Abstract
The presence of West Nile virus (WNV) was first documented in California, USA, during the summer of 2003, and subsequently the virus has become endemic throughout the state. Sequence analysis has demonstrated that the circulating strains are representative of the North American (WN02) genotype that has displaced the East Coast genotype (NY99). A recent study has indicated that enhanced vector competence at elevated temperatures may have played a role in the displacement of the East Coast genotype by WN02. In the current study, four WN02 strains from California, including an initial 2003 isolate (COAV997), were compared to strain NY99 in growth curve assays in mosquito and duck embryonic fibroblast (DEF) cell lines at differing, biologically relevant temperatures to assess the relative temperature sensitivities of these natural isolates. COAV997 was significantly debilitated in viral replication in DEF cells at 44 °C. Full-length sequence comparison of COAV997 against the NY99 reference strain revealed non-synonymous mutations in the envelope glycoprotein (V159A), non-structural protein 1 (NS1) (K110N) and non-structural protein 4A (NS4A) (F92L), as well as two mutations in the 3′ UTR: C→T at nt 10 772 and A→G at nt 10 851. These non-synonymous mutations were introduced into the NY99 viral backbone by site-directed mutagenesis. A mutant containing the NS1-K110N and NS4A-F92L mutations exhibited a debilitated growth phenotype in DEF cells at 44 °C, similar to that of COAV997. One explanation for the subsistence of this genotype is that COAV997 was obtained from an area of California where avian host species might not present elevated temperatures. These data indicate that the NS1 and NS4A mutations identified in some WN02 isolates could reduce thermal stability and impede replication of virus at temperatures observed in febrile avian hosts.
Introduction
West Nile virus (WNV; family Flaviviridae genus Flavivirus) is maintained and amplified in a two-host transmission cycle between Culex spp. mosquitoes and multiple bird species. When replicating in this two-host system, the virus is exposed to very different host environments, including disparate immune systems, host-cell receptors and temperatures, all of which can affect viral replication. The low genetic substitution rates described in flaviviruses, such as WNV, are generally attributed to the strong purifying selective effect of replication in the presence of such disparate factors (Coffey et al., 2008; Jerzak et al., 2008; Scott et al., 1994). Temperature plays an important role in viral replication rates and transmission, affecting the length of the extrinsic incubation, the seasonal phenology of mosquito host populations and geographical variation in human case incidence (Kilpatrick et al., 2008; Kinney et al., 2006; Reisen et al., 2006). West Nile virus is capable of replication across a wide range of temperatures, from as low as 14 °C in poikilothermic mosquitoes (Cornel et al., 1993; Reisen et al., 2006) to 45 °C in febrile avian hosts (Kinney et al., 2006). For transmission to occur, the virus must maintain a eurythermic replicative capacity, and retention of this thermal plasticity may act as an evolutionary constraint.
West Nile virus was first identified in North America in 1999 (Lanciotti et al., 1999), and the founding strain (NY99) was representative of the East Coast genotype (Lanciotti et al., 2002). NY99 is highly virulent for birds in the family Corvidae (Brault et al., 2007; Eidson et al., 2001; Wheeler et al., 2009) and has caused extensive morbidity and mortality in humans and equines (Hayes et al., 2005). Concurrent with the westward expansion of WNV, a new North American genotype (WN02) emerged, and by 2004 had largely displaced the East coast genotype in the United States (Davis et al., 2005). The WN02 genotype has been typified by the presence of a conserved valine-to-alanine mutation in the envelope glycoprotein (E) at amino acid 159 (E-V159A; Davis et al., 2005). The first isolation of WNV in California, USA, was made from a pool of Culex tarsalis collected during July 2003 in Imperial County, a desert biome adjacent to the Mexican border (Reisen et al., 2004). Strains associated with this initial introduction of WNV to California were found to contain the E-V159A mutation and phylogenetically clustered with other WN02 genotype viruses (Davis et al., 2005; Deardorff et al., 2006). Transmission resumed in 2004 with the northward spread of WNV into central and northern California, with a notably high incidence of morbidity and mortality in corvid populations (Hom et al., 2005), such as the yellow-billed magpie (Pica nuttalli) (Ernest et al., 2010).
In an effort to determine what phenotypic advantages facilitated WN02 displacement of the East Coast genotype, several in vitro and in vivo studies have compared isolates of the two genotypes (Ebel et al., 2004; Kilpatrick et al., 2008; Moudy et al., 2007). Kilpatrick et al. (2008) reported that warmer temperature disproportionately increased replication of WN02 virus, compared with NY99, in Culex spp. mosquitoes, thereby allowing WN02 to be transmitted more rapidly after infection, leading to eventual competitive displacement of NY99. The current study addressed the hypothesis that WN02 strains have a greater eurythermic capacity by comparing the replicative capacities of California WN02 strains with the founding North American NY99 genotype strain at temperature extremes in mosquito and avian cell lines. The strains utilized in these experiments vary by locality, year and type of host. Previous studies comparing WN02 and NY99 have been conducted either in vitro or in vivo at a single temperature (Ebel et al., 2004; Moudy et al., 2007), with one in vivo study investigating the effect of temperatures up to 32 °C on WNV infection and transmission by mosquitoes (Kilpatrick et al., 2008). The current study assessed WNV replication over a wider range of biologically relevant temperatures within vertebrate and invertebrate systems to assess the potential effect of host selection for restricting expansion of viruses with high fitness under specific temperature ranges. The Aedes albopictus-derived cell line (C6/36) was maintained at 22, 28 and 34 °C to simulate ambient temperatures during the spring/early summer, summer and late summer/early autumn in California. Duck embryonic fibroblasts (DEF) were maintained at 37, 41 and 44 °C to simulate the low, average and high avian body temperatures observed during terminal infection (Kinney et al., 2006). Mutations associated with altered replicative capacities at different temperatures were implicated by introducing them into an East Coast genotype, NY99, infectious cDNA clone by site-directed mutagenesis. These studies identified genetic modifications that are potentially associated with different replicative capacities at temperature extremes that could have mediated the evolution of WNV as it became established throughout California.
Results
Sequence analysis of the natural isolates
Full-length consensus sequences obtained from the natural isolates COAV997, IMP116, COAV689 and COAV2900 (Table 1) were analysed by comparing them with the NY99 reference genotype strain (Table 2). All four isolates contained non-synonymous mutations at genomic nucleotide position 1442 that associated with the predicted amino acid substitution of E-V159A, as well as the 3′ UTR A/G→G substitution at nt 10 851 (3′UTR-a/g10851g). The full-length sequence obtained from COAV997 revealed additional non-synonymous mutations that resulted in predicted amino acid substitutions in non-structural proteins 1 (NS1) (K110N) and 4A (NS4A) (F92L) (Table 2), as well as a nucleotide substitution in the 3′ UTR [C→T at nt 10 772 (3′UTR-c10772t)]. The IMP116 isolate exhibited predicted amino acid substitutions at NS1-K110N, NS4A-F92L/F and NS5-G845R (Table 2). The full-length sequence of the COAV689 isolate revealed amino acid substitutions at NS1-K110N, NS4A-F92L and NS4A-V135M, as well as nucleotide substitutions in the 3′ UTR [3′UTR-c10772t and T→C at nt 11 009 (3′UTR-t11009c); Table 2], whereas analysis of the COAV2900 isolate demonstrated amino acid substitutions at capsid (C)-V121A, NS4A-A85T, NS4B-N26S, NS5-K314R, NS5-N867D and NS5-D902N, as well as multiple nucleotide substitutions in the 3′ UTR (T→C at nt 10403, C→T at nt 10435, T→C at nt 10459 and G→A at nt 10682; Table 2).
Table 1. Origin and passage history of California natural isolates.
| GenBank accession no. | Strain | Location* | Year | Host | Passage history |
| AF196835 | 382-99 (IC) (NY99-IC) | New York | 1999 | Phoenicopterus chilensis | Vero-1 (BHK-transfection) |
| JF703162 | CA-03 COAV997 (COAV997) | Imperial Valley | 2003 | Cx. tarsalis | Vero-1 |
| DQ080055 | CA-03 IMPR 102 (IMP102) | Imperial Valley | 2003 | Cx. tarsalis | Vero-1 |
| JF703164 | CA-03 IMPR 116 (IMP116) | Imperial Valley | 2003 | Cx. tarsalis | Vero-1 |
| DQ080056 | CA-03 IMPR 1075 (IMP1075) | Imperial Valley | 2003 | Cx. tarsalis | Vero-1 |
| DQ080059 | CA-04 04-7168 (CA-04) | Sacramento County | 2004 | P. nuttalli | Vero-4 |
| JF703161 | CA-04 COAV689 (COAV689) | Coachella Valley | 2004 | Cx. tarsalis | Vero-1 |
| JF703163 | CA-05 COAV2900 (COAV2900) | Coachella Valley | 2005 | Cx. tarsalis | Vero-1 |
All locations are in California, USA, with the exception of New York, USA.
Table 2. Comparison of non-synonymous and non-coding mutations of California natural isolates as compared to strain NY99.
Virus strains in bold were sequenced in the current study, while others were published previously [NY99 strain 382-99 (Lanciotti et al., 1999) and strains IMP102, IMP1075 and CA-04 (Deardorff et al., 2006)]. Italics indicate the nucleotide or amino acid substitutions in the COAV997 strain that were introduced into the NY99 virus backbone by site-directed mutagenesis. Mutations marked with lower case letters indicate a change in nucleotide, upper case letters indicate a change in amino acid.
| Nucleotide position | Amino acid position | Virus strain | |||||||
| NY99 | COAV997 | IMP102 | IMP116 | IMP1075 | CA-04 | COAV689 | COAV2900 | ||
| 19 | 5′ UTR | g | a | ||||||
| 458 | C-121 | V | A | ||||||
| 1442 | E-159 | V | A | A | A | A | A | A | A |
| 2799 | NS1-110 | K | N | N | N | N | N | ||
| 6263 | NS3-551 | V | A | ||||||
| 6721 | NS4A-85 | A | T | ||||||
| 6742 | NS4A-92 | F | L | L/F | L | L | |||
| 6871 | NS4A-135 | V | M | M | |||||
| 6901 | NS4A-145 | S | G | ||||||
| 6992 | NS4B-26 | N | S | ||||||
| 8620–21 | NS5-314 | K | N | R | |||||
| 10 213 | NS5-845 | G | R | ||||||
| 10 279 | NS5-867 | N | D | ||||||
| 10 384 | NS5-902 | D | N | ||||||
| 10 403 | 3′ UTR | t | c | ||||||
| 10 435 | 3′ UTR | c | t | ||||||
| 10 459 | 3′ UTR | t | c | ||||||
| 10 682 | 3′ UTR | g | a | ||||||
| 10 772 | 3′ UTR | c | t | t | t | t | t | ||
| 10 851 | 3′ UTR | a/g | g | g | g | g | g | g | g |
| 10 980 | 3′ UTR | c | a | ||||||
| 11 009 | 3′ UTR | t | c | ||||||
Mosquito cell (C6/36) temperature-specific growth profiles of California WNV strains
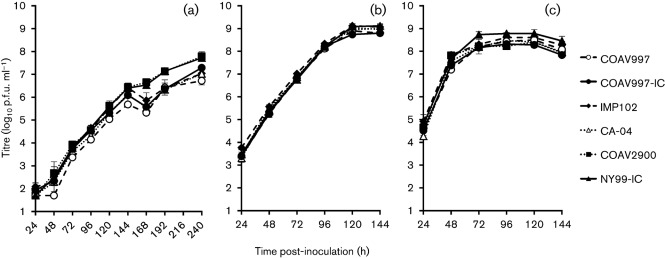
Viral growth was assessed in C6/36 cells at 22, 28 and 34 °C (Fig. 1) for a subset of the California strains (COAV997, IMP102, CA-04 and COAV2900) and infectious-clone-derived NY99 (NY99-IC) viruses (Table 1). COAV997 was the initial isolate of WNV from California and therefore was selected initially to represent the founding genotype. An infectious-clone-derived COAV997 strain (COAV997-IC) was also generated for use as a California reference strain and was utilized for comparisons with the corresponding natural COAV997 isolate. A repeated measures analysis of variance (ANOVA) with viral strains and temperatures as main effects and days after inoculation as the repeated measure indicated that there was no significant difference in the interaction between virus strains and temperature (P = 0.999). The ranges of the overall mean titres at each temperature were as follows: 4.5–5.1 log10 p.f.u. ml−1 at 22 °C, 6.9–7.1 log10 p.f.u. ml−1 at 28 °C and 7.4–7.9 log10 p.f.u. ml−1 at 34 °C.
Fig. 1.
Growth curves of natural California isolates in C6/36 cells at (a) 22, (b) 28 and (c) 34 °C. Error bars represent sd and the limit of detection is 1.7 log10 p.f.u. ml−1.
Avian cell (DEF) temperature-specific growth profiles of California WNV strains
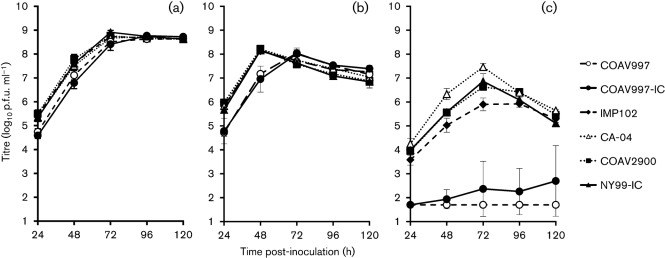
A repeated measures ANOVA was used to compare viral growth profiles of NY99-IC, COAV997, COAV997-IC, IMP102, CA-04 and COAV2900 in DEF cells at 37, 41 and 44 °C (Fig. 2). There was a significant interaction between virus strain and temperature (P<0.001), and most of the variance could be ascribed to temperature and time (P<0.001). Post-hoc analysis of the datasets for each individual temperature revealed that the overall mean titres for COAV997 and COAV997-IC were in a unique group that did not contain IMP102, CA-04, COAV2900 and NY99-IC at 37 °C or 44 °C (Tukey–Kramer multiple comparisons, α = 0.05), with overall mean titres correspondingly ranging from 7.5–7.9 and 1.7–6.0 log10 p.f.u. ml−1 at 37 and 44 °C, respectively (Table 3). No significant differences were found at 41 °C by this pairwise comparison of the overall means (Table 3); however, COAV997 and COAV997-IC peaked on day 3 post-inoculation (p.i.), while the other strains peaked on day 2 p.i. (Fig. 2b). Peak titres at 44 °C occurred on day 3 p.i. for IMP102, CA-04, COAV2900 and NY99-IC. Both COAV997 and COAV997-IC were debilitated at 44 °C, with a detectible virus titre (>1.7 log10 p.f.u. ml−1) being observed in none of the COAV997 replicates and in only one of three replicates for COAV997-IC. The peak titre for that replicate was 4.4 log10 p.f.u. ml−1 on day 5 p.i. As virus was detected in only one flask of the infectious-clone-derived virus and not in the COAV997 natural isolate, plaques were analysed to determine whether any compensatory mutations were generated in this single replicate. Full-length consensus sequencing of an individual plaque showed no additional mutations. Notably, since the COAV997-IC was engineered by site-directed mutagenesis using the two-plasmid system of NY99 as a template, it therefore contained additional non-synonymous mutations, C→T at nt 3880 (NS2A-H118Y) and A→G at nt 4922 (NS3-K104R) (Beasley et al., 2005; Kinney et al., 2006), that were present within the NY99 sequence. These additional non-synonymous mutations may have contributed to the slight replicative capacity of COAV997-IC at 44 °C.
Fig. 2.
Growth curves of natural California isolates in DEF cells at (a) 37, (b) 41 and (c) 44 °C. Error bars represent the sd and the limit of detection is 1.7 log10 p.f.u. ml−1.
Table 3. Summary of overall mean titres in DEF cells at 37, 41 and 44 °C.
Horizontal lines indicate the three groups of data from independent experiments.
| Virus strain | Overall mean virus titre (p.f.u. ml−1) | |||||
| 37 °C* | 41 °C* | 44 °C* | Δ37–41† | Δ41–44† | Δ37–44† | |
| COAV997 | 7.5a | 6.9a | 1.7a | 0.6 | 5.2 | 5.8 |
| COAV997-IC | 7.5a | 6.9a | 2.2a | 0.6 | 4.7 | 5.3 |
| IMP102 | 7.8b | 7.3a | 5.1b | 0.5 | 2.2 | 2.7 |
| CA-04 | 7.8b | 7.2a | 6.0b | 0.6 | 1.2 | 1.8 |
| COAV2900 | 7.9b | 7.2a | 5.6b | 0.7 | 1.6 | 2.3 |
| NY99 | 7.8b | 7.1a | 5.5b | 0.7 | 1.6 | 2.3 |
| COAV997 | 7.7ab | 7.5a | 1.9a | 0.2 | 5.6 | 5.8 |
| COAV997-IC | 7.5a | 7.3a | 2.9b | 0.2 | 4.4 | 4.6 |
| E | 7.6a | 7.5a | 6.0c | 0.1 | 1.5 | 1.6 |
| NS1 | 7.4a | 7.4a | 5.5c | 0 | 1.9 | 1.9 |
| E.NS1 | 7.5a | 7.5a | 5.7c | 0 | 1.8 | 1.8 |
| 3′ UTR | 8.0b | 7.6a | 5.9c | 0.4 | 1.7 | 2.1 |
| NY99 | 7.9b | 7.4a | 6.0c | 0.5 | 1.4 | 1.9 |
| COAV997 | 8.1a | 7.4b | 1.7a | 0.7 | 5.7 | 6.4 |
| COAV997-IC | 8.0a | 7.2ab | 2.6a | 0.8 | 4.6 | 5.4 |
| NS1 | 7.8a | 7.1a | 5.9c | 0.7 | 1.2 | 1.9 |
| NS4A | 8.1a | 7.0a | 5.4bc | 1.1 | 1.6 | 2.7 |
| NS1.NS4A | 7.9a | 7.0a | 1.9a | 0.9 | 5.1 | 6.0 |
| E.NS1.3′UTR | 8.0a | 7.4b | 4.3b | 0.6 | 3.1 | 3.7 |
| NY99 | 8.1a | 7.2ab | 6.3c | 0.9 | 0.9 | 1.8 |
Means followed by the same superscripts (a, b and c) were not significantly different by pairwise analysis (Tukey–Kramer multiple comparisons test, α<0.05).
The difference in overall mean titre between 37–41 °C, 41–44 °C and 37–44 °C, as indicated.
Avian cell (DEF) temperature-specific growth profiles of engineered WNV mutants
Non-synonymous and non-coding mutations observed in COAV997 (relative to the NY99 reference strain) were introduced into the NY99 viral backbone by site-directed mutagenesis in order to assess the phenotypic contribution of specific mutations either individually or in combination. The following mutants were engineered and utilized in growth curve assessments: (i) E-V159A (E), (ii) NS1-K110N (NS1), (iii) NS4A-F92L (NS4A), (iv) 3′ UTR-c10772t (3′UTR), (v) E-V159A with NS1-K110N (E.NS1), (vi) NS1-K110N with NS4A-F92L (NS1.NS4A), and (vii) E-V159A with NS1-K110N and 3′UTR-c10772t (E.NS1.3′UTR).
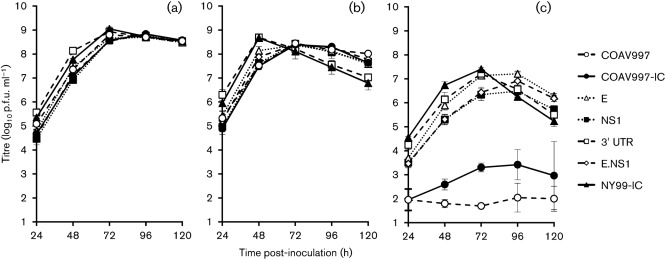
The first of two separate experiments utilized the E, NS1, E.NS1 and 3′UTR mutants with NY99-IC, COAV997 and COAV997-IC as controls (Fig. 3). A repeated measures ANOVA was used to compare growth profiles of these viruses in DEF cells at 37, 41 and 44 °C; a significant interaction between virus strain and temperature was observed, with most of the variance ascribed to temperature and time (P<0.001). Therefore, post-hoc analyses were performed on the data for each individual temperature. Pairwise comparisons of the overall means for each temperature resulted in overlapping groups at 37 °C and no differences at 41 °C, but significant differences were observed at 44 °C, with mean titres ranging from 1.9 to 6.0 log10 p.f.u. ml−1 (Table 3, Tukey–Kramer multiple comparisons, α = 0.05). The peak titre at 44 °C occurred on day 3 p.i. for NY99-IC and the 3′UTR mutant, while the E, NS1 and E.NS1 mutants peaked at day 4 p.i. (Fig. 3c). Detectible titres were found in all three replicates of the COAV997-IC in DEF at 44 °C, and peak titre occurred at day 4 p.i. COAV997 was debilitated in viral replication at 44 °C, with virus detected in one of three replicates on day 1 p.i. (2.0 log10 p.f.u. ml−1); a second replicate demonstrated detectible virus peaking on day 4 p.i. (2.7 log10 p.f.u. ml−1).
Fig. 3.
Growth curves of engineered mutants in DEF at (a) 37, (b) 41 and (c) 44 °C. Error bars represent sd and the limit of detection is 1.7 log10 p.f.u. ml−1.
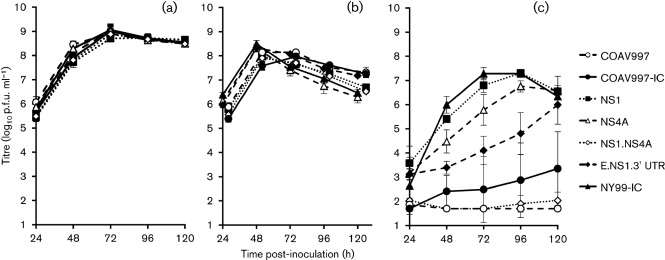
The second experiment utilized the NS1, NS4A, NS1.NS4A and E.NS1.3′UTR mutants with NY99-IC, COAV997 and COAV997-IC as controls (Fig. 4). A repeated measures ANOVA compared growth profiles of these viruses in DEF cells at 37, 41 and 44 °C, and again there was a significant interaction between virus strain and temperature, with most of the variance being ascribed to temperature and time (P<0.001). Therefore, a post-hoc analysis was performed on the data for each temperature. Pairwise comparison of the overall means resulted in no significant differences in DEF at 37 or 41 °C; however, significant differences were observed at 44 °C, with mean titres ranging from 1.7–6.3 log10 p.f.u. ml−1 (Table 3; Tukey–Kramer multiple comparisons, α = 0.05). At 44 °C, the NY99-IC strain elicited a peak viral titre on day 3 p.i. and detectible titre was found in two of the three replicates of the COAV997-IC strain, with the average occurring on day 5 p.i. COAV997 was debilitated in viral replication at 44 °C, with a detectible titre occurring in only one replicate on day 1 p.i. (2.2 log10 p.f.u. ml−1). The NS1 and NS4A individual mutants both peaked on day 4 p.i. at 44 °C, whereas the E.NS1.3′UTR mutant did not peak within 5 days p.i. (Fig. 4c). Detectable titres were found in two of the three replicates of the NS1.NS4A mutant, with peak titres of 2.7 log10 p.f.u. ml−1 on day 1 p.i. in one flask and 2.4 log10 p.f.u. ml−1 on day 5 p.i. in the other. Analysis of the overall mean titres for NS1.NS4A, COAV997 and COAV997-IC revealed that these viruses were not significantly different at 44 °C (Table 3), implicating the NS1-K110N and NS4A-F92L mutations as genetic determinants of the highly temperature sensitive (ts) phenotype observed for the COAV997 strain.
Fig. 4.
Growth curves of additional engineered mutants in DEF at (a) 37, (b) 41 and (c) 44 °C. Error bars represent sd and the limit of detection is 1.7 log10 p.f.u. ml−1 . Data points on Fig. 4b have been slightly offset for presentation purposes.
Comparative Vero cell plaque morphologies
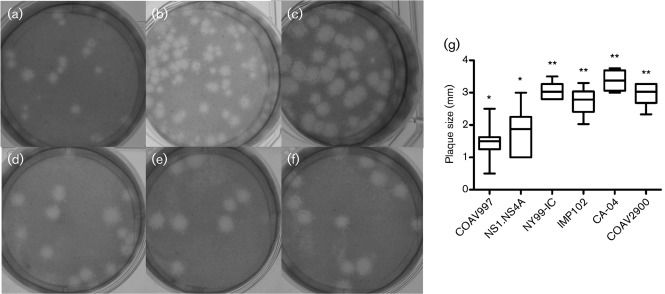
Plaque morphologies were described for a subset of the California strains (COAV997, IMP102, CA-04 and COAV2900), as well as NY99IC and the NS1.NS4A mutant, on Vero cells at 37 and 41 °C (Fig. 5). A comparison could not be made in Vero cells at 44 °C owing to the intolerance of high temperature by these mammalian cells. At 37 °C, the viruses all exhibited similar plaque phenotypes (not shown); however, when compared at 41 °C, differences in plaque size became apparent. The viruses assessed exhibited the following mean plaque sizes±sd: COAV997, 1.5±0.6 mm; IMP102, 2.7±0.4 mm; CA-04, 3.4±0.3 mm; COAV2900, 3.0±0.3 mm; NY99-IC, 3.1±0.3 mm; and NS1.NS4A, 1.8±0.7 mm. The mean plaque size for COAV997 and NS1.NS4A were not significantly different from each other; however, they were significantly different than NY99-IC, IMP-102, CA-04 and COAV2900 (Tukey’s multiple comparison test, P<0.05). These data lend support to the contention that of the NS1-K110N and NS4A-F92L mutations contribute to the ts phenotype of the COAV997 isolate.
Fig. 5.
Plaque phenotypes in Vero cells at 41 °C on day 3 p.i. for comparison of the California strains, NY99-IC and NS1.NS4A mutant: (a) COAV997, (b) NS1.NS4A, (c) NY99-IC, (d) IMP102, (e) CA-04 and (f) COAV2900. (g) Box plot of plaque sizes (mm) with the median value represented as the bar within the box. The 25 % quartiles are represented by the edges of the box, and the error bars span from the highest to the lowest measurements. Datasets labelled with the same number of asterisks (*) were not significantly different from one another as determined by Tukey’s multiple comparisons test (P<0.05).
Discussion
Previous studies have demonstrated a more rapid dissemination of the WN02 strain in Culex spp. mosquitoes held at elevated temperatures when compared with the NY99 North American founding genotype (Kilpatrick et al., 2008), thus suggesting a selective role for temperature in the adaptive landscape of WNV. In the current study, West Coast representatives of the WN02 genotype were compared to NY99 at a range of biologically relevant mosquito and avian temperatures, and, although most strains assayed were tolerant to replication at the extreme, high temperature (44 °C), one strain, COAV997, was found to be non-permissive for replication at this temperature. Comparison of growth profiles for these strains in the C6/36 mosquito cell line at lower temperatures revealed no statistical differences (Fig. 1). COAV997 and NY99-IC were previously assayed in a Cx. tarsalis-derived cell line (CxtarD1), which also resulted in no significant virus versus temperature interaction (Andrade et al., 2010). The COAV997 strain originated from a Cx. tarsalis mosquito pool collected in Imperial County, California, USA, during July 2003. IMP102, the first positive pool recorded in California, was obtained at nearly the same time (July 2003) and in close proximity (within 40 km) of COAV997, yet IMP102 did not show the severe ts effect observed with COAV997, indicating that California was invaded by multiple strains of WNV. Full-length sequence comparison of these two strains revealed that they both contained NS1-K110N and 3′UTR-c10772t, and only differed by two non-synonymous mutations, NS4A-F92L and a C→A change in the 3′ UTR at nt 10 980 (Table 2).
To determine which mutations contributed to the debilitated replicative phenotype in DEF at 44 °C, point mutants were engineered into the NY99 backbone by using site-directed mutagenesis, and growth curves were analysed for the various temperatures. The overall means of E-V159A, NS1-K110N and 3′UTR mutants were not statistically different from the NY99-IC strain at 44 °C, excluding the likelihood of these individual mutations contributing to the observed ts phenotype of COAV997. The double mutant containing both the E-V159A and NS1-K110N mutations similarly failed to demonstrate a significantly debilitated ts phenotype at 44 °C. However, the triple mutant, E.NS1.3′UTR, did exhibit a significant decrease in overall mean titre at 44 °C, as compared with NY99-IC, thus suggesting a cumulative ts effect. It is of interest that the ts phenotype was not observed without the addition of the 3′UTR mutant. Davis et al. examined ts WNV strains isolated in Texas in 2003 and found that their observed ts phenotype required either three 3′ UTR mutations, which included 3′UTR-a/g10851g, or several non-structural amino acid substitutions in concert with the 3′ UTR mutations (Davis et al., 2004, 2007). In the current study, there were no observed phenotypic differences between NY99-IC and E-V159A mutants, E-V159A being the genetic marker for the WN02 genotype; comparisons should be made in vivo to further understand the role of this particular non-synonymous mutation, especially in combination with mutants NS1-K110N and 3′UTR-c10772t.
The mean overall titre of the WNV mutant containing the NS4A-F92L individual point mutant, which is the only predicted amino acid variation between COAV997 and IMP102, was not statistically different from NY99-IC, and this served as experimental evidence that the highly ts phenotype could not be attributed to this single mutation. However, a recombinant WNV containing both NS1-K110N and NS4A-F92L mutations did demonstrate the complete COAV997 ts phenotype at 44 °C, indicating the combined role of NS1-K110N and NS4A-F92L in serving as the genetic determinants mediating this phenotype. This genetic synergism is not completely unexpected as previous trans-complementation studies have demonstrated a direct interaction between NS1 and NS4A proteins for the production of a viable replication complex (Lindenbach & Rice, 1999).
The NS1 protein plays a role in flavivirus RNA replication (Brinton, 2002; Lindenbach & Rice, 1999; MacKenzie et al., 1996; Muylaert et al., 1997), and a study utilizing conditional NS1 yellow fever virus mutants identified a highly ts virus that was deficient in accumulation of both positive- and negative-strand RNA at the non-permissive temperature (Muylaert et al., 1997). The function of the flaviviral NS4A transmembrane protein is not fully understood, but this small, hydrophobic protein (Brinton, 2002) is known to associate with membranes and has been associated with viral replication complexes (Miller et al., 2007) and evasion of the innate immune response of hosts (Lin et al., 2008). Research with related flaviviruses supports the proposition that the NS4A protein may be involved in mediating host innate immune responses by acting as an interferon antagonist and suppressing the phosphorylation of STAT during infection (Lin et al., 2008). The NS4A protein may also act as an NS3 cofactor involved in sustaining helicase activity during periods of ATP depletion (Shiryaev et al., 2009). A direct genetic interaction between NS1 and NS4A, potentially critical for RNA replication, was proposed based on the identification of an NS4A amino acid substitution in a yellow fever virus NS1 deletion mutant that allowed for successful complementation with dengue virus NS1 provided in trans (Lindenbach & Rice, 1999). The temperature sensitivity observed in COAV997 at 44 °C is potentially because such an interaction between NS1-K110N and NS4A-F92L results in instability of the RNA replication complex at elevated temperatures.
Full-length sequences from additional California strains revealed that two ancillary 2003 mosquito-pool isolates (IMP116 and IMP1075; Table 2) contained the same NS1-K110N and NS4A-F92L mutations that were identified in COAV997. This observation, in a limited number of sequences of isolates made following WNV introduction to California, could indicate the prevalence of these two mutations in introduced WNV strains circulating in the Imperial Valley or as being a possible result of a founder effect as the virus was initially introduced into the desert biome, probably from Arizona (Deardorff et al., 2006). An additional California strain (COAV689), containing the NS1-K110N and NS4A-F92L mutations, was identified from the 2004 transmission season, indicating that viruses containing these two mutations persisted overwinter or were reintroduced the following spring. COAV689 contained additional non-synonymous mutations in the NS4A (NS4A-V135M) and the 3′ UTR (3′UTR-t11009c) (Table 2). The NS4A-V135M mutation was also identified in strain IMP1075 from 2003 (Table 2). This additional NS4A mutation may have played a role in stabilizing any in vivo phenotypic effect resulting from the NS1-K110N and NS4A-F92L mutations; however, the validity of this assertion would require additional investigation.
The CA-04 strain was isolated in Northern California where the NS1-K110N and NS4A-F92L mutations have not been identified in any published sequences. The COAV2900 strain isolated in the Coachella Valley during 2005 does not contain the NS1-K110N and NS4A-F92L mutations; however, this strain has extensive non-synonymous and non-coding mutations compared with NY99, as well as sequence commonalities with isolates from Colorado and Arizona from previous transmission seasons, thus suggesting a possible common progenitor for these strains. The NS4A-A85T amino acid substitution (present in the COAV2900 virus described here) was also found in strains from Colorado in 2003 (CO2003-2) and Arizona in 2004 (AZ2004) (Davis et al., 2005). AZ2004 has an additional amino acid substitution in common with COAV2900 at NS5-K314R (Davis et al., 2005).
Previous studies reported WNV strains exhibiting ts phenotypes in avian cells at extreme temperatures that were associated with debilitated viral titres in vivo in either avian or mosquito hosts (Jia et al., 2007; Kinney et al., 2006). Characterization of a small-plaque variant of a New York strain WNV isolated in 2000 that contained non-synonymous mutations in the membrane glycoprotein (prM) and NS2A proteins similarly showed no significant differences when assessed at different temperatures (26, 30 and 34 °C) in C6/36; however, the small plaque variant exhibited significantly lower titres in Cx. pipiens when maintained at 15, 28 and 34 °C (Jia et al., 2007). In PDE cells (synonymous with DEF cells) maintained at various temperatures, the small-plaque variant did not produce detectable virus at the high temperature (42.5 °C), as compared with the large-plaque variant (Jia et al., 2007). Kinney et al. previously reported that a Kenyan WNV strain with reduced avian virulence potential was also highly ts in DEF at 44 °C, a temperature observed in WNV-infected viraemic American crows (Kinney et al., 2006). Interestingly, although only 11 non-synonymous mutations were observed between the Kenyan and NY99 viruses, there was both an amino acid change in NS1 as well as an NS4A-A85V substitution, which is similar to the COAV2900 NS4A-A85T mutation, and only 7 aa upstream of the critical NS4A-92 residue identified as being associated with retarded high temperature replication of the COAV997 strain. The highly ts phenotype observed at 44 °C with the COAV997 strain and NS1.NS4A mutant may be associated with a similar low-level avian virulence phenotype or decreased replication in mosquitoes. Investigation of COAV997 and the NS1.NS4A mutant in vivo is warranted in order to further elucidate the effect of naturally occurring WNV ts strains on replication in mosquito vectors as well as in avian hosts. Unlike mosquito studies that have demonstrated increased dissemination of the WN02 genotype at elevated temperatures (Ebel et al., 2004; Kilpatrick et al., 2008), the results presented herein have failed to identify a temperature-versus-strain interaction for in vitro C6/36 growth. One possible factor that could dictate this difference is the lack of a functional RNAi system in C6/36 cells (Brackney et al., 2010). This fact, coupled with a potential association of temperature in mediating RNAi responses (Szittya et al., 2003), could indicate that temperature and RNA interference pathways might be coupled and that differential antagonistic capacities of WNV strains could be critical for some of the observed in vivo differences that are mediated at elevated temperatures. Similar mosquito competence studies will need to be performed to assess this theory further. However, results from this study have allowed the independent assessment of temperature as a direct factor modulating growth of different strains of WNV. Interestingly, corvids are rare in south-eastern California and the primary maintenance and amplification hosts, house finches and house sparrows (Reisen et al., 2008), did not produce elevated temperatures, even following infection with NY99 virus (G. Worwa, unpublished data). The proliferation of viruses carrying the ts mutations, NS1-K110N and NS4A-F92L, could have been because of the fact that circulation in areas essentially devoid of corvids removed a selective force present in alternative landscapes where other passeriforms serve as important amplifying hosts. As previously mentioned, WNV isolates from Northern California, with robust corvid populations, have not been observed to express either ts mutation identified in this study.
Methods
Cells and virus strains.
DEF (ATCC accession no. CCL-141) and Ae. albopictus-derived mosquito cell (C6/36) cultures were used for temperature-dependent assays. Viral titres were determined with African green monkey kidney cells (Vero cells; ATCC no. CCL-81). Baby hamster kidney cells (BHK-21, clone 13; ATCC no. CCL-10) were utilized during transfection protocols for rescuing infectious-clone-derived viruses. Media used to maintain cell lines: Eagle’s minimum essential medium (ATCC) for DEF cells, Schneider’s Drosophila medium (Gibco) for C6/36 cells, Dulbecco’s minimal essential medium with low glucose (Gibco) for Vero cells and minimal essential medium (Gibco) for BHK cells. All media were routinely supplemented with 10 % FBS and penicillin/streptomycin (100 U ml−1; Gibco).
An infectious clone-derived virus based on NY99 strain 382-99 was generated as previously described (Beasley et al., 2005; Kinney et al., 2006). The COAV997 and CA-03 IMPR 102 (IMP102) isolates were made from Cx. tarsalis mosquito pools collected in 2003 from the Imperial Valley. CA-04 04-7168 (CA-04) was isolated from a yellow-billed magpie (P. nuttalli) that died in Sacramento County in 2004. The COAV2900 isolate was generated from a Cx. tarsalis mosquito pool made in 2005 having been collected from the Coachella Valley. All California isolates were made in Vero cell culture (Table 1).
Full-length sequencing of natural isolates.
Viral RNA was obtained from isolates after passage in Vero cells with a QIAamp Viral RNA Mini kit (Qiagen), following the manufacturer’s protocols. Primers were designed based on North American genomic sequences. SuperScript II Reverse Transcriptase (Invitrogen) and PfuUltra (Stratagene) were used to generate cDNA and PCR amplicons, respectively. To amplify the 5′ and 3′ ends of the genome, 5′ RACE and 3′ RACE Systems (Invitrogen) were used. The poly(A) tail necessary for the 3′ RACE amplification was engineered with a Poly-A Plus kit (Epicentre), following the manufacturer’s protocol. A Qiaquick gel extraction kit (Qiagen) was used to clean RT-PCR products, and sequencing was performed on an ABI capillary sequencer.
Construction and rescue of clone-derived infectious viruses.
A two-plasmid system based on NY99 (Kinney et al., 2006) was utilized to engineer an infectious-clone-derived version of the COAV997 natural isolate (COAV997-IC) containing E-V159A, NS1-K110N, NS4A-F92L and 3′UTR-c10772t mutations. Additionally, mutant viruses were engineered to insert individual or combinations of non-synonymous COAV997 point mutants into the NY99 virus backbone. The following clone-derived viruses containing individual or combinations of COAV997 mutations were engineered: (i) E-V159A, (ii) NS1-K110N, (iii) NS4A-F92L, (iv) 3′UTR-c10772t, (v) E-V159A and NS1-K110N, (vi) NS1-K110N and NS4A-F92L, and (vii) E-V159A, NS1-K110N and 3′UTR-c10772t. Of note, the NY99-IC was engineered with 3′UTR-a/g10851g and therefore all viruses derived from this strain contain this nucleotide configuration. Briefly, NY99-plasmid was mutagenized by using a QuikChange site-directed mutagenesis kit (Stratagene), and the presence of the desired mutations was confirmed by direct sequencing. Specific mutagenesis primers were designed to insert each individual point mutation on either the 5′ (5′UTR-NS1) or 3′ (NS1-3′UTR) plasmids by using wrap-around PCR amplification. Mutants containing more than one point mutant were engineered by using multiple rounds of mutagenesis, with altered genome sequences being confirmed between rounds. The 5′ and 3′ plasmids were digested and ligated to serve as template for RNA transcription. The viral RNA generated was transfected into BHK-21 cells (Kinney et al., 2006). Cells were observed daily for cytopathic effect, as compared with a negative control. Virus was harvested 3–5 days p.i. when 2+ cytopathogenicity was observed. Harvested supernatants were titrated on Vero cells and full-length sequencing performed from amplicons generated to assess the sequence identity of all derived viruses.
Temperature-dependent replication.
C6/36 and DEF cultures were grown in 25 cm2 flasks each inoculated in triplicate at an m.o.i. of 0.1. Initial inocula were later back-titrated to confirm m.o.i. Viruses were adsorbed for 90 min at 28 °C (C6/36) or 37 °C (DEF) in a 5 % CO2 atmosphere. After the adsorption period, the monolayer was washed twice with PBS (pH 7.4), 6 ml of maintenance medium containing 10 % FBS was added and the cultures were incubated at the selected temperatures. Inoculated cultures of C6/36 were incubated at 22, 28 or 34 °C without CO2, while DEF cultures were incubated at 37, 41 and 44 °C in a 5 % CO2 atmosphere. Samples were collected at 24 h intervals, diluted 1 : 10 in medium supplemented with 20 % FBS, and stored at –80 °C until being assayed for infectious titre.
Viral titration.
Infectious viral titres were quantified by serial tenfold dilutions of virus samples on Vero cells as previously described (Brault et al., 2004). Briefly, virus dilutions were allowed to adsorb for 1 h at 37 °C before the addition of a nutrient overlay containing 0.5 % agarose. A second overlay containing 0.005 % neutral red was added at day 2 p.i. P.f.u. were enumerated at day 3 p.i. and used to calculate the titre (log10 p.f.u. ml−1) of the sample. In order to document plaque morphology and size at the experimental temperatures, the viral titration protocol was performed as described above, except that after application of the overlay the cells were maintained at 37 °C or 41 °C (Vero cells did not tolerate temperatures >41 °C). Digital images and measurements were made on day 3 p.i. Vero cells were used as a neutral cell line for plaque morphology comparisons owing, in part, to poor plaque resolution in DEF cells, especially at 44 °C where non-virus-induced cytopathic effect has been observed. Additionally, plaque morphology measurements were taken at 41 °C in order to minimize the effects of agar overlay viscosity that have been observed at 44 °C (unpublished results).
Statistical analysis.
A repeated measures ANOVA (Hintze, 1998) was utilized to determine statistical significance between viral strains and temperatures, with days p.i. as the within repeated flask factor. If a significant virus strain versus temperature interaction was observed, then the repeated measures ANOVA was repeated with data specific for each temperature. Post-hoc analysis was performed by using the Tukey–Kramer test for all pairwise multiple comparisons between the means with α = 0.05.
Plaque measurements were analysed by using the Graph Pad Prism (version 5) software. A one-way ANOVA was used to determine whether there was statistically significant variation in the plaque size of the viruses. If a significant difference was observed, then a Tukey’s multiple comparisons test was used as a post-hoc analysis with P<0.05.
Acknowledgements
We thank Kelly Anderson and Pooja Maharaj for technical assistance. C. Andrade was supported by National Institutes of Health (NIH) training grant T32 AI074550. W. K. Reisen was supported, in part, by the Research and Policy in Infectious Disease Dynamics (RAPIDD) Program of the Science and Technology Directorate, Department of Homeland Security and Fogarty International Center, NIH. This research was funded, in part, by grants from the National Institutes of Allergy and Infectious Diseases, RO1-AI55607, using American Recovery and Reinvestment Act Support as well as funding from the Pacific South-west Regional Center for Excellence U54 AI065359 and Centers for Disease Control and Prevention CI000235.
References
- Andrade C. C., Maharaj P. D., Reisen W. K., Brault A. C. (2010). Effect of temperature on West Nile virus replication in different host cell types: potential for altered transmission cycles in California. Proc Annu Conf Mosq Vector Control Assoc Calif 78, 12–15 [Google Scholar]
- Beasley D. W., Whiteman M. C., Zhang S., Huang C. Y., Schneider B. S., Smith D. R., Gromowski G. D., Higgs S., Kinney R. M., Barrett A. D. (2005). Envelope protein glycosylation status influences mouse neuroinvasion phenotype of genetic lineage 1 West Nile virus strains. J Virol 79, 8339–8347 10.1128/JVI.79.13.8339-8347.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackney D. E., Scott J. C., Sagawa F., Woodward J. E., Miller N. A., Schilkey F. D., Mudge J., Wilusz J., Olson K. E., et al. (2010). C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl Trop Dis 4, e856 10.1371/journal.pntd.0000856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault A. C., Langevin S. A., Bowen R. A., Panella N. A., Biggerstaff B. J., Miller B. R., Komar N. (2004). Differential virulence of West Nile strains for American crows. Emerg Infect Dis 10, 2161–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault A. C., Huang C. Y., Langevin S. A., Kinney R. M., Bowen R. A., Ramey W. N., Panella N. A., Holmes E. C., Powers A. M., Miller B. R. (2007). A single positively selected West Nile viral mutation confers increased virogenesis in American crows. Nat Genet 39, 1162–1166 10.1038/ng2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinton M. A. (2002). The molecular biology of West Nile Virus: a new invader of the western hemisphere. Annu Rev Microbiol 56, 371–402 10.1146/annurev.micro.56.012302.160654 [DOI] [PubMed] [Google Scholar]
- Coffey L. L., Vasilakis N., Brault A. C., Powers A. M., Tripet F., Weaver S. C. (2008). Arbovirus evolution in vivo is constrained by host alternation. Proc Natl Acad Sci U S A 105, 6970–6975 10.1073/pnas.0712130105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornel A. J., Jupp P. G., Blackburn N. K. (1993). Environmental temperature on the vector competence of Culex univittatus (Diptera: Culicidae) for West Nile virus. J Med Entomol 30, 449–456 [DOI] [PubMed] [Google Scholar]
- Davis C. T., Beasley D. W., Guzman H., Siirin M., Parsons R. E., Tesh R. B., Barrett A. D. (2004). Emergence of attenuated West Nile virus variants in Texas, 2003. Virology 330, 342–350 10.1016/j.virol.2004.09.016 [DOI] [PubMed] [Google Scholar]
- Davis C. T., Ebel G. D., Lanciotti R. S., Brault A. C., Guzman H., Siirin M., Lambert A., Parsons R. E., Beasley D. W., Novak R. (2005). Phylogenetic analysis of North American West Nile virus isolates, 2001–2004: evidence for the emergence of a dominant genotype. Virology 342, 252–265 10.1016/j.virol.2005.07.022 [DOI] [PubMed] [Google Scholar]
- Davis C. T., Galbraith S. E., Zhang S., Whiteman M. C., Li L., Kinney R. M., Barrett A. D. (2007). A combination of naturally occurring mutations in North American West Nile virus nonstructural protein genes and in the 3′ untranslated region alters virus phenotype. J Virol 81, 6111–6116 10.1128/JVI.02387-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff E., Estrada-Franco J., Brault A. C., Navarro-Lopez R., Campomanes-Cortes A., Paz-Ramirez P., Solis-Hernandez M., Ramey W. N., Davis C. T., et al. (2006). Introductions of West Nile virus strains to Mexico. Emerg Infect Dis 12, 314–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel G. D., Carricaburu J., Young D., Bernard K. A., Kramer L. D. (2004). Genetic and phenotypic variation of West Nile virus in New York, 2000–2003. Am J Trop Med Hyg 71, 493–500 [PubMed] [Google Scholar]
- Eidson M., Komar N., Sorhage F., Nelson R., Talbot T., Mostashari F., McLean R., West Nile Virus Avian Mortality Surveillance Group (2001). Crow deaths as a sentinel surveillance system for West Nile virus in the northeastern United States, 1999. Emerg Infect Dis 7, 615–620 10.3201/eid0704.010402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernest H. B., Woods L. W., Hoar B. R. (2010). Pathology associated with West Nile virus infections in the yellow-billed magpie (Pica nuttalli): a California endemic bird. J Wildl Dis 46, 401–408 [DOI] [PubMed] [Google Scholar]
- Hayes E. B., Sejvar J. J., Zaki S. R., Lanciotti R. S., Bode A. V., Campbell G. L. (2005). Virology, pathology, and clinical manifestations of West Nile virus disease. Emerg Infect Dis 11, 1174–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintze, J. (1998). NCSS Statistical Software.
- Hom A., Marcus L., Kramer V. L., Cahoon B., Glaser C., Cossen C., Baylis E., Jean C., Tu E., et al. (2005). Surveillance for mosquito-borne encephalitis virus activity and human disease, including West Nile virus, in California, 2004. Proc Pap Annu Conf Calif Mosq Control Assoc 73, 66–77 [Google Scholar]
- Jerzak G. V., Brown I., Shi P. Y., Kramer L. D., Ebel G. D. (2008). Genetic diversity and purifying selection in West Nile virus populations are maintained during host switching. Virology 374, 256–260 10.1016/j.virol.2008.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Moudy R. M., Dupuis A. P., II, Ngo K. A., Maffei J. G., Jerzak G. V., Franke M. A., Kauffman E. B., Kramer L. D. (2007). Characterization of a small plaque variant of West Nile virus isolated in New York in 2000. Virology 367, 339–347 10.1016/j.virol.2007.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick A. M., Meola M. A., Moudy R. M., Kramer L. D. (2008). Temperature, viral genetics, and the transmission of West Nile virus by Culex pipiens mosquitoes. PLoS Pathog 4, e1000092 10.1371/journal.ppat.1000092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney R. M., Huang C. Y., Whiteman M. C., Bowen R. A., Langevin S. A., Miller B. R., Brault A. C. (2006). Avian virulence and thermostable replication of the North American strain of West Nile virus. J Gen Virol 87, 3611–3622 10.1099/vir.0.82299-0 [DOI] [PubMed] [Google Scholar]
- Lanciotti R. S., Roehrig J. T., Deubel V., Smith J., Parker M., Steele K., Crise B., Volpe K. E., Crabtree M. B., et al. (1999). Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286, 2333–2337 10.1126/science.286.5448.2333 [DOI] [PubMed] [Google Scholar]
- Lanciotti R. S., Ebel G. D., Deubel V., Kerst A. J., Murri S., Meyer R., Bowen M., McKinney N., Morrill W. E., et al. (2002). Complete genome sequences and phylogenetic analysis of West Nile virus strains isolated from the United States, Europe, and the Middle East. Virology 298, 96–105 10.1006/viro.2002.1449 [DOI] [PubMed] [Google Scholar]
- Lin C. W., Cheng C. W., Yang T. C., Li S. W., Cheng M. H., Wan L., Lin Y. J., Lai C. H., Lin W. Y., Kao M. C. (2008). Interferon antagonist function of Japanese encephalitis virus NS4A and its interaction with DEAD-box RNA helicase DDX42. Virus Res 137, 49–55 10.1016/j.virusres.2008.05.015 [DOI] [PubMed] [Google Scholar]
- Lindenbach B. D., Rice C. M. (1999). Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol 73, 4611–4621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie J. M., Jones M. K., Young P. R. (1996). Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 220, 232–240 10.1006/viro.1996.0307 [DOI] [PubMed] [Google Scholar]
- Miller S., Kastner S., Krijnse-Locker J., Bühler S., Bartenschlager R. (2007). The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J Biol Chem 282, 8873–8882 10.1074/jbc.M609919200 [DOI] [PubMed] [Google Scholar]
- Moudy R. M., Meola M. A., Morin L. L., Ebel G. D., Kramer L. D. (2007). A newly emergent genotype of West Nile virus is transmitted earlier and more efficiently by Culex mosquitoes. Am J Trop Med Hyg 77, 365–370 [PubMed] [Google Scholar]
- Muylaert I. R., Galler R., Rice C. M. (1997). Genetic analysis of the yellow fever virus NS1 protein: identification of a temperature-sensitive mutation which blocks RNA accumulation. J Virol 71, 291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisen W., Lothrop H., Chiles R., Madon M., Cossen C., Woods L., Husted S., Kramer V., Edman J. (2004). West Nile virus in California. Emerg Infect Dis 10, 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisen W. K., Fang Y., Martinez V. M. (2006). Effects of temperature on the transmission of West Nile virus by Culex tarsalis (Diptera: Culicidae). J Med Entomol 43, 309–317 10.1603/0022-2585(2006)043[0309:EOTOTT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Reisen W. K., Lothrop H. D., Wheeler S. S., Kennsington M., Gutierrez A., Fang Y., Garcia S., Lothrop B. (2008). Persistent West Nile virus transmission and the apparent displacement St. Louis encephalitis virus in southeastern California, 2003–2006. J Med Entomol 45, 494–508 10.1603/0022-2585(2008)45[494:PWNVTA]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott T. W., Weaver S. C., Mallampalli V. L. (1994). Evolution of mosquito-borne viruses. In The Evolutionary Biology of Viruses, pp. 293–324 Edited by Morse S. S. New York: Raven Press [Google Scholar]
- Shiryaev S. A., Chernov A. V., Aleshin A. E., Shiryaeva T. N., Strongin A. Y. (2009). NS4A regulates the ATPase activity of the NS3 helicase: a novel cofactor role of the non-structural protein NS4A from West Nile virus. J Gen Virol 90, 2081–2085 10.1099/vir.0.012864-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya G., Silhavy D., Molnár A., Havelda Z., Lovas A., Lakatos L., Bánfalvi Z., Burgyán J. (2003). Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J 22, 633–640 10.1093/emboj/cdg74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler S. S., Barker C. M., Fang Y., Armijos M. V., Carroll B. D., Husted S., Johnson W. O., Reisen W. K. (2009). Differential impact of West Nile virus on California birds. Condor 111, 1–20 10.1525/cond.2009.080013 [DOI] [PMC free article] [PubMed] [Google Scholar]