Abstract
Estimating genetic connectivity in disturbed riverine landscapes is of key importance for river restoration. However, few species of the disturbed riverine fauna may provide a detailed and basin-wide picture of the human impact on the population genetics of riverine organisms. Here we used the most abundant native fish, the three-spined stickleback (Gasterosteus aculeatus L.), to detect the geographical determinants of genetic connectivity in the eastern part of the Scheldt basin in Belgium. Anthropogenic structures came out as the strongest determinant of population structure, when evaluated against a geographically well-documented baseline model accounting for natural effects. These barriers not only affected genetic diversity, but they also controlled the balance between gene flow and genetic drift, and therefore may crucially disrupt the population structure of sticklebacks. Landscape models explained a high percentage of variation (allelic richness: adjusted R2 = 0.78; pairwise FST: adjusted R2 = 0.60), and likely apply to other species as well. River restoration and conservation genetics may highly benefit from riverine landscape genetics, including model building, the detection of outlier populations, and a specific test for the geographical factors controlling the balance between gene flow and genetic drift.
Keywords: conservation genetics, gene flow, genetic drift, GIS, isolation-by-distance, landscape genetics, river restoration, riverscapes
Introduction
Riverine ecosystems are vulnerable to anthropogenic perturbation. Pollution, barriers, and habitat loss through canalization directly affect ecosystem dynamics and population characteristics such as migration, reproductive success, and survival of various organisms (Vrijenhoek 1998; Fausch et al. 2002; Wiens 2002; Maes et al. 2005). In the long term, anthropogenic structures fragmenting rivers may enhance the polarity in population size, migration, and potential for local extinction that naturally characterizes river systems. From a genetic perspective, fragmentation may isolate populations, reduce gene flow, and decrease genetic diversity through the processes of genetic drift and inbreeding (Saccheri et al. 1998; Morita and Yamamoto 2002). The impact of migration barriers on genetic diversity and genetic connectivity in rivers has been demonstrated in fish populations from temperate regions. Examples include the migratory grayling (Thymallus thymallus; Meldgaard et al. 2003) and brown trout (Salmo trutta; Van Houdt et al. 2005; Heggenes and Røed 2006), coastal cutthroat trout (Oncorhynchus clarki clarki; Wofford et al. 2005) and the residential bullhead (Cottus gobio; Hänfling et al. 2002; Hänfling and Weetman 2006).
Estimating genetic connectivity is a key to understanding human impact on river systems, and may improve restoration and conservation strategies. However, detecting which forces crucially affect genetic connectivity in rivers may be complicated. Population genetics of riverine vertebrates and invertebrates has been correlated with a number of geographical and environmental features (Hughes et al. 1996; Heath et al. 2001; Hänfling et al. 2002; Kinnison et al. 2002; Taylor et al. 2003; Kelly and Rhymer 2005; Koizumi et al. 2006; Wilcock et al. 2007). However, geographical and environmental information tend to be highly correlated, complicating the detection of the causality of genetic structure. Evolutionary biologists and ecological geneticists have been considering a range of scenarios explaining riverine population genetics, including the role of geography versus natural selection (Crispo et al. 2006), recent versus historical river landscapes (Castric et al. 2001; Poissant et al. 2005), isolation-by-distance (IBD) versus long-term divergence (Raeymaekers et al. 2005), active versus passive dispersal (Michels et al. 2001), and landscape versus life-history processes (Neville et al. 2006). Likewise, it may be particularly challenging to distinguish the human versus natural impact on genetic connectivity.
There are two main challenges when quantifying the impact of man-made migration barriers on the genetic connectivity of riverine fishes (see Hänfling and Weetman 2006). First, historical processes (e.g. past upstream colonization) must be ruled out. Genetic signals of past colonization are expected to disappear over time as populations approach migration-drift equilibrium. Secondly, the effect of barriers must be distinguished from natural processes (e.g. downstream-biased gene flow). Neutral genetic diversity and population structure depend on the interplay between genetic drift and gene flow (Hutchison and Templeton 1999). It is assumed that the size of a habitat patch is a suitable indicator for effective population size (Ne) (Frankham 1996), and hence genetic drift (Hartl and Clark 1997). Geographical distance is a good indicator for gene flow according to a stepping-stone model leading to an IBD pattern at equilibrium (Hutchison and Templeton 1999). The contribution of anthropogenic barriers to genetic structure can therefore be assessed after control for habitat size or geographical distance. However, the number of barriers may be strongly correlated with habitat size and geographical distance; long rivers tend to have more barriers, and man tends to build more barriers on small rivers. The effect of man-made barriers on genetic diversity and population structure in a river system must therefore be evaluated against a multivariate model accounting for the system’s natural levels of genetic drift and gene flow simultaneously.
Multivariate geographical modeling of genetic data belongs to the field of landscape genetics (Manel et al. 2003; Spear et al. 2005; Broquet et al. 2006). The field asserts that landscape and habitat features largely determine dispersal and gene flow. The value of detailed consideration of landscape variables for understanding the process of population differentiation has been recognized in river landscapes as well (Koizumi et al. 2006; Neville et al. 2006). In this study, we model the genetic connectivity between three-spined stickleback (Gasterosteus aculeatus L.) populations from the Scheldt basin in Flanders (Belgium), to delineate guidelines for river restoration. The western European aquatic fauna has suffered from extensive anthropogenic pollution and habitat destruction during the 20th century. Twenty percent of all native fish species have been lost in Flanders (Vandelannoote and Coeck 1998). Current programs for water treatment have lowered pollution loads, opening perspectives for the restoration of the native fauna. However, man-made barriers remain a considerable obstacle to achieve this goal. Anthropogenic structures have been intensively monitored on the Flemish rivers (Monden et al. 2004), resulting in a public database implemented in a GIS environment (http://www.vismigratie.be). The Flemish government is using this database to assign restoration priorities to each tributary and barrier, based on ecological and economical criteria. However, an evolutionary perspective, taking into account the basin-wide genetic connectivity and local evolutionary potential of fish species, may greatly contribute to the river restoration program.
The three-spined stickleback represents an excellent organism for monitoring the influence of anthropogenic disturbance (Katsiadaki et al. 2002). In Belgium, its resistance to pollution, its high abundance and the absence of a stocking policy make it the only species that can provide an accurate, detailed and basin-wide picture of the impact of man-made barriers on population genetics. Because of its small size it should be sensitive to the smallest barrier. Therefore, we expect a high resolution to detect barrier-related patterns, possibly stronger than for salmonids which are larger and much better swimmers, and than for the bullhead (Cottus) which may be too sessile and naturally fragmented. Also, the generation time of sticklebacks is short, and evolutionary change may be great over a given number of years of human impact. From the perspective of river restoration, inferring guidelines from a species with a high sensitivity to barriers is preferable as it may generate a more detailed picture of the potential connectivity. We evaluated the average effect of a set of barriers on genetic diversity and differentiation in a network of stickleback populations. We opted for a river system with a high restoration priority, and a regular distribution of barriers. Both genetic diversity within and genetic differentiation among populations was modeled to compare the contribution of barriers to the contribution of natural effects. The available information also allowed evaluating the effect of barrier type and barrier height. In addition, we extended our landscape genetic analyses with two innovative aspects. First, we adapted a method of Hutchison and Templeton (1999), not only allowing us to detect the geographical factor most strongly limiting gene flow, but also the one allowing most genetic drift, which tends to be neglected in landscape models. Secondly, we not only determined the most influential landscape variables, but also evaluated the predictive power of our models, including the detection of populations that strongly deviate from geographical expectations.
Materials and methods
Sample collection
Three-spined sticklebacks were sampled from 21 sites in the eastern sub basins of the Scheldt River in Flanders (Belgium) in spring 2002 (Fig. 1; Table 1). Sticklebacks in this area belong to the low-plated upland freshwater ecotype (Raeymaekers et al. 2007). Eight sites (coded S4a–S13a, S14) were chosen at regular distances in, or as close as possible to, the main channel (Nete, Dijle and Demer). The remaining 13 sites (S5b, S9b–S9l, S13b) were chosen to be downstream and upstream on principal tributaries. Fifty adults per site were caught with a dip net or by electrofishing, and flash frozen in dry ice.
Figure 1.
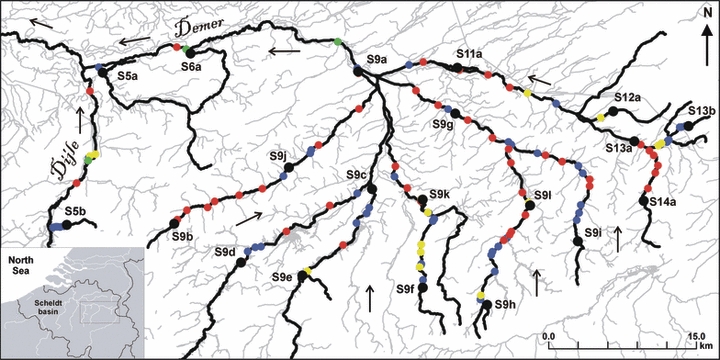
Sampling locations of 20 freshwater populations of the three-spined stickleback (Gasterosteus aculeatus) in the eastern sub-basins (Dijle and Demer) of the Scheldt River in Belgium (see inset). The most downstream population (S4a; not shown) and one more barrier are located 37 km east of population S5a. Red, blue, yellow and green dots represent water mills (n = 46), weirs (n = 57), tunnels (n = 14) and sluices (n = 4), respectively. Small arrows mark flow direction. Codes as in Table 1.
Table 1.
Characteristics of 21 sampling locations of three-spined stickleback (Gasterosteus aculeatus) populations in Belgium, genotyped at six microsatellite loci.
| Population | Code | Basin | Latitude | Longitude | HE | HO | AR |
|---|---|---|---|---|---|---|---|
| Mechelen | S4a | Nete | 51°03.734′ | 4°27.588′ | 0.83 | 0.83 | 10.34 |
| Werchter | S5a | Dijle | 50°57.806′ | 4°43.276′ | 0.78 | 0.77 | 9.46 |
| Vaalbeek | S5b | Dijle | 50°49.466′ | 4°40.105′ | 0.65 | 0.63 | 6.22 |
| Aarschot | S6a | Demer | 50°58.831′ | 4°50.898′ | 0.79 | 0.76 | 8.20 |
| Zelem | S9a | Demer | 50°57.761′ | 5°05.431′ | 0.73 | 0.72 | 7.66 |
| Boutersem | S9b | Demer | 50°49.506′ | 4°49.405′ | 0.66 | 0.68 | 5.78 |
| Zoutleeuw | S9c | Demer | 50°51.310′ | 5°6.531′ | 0.78 | 0.75 | 8.89 |
| Hoegaarden | S9d | Demer | 50°47.355′ | 4°55.150′ | 0.79 | 0.77 | 9.471 |
| Landen | S9e | Demer | 50°46.613′ | 5°00.441′ | 0.74 | 0.76 | 6.50 |
| Gingelom | S9f | Demer | 50°45.882′ | 5°10.842′ | 0.69 | 0.74 | 5.35 |
| Stevoort | S9g | Demer | 50°55.393′ | 5°13.823′ | 0.78 | 0.80 | 7.02 |
| Mechelen-Bovelingen | S9h | Demer | 50°44.904′ | 5°16.350′ | 0.55 | 0.56 | 3.67 |
| Borgloon | S9i | Demer | 50°48.318′ | 5°24.287′ | 0.44 | 0.43 | 3.46 |
| Kortenaken | S9j | Demer | 50°52.513′ | 4°59.953′ | 0.75 | 0.74 | 7.74 |
| St-Truiden | S9k | Demer | 50°50.702′ | 5°10.900′ | 0.80 | 0.81 | 7.32 |
| Wellen | S9l | Demer | 50°50.312′ | 5°20.196′ | 0.77 | 0.78 | 7.28 |
| Kermt | S11a | Demer | 50°57.900′ | 5°14.043′ | 0.73 | 0.72 | 7.56 |
| Diepenbeek | S12a | Demer | 50°55.409′ | 5°27.509′ | 0.79 | 0.73 | 7.59 |
| Bilzen | S13a | Demer | 50°53.749′ | 5°29.290′ | 0.81 | 0.83 | 7.73 |
| Zutendaal | S13b | Demer | 50°54.539′ | 5°34.006′ | 0.64 | 0.66 | 4.37 |
| Alt-Hoeselt | S14 | Demer | 50°50.479′ | 5°30.031′ | 0.79 | 0.71 | 7.39 |
HE, expected (unbiased) heterozygosity; HO, observed heterozygosity; AR, allelic richness standardized for 18 diploid individuals.
DNA extraction and microsatellite amplification
Genomic DNA was extracted from fin clips using a silica-based purification method (Elphinstone et al. 2003). Allelic variation was assessed at six microsatellite loci (Gac5196, Gac2111, Gac4170, Gac1097, Gac7033, Gac1125) developed by Largiadèr et al. (1999). All loci could be amplified simultaneously with the Qiagen® Multiplex PCR Kit (Qiagen, Venlo, The Netherlands). The 12.5 μL PCR contained 1–100 ng genomic DNA, 0.050 μm (Gac5196, Gac2111, Gac7033, Gac1125), 0.75 μm (Gac4170), 1 μm (Gac1097) forward and reverse primer, 1× Qiagen Multiplex PCR master Mix (3 mm MgCl2) and RNase-free water. The reaction consisted of an initial activation step of 15 min at 95°C, followed by 30 cycles of 30 s at 94°C, 90 s at 55°C and 1 min at 72°C. A final elongation step of 10 min at 72°C was performed. PCR products were visualized on an ABI3130 Avant Genetic analyzer (Applied Biosystems, Foster City, CA). Allele sizes were determined by means of an internal GeneScan 500-LIZ size standard and genotypes were obtained using genemapper 3.7 (Applied Biosystems). Genotypes were checked for scoring errors that might be attributable to stutter-products, large allele dropout or to the presence of null-alleles, using the software micro-checker 2.3 (Van Oosterhout et al. 2004).
Genetic data analysis
Genetic diversity was evaluated based on genotype and allele frequencies, the level of polymorphism, and the observed and unbiased expected heterozygosity (HO and HE) using genetix 4.04 (Belkhir et al. 2002). Allelic richness (AR) was quantified in fstat 2.9.3.2 (Goudet 1995) and averaged over loci. Departures from Hardy–Weinberg equilibrium (HWE) were calculated and tested with genepop 3.4 (Raymond and Rousset 1995). Tests for linkage disequilibrium (LD) were performed according to a permutation method implemented in genetix. Population differentiation was quantified in genetix using the standardized allelic variance FST, estimated as θ (Weir and Cockerham 1984). Overall and pairwise FST values were tested for significance against 104 random permutations of the data in genetix. Genetic and geographical distance matrices were visualized by nonmetric multidimensional scaling (NMDS) plots with the function isomds in s-plus (StatSoft, Tulsa, OK).
Hierarchical dendritic habitats like rivers deviate in multiple ways from the infinite island model (Wright 1951), including marked asymmetries in gene flow and subpopulation sizes. In particular, migration-drift equilibrium, a critical assumption for the most analytical methods if gene flow estimates are to reflect ongoing rather than historical processes, might never be attained when populations are prone to cycles of decline and recovery. Reaching equilibrium conditions under a stepping-stone model, which may be more applicable to river populations (Vrijenhoek 1998; Hänfling and Weetman 2006), is expected to take an extremely long period of time (Slatkin 1993; Efremov 2004). To investigate whether our stickleback populations had attained regional migration-drift equilibrium, we used the graphical IBD method of Hutchison and Templeton (1999). This method is based on the changing relative influences of gene flow and genetic drift as populations become more geographically separated. Hutchison and Templeton (1999) predicted that, assuming a stepping-stone model of regional population structure, a strong IBD relationship throughout the sampled range, and increasing variability in genetic differentiation with geographical distance, is compatible with regional migration-drift equilibrium. We tested this prediction with the regression of pairwise FST on river distance (i.e. IBD), and with the correlation of the absolute value of the residuals of this relationship with river distance.
Geographical information
Geographical information was obtained from a digital map of the river system (Aminal Section Water 2000), and from a digital map containing the migration barriers on the main river channels (Monden et al. 2004; Fig. 1). We carried out complementary field surveys with a Global Positioning System (Etrex, Garmin) to locate sampling sites, and to type and digitize additional barriers on some unexplored river sections between sampling sites. We also recorded the width of the stream at each sampling site, calculated as the mean value of two independent measurements. Migration barriers consisted of several types but were classified in three main categories. The first category included water mills, which can be considered as constructions with a long history (100–500 years). The second category (weirs) consisted of more recent (<100 years) hydraulic artifacts like small dams and inappropriately constructed culverts creating waterfalls. The third category included tunnels, which are not physical barriers but which can be up to several kilometers long, and sluices (water channels with a gate controlling water levels), which are temporary barriers only. We used the vertical height of a barrier as a measure of barrier strength. These data were obtained from Monden et al. (2004), or were measured during our own field surveys. As the Scheldt basin is a shallow watershed, there are no natural rapids hindering dispersal or gene flow.
All geographical information was combined in a Geographical Information System (geomedia professional 5.2, Intergraph Co., Huntsville, AL) and rasterized in geomedia grid 5.2 (Intergraph Co.). Using the standard cost analysis tool in geomedia grid we calculated watershed position, defined as the maximal upstream river distance from a sampling site. Note that high values reflect a low degree of isolation. A custom command was developed in the geomedia grid environment to automate the calculation of all pairwise distances along waterways between sampling sites. This function (available from the authors) was based on the standard cost analysis tool and implemented river distance (km), total number of barriers, number of each type of barrier or total vertical barrier height (m) as friction. These calculations revealed that populations were separated by up to 116 km, 23 mills, eight tunnels and sluices, 20 weirs and 31 m vertical height. We also calculated pairwise average habitat width (generally enabling high Ne) and pairwise average watershed position (corresponding to a low degree of isolation), which must be seen as geographical estimates of genetic similarity among each pair of sampling sites (pairs with high averages should be genetically more similar).
Landscape genetics
Analyses focused on genetic diversity within sites, and genetic differentiation between sites. For all tests, variables were inspected for normality and log10-transformed when necessary. First, we tested the impact of river distance, barrier characteristics, watershed position and habitat width on AR within sites. River distance, total number of barriers, total barrier height and the number of each barrier type were calculated starting from the most downstream population (S4a). Pearson correlations with AR (based on n = 21 populations) were tested in statistica 6.0. Secondly, we tested the impact of pairwise river distance, barrier characteristics, log-transformed average watershed position, and log-transformed average habitat width on genetic differentiation based on pairwise matrix correlations. Mantel correlations (Mantel 1967) with pairwise FST (n = 210 pairwise combinations) were calculated and tested with a matrix permutation method programmed in s-plus.
To identify the anthropogenic effects of barriers on genetic diversity and genetic differentiation, we carried out multiple regression analyses, evaluating the effect of barriers against multiple geographical features accounting for the natural levels of genetic drift and gene flow. Response variables were AR and pairwise FST. Multicollinearity among variables that are geographically linked may be considerable and may interfere with the detection of the most relevant ones. Therefore we tested for multicollinearity among explanatory variables using the variance inflation factor (VIF), which should be <10 (Neter et al. 1996, p. 387). Significance of geographical features was assessed with a parametric regression model for AR in statistica, and with a nonparametric regression model for pairwise FST in fstat. Model fit was compared based on AICC criteria following Koizumi et al. (2006). We examined the predictive power of the models with the coefficient of determination (R2 and adjusted R2) and by identifying studentized residuals with absolute values larger than two, pointing to observations that are strongly under- or overestimated. To improve the predictive power, models were extended with detailed barrier characteristics. Here we neglected multicollinearity, as this does not affect the precision of predictions if the predicted variable follows the same multicollinearity pattern (Neter et al. 1996, p. 410). Predictive power among extended models was compared based on adjusted R2.
Finally, we adapted the method of Hutchison and Templeton (1999) to detect the strongest genetic barrier. The method relies on IBD plots to assess if the stochastic effect of drift gradually becomes more important than the homogenizing effect of gene flow as populations become geographically more separated. Conversely, under or close to migration-drift equilibrium, it should be possible to use isolation-by-geographical feature plots to detect the geographical feature that strongest determines genetic isolation. We calculated the correlation of each geographical feature with pairwise FST to determine the strongest barrier to gene flow. In addition, we correlated the absolute values of the residuals of each isolation-by-geographical feature plot with the corresponding geographical feature. As these residuals account for the variability in genetic differentiation, these correlations should reveal the geographical feature allowing most genetic drift. The analyses based on correlations (gene flow) and scatter (genetic drift) of the isolation-by-geographical feature plots should corroborate each other and point to the most isolating geographical factor.
Results
Genetic diversity, genetic differentiation, and equilibrium conditions
Mean AR was maximal (10.34) in S4a, the most downstream population, and minimal (3.46) in S9i, one of the most upstream populations (Table 1). Observed and expected heterozygosity ranged between 0.43 and 0.83. From a total of 315 tests (15 locus pairs in 21 populations), LD was detected nine times after Bonferroni correction. This was observed three times in population S13a, and two times in population S6a and S13b. Physical linkage is unlikely, because each of the nine cases occurred in a different locus pair, and because four of six loci are on different linkage groups in other populations (Peichel et al. 2001). Deviations from HWE were significant in populations S6a and S14 because of few homozygotes with rare alleles. There was no evidence for systematic scoring errors according to micro-checker.
Allelic richness (Table 2A) strongly decreased with the river distance (r = −0.62; P = 0.0029; Fig. 2A) and with the total number of barriers from site S4a (r = −0.86; P < 0.0001; Fig. 2C). Among barrier types, the correlation was strongest with the number of weirs (r = −0.82; P < 0.0001), and weakest with the number of tunnels and sluices (r = −0.63; P = 0.0022). Watershed position (r = 0.75; P < 0.0001) and habitat width (r = 0.60; P = 0.0038) were positively correlated with AR. Overall genetic differentiation was high (FST = 0.15; P < 0.0001). Only two of the 210 pairwise FST values were not significant after Bonferroni correction (S9a vs S9j –FST = 0.010; S12a vs S13a –FST = 0.008). A NMDS plot of pairwise FST values (Fig. 3A) showed that each of the upstream population S5b, S9h, S9i and S13b was highly differentiated from a cluster of downstream populations (S4a, S5a and S6a) and a cluster of downstream and upstream populations. Populations showed a significant IBD relationship (Table 2B; Fig. 2B; r = 0.46; P = 0.0039). Other geographical features and barrier types, except the number of tunnels and sluices, also correlated with pairwise FST (Table 2B). The strongest correlation here was with the total number of barriers (r = 0.70; P = 0.0003).
Table 2.
Correlations of landscape variables with genetic diversity and genetic differentiation obtained from 21 three-spined stickleback populations. (A) Pearson correlations of geographical features with allelic richness (AR); (B) Mantel matrix correlations of pairwise geographical features with pairwise FST, and with the absolute values of residual pairwise FST.
| (A) AR | (B) †Pairwise FST | ||
|---|---|---|---|
| Geographical feature | R | R | Residual |
| River distance (km) | −0.62** | 0.46** | 0.13 |
| Mills | −0.67*** | 0.53** | 0.26* |
| Tunnels and sluices | −0.63** | 0.35 | −0.08 |
| Weirs | −0.82*** | 0.68*** | 0.32* |
| All barriers | −0.86*** | 0.70*** | 0.17 |
| Barrier height (m) | −0.78*** | 0.68*** | 0.24* |
| Log10(habitat width) | 0.60** | −0.57*** | −0.21* |
| Log10(watershed position) | 0.75*** | −0.37* | −0.16 |
Underlined correlations are plotted in Fig. 2.
P < 0.05;
P < 0.01;
P < 0.001.
P-values are given after 10 000 randomisations.
Figure 2.
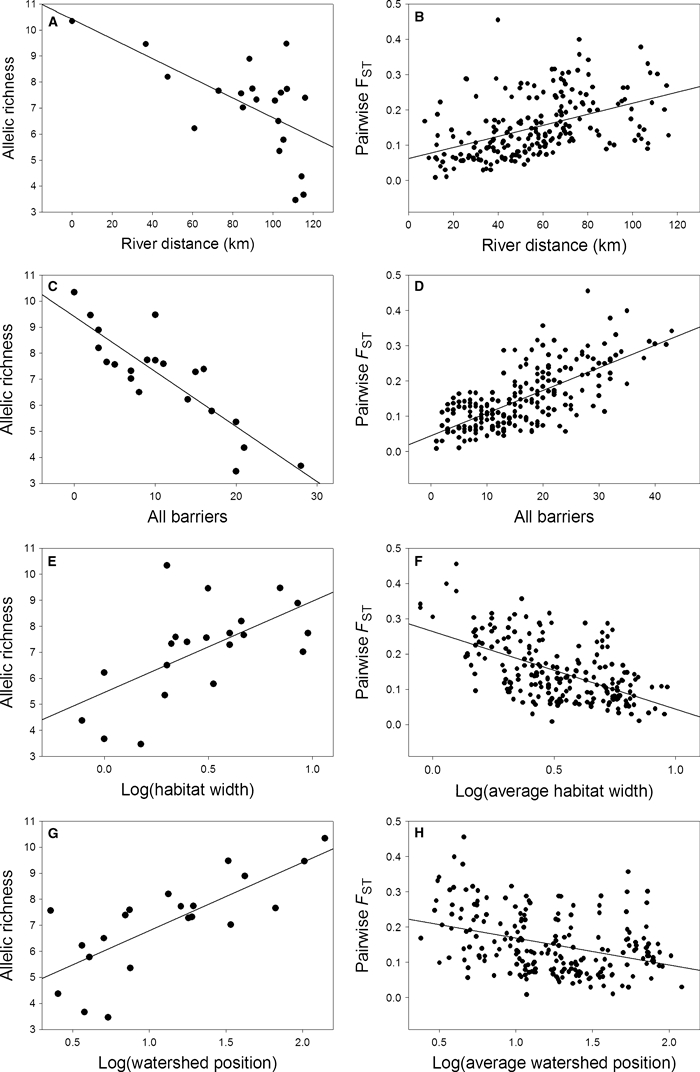
Relationship between geographical features and genetic diversity (left) and genetic differentiation (right) based on six microsatellite loci in 21 three-spined stickleback populations. Predictors are (A,B) river distance; (C,D) total number of barriers; (E,F) log10 transformed habitat width (F: pairwise averaged) and (G,H) log10 transformed watershed position (H: pairwise averaged).
Figure 3.
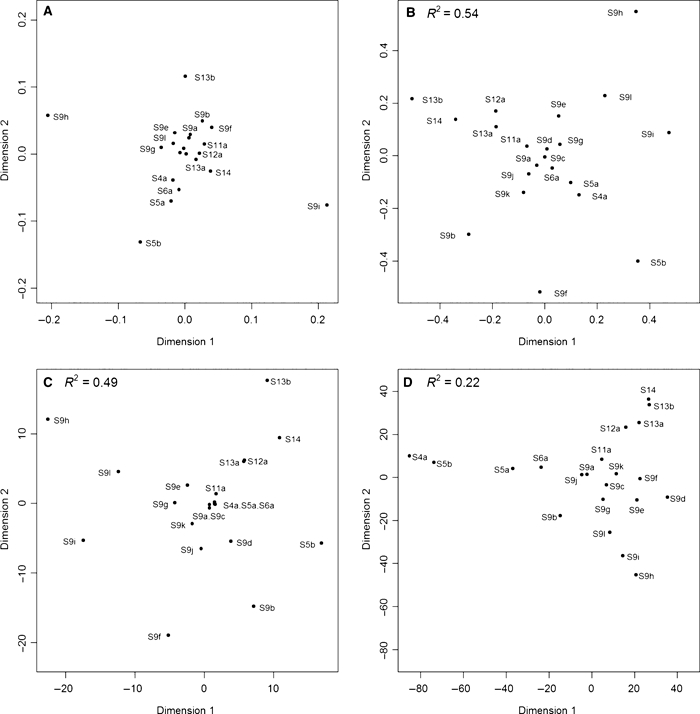
Comparison of Nonmetric Multidimensional Scaling plots based on (A) observed pairwise FST (stress: 0.11); four central points (S9c, S9d, S9j and S9k) are left unlabelled; (B) pairwise FST predicted from the model in Table 3B (stress: 0.19); (C) pairwise number of barriers (stress: 0.15) and (D) pairwise river distance (stress: 0.18) among 21 three-spined stickleback populations. R2 values (panel B–D) refer to the explained variation in observed pairwise FST (panel A).
Despite the significant IBD pattern, the variability in FST (as revealed by the absolute value of the residuals) did not increase significantly with distance (Table 2B; r = 0.13; P = 0.1720), suggesting a deviation from migration-drift equilibrium. However, the absolute value of residual FST increased significantly with barrier height and the number of weirs and mills, and decreased significantly with habitat width (Table 2B). This suggests that migration-drift equilibrium is associated with geographical features other than river distance.
Modeling genetic connectivity in riverine landscapes
A multiple regression of the number of barriers, habitat width and watershed position explained 76% of the variation in AR (adjusted R2 = 0.76; F3,17 = 22.30; P < 0.0001; VIFMAX = 2.09; Tables 3A and 4A). The relationship between AR and each of the predictors is represented in Fig. 2C,E,G. The number of barriers was most strongly related to AR and was the only significant effect in the model (P = 0.0012). Among the model from Table 3A and all its subsets, the AICC criterion (see Koizumi et al. 2006) supported a model that included both barriers and watershed position (AICC = −1.40), followed by the model including only barriers (AICC = −0.43). Other models were not supported by the AICC criterion (i.e. ΔAICC > 2). A studentized residual larger than two indicated that AR was underestimated in population S9d (observed AR = 9.47; predicted AR = 7.84). An extremely negative residual revealed that population S9i was much less genetically diverse than expected (observed AR = 3.46; predicted AR = 5.20). Model extensions replacing the total number of barriers by detailed barrier characteristics maximally explained 78% of the variation as revealed by the adjusted R2.
Table 3.
(A) Multiple regression analysis of number of barriers, log10(habitat width) and log10(watershed position) on allelic richness (AR; R2 = 0.80; adjusted R2 = 0.76; F3,17 = 22.30; P < 0.0001). (B) Nonparametric multiple regression of number of barriers, log10(pairwise average habitat width), log10(pairwise average watershed position) and river distance on pairwise FST (R2 = 0.54; adjusted R2 = 0.53).
| (A) AR | (B) †Pairwise FST | |||||
|---|---|---|---|---|---|---|
| Effect | df | MS | F | P-value | β | P-value |
| Intercept | 1 | 44.60 | 56.51 | <0.0001 | – | – |
| All barriers | 1 | 11.98 | 15.17 | 0.0012 | 0.00490 | 0.0001 |
| Log10(habitat width) | 1 | 0.39 | 0.49 | 0.4935 | −0.09611 | 0.0048 |
| Log10(watershed position) | 1 | 2.51 | 3.18 | 0.0926 | 0.00551 | 0.7742 |
| River distance | – | – | – | – | 0.00027 | 0.3776 |
| Error | 17 | 0.79 | ||||
Significant P-values are in bold.
P-values are given after 10 000 randomizations.
Table 4.
Correlations among the explanatory variables used in the landscape models from Table 3. (A) Pearson correlations among the geographical features from Table 3A. (B) Mantel matrix correlations among the pairwise geographical features from Table 3B.
| (A) | All barriers | Log10(habitat width) | Log10(watershed position) |
|---|---|---|---|
| All barriers | – | −0.5701 | −0.6858 |
| Log10(habitat width) | 0.007 | – | 0.5592 |
| Log10(watershed position) | 0.001 | 0.008 | – |
| (B)† | All barriers | Log10(habitat width) | Log10(watershed position) | River distance |
|---|---|---|---|---|
| All barriers | – | −0.5553 | −0.5447 | 0.5170 |
| Log10(habitat width) | 0.0008 | – | 0.4261 | −0.4390 |
| Log10(watershed position) | 0.0004 | 0.02310 | – | 0.0146 |
| River distance | 0.0004 | 0.0024 | 0.4502 | – |
Correlation coefficients are given above the diagonal, P-values below the diagonal. Significant P-values are in bold.
P-values are given after 10 000 randomizations.
A nonparametric multiple regression of the number of barriers, pairwise average habitat width, pairwise average watershed position and river distance explained 53% of the variation in pairwise FST (adjusted R2 = 0.53; VIFMAX = 2.41; Tables 3B and 4B). The relationship between pairwise FST and each of the predictors is represented in Fig. 2B,D,F,H. Genetic differentiation was strongly linked to the total number of barriers (β = 0.005; P = 0.0001). The relation with pairwise average habitat width was significantly negative (β = −0.096; P = 0.0048). The effect of pairwise average watershed position and river distance was not significant. Among the model from Table 3B and all its subsets, the model including only barriers was the best (AICC = −64.81), followed by the model including barriers and pairwise average habitat width (AICC = −64.09). Other models were not supported by the AICC criterion. Models extended with all barrier characteristics maximally explained 60% of the variation as revealed by the adjusted R2.
Nonmetric multidimensional scaling plots of observed pairwise FST values (Fig. 3A) and pairwise FST values predicted from the model (Fig. 3B; Table 3B) display a similar ordination of the stickleback populations, confirming the predictive power of the model: upstream populations, in particular S5b, S9h, S9i and S13b, were in general more divergent than downstream populations. Figure 3 also shows that barrier distances (Fig. 3C) contributed much more to the model predictions than river distances (Fig. 3D). At the same time, much more scatter in Fig. 3B versus Fig. 3A illustrates why the predictive power of the model is not higher. Studentized residuals with absolute values larger than two indicated that the divergence among some population pairs was considerably under- (e.g. S9i vs S9h) or overestimated (S9f vs S9d and S9f vs S9b), respectively. For instance, the divergence between S9i and S9h on a neighboring tributary (FST = 0.45) largely exceeded the predicted value (0.24). Among all geographical features, this outlier was particularly insufficiently explained by river distance, as can be noticed from Fig. 2B. Residual pairwise FST was negatively associated with pairwise mean AR (r = −0.36; P = 0.0320), indicating that the model from Table 3B lacks sufficient geographical information to predict low FST among population pairs with high mean genetic variability and vice versa. For instance, adding pairwise mean AR to the model explained 30% more variability (R2 = 0.83). Absolute values of residual pairwise FST were also negatively associated with pairwise mean AR (r = −0.28; P = 0.0287), indicating that, because of genetic drift, predicting FST from geography is more difficult among population pairs with low mean genetic variability than among population pairs with high mean genetic variability.
Discussion
Restoring the connectivity of river systems is central to many river restoration programs. European and Flemish legislation postulates that human activities limiting the free migration of riverine organisms should be minimized (Monden, 2007). Potential barriers to migration have been intensively monitored on the Flemish rivers and streams. As a basis for river restoration, we analyzed the genetic structure of the most abundant native fish in the region, the three-spined stickleback, to obtain a basin-wide picture of the geographical features that affect connectivity. Population genetics emphasizes a concern for conserving genetic diversity in restoration projects. Stickleback population genetics provided connectivity estimates on a geographical scale and with a resolution that may be difficult to achieve with other methods or with other (rare or endangered) fish species.
Geographical determinants of genetic diversity and genetic differentiation
Levels of genetic differentiation between stickleback populations of the eastern sub-basins of the Scheldt River were high, consistent with earlier studies indicating marked differentiation in freshwater stickleback within and among watersheds (Taylor and Mcphail 2000; Reusch et al. 2001; Hendry et al. 2002; Hendry and Taylor 2004; Raeymaekers et al. 2005, 2007; Mäkinen et al. 2006; Moore et al. 2007). Within this network of upland rivers, watershed position was accompanied by a strong cline in genetic diversity. Genetic diversity can often be reliably predicted from the position in the river system, as evidenced in bullhead (Hänfling et al. 2002; Hänfling and Weetman 2006) and guppies (Crispo et al. 2006).
Barrier characteristics revealed strong univariate relationships with genetic diversity and genetic differentiation (Table 2). Among barrier types, weirs were more influential than water mills. This is against expectations, because mills are thought to have affected stickleback genetic structure during a longer time period. However, weirs were more numerous than mills on small tributaries which may be easier to obstruct, resulting in a larger overall effect. From the perspective of river restoration, it should be realized that not only ancient mills, but also modern constructions, can have a large impact on the genetic connectivity of fish populations, and that this impact may be location-dependent. Unfortunately, this category of barriers also includes some fish passages with a poor design. The influence of tunnels and sluices on genetic diversity was lowest among barrier types, and there was no significant correlation with genetic differentiation. This corresponds to recent tagging studies (H. Verbiest, personal communication) and genetic simulations (Knaepkens et al. 2004) demonstrating that tunnels are passable for a number of fish species, and justifies the low restoration priority the Flemish river restoration program has assigned to these constructions (Monden et al. 2004). Barrier height was a good predictor for genetic diversity and genetic differentiation, but did not perform better than the total number of barriers. This may be due to measurement error associated with seasonal variation in water levels.
Significant correlations between AR and watershed position (this study) or upstream size (e.g. Hänfling et al. 2002; Hänfling and Weetman 2006) show that these measures can be considered indicative for the geographical range of riverine populations, accounting for effective population size, drift, or higher rates of immigration in the downstream populations (Frankham 1996; Hänfling et al. 2002). Allelic richness was also correlated with habitat width, which may represent an alternative measure for effective population size or drift. It is also possible that habitat size or watershed position just reflect to what extent the position within the river system is isolated by distance. Habitat width and watershed position, averaged over population pairs, were negatively correlated with pairwise FST, suggesting that small – or geographically isolated – populations are more differentiated.
In summary, the dependence of genetic diversity and genetic differentiation on gene flow and genetic drift is apparent in our data set. Less clear is which landscape variable accounts best for the interplay between gene flow and genetic drift, and which variable is the strongest determinant of genetic structure. First, we extend Hutchison and Templeton’s (1999) rule towards other geographical features, acknowledging that river distance is not necessarily a good determinant of migration-drift equilibrium. Evidence from slopes (gene flow) and scatter (drift) derived from the isolation-by-river distance plot did not corroborate each other. Secondly, it seems unwise to leave part of the natural variation unexplained by relying on just one variable, as anthropogenic barriers may pick up this variation. The suggested impact of barriers should therefore appear from a multivariate landscape model containing information based on multiple geographical features.
Geographical determinants of migration-drift equilibrium
Throughout a sampled range of maximally 116 km along waterways, we observed a significant IBD pattern, consistent with regional migration-drift equilibrium (Slatkin 1993; Hutchison and Templeton 1999). On the other hand, the increase of genetic drift (as quantified with absolute values of the residuals) with river distance was not significant. According to Hutchison and Templeton (1999), this suggests that the equilibrium may be only partially attained. FST may still perform as a useful inverse index of gene flow when equilibrium conditions are not fully met (Hänfling and Weetman 2006), although gene flow estimates may not reflect exclusively ongoing population dynamics. Interestingly, our extension of the method of Hutchison and Templeton (1999) showed that other geographical features, in particular weirs and mills, were significant determinants of the balance between genetic drift and gene flow. This suggests that these barriers have disturbed the natural balance between genetic drift and gene flow, such that migration-drift equilibrium with respect to river distance does no longer exist in the current riverscape. An equivalent explanation is that a small number of outlier (isolated) populations biased the IBD pattern (see Koizumi et al. 2006), while fitting the predictions of an ‘isolation-by-barrier’ pattern. Because of genetic drift, predicting FST becomes particularly more difficult when more barriers separate populations.
Modeling genetic connectivity in riverine landscapes
Geography was an important determinant of stickleback genetic structure. The model fit for genetic diversity (max. 78%) was higher than for genetic differentiation (max. 60%). This is expected because at neutral loci the latter is much more influenced by the stochastic outcome of genetic drift (Hartl and Clark 1997). Geography may be a good predictor of the magnitude of drift, determining AR, but it cannot predict the outcome of drift, determining which alleles will be lost or preserved.
The models for both genetic measures (diversity and differentiation) pointed to a major effect of barriers, reducing AR and long-term gene flow. In the case of genetic diversity, the effect of barriers was substantial and likely represented a true anthropogenic effect, as two covariates in the model accounted for natural patterns (watershed position and habitat width). In the case of genetic differentiation, confidence that barriers truly affect population structure was obtained after incorporation of river distance, habitat width and watershed position. Habitat width also appeared to influence genetic differentiation, suggesting that smaller habitat patches have an isolating effect as well.
R2 values of population-based models appear to be fairly consistent across riverscapes. Crispo et al. (2006) found an almost identical maximal R2 value of 60% for a model explaining pairwise FST values among riverine guppy (Poecilia) populations by a combination of distance, number of waterfalls, predation regime and habitat characteristics. Koizumi et al. (2006) detected a maximal R2 value of 54% for a model explaining the [FST/(1−FST)] matrix in stream-dwelling Dolly Varden charr (Salvelinus) by geographical distance, potential anthropogenic disturbance, habitat size and the occurrence of a physical barrier. These percentages are high in an ecological context, making population-based riverscape models useful to quantify the effect of geography or environment. It is worth mentioning here that the performance of individual-based landscape models has been found much lower (R2 values lower than 1%; Broquet et al. 2006). Instead, in a case study of the blotched tiger salamander (Ambystoma; Spear et al. 2005), population-based landscape models explained up to 80% of the variation in pairwise FST. Such high percentage is remarkable, because of the uncertainty associated with the multiple pathways salamanders may use to disperse through the terrestrial matrix. In contrast, uncertainty with respect to the migratory pathways is low in riverine organisms like guppies (Crispo et al. 2006), Dolly Varden charr (Koizumi et al. 2006) and sticklebacks (this study). However, our models predicted that, in some populations, neither barriers, habitat width nor watershed position could account for the high or low AR. This also translated in cases of severe over- or underestimation of genetic differentiation. Hence, our models for pairwise FST performed poorly, in the sense that there is potential for improvement by the incorporation of information sufficiently linked to genetic diversity. The marginally higher R2 values of extended models indicated that improvement based on geographical information might be difficult to achieve. Compared with terrestrial population-based models, models in riverscapes may leave more variation unexplained because of demographic events (e.g. local extinctions) that naturally characterizes rivers (Fagan 2002).
Conclusions and applicability
Anthropogenic barriers have a severe impact on three-spined stickleback population structure in the eastern sub-basins of the Scheldt River. We showed that barriers not only affect genetic diversity, but that they also control the balance between gene flow and genetic drift. Therefore, barriers may disrupt stickleback population structure. First, physical isolation increases the risk of stochastic population extinction, which is particularly high in rivers (Fagan 2002). Secondly, inbreeding depression caused by genetic isolation might lower survival and population sizes (Saccheri et al. 1998; Brook et al. 2002). Given the marine ancestry of sticklebacks and their natural history of extinction and fast recolonization (Wootton 1976; Bell and Foster 1994; Raeymaekers et al. 2005), this may not harm the species in an evolutionary perspective. However, it may result in temporarily impoverished populations, in particular those inhabiting small, shallow tributaries or upstream rivers stretches (width: 1–2 m).
These results provide a basin-wide picture of the potential connectivity of fish populations, and of the relative impact of various barrier types in a system with a highly disturbed aquatic fauna. This knowledge is particularly helpful to guide general management strategies, including stocking policy and ecologically effective restoration projects (Giller 2005). We provide three specific recommendations for the restoration program for the system under study. First, river sections connecting S5b, S9h, S9i and S13b should receive a higher restoration priority. Secondly, weirs and mills should be treated with high priority, while it is acceptable to assign a low priority to the removal of tunnels and sluices. Thirdly, small tributaries and upstream rivers stretches should be monitored for barriers more intensively, as connectivity problems in these sections are likely to have a larger impact on the population genetics of riverine fish. Importantly, these guidelines apply to river restoration and to the whole fish community, rather than to specific conservation goals. This is because stickleback population structure may not be entirely comparable to the genetic structure of specific conservation target species in the system. In addition, barriers may be advantageous to some species by protecting populations from introgression with exotic lineages (Van Houdt et al. 2005), requiring a good knowledge of the indigenous nature of the populations under study before removing barriers.
We assessed the complexity of the natural component of the variation in genetic diversity and differentiation using a landscape genetics approach, to estimate the contribution of barriers with high confidence. Our method also detected outlier populations of low evolutionary potential, requiring special attention from conservation managers to avoid the risk of inbreeding. Extensive modeling of genetic diversity and genetic differentiation in riverscapes can maximize the detection of meaningful links between genetic structure and specific geographical or environmental components. Geographical modeling of genetic information may therefore greatly contribute to our understanding of landscape processes, population dynamics, adaptation and evolutionary potential in river systems.
Acknowledgments
We thank B. Christiaen and N. Leysen for field support, and I. Koizumi, A. P. Hendry, C. W. Epps and three anonymous referees for valuable comments. S. Monden (INBO) and the Ministry of the Flemish Community (VMM Section Water) kindly provided the digital maps. J. Spillemaeckers (InterGraph Benelux) and E. Michels provided useful and funny spatial solutions, respectively. A. Ghesquiere and J. Tosh were gentle proofreaders. J. A. M. R. received a PhD fellowship from the Fund for Scientific Research – Flanders (FWO-Vlaanderen) and was kindly hosted by A. P. Hendry during the finalization of this study. G. E. M. received a postdoctoral fellowship from FWO-Vlaanderen. Research was sponsored by FWO-Vlaanderen (project G.0142.03).
Literature cited
- Aminal Section Water. De Vlaamse Hydrografische Atlas. Vlaamse Gemeenschap, Administratie voor Milieu-, Natuur, Land- en Waterinrichting; 2000. 29-05-2000. http://www.vmm.be/water. [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. Genetix 4.04: Logiciel Sous Windows TM pour la génétique des populations. Montpellier, France: Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II; 2002. [Google Scholar]
- Bell MA, Foster SA. The Evolutionary Biology of the Threespine Stickleback. Oxford: Oxford Science Publications, Oxford University Press; 1994. [Google Scholar]
- Brook BW, Tonkyn DW, O’Grady JJ, Frankham R. Contribution of inbreeding to extinction risk in threatened species. Conservation Ecology. 2002;6:16. [Google Scholar]
- Broquet T, Ray N, Petit E, Fryxell JM, Burel F. Genetic isolation by distance and landscape connectivity in the American marten (Martes americana. Landscape Ecology. 2006;21:877–889. [Google Scholar]
- Castric V, Bonney F, Bernatchez L. Landscape structure and hierarchical genetic diversity in the brook charr, Salvelinus fontinalis. Evolution. 2001;55:1016–1028. doi: 10.1554/0014-3820(2001)055[1016:lsahgd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Crispo E, Bentzen P, Reznick DN, Kinnison MT, Hendry AP. The relative influence of natural selection and geography on gene flow in guppies. Molecular Ecology. 2006;15:49–62. doi: 10.1111/j.1365-294X.2005.02764.x. [DOI] [PubMed] [Google Scholar]
- Efremov VV. The rate of approach to the equilibrium value of FST in island and one-dimensional stepping-stone models of migration. Russian Journal of Genetics. 2004;40:1041–1045. [PubMed] [Google Scholar]
- Elphinstone MS, Hinten GN, Anderson MJ, Nock CJ. An inexpensive and high-throughput procedure to extract and purify total genomic DNA for population studies. Molecular Ecology Notes. 2003;3:317–320. [Google Scholar]
- Fagan WF. Connectivity, fragmentation, and extinction risk in dendritic metapopulations. Ecology. 2002;83:3243–3249. [Google Scholar]
- Fausch KD, Torgersen CE, Baxter CV, Li HW. Landscapes to riverscapes: bridging the gap between research and conservation of stream fishes. Bioscience. 2002;52:483–498. [Google Scholar]
- Frankham R. Relationship of genetic variation to population size in wildlife. Conservation Biology. 1996;10:1500–1508. [Google Scholar]
- Giller PS. River restoration: seeking ecological standards. Editor’s introduction. Journal of Applied Ecology. 2005;42:201–207. [Google Scholar]
- Goudet J. Fstat (version 1.2): a computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Hänfling B, Weetman D. Concordant genetic estimators of migration reveal anthropogenically enhanced source-sink population structure in the river sculpin, Cottus gobio. Genetics. 2006;173:1487–1501. doi: 10.1534/genetics.105.054296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänfling B, Hellemans B, Volckaert FAM, Carvalho GR. Late glacial history of the cold-adapted freshwater fish Cottus gobio, revealed by microsatellites. Molecular Ecology. 2002;11:1717–1729. doi: 10.1046/j.1365-294x.2002.01563.x. [DOI] [PubMed] [Google Scholar]
- Hartl DL, Clark AG. Principles of Population Genetics. Sunderland: Sinauer Associates, Inc. Publishers; 1997. [Google Scholar]
- Heath DD, Pollard S, Herbinger C. Genetic structure and relationships among steelhead trout (Oncorhynchus mykiss) populations in British Columbia. Heredity. 2001;86:618–627. doi: 10.1046/j.1365-2540.2001.00867.x. [DOI] [PubMed] [Google Scholar]
- Heggenes J, Røed KH. Do dams increase genetic diversity in brown trout (Salmo trutta)? Microgeographic differentiation in a fragmented river. Ecology of Freshwater Fish. 2006;15:366–375. [Google Scholar]
- Hendry AP, Taylor EB. How much of the variation in adaptive divergence can be explained by gene flow? - An evaluation using lake-stream stickleback pairs. Evolution. 2004;58:2319–2331. doi: 10.1111/j.0014-3820.2004.tb01606.x. [DOI] [PubMed] [Google Scholar]
- Hendry AP, Taylor EB, Mcphail JD. Adaptive divergence and the balance between selection and gene flow: lake and stream stickleback in the Misty system. Evolution. 2002;56:1199–1216. doi: 10.1111/j.0014-3820.2002.tb01432.x. [DOI] [PubMed] [Google Scholar]
- Hughes JM, Bunn SE, Hurwood DA, Choy S, Pearson RG. Genetic differentiation among populations of Caridina zebra (Decapoda: Atyidae) in tropical rainforest streams, northern Australia. Freshwater Biology. 1996;36:289–296. [Google Scholar]
- Hutchison DW, Templeton AR. Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution. 1999;53:1898–1914. doi: 10.1111/j.1558-5646.1999.tb04571.x. [DOI] [PubMed] [Google Scholar]
- Katsiadaki I, Scott AP, Mayer I. The potential of the three-spined stickleback (Gasterosteus aculeatus L.) as a combined biomarker for oestrogens and androgens in European waters. Marine Environmental Research. 2002;54:725–728. doi: 10.1016/s0141-1136(02)00110-1. [DOI] [PubMed] [Google Scholar]
- Kelly MW, Rhymer JM. Population genetic structure of a rare unionid (Lampsilis cariosa) in a recently glaciated landscape. Conservation Genetics. 2005;6:789–802. [Google Scholar]
- Kinnison MT, Bentzen P, Unwin MJ, Quinn TP. Reconstructing recent divergence: evaluating nonequilibrium population structure in New Zealand chinook salmon. Molecular Ecology. 2002;11:739–754. doi: 10.1046/j.1365-294x.2002.01477.x. [DOI] [PubMed] [Google Scholar]
- Knaepkens G, Verheyen E, Galbusera P, Eens M. The use of genetic tools for the evaluation of a potential migration barrier for the bullhead. Journal of Fish Biology. 2004;64:1737–1744. [Google Scholar]
- Koizumi I, Yamamoto S, Maekawa K. Decomposed pairwise regression analysis of genetic and geographic distances reveals a metapopulation structure of stream-dwelling Dolly Varden charr. Molecular Ecology. 2006;15:3175–3189. doi: 10.1111/j.1365-294X.2006.03019.x. [DOI] [PubMed] [Google Scholar]
- Largiader CR, Fries V, Kobler B, Bakker TCM. Isolation and characterization of microsatellite loci from the three-spined stickleback (Gasterosteus aculeatus L. Molecular Ecology. 1999;8:342–344. [PubMed] [Google Scholar]
- Maes GE, Raeymaekers JAM, Pampoulie C, Seynaeve A, Goemans G, Belpaire C, Volckaert FAM. The catadromous European eel Anguilla anguilla (L.) as a model for freshwater evolutionary ecotoxicology: relationship between heavy metal bioaccumulation, condition and genetic variability. Aquatic Toxicology. 2005;73:99–114. doi: 10.1016/j.aquatox.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Mäkinen HS, Cano JM, Merilä J. Genetic relationships among marine and freshwater populations of the European three-spined stickleback (Gasterosteus aculeatus) revealed by microsatellites. Molecular Ecology. 2006;15:1519–1534. doi: 10.1111/j.1365-294X.2006.02871.x. [DOI] [PubMed] [Google Scholar]
- Manel S, Schwartz MK, Luikart G, Taberlet P. Landscape genetics: combining landscape ecology and population genetics. Trends in Ecology & Evolution. 2003;18:189–197. [Google Scholar]
- Mantel N. The detection of disease clustering and generalised regression approach. Cancer Research. 1967;27:209–220. [PubMed] [Google Scholar]
- Meldgaard T, Nielsen EE, Loeschcke V. Fragmentation by weirs in a riverine system: a study of genetic variation in time and space among populations of European grayling (Thymallus thymallus) in a Danish river system. Conservation Genetics. 2003;4:735–747. [Google Scholar]
- Michels E, Cottenie K, Neys L, De Gelas K, Coppin P, De Meester L. Geographical and genetic distances among zooplankton populations in a set of interconnected ponds: a plea for using GIS modelling of the effective geographical distance. Molecular Ecology. 2001;10:1929–1938. doi: 10.1046/j.1365-294x.2001.01340.x. [DOI] [PubMed] [Google Scholar]
- Monden S. Vismigratie en het oplossen van vismigratieknelpunten. Water. 2007;30:1–4. [Google Scholar]
- Monden S, Van Liefferinge C, Vandenauweele I, Simoens I, Beyens J, Denayer B, Yseboodt R, et al. 2004. Databank vismigratieknelpunten op prioritaire waterlopen in het Vlaamse gewest. IBW-UIA databank, http://www.vismigratie.be.
- Moore JS, Gow JL, Taylor EB, Hendry AP. Quantifying the constraining influence of gene flow on adaptive divergence in the lake-stream threespine stickleback system. Evolution. 2007;61:2015–2026. doi: 10.1111/j.1558-5646.2007.00168.x. [DOI] [PubMed] [Google Scholar]
- Morita K, Yamamoto S. Effects of habitat fragmentation by damming on the persistence of stream-dwelling charr populations. Conservation Biology. 2002;16:1318–1323. [Google Scholar]
- Neter J, Kutner MH, Nachtsheim CJ, Wasserman W. Applied Linear Statistical Models. 4th edn. New York: WCB/McGraw-Hill; 1996. [Google Scholar]
- Neville HM, Dunham JB, Peacock MM. Landscape attributes and life history variability shape genetic structure of trout populations in a stream network. Landscape Ecology. 2006;21:901–916. [Google Scholar]
- Peichel CL, Nereng KS, Ohgi KA, Cole BLE, Colosimo PF, Buerkle CA, Schluter D, Kingsley DM. The genetic architecture of divergence between threespine stickleback species. Nature. 2001;414:901–905. doi: 10.1038/414901a. [DOI] [PubMed] [Google Scholar]
- Poissant J, Knight TW, Ferguson MM. Nonequilibrium conditions following landscape rearrangement: the relative contribution of past and current hydrological landscapes on the genetic structure of a stream-dwelling fish. Molecular Ecology. 2005;14:1321–1331. doi: 10.1111/j.1365-294X.2005.02500.x. [DOI] [PubMed] [Google Scholar]
- Raeymaekers JAM, Maes GE, Audenaert E, Volckaert FAM. Detecting Holocene divergence in the anadromous-freshwater three-spined stickleback (Gasterosteus aculeatus) system. Molecular Ecology. 2005;14:1001–1014. doi: 10.1111/j.1365-294X.2005.02456.x. [DOI] [PubMed] [Google Scholar]
- Raeymaekers JAM, Van Houdt JKJ, Larmuseau MHD, Geldof S, Volckaert FAM. Divergent selection as revealed by PSTand QTL-based FST in three-spined stickleback (Gasterosteus aculeatus) populations along a coastal-inland gradient. Molecular Ecology. 2007;16:891–905. doi: 10.1111/j.1365-294X.2006.03190.x. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. Genepop (version 1.2) – population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Reusch TBH, Wegner KM, Kalbe M. Rapid genetic divergence in postglacial populations of threespine stickleback (Gasterosteus aculeatus): the role of habitat type, drainage and geographical proximity. Molecular Ecology. 2001;10:2435–2445. doi: 10.1046/j.0962-1083.2001.01366.x. [DOI] [PubMed] [Google Scholar]
- Saccheri I, Kuussaari M, Kankare M, Vikman P, Fortelius W, Hanski I. Inbreeding and extinction in a butterfly metapopulation. Nature. 1998;392:491–494. [Google Scholar]
- Slatkin M. Isolation by distance in equilibrium and nonequilibrium populations. Evolution. 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- Spear SF, Peterson CR, Matocq MD, Storfer A. Landscape genetics of the blotched tiger salamander (Ambystoma tigrinum melanostictum. Molecular Ecology. 2005;14:2553–2564. doi: 10.1111/j.1365-294X.2005.02573.x. [DOI] [PubMed] [Google Scholar]
- Taylor EB, Mcphail JD. Historical contingency and ecological determinism interact to prime speciation in sticklebacks, Gasterosteus. Proceedings of the Royal Society of London Series B-Biological Sciences. 2000;267:2375–2384. doi: 10.1098/rspb.2000.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor EB, Stamford MD, Baxter JS. Population subdivision in westslope cutthroat trout (Oncorhynchus clarki lewisi) at the northern periphery of its range: evolutionary inferences and conservation implications. Molecular Ecology. 2003;12:2609–2622. doi: 10.1046/j.1365-294x.2003.01937.x. [DOI] [PubMed] [Google Scholar]
- Van Houdt JKJ, Flamand MC, Briquet M, Dupont E, Volckaert FAM, Baret PV. Migration barriers protect indigenous brown trout (Salmo trutta) populations from introgression with stocked hatchery fish. Conservation Genetics. 2005;6:175–191. [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- Vandelannoote A, Coeck J. Rode lijst van de inheemse en ingeburgerde zoet- en brakwatersoorten en van de rondbekken in Vlaanderen. In: Vandelannoote A, Yseboodt R, Bruylants B, Verheyen RF, Coeck J, Maes J, Belpaire C, Van Thuyne G, Denayer B, Beyens J, De Charleroy D, Vandenabeele P, editors. Atlas van de Vlaamse beek- en riviervissen. Wijnegem: Water-Energik-vLario; 1998. pp. 259–264. [Google Scholar]
- Vrijenhoek RC. Conservation genetics of freshwater fish. Journal of Fish Biology. 1998;53:394–412. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wiens JA. Riverine landscapes: taking landscape ecology into the water. Freshwater Biology. 2002;47:501–515. [Google Scholar]
- Wilcock HR, Bruford MW, Nichols RA, Hildrew AG. Landscape, habitat characteristics and the genetic population structure of two caddisflies. Freshwater Biology. 2007;52:1907–1929. [Google Scholar]
- Wofford JEB, Gresswell RE, Banks MA. Influence of barriers to movement on within-watershed genetic variation of coastal cutthroat trout. Ecological Applications. 2005;15:628–637. [Google Scholar]
- Wootton RJ. The Biology of the Sticklebacks. London: Academic Press; 1976. [Google Scholar]
- Wright S. The genetical structure of populations. Annals of Eugenics. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]


