canine parvovirus. Vet Microbiol 69, 47–50 10.1016/S0378-1135(99)00086-3 [DOI] [PubMed] [Google Scholar]
- Tsugawa T., Hoshino Y. (2008). Whole genome sequence and phylogenetic analyses reveal human rotavirus G3P[3] strains Ro1845 and HCR3A are examples of direct virion transmission of canine/feline rotaviruses to humans. Virology 380, 344–353 10.1016/j.virol.2008.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usuku S., Kumazaki M., Kitamura K., Tochikubo O., Noguchi Y. (2008). An outbreak of food-borne gastroenteritis due to sapovirus among junior high school students. Jpn J Infect Dis 61, 438–441 [PubMed] [Google Scholar]
- Victoria J. G., Kapoor A., Li L., Blinkova O., Slikas B., Wang C., Naeem A., Zaidi S., Delwart E. (2009). Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol 83, 4642–4651 10.1128/JVI.02301-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis A. M. (2000). Canine viral infections. Vet Clin North Am Small Anim Pract 30, 1119–1133 [DOI] [PubMed] [Google Scholar]
- Wright N. G. (1973). Recent advances in canine virus research. J Small Anim Pract 14, 241–250 10.1111/j.1748-5827.1973.tb06455.x [DOI] [PubMed] [Google Scholar]
- Yamashita T., Sakae K., Tsuzuki H., Suzuki Y., Ishikawa N., Takeda N., Miyamura T., Yamazaki S. (1998). Complete nucleotide sequence and genetic organization of Aichi virus, a distinct member of the Picornaviridae associated with acute gastroenteritis in humans. J Virol 72, 8408–8412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T., Ito M., Kabashima Y., Tsuzuki H., Fujiura A., Sakae K. (2003). Isolation and characterization of a new species of kobuvirus associated with cattle. J Gen Virol 84, 3069–3077 10.1099/vir.0.19266-0 [DOI] [PubMed] [Google Scholar]
- Yang S., Zhang W., Shen Q., Huang F., Wang Y., Zhu J., Cui L., Yang Z., Hua X. (2009). Molecular characterization and phylogenetic analysis of the complete genome of a porcine sapovirus from Chinese swine. Virol J 6, 216 10.1186/1743-422X-6-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.-M., Jin M., Zhang Q., Li H.-Y., Li D.-D., Xu Z.-Q., Li J.-S., Cui S.-X., Yang S.-H., et al. (2009). Candidate porcine Kobuvirus, China. Emerg Infect Dis 15, 823–825 10.3201/eid1505.081518 [DOI] [PMC free article] [PubMed] [Google Scholar]