Abstract
When species hybridize, offspring typically exhibit reduced fitness and maladapted phenotypes. This situation has biosafety implications regarding the unintended spread of novel transgenes, and risk assessments of crop-wild hybrids often assume that poorly adapted hybrid progeny will not evolve adaptive phenotypes. We explored the evolutionary potential of early generation hybrids using nontransgenic wild and cultivated radish (Raphanus raphanistrum, Raphanus sativus) as a model system. We imposed four generations of selection for two weedy traits – early flowering or large size – and measured responses in a common garden in Michigan, USA. Under selection for early flowering, hybrids evolved to flower as early as wild lineages, which changed little. These early-flowering hybrids also recovered wild-type pollen fertility, suggesting a genetic correlation that could accelerate the loss of crop traits when a short life cycle is advantageous. Under selection for large size at reproduction, hybrids evolved longer leaves faster than wild lineages, a potentially advantageous phenotype under longer growing seasons. Although early generation hybrid offspring have reduced fitness, our findings provide novel support for rapid adaptation in crop-wild hybrid populations. Biosafety risk assessment programs should consider the possibility of rapid evolution of weedy traits from early generations of seemingly unfit crop-wild hybrids.
Keywords: artificial selection, contemporary evolution, correlated evolution, crop-to-wild gene flow, extreme phenotypes, flowering phenology, introgression, plant size
The development and wide-scale adoption of genetically engineered crops have caused concern over the possibility for engineered traits (or transgenes) to ‘escape’ cultivation via hybridization with populations of wild relatives (Colwell et al. 1985; Ellstrand et al. 1999). Transgenes from domesticated plants that enhance yield or confer resistance to herbicides, disease, or insect pests may alter the survival and fecundity of noncultivated species, potentially making existing weeds more difficult to control or enhancing the weediness of species that are not currently problematic (Snow et al. 2003; Perez-Jones et al. 2006). In some cases, resulting economic and environmental damage could overshadow any benefits achieved through transgenic crop breeding (Wolfenbarger and Phifer 2000; Smyth et al. 2002). Consequently, it is helpful to understand the potential for crop-wild hybrid weeds to evolve and adapt to different ecological conditions before those crops are genetically engineered.
Although many crops and related weed taxa co-occur and hybridize (Ellstrand et al. 1999; Hails and Morley 2005), crop-to-wild gene flow does not necessarily result in persistent introgression. Whether the introduced alleles persist in the long-term may be governed by the nature of the introduced trait, the genetic and environmental background in which it is expressed, and levels of local selection, all of which influence the fitness of early generation hybrid offspring (Rissler and Mellon 1996; Arnold 1997; Piálek and Barton 1997; Whitney et al. 2006; Wu and Campbell 2006). In general, when most hybrid offspring have reduced fertility and maladaptive phenotypes for local environments, the risk of persistent introgression is assumed to be low (see Arnold et al. 1999 for a discussion of this assumption). Specifically, reduced pollen fertility in hybrid plants may be due to either genetic incompatibilities among the parents or the presence of chromosome structural heterozygosity leading to unbalanced meiotic products and the production of nonviable gametes (Heiser 1947; Bouck 2004; Arnold 2006). However, despite extremely low fertility and viability in early generation hybrids, extensive gene flow and the establishment of new evolutionary lineages have been repeatedly documented in natural systems via allopolyploidy (e.g., Otto and Whitton 2000) and, less frequently, in homoploid hybrid lineages in both plants (Hegarty and Hiscock 2005) and animals (Mallet 2007).
In general, evidence for new lineages has been inferred from the geographic occurrence of hybrid zones. The use of experimental hybridization and artificial selection to speed up natural processes allows us to explore ecological scenarios under which the fitness of hybrids lineages may equal or exceed the fitness of their wild parent, favoring the long-term persistence of crop alleles. However, this combination of evolutionary tools has not been used before in crop-wild hybrids. Furthermore, it is important to determine whether introgression can occur under a diversity of selective environments, some of which may eliminate many, if not most, maladaptive crop-derived traits (i.e., when selective filters are strong). Populations of crop-wild hybrids may generally experience purifying selection towards the wild phenotype (Hauser et al. 1998; Snow and Campbell 2005) and yet occasionally possess neutral or advantageous crop-derived traits, such as resistance to certain diseases or herbicides (e.g., Snow et al. 2003; Warren and James 2006; Baack et al. 2008; Warwick et al. 2008).
Rapid evolution of plants in natural populations in response to both anthropogenic and natural environmental disturbances has been documented repeatedly (reviewed in Thompson 1998; Bone and Farres 2001; Reznick and Ghalambor 2001; Hairston et al. 2005). Although rapid evolution via natural selection is well understood, we have a poor understanding of how often hybridization facilitates rapid evolution (Lewontin and Birch 1966; Rieseberg 1991; Ellstrand et al. 1999; Jarvis and Hodgkin 1999). Early generation hybrids often exhibit significant reductions in pollen fertility and seed viability (Dobzhansky 1937, p. 231; Arnold and Hodges 1995; Arnold 1997), perhaps due to disruption of co-adapted gene complexes (Dobzhansky 1937; Mayr 1963), chromosomal rearrangements (Panetsos and Baker 1967), or the introduction of maladapted genes (Waser and Price 1991; Arnold and Hodges 1995). Thus, interspecific hybridization has the potential to be an evolutionary dead-end.
Nevertheless, hybridization is increasingly recognized as a creative evolutionary force that sometimes produces ‘preadapted’ and highly novel genotypes and morphologies (Anderson and Stebbins 1954; Lewontin and Birch 1966; Rieseberg 1995; Arnold 1997). Furthermore, there is evidence that hybridization plays a significant role in the evolution of some weedy or invasive species (Ellstrand and Schierenbeck 2000; Arnold 2004). Ellstrand and Schierenbeck (2000) documented 28 examples of weedy, hybrid-derived taxa and suggested that their invasiveness may be a direct result of the effects of hybridization in many cases. However, in a global analysis, Whitney et al. (2008) found that vascular plant families with a higher propensity for hybridization were not more likely to produce more naturalized, weedy, or invasive species than families less prone to hybridization, suggesting that the link between hybridization and invasiveness is species-specific (e.g., Greenwood et al. 2004; Facon et al. 2005; Campbell et al. 2006; Whitney et al. 2006).
The rate of evolution after hybridization may depend on the degree of evolutionary divergence of the parental taxa, especially when considering hybridization among crops and their wild relatives. When a crop is recently derived from a wild relative (i.e., a wild progenitor), the crop should contain a subset of the alleles present in the wild species (e.g., sea beets and cultivated beets, Bartsch et al. 1999; wild and cultivated rice, Zhu et al. 2007) and hybridization is unlikely to dramatically alter the potential for phenotypic evolution in the wild relative. Alternatively, if the crop is distantly related to a sexually compatible wild species or the wild progenitor and crop lineages have evolved independently for many generations, the crop may contain unique traits that contribute to phenotypic evolution in weedy relatives via hybridization.
To measure rates of evolution in crop-wild hybrids and nonhybrid wild populations, we used the model ecological system of Raphanus raphanistrum (jointed charlock or wild radish) and its domesticated relative, Raphanus sativus (cultivated radish). Radish is an ancient crop that appears to have multiple origins from several wild species, including R. raphanistrum (Ellstrand and Marshall 1985; Crisp 1995; Yamagishi and Terachi 2003). Allozyme studies revealed that cultivated radishes have retained nearly as much genetic variation as R. raphanistrum and most of this variation is found within, rather than among, cultivars, similar to the wild relative (Ellstrand and Marshall 1985). In California, USA, a fertile, invasive hybrid has evolved and has supplanted R. raphanistrum within the region (Hegde et al. 2006). Known as wild R. sativus (or California wild radish), this hybrid has characteristics of both parental species – its flowering phenology and leaf length are intermediate to wild and crop radish (Hegde et al. 2006). Ridley et al. (2008) recently discovered that bi-directional hybridization among multiple cultivars and multiple European wild radish populations contributed to the diversity of cpDNA haplotypes within California. Essentially, we have attempted to recreate a simplified version of this invasive hybrid lineage by hybridizing a single radish cultivar with a single wild radish population.
An ideal weed has broad environmental tolerances for germination and seed production, rapid growth and early flowering, and continuous and high seed production in favorable environmental circumstances (Baker 1965). Although crop breeders often impose selection for these same traits (e.g., Chloupek and Hrstkova 2005), domestication has often resulted in crop life-history strategies that differ from those of their wild relatives (e.g., perennial teosinte versus annual maize, Doebley 1992; Doebley et al. 1997; determinate growth and a single capitulum versus indeterminate growth and multiple capitula in annual sunflowers, Fick and Miller 1997). When F1 hybrid phenotypes are intermediate to their parental taxa, early-generation hybrid phenotypes may be maladapted to parental environments (Arnold and Hodges 1995; but see Rieseberg et al. 1999), but hybrids could respond to selection and may ultimately produce novel and advantageous phenotypes.
Here, we used artificial selection to measure the rate of evolution of crop-wild hybrids compared to their wild relative under simulated ecological contexts that favored two traits found in a hybrid-derived weed: early flowering, a wild-type trait, or long leaf length, a crop-derived trait that is correlated with plant size and lifetime fecundity. Further, we monitored the correlated responses of hybrid pollen fertility and flower petal color, a simply inherited crop-derived trait, to selection for earlier flowering or longer leaves. We expected trait evolution to occur more rapidly in hybrid lineages than in wild ones because segregating hybrid populations contain more genetic diversity and may even generate extreme phenotypes relative to the parental taxa. The manipulative approach taken here affords us the opportunity to make strong causal inferences and provide unique perspectives on the potential role of hybridization in adaptive evolution.
Materials and methods
Study organism
Raphanus raphanistrum, known as wild radish or jointed charlock, is an economically important, annual weed that flowers early in the growing season (Warwick and Francis 2005; Campbell and Snow 2007), whereas cultivated radish (R. sativus) has been bred for delayed flowering to favor the production of an economically valuable, enlarged hypocotyl or root (Curtis 2003). The F1 hybrid of these taxa has an intermediate flowering phenology (Snow et al. 2001; Campbell 2007). Early flowering can be selectively advantageous for both wild and hybrid-derived radishes because plants can complete their life cycle before being out-competed or killed by frost, drought, herbivores, or crop harvest, even when germination is induced later in the growing season (Snow et al. 2001; Campbell and Snow 2007). Long leaf length and large size are favored for both wild and hybrid-derived radishes because larger plants tend to produce more flowers and ultimately more seeds (Campbell and Snow 2007). Radishes produce a basal rosette of leaves prior to flowering, and the length of the longest leaf is correlated with lifetime fecundity (Campbell 2007; Campbell and Snow 2007). Furthermore, a positive genetic correlation between age at flowering and leaf length was found in California wild radish populations (Mazer and Schick 1991a,b). Longer leaves were correlated with later flowering in these hybrid radishes.
Although R. sativus and R. raphanistrum are considered inter-fertile, early generation hybrids have reduced pollen fertility, commonly producing approximately 50–60% aborted pollen grains (Campbell et al. 2006). Reduced pollen fertility in Raphanus hybrids is due to heterozygosity for a reciprocal translocation that affects chromosome pairing during meiosis (Panetsos and Baker 1967). Yet, experimental populations of hybrid radish can recover relatively high pollen fertility within a few generations (Campbell et al. 2006; A. A. Snow, T. M. Culley, L. G. Campbell, P. M. Sweeney, S. G. Hegde, N. C. Ellstrand, unpublished data).
Wild radish and its cultivated relative have several advantages for studies of hybrid fitness and introgression. As annuals, they are easily grown in large quantities and can be hand-pollinated without emasculation due to self-incompatibility (Warwick and Francis 2005). Furthermore, flower color provides a useful indicator in advanced-generation hybrid populations of persistent introgression of crop R. sativus alleles. Raphanus sativus is homozygous for the dominant, white petal allele, whereas R. raphanistrum is homozygous for the recessive, yellow carotenoid pigment (Panetsos and Baker 1967; Kay 1976; Campbell et al. 2006). Therefore, the wild-type flower color could be detected in homozygotes but not heterozygotes at this locus. Inheritance of pinkish petal hues that blend with white or yellow colors is more variable and complex (Irwin et al. 2003). In the context of this study, pink-flowered plants were grouped with white-flowered ones, and bronze-flowered plants were grouped with yellows (as in Snow et al. 2001).
Seed sources and base generation
In 2001, we collected seeds from several hundred plants in a natural R. raphanistrum population near a recently abandoned potato field in Pellston, MI, USA (plants were homozygous for the yellow petal-color allele). In a greenhouse in Columbus, OH, 100 wild plants were hand-pollinated with either wild pollen to create F1 wild plants, or crop pollen to create F1 hybrid plants. Below, we refer to these radish biotypes as wild or hybrid based on pollinations in this first generation. We obtained crop pollen from 100 ‘Red Silk’R. sativus plants (homozygous for the white petal color allele; Harris-Moran Seed Co., Modesto, CA, USA). Plants were grown under controlled conditions, including a 16 h daylight schedule with a 23–28°C/20–25°C day/night temperature range. A long-day photoperiod encourages radish flowering (Erwin et al. 2002) and the temperature range discouraged the breakdown of the self-incompatibility system (el Murabaa 1957). These conditions were maintained during the artificial selection experiment described below. Maternal parents were randomly assigned to pollen donors from concurrently flowering plants. We used unpollinated flowers to confirm that no flowers were self- or cross-pollinated inadvertently.
The selection experiment started in the second generation. F2 seeds were produced from 100 F1 wild × F1 wild, and 100 F1 hybrid × F1 hybrid crosses in the same growth room and under similar environmental conditions as the parental generation. We randomly assigned F2 plants to one of three selection treatments (early flowering, long leaf length, and no selection control) and to one of three replicate lineages per selection treatment, for a total of 18 lineages (see Fig. 1). Three replicate lineages and the control treatment allowed us to exclude drift as the cause of trait evolution. There were 140 plants per F2 lineage.
Figure 1.
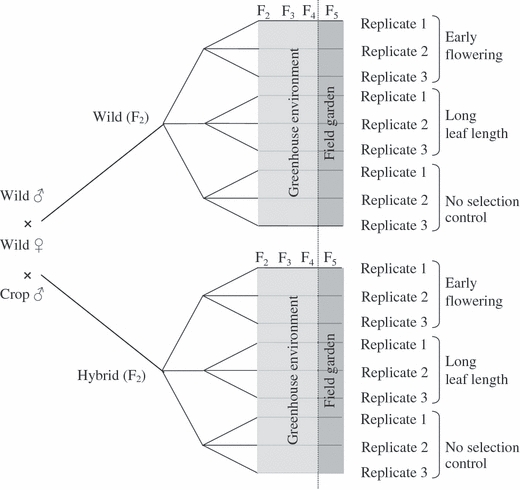
Design of the artificial selection experiment. Each F2 plant was randomly assigned to one of three selection treatments (early flowering, long leaf length, or the no-selection control) and one of three replicates within each treatment. Each lineage in each generation was initially composed of 130–160 plants and 10% of each cohort was selected to produce the following generation.
Undoubtedly, back-cross (BC) hybrids will be more commonly produced under natural conditions than F2 hybrids, because the hybrid offspring are often rare relative to their wild and crop progenitors. However, F2 hybrids possess more genetic variation than BC hybrids and therefore represent the lineage with the most evolutionary potential.
Artificial selection on age or leaf size at reproduction
We imposed truncation selection on the F2–F4 generations of nine wild and nine hybrid lineages, with a selection intensity of 10% (Fig. 1). Truncation selection is an efficient form of directional selection often used by plant breeders and evolutionary biologists. Individuals are sorted according to specific traits of interest and those with the most extreme phenotypes (in our case the earliest flowering or longest leaves) are selected to serve as parents. Hand-pollination was used to allow random mating among the subset of selected parents, which contributed equally to the following generation.
Over the course of the selection experiment, plants were grown in Cone-tainers (Stuewe and Sons, Corvallis, OR, USA), filled with standard potting soil (PRO-MIX BX peat, Premier Horticulture Ltd., Rivière-du-Loup, Canada), so that we could simultaneously raise several thousand plants in the greenhouse. The planting dates for the F2, F3, and F4 generations were February 13–18, 2003, November 10–17, 2003, and August 17–20, 2004, respectively. We randomly repositioned the plants every 2 weeks so as to reduce the effects of any variation in light, watering, and other environmental conditions of the greenhouse. Additionally, each replicate occupied two adjacent greenhouse benches. Each generation of selection was completed in approximately 5 months.
For the F2–F4 generations, each of the 18 lineages was initiated with at least 225 seeds. Two weeks after planting, the size of every lineage was reduced to the number of germinated plants in the smallest lineage, maintaining equally sized lineages. In the F2, F3, and F4 generations, each lineage included 160, 130, and 140 plants from 100, 16, and 13 parental plants, respectively. For the purposes of imposing selection and following trait evolution, we recorded dates of germination and anthesis, and leaf length at anthesis of each plant. Age at flowering was calculated as the difference, in days, between germination and anthesis. The length of the longest leaf on the first day of flowering served as a measure of plant size at the time of reproduction. Applying truncation selection, we selected 10% of the plants from each lineage that represented the earliest-flowering individuals for early lineages, 10% of the plants from each lineage that represented the longest-leafed individuals for long lineages, and randomly selected 10% of the plants from the control lineages to produce the following generation. Selected plants were cross-pollinated within a lineage in a complete diallel design. Summary statistics for the data recorded for the F2–F4 generations are reported in Appendix 1.
Phenotypes of F5 plants in a common garden
We measured the phenotypes of F5 plants in a common garden at the University of Michigan Biological Station (UMBS) in Pellston, MI (42°35′N, 84°42′W). The garden area was leveled and the sandy topsoil was thoroughly roto-tilled twice to remove vegetation. First, seeds were planted in 300 mL of Pro-mix BX peat in Jiffy fiber pots (May 3–May 10, 2005) with four oat seeds (Blaskowski's Feed and Seed, Cheboygan, MI, USA) in a greenhouse at UMBS. Cultivated spring oats were added to provide a uniform competitor and mimic natural conditions. Oat seedling density was reduced to two seedlings per pot. Once radish seedlings developed their first true leaves (May 19–26), each fiber pot was transplanted into a 2-L tube pot with 1.7 L of local topsoil that enveloped the fiber pot. Each wild and hybrid lineage was represented by 42 and 84 plants, respectively. Plants were arranged in a complete block design randomized with respect to selection treatment, biotype, and replicate. Plants were separated by 30 cm and the pots served to reduce root competition among neighbors. Seedlings that died within the first week after transplanting were replaced. Plants were watered daily for the first month and every other day until August 31. On June 18, 13 mg of fertilizer (Slow-release Osmocote 19-6-12) was added to each pot, as local soil was nutrient poor. Insect herbivory was kept at low levels by applying an insecticide three times during the first month (every 2 weeks) after transplantation, when herbivory was highest (Ortho® Bug-B-Gon® Garden and Landscape Insect Killer Concentrate, 0.0033% esfenvalerate and Bt, 20 g/2.5 gal; Scotts Miracle-Gro Co., Marysville, OH, USA). Aphids were present at low densities later in the season but did not colonize any plant heavily.
We recorded the dates of germination and anthesis, flower petal color, and maximum leaf length at anthesis for each plant, as described above. We also collected pollen from 40 plants per lineage to estimate pollen fertility. After staining (Alexander 1969), pollen fertility was assessed using a compound microscope to count the proportion of aborted grains in samples of at least 100 grains per plant. Three plants did not flower before the first hard frost (September 16–20) and were excluded from the experiment. We recognize that measuring the phenotype of the plants in a different environment than the selection environment requires the assumption that gene × environment interactions are rare. However, additional data collected from these plants were required for related experiments (Campbell et al. 2006; Campbell and Snow, in press; Campbell 2007), and we wanted to evaluate their phenotypes under semi-natural conditions. Therefore, we collected the phenotypic data in the F5 generation outdoors with the caveat that gene × environment interactions might affect our results.
Analysis
To determine if lineages responded to direct or correlated selection for early flowering and long leaf length, and whether biotypes differed in their response to selection, we compared the phenotypes of F5 populations using an ANOVA for each quantitative trait (age at flowering, leaf length, pollen fertility). We compared the values for each biotype and selection treatment (fixed factors), replicate and block (random factors). If the ANOVA revealed significant differences among lineages for a particular trait, a series of planned nonorthogonal pair-wise comparisons was made. To determine if hybrid lineages differed in the strength of phenotypic correlations, we determined the slope of the regression between leaf length and age at flowering for each wild and hybrid control lineage. We then compared the standardized coefficients using an independent samples t-test. The frequency of white-flower petal plants was compared among selection treatments across replicate hybrid lineages using a loglinear analysis (Stokes et al. 2000). For the flower color analysis, only hybrid lineages were included, as wild lineages possess no genetic variation for flower petal color (i.e., they were all yellow).
Results
Responses to direct selection
Early flowering hybrid lineages evolved more rapidly than early flowering wild lineages (Fig. 2A, Appendix 2). These hybrid lineages flowered 11 days earlier than hybrid-control lineages, whereas wild-early lineages flowered 4 days earlier than wild-control lineages (Biotype × Selection: F1,3.8 = 13.56, P = 0.023). Hybrid-early lineages flowered almost synchronously with wild-early lineages and were similar to wild-control lineages (95% CI hybrid-early: 29.6–31.7 days; wild-control: 32.4–35.3 days; wild-early: 28.4–31.2 days). Overall, hybrid lineages flowered later than wild lineages (Biotype: F1,3.8 = 25.34, P = 0.009) and both biotypes responded to selection for early flowering (Selection: F1,4.2 = 21.15, P = 0.009). Age at flowering did not differ among replicates (P = 0.12) but did differ among blocks (F20,11.8 = 5.05, P = 0.009). Also, age at flowering of replicates within selection treatments differed significantly among biotypes (F4,80.4 = 6.41, P < 0.001). Thus, early flowering hybrid lineages rapidly evolved a wild-type flowering phenology and wild lineages evolved more slowly.
Figure 2.
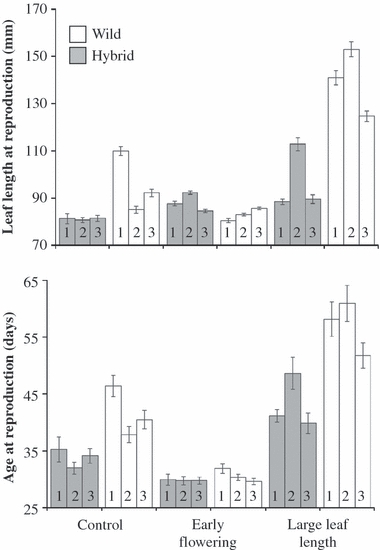
Effects of artificial selection for early flowering or large leaves (i.e., long leaf length) on (A) age at reproduction, and (B) leaf length at reproduction, in wild and hybrid F5 lineages in a common garden in Michigan. Numbers within bars indicate the replicate lineage within each biotype. Error bars represent ±1 SE of the mean; for each lineage nWild = 42, nHybrid = 84.
Long-leafed hybrid lineages also evolved more rapidly than long-leafed wild lineages (Fig. 2A, Appendix 2). Hybrid-long lineages grew leaves 43 mm longer than hybrid-control lineages, whereas wild-long lineages grew leaves 16 mm longer than wild-control lineages (Biotype × Selection: F1,4 = 10.6, P = 0.031). Overall, hybrid lineages grew longer leaves than wild lineages (Biotype: F1,3.7 = 43.8, P = 0.004) and both biotypes responded to selection for long leaves (Selection: F1,4.1 = 12.2, P = 0.024). Leaf length did not differ among replicates within selection treatments (P = 0.12) but did differ among blocks (F20,7.8 = 4.3, P = 0.021). Also, leaf length of replicates within selection treatments differed significantly among biotypes (F4,102.1 = 4.4, P = 0.002). Thus, long-leafed hybrid lineages rapidly evolved away from a wild-type size, whereas wild lineages evolved more slowly and did not evolve as extreme leaf lengths as the hybrid lineages.
Correlated Evolution
Hybridization allowed us to detect a strong life-history trade-off between age and size at reproduction (Fig. 3). Although wild control lineages exhibited a weak, correlation between age and size at first flower (β = −0.04, SD = 0.12, n = 3), hybrid control lineages exhibited a significantly stronger, positive correlation between age and size at first flower (β = 0.57, SD = 0.26, n = 3) (t-test: t = 3.67, unequal variances assumed, P = 0.04, Fig. 3). As we selected for longer leaf length, age at first flower became increasingly delayed.
Figure 3.
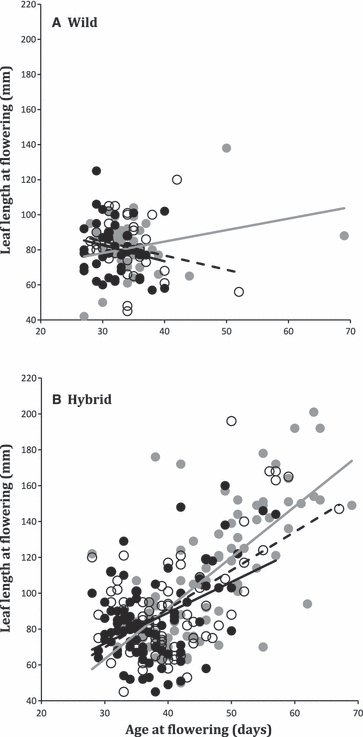
Phenotypic correlations between age and leaf length at reproduction for (A) wild control and (B) hybrid control lineages. Replicate 1 is represented by grey circles and a grey line; replicate 2 is represented by solid black circles and a solid black line; replicate 3 is represented by white circles and a dashed black line.
Earlier flowering in selected hybrid lineages coincided with a recovery of pollen fertility (Fig. 4). Hybrid-early lineages regained pollen fertility (average of 79.7%), similar to wild-control lineages (82.9%; pair-wise comparison: P = 0.323). In contrast, by the F5 generation, hybrid-control lineages continued to produce significantly fewer fertile pollen grains (70%) than wild-control lineages and hybrid-long lineages produced even fewer (63.8%; F2,3.6 = 8.01, P = 0.041). Consequently, hybrids that evolved a wild-type flowering phenology also rapidly regained wild-type fertility, whereas hybrids that evolved long leaf length did not regain wild-type fertility.
Figure 4.
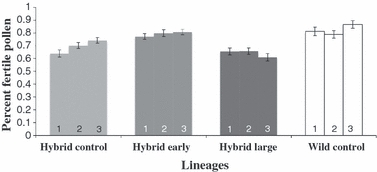
Effects of artificial selection for early flowering or large leaves (i.e., long leaf length) at reproduction on pollen fertility in wild and hybrid F5 lineages. Numbers within bars indicate the replicate lineage within each biotype. Error bars represent ±1 SE of the mean; for each lineage, nWild = 40, nHybrid = 40.
Persistence of a crop-derived allele
White flower petal color provided an easily assessed, dominant, crop-derived trait (wild lineages included only yellow-flowered plants). By the F5 generation, the frequency of white-flowered plants did not differ significantly among early and control lineages (Fig. 5, P = 0.99), but the frequency of white-flowered plants in long-leafed lineages was significantly greater than in control lineages (χ2 = 23.6, P < 0.0001). Therefore, changes in flower color frequencies demonstrated persistence of a crop trait under contrasting selection treatments, and suggest some degree of linkage between the white petal allele and genes that confer later flowering.
Figure 5.
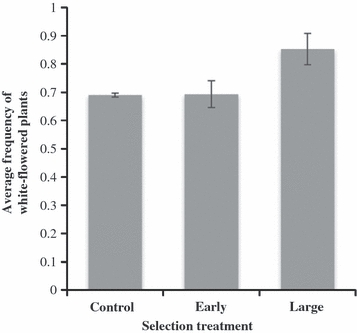
The frequency of white flower color, a crop-derived trait, in hybrid lineages after selection for early flowering or large leaves (i.e., long leaf length) relative to randomly mated controls. Error bars represent the mean of three replicate lineages ±1 SE (84 plants per lineage).
Discussion
Weedy plants are often introduced into foreign environments. To succeed, these plants must at least cope with, if not thrive in, diverse conditions by evolving adaptive phenotypes. Our results demonstrate that hybrids may evolve more rapidly, recover wild-type flowering phenology, and evolve more extreme leaf lengths than their wild relatives (Fig. 2), suggesting that crop-wild hybridization may facilitate rapid weed evolution. Hybrids exhibited a greater evolutionary response than wild plants under ecologically relevant artificial scenarios favoring early flowering or large size at reproduction. Further, hybridization revealed phenotypic trait correlations both in life-history trade-offs (size versus age at flowering) and pollen fertility (wild-type fertility in early-selected hybrids). Recovery of pollen fertility in early-flowering hybrids suggests that heterozygosity for the reciprocal translocation that causes low pollen fertility diminished in frequency, perhaps because the wild-type translocation is linked to genes that confer earlier flowering.
These experiments indicate that a substantial part of the measured differences in the flowering phenology and leaf length between wild and hybrid radishes and among selection treatments were of genetic origin. These genetic differences may have been produced by either random effects (drift, founder effects, etc.) or selection. Because we used control lineages and replicated each selection treatment three times, random effects are an unlikely cause. Therefore, the most probable explanation for the genetic differentiation of age and size at first flowering, pollen fertility, and flower color frequencies was selection. Several previous studies also demonstrated heritable variation in flowering times of California wild radish (Mazer and Schick 1991a,b) and R. raphanistrum (Conner et al. 2003).
Rapid, adaptive evolution
The potential to evolve earlier flowering may be fundamental for crop-wild hybrid success and, ultimately, for crop trait introgression. Many annual weeds have life histories similar to wild radish, exhibiting rapid growth and early flowering to be able to complete their life cycle before being out-competed or killed (Baker 1965). On the other hand, when the economically important crop trait is vegetative (such as the swollen radish hypotcotyl), the crop flowering phenology is often delayed, as in cultivated radish. In fact, our work in a temperate habitat with a short growing season showed that relatively early flowering radishes, wild or hybrid, are most fecund (Snow et al. 2001; Campbell and Snow 2007). Conversely, delayed flowering and even biennial life histories may be advantageous in locations with milder climates and longer growing seasons, such as on the coast of California (L. G. Campbell, unpublished data). Our results suggest that the capacity for rapid evolution of flowering phenology in crop-wild hybrids may allow more rapid demographic growth in weed populations than anticipated, although this should be confirmed under natural selective pressures in the field. In Michigan, our research on naturally evolving crop-wild hybrids supports this prediction (Campbell et al. 2006; Campbell 2007).
Phenotypic evolution proceeded more rapidly in hybrid than wild lineages, as indicated by the estimates of evolutionary rates in Table 1. We estimated average rates of evolution across replicates, acknowledging significant levels of variation among replicates. We can use relative rates of evolution via artificial selection as a priori expectations of relative evolutionary rates via natural selection (Bone and Farres 2001). In fact, we found that leaf length evolved twice as fast in hybrid lineages as in wild ones. Further, the rates of evolution we documented in the hybrid lineages (18–92 × 10−3 darwins) were faster than rates of evolution in natural hybrid sunflower populations over a 50-year period (6–20 × 10−3 darwins; Carney et al. 2000; calculated by Bone and Farres 2001). These differences could be attributed to our use of early generation lineages and strong, directional selection on single traits, whereas the rate of evolution in a field population of hybrid sunflowers was estimated for a suite of traits that likely experienced fluctuating natural selection over the 50-year period (Carney et al. 2000). In addition, the rates of evolution documented here in hybrid lineages in response to artificial selection were faster than those documented in crop lineages in response to artificial selection (Bone and Farres 2001 and references therein). Given that crop lineages are known to contain lower amounts of standing genetic variation and these studies were done over more generations (9–100), the faster rate of evolution of hybrid lineages is not surprising.
Table 1.
Estimated rates of evolution of plants experiencing artificial selection for early flowering (Early) or long leaf length at reproduction (Long) in hybrid and wild radish lineages
| Lineage | Trait | Darwins (×10−3)* |
|---|---|---|
| Hybrid Long | Leaf length | 92.5 |
| Hybrid Long | Age at flowering | 76.3 |
| Hybrid Early | Age at flowering | 75.9 |
| Wild Long | Age at flowering | 58.7 |
| Wild Long | Leaf length | 45.1 |
| Hybrid Early | Leaf length | 36.4 |
| Hybrid Early | Pollen fertility | 33.8 |
| Wild Early | Age at flowering | 31.3 |
| Wild Early | Leaf length | 20.7 |
| Hybrid Long | Pollen fertility | 18.8 |
Darwins = |(ln(x2) − ln(x1))/t| (Haldane, 1949) where x1 is the mean trait value for control lineages and x2 is the mean trait value of selected lineages, t is time in millions of years (4 years).
Beyond increased genetic variation, the genetic basis of faster phenotypic evolution in hybrid relative to wild lineages is likely due to linkage disequilibrium. Wild or nonhybrid species tend to show low phenotypic correlations (e.g., Murren et al. 2002), because the outcrossing wild populations would tend to exhibit random association of alleles in gametes, i.e., linkage equilibrium, and selection would be acting on each trait independently of other traits. In contrast, strong linkage disequilibrium in early generation hybrids in combination with strong selection would effectively lead to the fixation of an entire block of a chromosome, dragging along alleles controlling other traits within that genomic region. Although we have not demonstrated this phenomenon here, Rieseberg (1991) demonstrated that, in homoploid hybrid sunflowers, large chromosomal blocks from each parent were fixed in the hybrid species in response to strong selective pressures, reflecting (to some extent) the linkage disequilibrium that resulted from the original hybridization events.
Genetic correlations and constraints
Commonly, selection imposed on one trait results in correlated changes in other traits, revealing genetic correlations among traits. We detected a life-history trade-off between age and size at reproduction in hybrid lineages (Fig. 2). We also detected an increase in a crop-specific trait, white petal color, after selecting for long leaf length in hybrid lineages, and an increase in pollen fertility after selecting for early flowering in hybrid lineages (Figs 3 and 4). These results suggest that wild populations with high frequencies of the white petal allele may have experienced recent hybridization and/or selection for large plant size (e.g., Panetsos and Baker 1967; Kercher and Conner 1996; Campbell et al. 2006). The rapid recovery of pollen fertility documented in field-grown hybrid populations may be associated with selection for advanced flowering (e.g., Snow et al. 2001; Campbell et al. 2006).
Selection, natural or artificial, tends to create a state of linkage disequilibrium between selected traits and correlated traits. Under natural conditions, recombination tends to restore linkage equilibrium (or a random association among heritable traits), and so the linkage disequilibrium generated by correlated selection persists for a relatively short time (Przeworski 2002). However, if selection is sufficiently strong, as in truncation selection, or if matings occur between closely related individuals making recombination less effective, as in artificial selection experiments, the signature of selection on one trait can sometimes be detected in correlated traits (Hartl and Clark 2007). Therefore, our experimental design may have inflated the degree of correlated evolution in both hybrid and wild lineages, relative to what could be detected under more natural conditions and our results must be interpreted accordingly.
Here, we have determined that the phenotypic correlation between size and age at reproduction may also have a genetic basis and that this life-history correlation is stronger in hybrid than wild lineages. Murren et al. (2002) also found that three different hybrid combinations had stronger phenotypic correlations than their parental taxa even though mean phenotypic values of individual traits in hybrids were intermediate to parental taxa. Although hybridization increased phenotypic variation and heritability of age at first flower in our study, and thus increased their evolutionary potential, hybridization also may have constrained phenotypic evolution by increasing phenotypic correlations and producing life-history trade-offs. For instance, whereas selection for leaf length appears to continue to produce ever longer leaves, selection for early flowering in hybrid lineages appears to be limited to 4 days earlier than the wild control lineages. This may be a result of the correlation between age and size at reproduction. Although these strong phenotypic correlations may constrain the direction of evolution, they will also result in the rapid evolution of multiple traits simultaneously. Although we evaluated the evolution of early flowering and long leaf length independently, these traits have evolved concurrently in the California weedy radish, an unlikely combination from the results of this experiment. The diverse parentage of the California radish, with multiple cultivars and wild lineages (Ridley et al. in press) may have allowed these plants to escape the life-history trade-off detected here. In summary, early-generation crop-wild hybrid populations may quickly transition from high frequencies of relatively nonweedy phenotypes to high frequencies of a suite of adaptive traits, allowing them to be successful in a wide range of environments.
Correlated evolution in response to selection for a weedy phenotype may have a significant impact on the introgression of crop-specific traits (Gavrilets 1997; Barton 2001). Given that QTLs of domestication traits are not randomly or evenly distributed throughout the genome of many crops, but rather occur as linked clusters in certain chromosomal regions (reviewed in Ross-Ibarra 2005), we expect that the joint processes of crop-wild hybridization and strong selection could quickly eliminate domestication traits that are maladaptive or are linked to selectively deleterious traits. In this study, linkage disequilibrium did not eliminate all crop-derived traits, because white petal color persisted at high frequencies in all hybrid lineages. Indeed, field populations of hybrid radish vary widely in the frequency of white-flowered plants (6%–75%), suggesting the ease with which crop-derived alleles may introgress (Panetsos and Baker 1967; Kercher and Conner 1996; Snow et al. 2001; Campbell et al. 2006). Further, the crop version of the reciprocal translocation that led to low pollen fertility was maintained in hybrid, long-leaf lineages. This is similar to the apparent persistence of the crop version of the reciprocal translocation in California wild radish described in Panetsos and Baker (1967) and could be due to selection for large size and late flowering in wild populations.
Summary
Hybridization has undoubtedly played a role in the diversification of evolutionary lineages in general and the evolution of weed taxa specifically (Ellstrand and Schierenbeck 2000; Rosenthal et al. 2005). Our results reveal that hybridizing populations may evolve more rapidly than nonhybridizing populations and hybrid individuals may exhibit more extreme phenotypes than those of their parental taxa. Although one might underestimate the evolutionary potential of early generation hybrids (e.g., Mayr 1963; Stewart et al. 2003), in some cases these lineages can rapidly become adapted to environmental conditions and may even exhibit novel phenotypes (Schwarzbach et al. 2001; Rieseberg et al. 2003, 2007; Rosenthal et al. 2005). However, correlations and trade-offs among key life-history traits could limit this rapid evolution. To the authors’ knowledge, this study represents the first estimate of the response of quantitative traits to selection in crop-wild hybrid lineages and one of the few that has compared the response to selection of hybrid populations to nonhybrid populations (Lewontin and Birch 1966; Hercus and Hoffman 1999). By using hybridization, artificial and natural selection experiments, we will begin to appreciate the evolutionary potential and limitations of crop-wild hybrids and how to minimize any unwanted evolutionary and ecological effects associated with the release of crops with novel, transgenic traits.
Acknowledgments
We thank J. Leonard, S. Clark, H. Eisel, S. Pfingsten, the UMBS staff, T. Waite, and many student researchers for their help in the greenhouse, field, and laboratory. The United States Department of Agriculture (Grant #2002-03715), University of Michigan Biological Station, National Science Foundation Environmental Biology Program Doctoral Dissertation Improvement Grant (DEB-0508615), Ohio State University Presidential Fellowship, Nature Conservancy of Michigan, Janice Carson Beatley Endowment, and Sigma Xi financially supported this research. Many thanks to L. Gibbs, K. Mercer, K. Whitney and two anonymous reviewers for valuable comments.
Appendix 1
Summary statistics of fitness components for F2–F5 wild and hybrid plants that experienced one of three selection treatments: (i) to decrease time to flowering (Early), (ii) to increase leaf length (Long), and (iii) random mating (Control). Each selection treatment was represented by three replicated lineages. Plants were grown in a greenhouse at Ohio State University. Age is reported in days and leaf length in millimeters. Each lineage was represented by n plants each generation; n for pollen fertility = 14; n/a indicates not available.
| Selection treatment | Replicate | Trait | F2 base (SE) | F3 (SE) | F4 (SE) | F5 (SE) |
|---|---|---|---|---|---|---|
| Biotype: Wild | ||||||
| Early | 1 | Age at flowering | n/a | 39 (0.6) | 22 (0.2) | 30 (0.5) |
| Leaf length | n/a | 139 (4.9) | 142 (1.6) | 88 (3.3) | ||
| n | n/a | 130 | 140 | 42 | ||
| 2 | Age at flowering | n/a | 41 (0.6) | 20 (0.2) | 30 (0.4) | |
| Leaf length | n/a | 204 (5.7) | 129 (1.9) | 92 (3.0) | ||
| n | n/a | 130 | 140 | 42 | ||
| 3 | Age at flowering | n/a | 43 (0.9) | 21 (0.2) | 30 (0.3) | |
| Leaf length | n/a | 224 (5.1) | 140 (1.9) | 85 (2.7) | ||
| n | n/a | 130 | 140 | 42 | ||
| Avg | Age at flowering | n/a | 41 (0.4) | 21 (0.1) | 30 (0.2) | |
| Leaf length | n/a | 189 (3.5) | 137 (1.1) | 88 (1.7) | ||
| n | n/a | 3 | 3 | 3 | ||
| Control | 1 | Pollen fertility (%) | 87 (2.6) | 85 (4.7) | 82 (5.4) | 81 (3.3) |
| Age at flowering | 47 (1.1) | 41 (0.7) | 24 (0.3) | 35 (1.1) | ||
| Leaf length | 197 (4.5) | 115 (4.4) | 130 (2.2) | 81 (2.7) | ||
| n | 160 | 130 | 140 | 42 | ||
| 2 | Pollen fertility (%) | 92 (1.4) | 89 (4.7) | 81 (4.4) | 79 (2.9) | |
| Age at flowering | 45 (1.0) | 40 (0.5) | 23 (0.3) | 32 (0.5) | ||
| Leaf length | 196 (4.6) | 195 (6.7) | 130 (1.9) | 81 (2.4) | ||
| n | 160 | 130 | 140 | 42 | ||
| 3 | Pollen fertility (%) | 92 (1.6) | 94 (1.7) | 91 (1.5) | 87 (3.0) | |
| Age at flowering | 46 (1.0) | 44 (0.9) | 24 (0.3) | 34 (0.7) | ||
| Leaf length | 198 (4.4) | 194 (5.1) | 133 (2.2) | 81 (2.6) | ||
| n | 160 | 130 | 140 | 42 | ||
| Avg | Pollen fertility (%) | 90 (1.1) | 90 (1.9) | 85 (2.3) | 82 (1.8) | |
| Age at flowering | 41 (1.1) | 41 (0.4) | 24 (0.2) | 34 (0.5) | ||
| Leaf length | 190 (4.6) | 168 (3.6) | 131 (1.2) | 81 (1.5) | ||
| n | 3 | 3 | 3 | 3 | ||
| Long | 1 | Age at flowering | n/a | 53 (1.3) | 42 (1.8) | 41 (0.6) |
| Leaf length | n/a | 184 (6.7) | 150 (4.3) | 89 (3.2) | ||
| N | n/a | 130 | 140 | 42 | ||
| 2 | Age at flowering | n/a | 52 (1.1) | 36 (1.2) | 49 (1.4) | |
| Leaf length | n/a | 228 (6.5) | 150 (3.2) | 113 (4.0) | ||
| n | n/a | 130 | 140 | 42 | ||
| 3 | Age at flowering | n/a | 51 (1.0) | 30 (0.8) | 40 (0.9) | |
| Leaf length | n/a | 229 (5.5) | 157 (3.3) | 90 (3.9) | ||
| n | n/a | 130 | 140 | 42 | ||
| Avg | Age at flowering | n/a | 52 (0.7) | 36 (0.8) | 43 (0.7) | |
| Leaf length | n/a | 214 (3.7) | 153 (2.1) | 97 (2.4) | ||
| n | n/a | 3 | 3 | 3 | ||
| Biotype: Hybrid | ||||||
| Early | 1 | Pollen fertility (%) | n/a | 87 (3.4) | 87 (2.9) | 77 (2.2) |
| Age at flowering | n/a | 45 (0.7) | 24 (0.3) | 32 (0.4) | ||
| Leaf length | n/a | 133 (5.1) | 130 (1.6) | 80 (2.0) | ||
| n | n/a | 130 | 140 | 84 | ||
| 2 | Pollen fertility (%) | n/a | 72 (8.2) | 85 (0.04) | 80 (2.6) | |
| Age at flowering | n/a | 41 (0.5) | 22 (0.3) | 30 (0.3) | ||
| Leaf length | n/a | 99 (6.7) | 123 (1.9) | 83 (1.8) | ||
| N | n/a | 130 | 140 | 84 | ||
| 3 | Pollen fertility (%) | n/a | 89 (2.0) | 87 (3.3) | 81 (2.1) | |
| Age at flowering | n/a | 42 (0.4) | 20 (0.2) | 30 (0.3) | ||
| Leaf length | n/a | 214 (4.4) | 129 (1.9) | 86 (1.7) | ||
| n | n/a | 130 | 140 | 84 | ||
| Avg | Pollen fertility (%) | n/a | 84 (2.9) | 86 (2.0) | 79 (1.3) | |
| Age at flowering | n/a | 43 (0.3) | 22 (0.2) | 31 (0.2) | ||
| Leaf length | n/a | 149 (3.8) | 128 (1.0) | 83 (1.1) | ||
| n | n/a | 3 | 3 | 3 | ||
| Control | 1 | Pollen fertility (%) | 71 (3.9) | 69 (5.5) | 75 (3.7) | 64 (2.7) |
| Age at flowering | 58 (1.4) | 74 (2.1) | 45 (2.3) | 46 (1.0) | ||
| Leaf length | 245 (6.4) | 198 (8.0) | 168 (4.5) | 110 (4.0) | ||
| n | 160 | 130 | 140 | 84 | ||
| 2 | Pollen fertility (%) | 73 (3.9) | 70 (5.8) | 73 (5.6) | 70 (2.2) | |
| Age at flowering | 63 (1.6) | 73 (1.8) | 38 (1.8) | 38 (0.7) | ||
| Leaf length | 247 (6.3) | 256 (5.7) | 136 (2.9) | 85 (2.7) | ||
| n | 160 | 130 | 140 | 84 | ||
| 3 | Pollen fertility (%) | 76 | 67 (4.4) | 62 (8.3) | 74 (2.2) | |
| Age at flowering | 58 (1.4) | 66 (1.7) | 42 (2.0) | 41 (0.8) | ||
| Leaf length | 253 (6.4) | 206 (8.7) | 151 (3.3) | 92 (3.1) | ||
| n | 160 | 130 | 140 | 84 | ||
| Avg | Pollen fertility (%) | 73 (2.2) | 68 (2.9) | 70 (3.5) | 69 (1.4) | |
| Age at flowering | 67 (2.5) | 71 (1.1) | 42 (1.2) | 42 (0.5) | ||
| Leaf length | 240 (6.6) | 220 (4.5) | 152 (2.2) | 96 (2.0) | ||
| n | 3 | 3 | 3 | 3 | ||
| Long | 1 | Pollen fertility (%) | n/a | 65 (7.3) | 56 (6.2) | 65 (2.8) |
| Age at flowering | n/a | 82 (2.8) | 89 (2.9) | 58 (1.5) | ||
| Leaf length | n/a | 286 (9.2) | 187 (7.0) | 141 (4.3) | ||
| n | n/a | 130 | 140 | 84 | ||
| 2 | Pollen fertility (%) | n/a | 66 (4.8) | 67 (5.1) | 66 (2.6) | |
| Age at flowering | n/a | 84 (2.0) | 54 (2.7) | 61 (1.6) | ||
| Leaf length | n/a | 253 (7.9) | 164 (4.5) | 153 (5.1) | ||
| n | n/a | 130 | 140 | 84 | ||
| 3 | Pollen fertility (%) | n/a | 69 (5.2) | 64 (6.1) | 61 (2.9) | |
| Age at flowering | n/a | 73 (1.8) | 60 (2.8) | 52 (1.1) | ||
| Leaf length | n/a | 258 (6.4) | 169 (4.1) | 125 (4.4) | ||
| n | n/a | 130 | 140 | 84 | ||
| Avg | Pollen fertility (%) | n/a | 67 (3.1) | 62 (3.3) | 64 (1.6) | |
| Age at flowering | n/a | 79 (1.2) | 68 (1.8) | 57 (0.9) | ||
| Leaf length | n/a | 262 (4.5) | 174 (3.2) | 139 (2.7) | ||
| n | n/a | 3 | 3 | 3 | ||
Appendix 2
Summary statistics of fitness components for F5 wild and hybrid plants that experienced one of three selection treatments: (i) to decrease time to flowering (Early), (ii) to increase leaf length (Large), and (iii) random mating (Control). Each selection treatment was represented by three replicated lineages. Plants were grown in an outdoor common garden in Michigan, USA. Each lineage was represented by n plants each generation, as indicated, except for pollen fertility (n = 40); n/a indicates not available.
| Biotype | Selection treatment | Replicate | n | Days to flowering (days, SE) | Leaf length (mm, SE) | Pollen fertility (%) |
|---|---|---|---|---|---|---|
| Wild | Early | 1 | 42 | 30 (0.5) | 88 (3.3) | n/a |
| 2 | 42 | 30 (0.4) | 92 (3.0) | n/a | ||
| 3 | 42 | 30 (0.3) | 85 (2.7) | n/a | ||
| Avg | 3 | 30 (0.2) | 88 (1.7) | n/a | ||
| Control | 1 | 42 | 35 (1.1) | 81 (2.7) | 81 (3.3) | |
| 2 | 42 | 32 (0.5) | 81 (2.4) | 79 (2.9) | ||
| 3 | 42 | 34 (0.7) | 81 (2.6) | 87 (3.0) | ||
| Avg | 3 | 34 (0.5) | 81 (1.5) | 82 (1.8) | ||
| Large | 1 | 42 | 41 (0.6) | 89 (3.2) | n/a | |
| 2 | 42 | 49 (1.4) | 113 (4.0) | n/a | ||
| 3 | 42 | 40 (0.9) | 90 (3.9) | n/a | ||
| Avg | 3 | 43 (0.7) | 97 (2.4) | n/a | ||
| Hybrid | Early | 1 | 84 | 32 (0.4) | 80 (2.0) | 77 (2.2) |
| 2 | 84 | 30 (0.3) | 83 (1.8) | 80 (2.6) | ||
| 3 | 84 | 30 (0.3) | 86 (1.7) | 81 (2.1) | ||
| Avg | 3 | 31 (0.2) | 83 (1.1) | 79 (1.3) | ||
| Control | 1 | 84 | 46 (1.0) | 110 (4.0) | 64 (2.7) | |
| 2 | 84 | 38 (0.7) | 85 (2.7) | 70 (2.2) | ||
| 3 | 84 | 41 (0.8) | 92 (3.1) | 74 (2.2) | ||
| Avg | 3 | 42 (0.5) | 96 (2.0) | 69 (1.4) | ||
| Large | 1 | 84 | 58 (1.5) | 141 (4.3) | 65 (2.8) | |
| 2 | 84 | 61 (1.6) | 153 (5.1) | 66 (2.6) | ||
| 3 | 84 | 52 (1.1) | 125 (4.4) | 61 (2.9) | ||
| Avg | 3 | 57 (0.9) | 139 (2.7) | 64 (1.6) |
Literature cited
- Alexander MP. Differential staining of aborted and non-aborted pollen. Stain Technology. 1969;44:117–122. doi: 10.3109/10520296909063335. [DOI] [PubMed] [Google Scholar]
- Anderson E, Stebbins GL. Hybridization as an evolutionary stimulus. Evolution. 1954;8:378–388. [Google Scholar]
- Arnold ML. Natural Hybridization and Evolution. New York, NY, USA: Oxford University Press; 1997. [Google Scholar]
- Arnold ML. Natural hybridization and the evolution of domesticated, pest and disease organisms. Molecular Ecology. 2004;13:997–1007. doi: 10.1111/j.1365-294X.2004.02145.x. [DOI] [PubMed] [Google Scholar]
- Arnold ML. Evolution Through Genetic Exchange. New York, NY, USA: Oxford University Press; 2006. [Google Scholar]
- Arnold ML, Hodges SA. Are natural hybrids fit or unfit relative to their parents? Trends in Ecology & Evolution. 1995;10:67–71. doi: 10.1016/S0169-5347(00)88979-X. [DOI] [PubMed] [Google Scholar]
- Arnold ML, Bulger MR, Burke JM, Hempel AL, Williams JH. Natural hybridization: how low can you go and still be important? Ecology. 1999;80:371–381. [Google Scholar]
- Baack EJ, Sapir Y, Chapman MA, Burke JM, Rieseberg LH. Selection on domestication traits and quantitative trait loci in crop-wild sunflower hybrids. Molecular Ecology. 2008;17:666–677. doi: 10.1111/j.1365-294X.2007.03596.x. [DOI] [PubMed] [Google Scholar]
- Baker HG. Characteristics and modes of origin of weeds. In: Baker HG, Stebbins GL, editors. The Genetics of Colonizing Species. New York, NY, USA: Academic; 1965. pp. 147–172. [Google Scholar]
- Barton NH. The role of hybridization in evolution. Molecular Ecology. 2001;10:551–568. doi: 10.1046/j.1365-294x.2001.01216.x. [DOI] [PubMed] [Google Scholar]
- Bartsch D, Lehnen M, Clegg J, Pohl-Orf M, Schuphan I, Ellstrand NC. Impact of gene flow from cultivated beet on genetic diversity of wild sea beet populations. Molecular Ecology. 1999;8:1733–1741. doi: 10.1046/j.1365-294x.1999.00769.x. [DOI] [PubMed] [Google Scholar]
- Bone E, Farres A. Trends and rates of microevolution in plants. Genetica. 2001;112:165–182. [PubMed] [Google Scholar]
- Bouck AC. The Genetic Architecture of Reproductive Isolation in Louisiana Irises. Athens, GA, USA: University of Georgia; 2004. Ph.D. thesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LG. Rapid Evolution in a Crop-Weed Complex (Raphanus spp.) Columbus, OH, USA: The Ohio State University; 2007. p. 235. Ph.D. dissertation. [Google Scholar]
- Campbell LG, Snow AA. Competition alters life-history traits and increases the relative fecundity of crop-wild hybrids (Raphanus spp.) New Phytologist. 2007;173:648–660. doi: 10.1111/j.1469-8137.2006.01941.x. [DOI] [PubMed] [Google Scholar]
- Campbell LG, Snow AA. Radish dedomestication (Raphanus sativus; Brassicaceae) – will gene flow assist the evolution of feral weeds? American Journal of Botany. 2008 doi: 10.3732/ajb.0800054. In press. [DOI] [PubMed] [Google Scholar]
- Campbell LG, Snow AA, Ridley CE. Weed evolution after crop gene introgression: greater survival and fecundity of hybrids in a new environment. Ecology Letters. 2006;11:1198–1209. doi: 10.1111/j.1461-0248.2006.00974.x. [DOI] [PubMed] [Google Scholar]
- Carney SE, Gardner KA, Rieseberg LH. Evolutionary changes over the fifty-year history of a hybrid population of sunflowers (Helianthus. Evolution. 2000;54:462–474. doi: 10.1111/j.0014-3820.2000.tb00049.x. [DOI] [PubMed] [Google Scholar]
- Chloupek O, Hrstkova P. Adaptation of crops to environment. Theoretical and Applied Genetics. 2005;111:1316–1321. doi: 10.1007/s00122-005-0060-x. [DOI] [PubMed] [Google Scholar]
- Colwell RK, Norse EA, Pimentel D, Sharples FE, Simberloff D. Genetic-engineering in agriculture. Science. 1985;229:111–112. doi: 10.1126/science.229.4709.111. [DOI] [PubMed] [Google Scholar]
- Conner JK, Franks R, Stewart CN. Expression of additive genetic variances and covariances for wild radish floral traits: Comparison between field and greenhouse environments. Evolution. 2003;57:487–495. doi: 10.1111/j.0014-3820.2003.tb01540.x. [DOI] [PubMed] [Google Scholar]
- Crisp P. Radish, Raphanus sativus (Cruciferae) In: Smartt J, Simmonds NW, editors. Evolution of Crop Plants. 2nd edn. Essex, Harlow, UK: Longman Scientific & Technical; 1995. pp. 86–89. [Google Scholar]
- Curtis IS. The noble radish: past, present and future. Trends in Plant Sciences. 2003;8:305–307. doi: 10.1016/S1360-1385(03)00127-4. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics and the Origin of Species. New York, NY, USA: Columbia University Press; 1937. [Google Scholar]
- Doebley J. Mapping the genes that made maize. Trends in Genetics. 1992;8:302–307. doi: 10.1016/0168-9525(92)90261-2. [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- Ellstrand N, Marshall D. The impact of domestication on distribution of allozyme variation within and among cultivars of radish, Raphanus sativus L. Theoretical and Applied Genetics. 1985;69:393–398. doi: 10.1007/BF00570908. [DOI] [PubMed] [Google Scholar]
- Ellstrand NC, Schierenbeck KA. Hybridization as a stimulus for the evolution of invasiveness in plants? Proceedings of the National Academy of Science of the United States of America. 2000;97:7043–7050. doi: 10.1073/pnas.97.13.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellstrand NC, Prentice HC, Hancock JF. Gene flow and introgression from domesticated plants into their wild relatives. Annual Review of Ecology and Systematics. 1999;30:539–563. [Google Scholar]
- Erwin JE, Warner RM, Smith AG. Vernalization, photoperiod and GA3 interact to affect flowering of Japanese radish (Raphanus sativus Chinese Radish Jumbo Scarlet) Physiologia Plantarum. 2002;115:298–302. doi: 10.1034/j.1399-3054.2002.1150217.x. [DOI] [PubMed] [Google Scholar]
- Facon B, Jarne P, Pointier JP, David P. Hybridization and invasiveness in the freshwater snail Melanoides tuberculata: hybrid vigour is more important than increase in genetic variance. Journal of Evolutionary Biology. 2005;18:524–535. doi: 10.1111/j.1420-9101.2005.00887.x. [DOI] [PubMed] [Google Scholar]
- Fick GN, Miller JF. The genetics of sunflower. In: Schneiter AA, editor. Sunflower Technology and Production. Madison, WI, USA: Soil Science of America; 1997. pp. 395–440. [Google Scholar]
- Gavrilets S. Hybrid zones with Dobzhansky-type epistatic selection. Evolution. 1997;51:1027–1035. doi: 10.1111/j.1558-5646.1997.tb03949.x. [DOI] [PubMed] [Google Scholar]
- Greenwood H, O'Dowd DJ, Lake PS. Willow (Salix x rubens) invasion of the riparian zone in south-eastern Australia: reduced abundance and altered composition of terrestrial arthropods. Diversity and Distributions. 2004;10:485–492. [Google Scholar]
- Hails RS, Morley K. Genes invading new populations: a risk assessment perspective. Trends in Ecology & Evolution. 2005;20:245–252. doi: 10.1016/j.tree.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Hairston NG, Ellner SP, Geber MA, Yoshida T, Fox JA. Rapid evolution and the convergence of ecological and evolutionary time. Ecology Letters. 2005;8:1114–1127. [Google Scholar]
- Haldane JBS. Suggestions as to quantitative measurement of rates of evolution. Evolution. 1949;3:51–56. doi: 10.1111/j.1558-5646.1949.tb00004.x. [DOI] [PubMed] [Google Scholar]
- Hartl DL, Clark AG. Principles of Population Genetics. 4th edn. Sunderland, MA: Sinauer Associates, Inc; 2007. p. 481. [Google Scholar]
- Hauser T, Jørgensen RB, Østergård H. Fitness of backcross and F2 hybrids between weedy Brassica rapa and oilseed rape (B. napus. Heredity. 1998;81:436–443. [Google Scholar]
- Hegarty MJ, Hiscock SJ. Hybrid speciation in plants: new insights from molecular studies. New Phytologist. 2005;165:411–423. doi: 10.1111/j.1469-8137.2004.01253.x. [DOI] [PubMed] [Google Scholar]
- Hegde SG, Nason JD, Clegg JM, Ellstrand NC. The evolution of California's wild radish has resulted in the extinction of its progenitors. Evolution. 2006;60:1187–1197. [PubMed] [Google Scholar]
- Heiser CB., Jr Hybridization between the sunflower species Helianthus annuus and H. petiolaris. Evolution. 1947;1:249–262. [Google Scholar]
- Hercus MJ, Hoffman AA. Does interspecific hybridization influence evolutionary rates? An experimental study of laboratory adaptation in hybrids between Drosophila serrata and Drosophila birchii. Proceedings of the Royal Society, Series B. 1999;266:2195–2200. doi: 10.1098/rspb.1999.0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin RE, Strauss SY, Storz S, Emerson A, Guibert G. The role of herbivores in the maintenance of a flower color polymorphism in wild radish. Ecology. 2003;84:1733–1743. [Google Scholar]
- Jarvis DI, Hodgkin T. Wild relatives and crop cultivars: detecting natural introgression and farmer selection of new genetic combinations in agro-ecosystems. Molecular Ecology. 1999;8:S159–S173. [Google Scholar]
- Kay QON. Preferential pollination of yellow-flowered morphs of Raphanus raphanistrum by Pieris and Eristalis spp. Nature. 1976;261:230–232. [Google Scholar]
- Kercher S, Conner JK. Patterns of genetic variability within and among populations of wild radish, Raphanus raphanistrum (Brassicaceae) American Journal of Botany. 1996;83:1416–1421. [Google Scholar]
- Lewontin RC, Birch LC. Hybridization as a source of variation for adaptation to new environments. Evolution. 1966;20:315–336. doi: 10.1111/j.1558-5646.1966.tb03369.x. [DOI] [PubMed] [Google Scholar]
- Mallet J. Hybrid speciation. Nature Reviews. 2007;446:279–283. doi: 10.1038/nature05706. [DOI] [PubMed] [Google Scholar]
- Mayr E. Animal Species and Evolution. Cambridge: Harvard University Press; 1963. [Google Scholar]
- Mazer SJ, Schick CT. Constancy of population parameters for life history and floral traits in Raphanus sativus L. I. Norms of reaction and the nature of genotype by environmental interactions. Heredity. 1991a;67:143–156. [Google Scholar]
- Mazer SJ, Schick CT. Constancy of population parameters for life-history and floral traits in Raphanus sativus L. 2. Effects of planting density on phenotype and heritability estimates. Evolution. 1991b;45:1888–1907. doi: 10.1111/j.1558-5646.1991.tb02694.x. [DOI] [PubMed] [Google Scholar]
- El Murabaa AIM. Effect of high temperature on incompatibility in radish. Euphytica. 1957;6:268–270. [Google Scholar]
- Murren CJ, Pendleton N, Pigliucci M. Evolution of phenotypic integration in Brassica (Brassicaceae) American Journal of Botany. 2002;89:655–663. doi: 10.3732/ajb.89.4.655. [DOI] [PubMed] [Google Scholar]
- Otto SP, Whitton J. Polyploid incidence and evolution. Annual Review of Genetics. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Panetsos CA, Baker HG. The origin of variation in “wild”Raphanus sativus (Cruciferae) in California. Genetica. 1967;38:243–274. [Google Scholar]
- Perez-Jones A, Mallory-Smith CA, Hansen JL, Zemetra RS. Introgression of an imidazolinone-resistance gene from winter wheat (Triticum aestivum L.) into jointed goatgrass (Aegilops cylindrica Host) Theoretical and Applied Genetics. 2006;114:177–186. doi: 10.1007/s00122-006-0421-0. [DOI] [PubMed] [Google Scholar]
- Piálek J, Barton NH. The spread of an advantageous allele across a barrier: The effects of random drift and selection against heterozygotes. Genetics. 1997;145:493–504. doi: 10.1093/genetics/145.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M. The signature of positive selection at randomly chosen loci. Genetics. 2002;160:1179–1189. doi: 10.1093/genetics/160.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick DN, Ghalambor CK. The population ecology of contemporary adaptations: what empirical studies reveal about the conditions that promote adaptive evolution. Genetica. 2001;112-113:183–198. [PubMed] [Google Scholar]
- Ridley CE, Kim SC, Ellstrand NC. Bi-directional history of hybridization in California wild radish Raphanus sativus (Brassicaceae) as revealed by chloroplast DNA. American Journal of Botany. 2008;95:1437–1442. doi: 10.3732/ajb.0800119. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. Homoploid reticulate evolution in Helianthus (Asteraceae): evidence from ribosomal genes. American Journal of Botany. 1991;78:1218–1237. [Google Scholar]
- Rieseberg LH. The role of hybridization in evolution – old wine in new skins. American Journal of Botany. 1995;82:944–953. [Google Scholar]
- Rieseberg LH, Archer MA, Wayne RK. Transgressive segregation, adaptation and speciation. Heredity. 1999;83:363–372. doi: 10.1038/sj.hdy.6886170. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Widmer A, Arntz MA, Burke JM. The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Philisophical Transactions of the Royal Society of London. 2003;358:1141–1147. doi: 10.1098/rstb.2003.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Kim S-C, Randell RA, Whitney KD, Gross BL, Lexer C, Clay K. Hybridization and the colonization of novel habitats by annual sunflowers. Genetica. 2007;129:149–165. doi: 10.1007/s10709-006-9011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissler J, Mellon M. The Ecological Risks of Engineered Crops. Cambridge, MA: The MIT Press; 1996. [Google Scholar]
- Rosenthal DM, Rieseberg LH, Donovan LA. Re-creating ancient hybrid species’ complex phenotypes from early-generation synthetic hybrids: Three examples using wild sunflowers. American Naturalist. 2005;166:26–41. doi: 10.1086/430527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Ibarra J. Quantitative trait loci and the study of plant domestication. Genetica. 2005;123:197–204. doi: 10.1007/s10709-004-2744-6. [DOI] [PubMed] [Google Scholar]
- Schwarzbach AE, Donavan LA, Rieseberg LH. Transgressive character expression in a hybrid sunflower species. American Journal of Botany. 2001;88:270–277. [PubMed] [Google Scholar]
- Smyth S, Khachatourians GG, Phillips PWB. Liabilities and economics of transgenic crops. Nature, Biotechnology. 2002;20:537–541. doi: 10.1038/nbt0602-537. [DOI] [PubMed] [Google Scholar]
- Snow AA, Campbell LG. Can feral radishes become weeds? In: Gressel J, editor. Crop Ferality and Volunteerism. Boca Raton, FL, USA: CRC Press; 2005. pp. 193–208. [Google Scholar]
- Snow AA, Uthus KL, Culley TM. Fitness of hybrids between weedy and cultivated radish: implications for weed evolution. Ecological Applications. 2001;11:934–943. [Google Scholar]
- Snow AA, Pilson D, Rieseberg LH, Paulsen MJ, Pleskac N, Reagon MR, Wolf DE, et al. A Bt transgene reduces herbivory and enhances fecundity in wild sunflowers. Ecological Applications. 2003;13:279–286. [Google Scholar]
- Stewart CN, Halfhill MD, Warwick SI. Transgene introgression from genetically modified crops to their wild relatives. Nature Reviews, Genetics. 2003;4:806–817. doi: 10.1038/nrg1179. [DOI] [PubMed] [Google Scholar]
- Stokes ME, Davis CS, Koch GG. Categorical Data Analysis Using the SAS System. 2nd edn. Cary, NC, USA: SAS Institute Inc; 2000. [Google Scholar]
- Thompson JN. Rapid evolution as an ecological process. Trends in Ecology & Evolution. 1998;13:329–332. doi: 10.1016/s0169-5347(98)01378-0. [DOI] [PubMed] [Google Scholar]
- Warren J, James P. The ecological effects of exotic disease resistance genes introgressed into British gooseberries. Oecologia. 2006;147:69–75. doi: 10.1007/s00442-005-0257-3. [DOI] [PubMed] [Google Scholar]
- Warwick SI, Francis A. The biology of Canadian weeds. 132. Raphanus raphanistrum, L. Canadian Journal of Plant Sciences. 2005;85:709–733. [Google Scholar]
- Warwick SI, Légère A, Simard MJ, James T. Do escaped transgenes persist in nature? The case of an herbicide resistance transgene in a weedy Brassica rapa population. Molecular Ecology. 2008;17:1387–1395. doi: 10.1111/j.1365-294X.2007.03567.x. [DOI] [PubMed] [Google Scholar]
- Waser NM, Price MK. Outcrossing distance effects in Delphinium nelsonii: pollen loads, pollen tubes, and seed set. Ecology. 1991;72:171–179. [Google Scholar]
- Whitney KD, Randell RA, Rieseberg LH. Adaptive introgression of herbivore resistance traits in the weedy sunflower Helianthus annuus. American Naturalist. 2006;167:794–807. doi: 10.1086/504606. [DOI] [PubMed] [Google Scholar]
- Whitney KD, Ahern JR, Campbell LG. Hybridization frequencies do not predict numbers of invasives across plant families. Biological Invasions. 2008 DOI: 10.1007/s10530-008-9390-3. [Google Scholar]
- Wolfenbarger LL, Phifer PR. Biotechnology and ecology – The ecological risks and benefits of genetically engineered plants. Science. 2000;290:2088–2093. doi: 10.1126/science.290.5499.2088. [DOI] [PubMed] [Google Scholar]
- Wu CA, Campbell DR. Environmental stressors differentially affect leaf ecophysiological responses in two Ipomopsis species and their hybrids. Oecologia. 2006;148:202–212. doi: 10.1007/s00442-006-0363-x. [DOI] [PubMed] [Google Scholar]
- Yamagishi H, Terachi T. Multiple origins of cultivated radishes as evidenced by a comparison of the structural variations in mitochondrial DNA of Raphanus. Genome. 2003;46:89–94. doi: 10.1139/g02-110. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Zheng X, Luo J, Gaut BS, Ge S. Multilocus analysis of nucleotide variation of Oryza sativa and its wild relatives: severe bottleneck during domestication of rice. Molecular Biology and Evolution. 2007;24:875–888. doi: 10.1093/molbev/msm005. [DOI] [PubMed] [Google Scholar]


