Abstract
Infectious disease is a major causal factor in the demography of human, plant and animal populations. While it is generally accepted in medical, veterinary and agricultural contexts that variation in host resistance and pathogen virulence and aggressiveness is of central importance to understanding patterns of infection, there has been remarkably little effort to directly investigate causal links between population genetic structure and disease dynamics, and even less work on factors influencing host–pathogen coevolution. The lack of empirical evidence is particularly surprising, given the potential for such variation to not only affect disease dynamics and prevalence, but also when or where new diseases or pathotypes emerge. Increasingly, this lack of knowledge has led to calls for an integrated approach to disease management, incorporating both ecological and evolutionary processes. Here, we argue that plant pathogens occurring in agro-ecosystems represent one clear example where the application of evolutionary principles to disease management would be of great benefit, as well as providing model systems for advancing our ability to generalize about the long-term coevolutionary dynamics of host–pathogen systems. We suggest that this is particularly the case given that agro-ecological host–pathogen interactions represent a diversity of situations ranging from those that only involve agricultural crops through to those that also include weedy crop relatives or even unrelated native plant communities. We begin by examining some of the criteria that are important in determining involvement in agricultural pathogen evolution by noncrop plants. Throughout we use empirical examples to illustrate the fact that different processes may dominate in different systems, and suggest that consideration of life history and spatial structure are central to understanding dynamics and direction of the interaction. We then discuss the implications that such interactions have for disease management in agro-ecosystems and how we can influence those outcomes. Finally, we identify several major gaps where future research could increase our ability to utilize evolutionary principles in managing disease in agro-ecosystems.
Keywords: coevolution, epidemiology, life history, resistance, spatial, virulence
Introduction
Increased human impacts on all levels of biological organization (e.g. fragmentation of natural systems and changing patterns of landuse, global movement of species), and in many novel ways (e.g. genetically modified organisms, introduction of new resistance genes into crops) has led to the explicit recognition of the value of an applied science of coevolutionary biology in management and planning contexts (Thompson 2005). Ultimately, the development of such a discipline will increase our ability to predict the outcomes of different types of human intervention (e.g. antibiotics, species introductions). Furthermore, our ability to both disrupt natural coevolutionary interactions as well as shift them in new and as yet unpredictable directions (e.g. emerging diseases) emphasizes the need for this predictive capability.
One arena where coevolutionary principles can be broadly applied lies in understanding dynamical processes and feedbacks across the interface between agricultural production systems and natural plant communities. Increasingly, as agricultural production intensifies or is altered to meet new demands (e.g. environmental forestry) in response to a range of external drivers, there are corresponding changes in the potential for contact with native elements in these landscapes. Of particular interest in this context are the ecological and evolutionary dynamics of plant pathogens that may move across this interface to varying degrees. A key question is the extent to which interactions between agriculture and native ecosystems may alter disease epidemiology and the potential for emergence of new plant pathogens. More generally, host co-infection by different pathogens and conversely the infection of different hosts by a given pathogen may be especially prevalent in agro-ecosystems, leading to novel coevolutionary dynamics. In this article, we specifically focus on existing evidence for such impacts, and highlight areas for future investigation.
The temporal and spatial nature of interactions between host plants and their fungal pathogens are forever shifting. Heterogeneity in environmental conditions across the distributional range of pathogens affects their ability to thrive and hence the size, frequency and severity of the impacts they have on their hosts. Against this backdrop though, the interactions between most hosts and their pathogens have an added level of subtle complexity. As they respectively place strong selection pressure on each other this can affect the frequency of resistance and avirulence genes within and among different populations, and indeed aspects of their respective life histories.
That pathogens evolve in response to changes in their hosts has been broadly documented in all types of interactions including human health, veterinary and wildlife diseases, plant-based agriculture and natural communities. Of these, changes in pathogen population structure following the deployment of novel cultivars with new resistance combinations are probably among the best examples of evolution in action. Indeed, the sequential deployment of single resistance genes in wheat in the 1950s and 1960s to counter stem rust, and the extremely rapid response of increased virulence in the previously avirulent pathogen population was summarized in the phrase ‘man-guided evolution of the rusts’ (Johnson 1961). However, this phenomenon has not been restricted to this particular interaction, occurring instead in virtually all interactions in which single major genes for resistance have been deployed in crops grown under high density and over large areas (e.g. all rusts and mildew of all cereals, mildews of lettuce).
In some cases, interactions between agricultural crops and their pathogens are largely self-contained (e.g. cereal smuts); in other cases though, crop volunteers (self-sown individual crop plants), wild weedy relatives or even unrelated native plants that are host to the same pathogen may play a significant role in shaping the dynamics and direction of the interaction (e.g. barley scald on volunteer plants and weedy Hordeum species; wheat rust parasitizing alternate hosts). What criteria are important in determining involvement in agricultural pathogen evolution by noncrop plants?; what implications do such interactions have for disease management in agro-ecosystems?; and how can we influence those outcomes?
Spatial and temporal aspects of host and pathogen life-history characteristics are crucial determinants of whether or not hosts and pathogens interact, and if they do, the extent and nature of those interactions. The magnitude and extent of influence wielded by noncrop components on disease will strongly depend on such factors. For example, spatially explicit simulation models have shown that the level of pathogen virulence which most strongly reduces host population size is dependent on host longevity, as well as whether pathogen impacts are felt through reduced fecundity or increased mortality (Thrall and Burdon 2004). Other life-history features of the interaction such as the relative spatial scales of host and pathogen dispersal can also profoundly affect patterns of disease incidence and prevalence, as well as the dynamical behaviour of the system (i.e. endemic versus epidemic patterns of disease increase; Thrall and Burdon 1999; Carlsson-Granér and Thrall 2002).
From an evolutionary perspective, among-population connectivity (as determined by both population structure and dispersal) influences the evolution and maintenance of genetic diversity in host resistance and pathogen virulence genes (Thrall and Burdon 2002). Importantly, the relative migration rates of hosts and pathogens play a key role in determining patterns of local adaptation (Gandon et al. 1996), which can in turn influence spatial patterns of disease. Relatively little work has been performed on the consequences of host specificity, although clearly, plant pathogens vary from those that are highly host specific (e.g. many rusts) through to those that are able to attack a broad diversity of host species (e.g. many soil-borne pathogens like Rhizoctonia). This may be of particular importance in the context of host–pathogen interactions across the agro-ecological interface. For example, recent theoretical work (Gandon 2004) suggests that variation in both the intensity and direction of selection pressures across different host species can strongly impact on the evolution of pathogen transmission, and patterns of host exploitation. In turn this feeds back on the epidemiological behaviour of pathogens.
Epidemiology of the pathogen
That crop and noncrop species (whether the latter is an introduced weed or native plant) are attacked by the same species of pathogen or pathogens is, a necessary, but not sufficient, condition for crop pathogen evolution to be significantly affected by the presence of a weedy host. It is the relative phenology of pathogen development on crop and noncrop that is important in determining whether the two cohorts are indeed just different parts of the one population or whether they are essentially isolated from each other despite spatial proximity.
Wild or weedy species that act as a ‘reservoir’ of inoculum providing a ‘green bridge’ between the maturity of one crop and the appearance of susceptible material in the next (Fig. 1A), certainly may prevent populations of obligate biotrophic fungi (e.g. rusts and mildews) with no effective resting stage from crashing precipitously with a consequent potential loss of variation through genetic drift or even local extinction. Thus in Australia, weedy Hordeum species have been implicated together with volunteer wheat plants as off-season (over-summering) hosts for Puccinia striiformis allowing this pathogen, that has no protected spore stage, to survive for the several months between maturity of one year’s wheat crop and emergence of the next (Wellings 2007). Similarly, weedy species often act as alternative hosts for many soil pathogens, maintaining pathogen populations at high levels in the absence of the crop host (Fig. 1B). A good example of this effect is seen in the importance of weedy species in contributing to levels of Fusarium spp. in agricultural soil (Jenkinson and Parry 1994; Hennessy et al. 2005). Recognition of this potential impact is at the heart of many crop sanitization programmes. Finally, weedy species that are only present at the same time as the crop are unlikely to have a reservoir effect although they may still play an important evolutionary role in affecting the genetic structure of the pathogen population.
Figure 1.
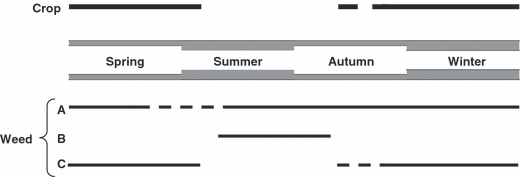
Hypothetical phenological cycles of a crop and associated weeds that are host to the same pathogens.
Genetic interactions
While epidemiological impacts may be the most obvious way in which crops and noncrop species affect one another, the most significant and far-reaching effects flow from more discreet interactions that generate genetic change in pathogen populations. What is it about the agro-ecological interface that makes it potentially such a hot-bed for evolutionary change? This boundary is essentially a breakpoint in a continuum in a broad range of ecological, environmental and species diversity measures. On the one hand, are crop communities that, notwithstanding the use of mixtures, alley cropping and other mixed approaches in horticulture, are essentially high density – low diversity and genetically controlled by man’s direct intervention. On the other hand, nearly all natural or even semi-natural plant communities are characterized by considerable inter and intra-specific diversity and lower individual species densities. Adding to this complexity, in many cases noncrop plant communities are composed of mixtures of native and exotic plant species, the latter ranging from introduced weeds with low diversity to those that are quite diverse (e.g. due to multiple introductions). These will vary in the suite of life histories represented and the degree of susceptibility to pathogen attack (with direct consequences for pathogen evolution).
Moreover, the potential for contact between agricultural and noncrop communities will itself vary across landscapes depending on the extent and configuration of cropping systems. For example, in the Middle East, there are a broad range of wild Cicer species which grow in varying degrees of proximity to cultivated chickpea (Cicer arietinum) and other crop legumes. Recent work suggests that the occurrence and severity of ascochyta diseases on the crops may be related to the proximity of wild hosts (Frenkel et al. 2007).
However, this is not to imply that evolution only or even predominantly, occurs in the agro-ecological zone. There are many examples of evolutionary change occurring within both agricultural and natural ecosystems. Evolutionary changes in pathogen populations in natural systems have often been assumed rather than demonstrated but clear examples exist of the appearance of novel pathotypes in populations (migration), year-to-year fluctuations in pathotype frequency (drift), changes within seasons (selection) and even much more significant changes resulting from genetic recombination between distinctly different genetic linkages or different species.
Evolutionary change occurring in pathogen populations attacking agricultural crops is also a well-recognized phenomenon but these changes typically reflect within-species changes – the evolution of novel virulence in response to the deployment of crop varieties carrying new resistance genes (mutation) or the re-assortment of characters through sexual recombination or the merging of closely related lineages. Not surprisingly, given the highly selected nature of most crops, the opportunities for major recombination between distinctly different but related pathogen species is very limited as they are rarely found on the one host.
The agro-ecological interface sits like a ‘hybrid’ zone between these two extremes, and like a hybrid zone is a site of overlap where crop species (whether cereals, horticultural crops or trees) will be in closer proximity to other related species (whether weedy or wild) than elsewhere, and where pathogens of these crops and related species (and hence more closely related pathogens) are also in closer contact. This situation is ripe with opportunities for pathogen evolution as a consequence of an increased diversity of selective pressures – different host species with potentially different resistance genes or mechanisms; novel encounters by related pathogen species. Indeed, in the agro-ecological zone the full gamut of potential changes to pathogens may occur, single virulence mutations to massive whole genome recombination, immediately adjacent to large uniform testing grounds – the agricultural crop! It is worth noting that such situations are not unique to plant–pathogen interactions. In the human context, similar ‘testbeds for disease’ exist in hospitals which are well-known hotspots for disease emergence and evolution (Ewald 1994). Basically, any situation which brings together variation (and enhances disease transmission potential) will clearly provide more opportunities for evolution.
Sources of variation
As with all other species, mutation and recombination are the ultimate source of genetically based variation in pathogen populations. Selection, random drift and migration (gene-flow) then acts on this basic variation to shape the structure of individual populations and hence the way they interact with their environment. Pathogen populations in agricultural crops experience a substantially different selective environment to those in natural ecosystems. The large and genetically uniform plant populations of agriculture provide a relatively uniform/predictable selective environment. Furthermore, notwithstanding the deployment of multiple varieties in the same general area, their use in large uniform blocks ensures that the fraction of the pathogen population that moves from one selective environment to another (e.g. one variety to another) is lower than in comparable wild situations where small, mixed host populations with multiple resistance gene combinations are the norm rather than the exception. The overall numerical and genetical dynamics of pathogen populations at the agro-ecological interface are therefore much better viewed as complex multi-population and multi-species interactions (i.e. metapopulations or even metacommunities) in which environmental, temporal and spatial heterogeneity, and marked differences in life-history features of hosts and pathogens may exert significant evolutionary influence.
Mutation – the wild host as a selective agent
Random mutation to virulence occurs in all pathogen populations. However, the probability that any such mutation amounts to any more than an ephemeral spark of evolution is dependent on a number of factors, not least of which will be any selective advantage such a mutation may gain through an ability to overcome resistances within the host population. In a genetically uniform crop pathogen population the appearance of a novel virulent pathotype capable of overcoming the deployed resistance may easily lead to a selective sweep culminating in the presence of a single clone or clonal lineage (this is particularly the case in asexual foliar pathogen populations). In diverse host populations, however, such a loss of diversity is far less likely to occur as different hosts with different resistance genes exert different selection pressures on the pathogen population. While it has been argued that over time repeated selection on hosts with different resistance gene combinations would lead to dominance by a single ‘super-race’, in reality in both agricultural mixtures and natural systems this tendency appears to be counter-balanced by greater fitness (increased fecundity) of isolates with lower virulence (Chin and Wolfe 1984; Thrall and Burdon 2002, 2003). A potential consequence of these interactions is then, the possibility that genetically diverse weedy or wild host populations existing at the agro-ecological interface may be effective generators of pathogen diversity (Fig. 2).
Figure 2.
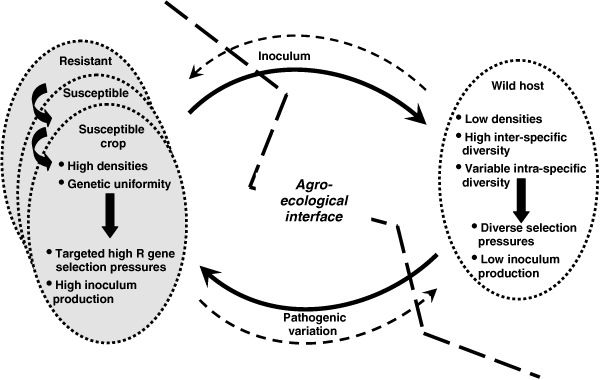
Interactions at the agro-ecological interface between crops and wild and weedy plants leading to an interchange of inoculum and pathogen variation.
Although there are good examples of no resistance being present, intra and inter-population diversity in resistance phenotypes is the norm rather than the exception in wild and weedy plant species that are hosts of agricultural pathogens (for example, resistance to Rhynchosporium secalis in Hordeum leporinum (Jarosz and Burdon 1996), to Puccinia graminis avenae and Puccinia coronata in Avena spp. (Dinoor 1970; Burdon et al. 1983), and to Bremia lactucae in Lactuca spp. (Lebeda and Boukema 1991; Lebeda and Zinkernagel 2003). However, the extent of such variation will be affected by a range of issues surrounding the history of the species involved. Thus in the case of recent weedy immigrants to a new environment, the number and magnitude of introductions, the length of time that has elapsed since such introductions, and the disease pressure experienced in the new habitat will all affect the extent of resistance gene diversity. Examples of the impact of these factors in weedy crop relatives that are host to agricultural pathogens have not been documented, but the marked difference in diversity of resistance found in Chondrilla juncea to its rust pathogen, Puccinia chondrillina, between populations in its native range (Turkey: Espiau et al. 1998) and in Australia (Burdon et al. 1982) illustrates this effect.
In all these cases, the ‘matching’ crop species typically occurs in large uniform stands in which resistance diversity within individual stands is very limited. Unfortunately, while screening plant populations for resistance patterns may be slow, assessing the virulence structure of matching pathogen populations is extremely laborious and has been performed very infrequently. As a consequence, evidence available for the agro-ecological interface as a major source of novel pathogen phenotypes is largely circumstantial. However, the most parsimonious explanation for the occurrence of increased pathotypic diversity found on wild and weedy plant populations at the A–E interface is selection on the range of resistance phenotypes found in those diverse host populations.
Below we illustrate these general points with evidence from a number of specific empirical examples:
Gene flow between pathogens of weeds and crops
The interaction occurring between oats [both cultivated (Avena sativa) and wild (Avena barbata, Avena fatua)] and its stem and crown rust pathogens (P. graminis f.sp. avenae and P. coronata respectively) in Australia provides good circumstantial evidence for the role that wild hosts play in both selectively favouring novel mutations as well as contributing variation to the pathogen population on a cultivated crop through gene flow (Burdon et al. 1983, 1992; Oates et al. 1983). In Australia, the sexual hosts of both of these pathogens are absent and although a number of different cultivated oat varieties are typically grown in the same geographic region, each cultivar is essentially a uniform line. In contrast, the wild oat populations show significant diversity in their resistance. Thus, studies of the occurrence of resistance to these two rust pathogens in 21 wild oat populations in eastern Australia found marked differences in the frequency of different seedling resistance phenotypes within and among different host populations (Burdon et al. 1983). Furthermore, within individual wild oat populations differences were found in both the response of individual host lines to different pathotypes of either pathogen, and between different host lines in their response to any one pathotype. This stands in clear contrast to the pattern of resistance in individual cultivated oat populations where essentially all individuals have the same resistance phenotype.
Against this backdrop of marked differences in resistance, comparisons of the diversity and virulence structure of pathogen populations occurring on either cultivated or wild oat populations from the same areas found no significant differences. However, spatial differences in the resistance structure of the wild oat populations (southern versus northern populations) was positively correlated with the complexity of the virulence structure of pathogen populations in the area (Oates et al. 1983; Burdon et al. 1992;) – a result that provides strong circumstantial support for a major role for the wild oat populations in driving virulence evolution in the pathogen populations.
It is also relevant to note here the possibility of gene flow between cultivated and wild host plant populations as this may alter the dynamics of any particular host–pathogen interaction. Naturally occurring introgressive hybridization leading to acquisition of novel genes or alleles in wild plants has been documented for a range of traits including, neutral isozyme markers in cultivated–wild oat associations (Burdon et al. 1992), herbicide resistance flowing from wheat to jointed goat grass (Aegilops cylindrica) (Hanson et al. 2005) and mildew resistance genes from exotic to native gooseberry species (Warren and James 2006). Where such introgression results in the flow of novel resistance genes introduced by breeders from newly released crop varieties into closely related weedy species this may ultimately reduce the range of disease control options available. For example, disease control strategies based around the controlled spatial or temporal use of resistance genes would become less efficient if such resistances remained in the environment (and hence continued to apply selection pressure) after crop varieties containing those genes were removed.
Recombination in pathogen populations
Sexual recombination
Classic examples of this phenomenon where there is a clear role played by a noncrop host are found amongst heteroecious cereal rust fungi where an indeterminate number of asexual pathogen generations on cereals alternates with a sexual stage occurring on either herbaceous or woody wild species. Thus, the occurrence of barberry (Berberis spp.) or buckthorn (Rhamnus spp.) bushes in hedgerows or wooded areas adjacent to cereal crops has been repeatedly shown to be a source of significant pathogen variation in wheat stem, and oat crown rust. Indeed, it is widely accepted that the barberry eradication campaigns that were waged in the Great Plains of the USA in the first third of the 20th Century were responsible for a significant reduction in the overall pathogenic diversity of the P. graminis tritici population (Roelfs and Groth 1980) and in the frequency of major stem rust epidemics. In that situation, eradication of the barberry not only reduced the diversity of pathotypes locally available to attack newly deployed wheat varieties, but also very substantially reduced the level of inoculum to which the developing crop was subject early in the spring. The lack of sexual recombination for more than 70 years has resulted in a population composed of a limited number of pathotypes that represent a few distinct clonal lineages. Each lineage is composed of a number of closely related pathotypes with one or two virulence differences, while among lineage differences are typically in the order of eight to 10 virulences (Burdon and Roelfs 1985a). Further evidence for the powerful effect of recombination in generating pathogenic diversity is seen in comparisons of the Great Plains populations with those populations from the Washington/Oregon area where the sexual stage of P. graminis tritici occurs annually. In the latter area, the pathogen population is composed of a very broad array of different pathotypes with no evidence of linkage disequilibrium (Burdon and Roelfs 1985b). Where the alternate host is herbaceous, as occurs in the interaction between wheat, Ornithogalum umbellatum and Puccinia hordei the interaction can be even more spatially intimate with the Ornithogalum sexual host growing within the cereal stand itself (Wallwork et al. 1992).
Somatic recombination
A feature common to many newly emerging diseases – whether they attack plants or animals (including humans) is their appearance in situations where changes in agricultural practices and human activities have either moved existing pathogens large distances around the world or brought species into new proximity (Anderson et al. 2004; Woolhouse et al. 2005). In the world of fungal plant pathogens, there is a steady increasing number of such examples which provides the basic minimum conditions needed (intimate proximity) for the creation of new species through horizontal gene transfer. In this process, genetic exchange occurring via anastomosis of vegetative hyphae (Little and Manners 1969) is widely regarded as the most likely mechanism, although, given that direct observation of this process has rarely occurred and the existence of fungal nonself recognition systems, the precise mechanism whereby exchange occurs is still in question (Glass et al. 2000). Exchange may result in transfer of nuclear material – whole nuclei (Burdon et al. 1982), individual chromosomes (Manners et al. 1992) and or of other cytoplasmic components including plasmids and viruses (Lawrence et al. 1988).
Regardless of the actual mechanism, increasingly, evidence provided by sequence data (e.g. Friesen et al. 2006) is providing confirmatory support for claims, based on changed pathogenicity patterns, of horizontal gene transfer in fungi. Indeed, with the growing use of sophisticated molecular markers it has become increasingly apparent that such somatic hybrids are more common than previously thought. For example, a number of novel pathogens of trees of agricultural landscapes have arisen in this way. Thus novel hybrid poplar rusts with a broader host range within Populus spp. has resulted from the transfer of pathogenicity genes between different species of Melampsora infecting poplars (Spiers & Hopcroft 1994; Newcombe et al. 2000) while a hybrid species of Phytophthora responsible for the deaths of alder trees in Europe has arisen from horizontal gene transfer between Phytophthora cambivora (a pathogen of hardwood trees) and Phytophthora fragariae (a pathogen of strawberries and raspberries) (Brasier et al. 1999; Brasier 2000). Although these examples do not necessarily relate to interactions at the agro-ecological interface, they do provide strong evidence for the potential for such events and their possible implications.
An example that truly sits along the ‘fence-line’ between agricultural crops and wild and weedy plants is the Australian ‘scabrum’ rust story, where a native wild wheat relative (Agropyrum scabrum) that is host to both P. graminis f.sp. tritici and P. graminis f.sp. secalis (stem rust of wheat and rye respectively) provided a common host on which a somatic hybrid naturally developed. The hybrid rust, generated through the exchange of haploid nuclei between the two dikaryotic parent formae speciales (Burdon et al. 1985a,b), has a different host range to either parent, having acquired through this recombination process virulence on a range of Hordeum vulgare lines that are immune to either parent (Luig and Watson 1977).
Implications for crop management
The role that weeds and native plants play as reservoirs of crop pathogens influencing the incidence and timing of disease in agricultural crops has been widely recognized (Wisler and Norris 2005). However, even in these seemingly simple numerical interactions not everything is as it might seem. Thus cryptic speciation is common among plant pathogens and one cannot assume that because the same species sensu lato occurs on a crop and on wild species nearby there is necessarily any interaction between them. Morphologically identical ‘species’ may indeed to be biologically separate entities with no effective genetic exchange.
When consideration is turned to the processes of evolutionary change in plant pathogens and how this may be affected by the spatial, temporal and biological complexities introduced at the agro-ecological interface, it becomes increasingly clear that provided the pathogen populations occurring on crop and wild plants are part of the same metapopulation, even relatively uncommon events occurring off the crop may be of great agricultural consequence. The important message is that plant pathogens are amazingly labile organisms. Here, we have provided a range of examples in which the evolution of those typically regarded as crop pathogens may be altered by interactions occurring on other hosts that are wild or weedy species growing in the same general vicinity.
The example provided by the North American experience with Berberis and its impact on wheat rust epidemics – both through a reservoir effect leading to early epidemic development, but more importantly on the evolutionary flexibility of the pathogen in the face of disease resistance breeding strategies is a classic demonstration of the potential of such interactions. In that example, knowledge of the life cycle of the pathogen, and the role of the sexual stage in generating new variation provided the scientific support for the massive eradication campaigns in the American Mid-West in the 1920s and 1930s. Similarly knowledge of the role of wild oats in generating pathogenic variation in oat crown rust has highlighted the futility of a standard major gene resistance approach to rust in oats if no attempt is made to simultaneously tackle wild oat populations.
Major issues for future research
Connectivity between production and natural components of agricultural landscapes has the potential to significantly influence disease dynamics and evolution, and there are clear examples where such interactions may have had significant economic consequences. Moreover, it seems highly likely that heterogeneity in agro-ecological interactions across geographical ranges could result in novel disease dynamics (e.g. shifts from ‘boom-and-bust’ to more endemic situations and vice versa) as well as spatial variation in the likelihood of disease emergence and the evolution of new virulence. From an applied evolution perspective, therefore one research issue of clear importance has to do with understanding how agricultural management (e.g. crop spatial arrangement and extent, rotational sequences) in rural landscapes might influence host–pathogen population dynamics. Notwithstanding work on within-field spatial patterning of cereals (Brophy and Mundt 1991; Mundt et al. 1996), we currently have little ability to predict the consequences of different spatial arrangements of production enterprises for disease dynamics and pathogen evolution. Even basic information relating to whether the relative proximity of wild host populations is correlated with crop disease epidemiology or the rate at which new pathotypes appear (or where they first emerge) is generally lacking. For example, cotton Fusarium wilt first appeared in Australian cotton-growing regions in the early 1990s. Recent molecular studies have shown that pathogenic strains, rather than being introduced, were derived from populations of related nonpathogenic Fusarium associated with a range of native Gossypium species (Wang et al. 2004) although it is still unclear as to the full range of factors involved in facilitating the evolution of pathogenicity on the crop.
It is also worth considering that evolutionary changes are not restricted to pathogen populations. For example it is also possible that pathogens may act as a bridge between cultivated and wild hosts. Indeed, studies of the cultivated–wild oat interaction in Australia have demonstrated gene flow between the crop and wild species which has clear implications for disease control strategies based on resistance gene deployment (Burdon et al. 1992). To what extent does the co-occurrence of crops and wild plants influence ecological and evolutionary processes in the natural hosts (e.g. resistance variation, maintenance of sex)? Can the incursion of exotic weeds influence the invasion and persistence of pathogens? Is spatial structure the most important factor determining host–pathogen dynamics in agro-ecosystems (can we detect the effects of variation in crop management and native community composition)? Is it possible to use mosaic management approaches to landscapes to control disease? Overall, it is becoming increasingly clear that managing biological interactions in fragmented landscapes requires studying coevolution in a community context (Thrall et al. 2007); this includes plant–pathogen interactions that span agricultural and native ecosystems.
Literature cited
- Anderson PK, Cunningham AA, Patel NG, Morales FJ, Epstein PR. Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends in Ecology & Evolution. 2004;19:535–544. doi: 10.1016/j.tree.2004.07.021. [DOI] [PubMed] [Google Scholar]
- Brasier CM. The rise of the hybrid fungi. Nature. 2000;405:134–135. doi: 10.1038/35012193. [DOI] [PubMed] [Google Scholar]
- Brasier CM, Cooke DEL, Duncan JM. Origin of a new Phytophthora pathogen through interspecific hybridization. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:5878–5883. doi: 10.1073/pnas.96.10.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brophy LS, Mundt CC. Influence of plant spatial patterns on disease dynamics, plant competition and grain-yield in genetically diverse wheat populations. Agriculture, Ecosystems and Environment. 1991;35:1–12. [Google Scholar]
- Burdon JJ, Roelfs AP. Isozyme and virulence variation in asexually reproducing populations of wheat leaf and stem rust. Phytopathology. 1985a;75:907–913. [Google Scholar]
- Burdon JJ, Roelfs AP. The effect of sexual and asexual reproduction on the isozyme structure of wheat stem rust populations. Phytopathology. 1985b;75:1068–1073. [Google Scholar]
- Burdon JJ, Groves RH, Cullen JM. The impact of biological control on the distribution and abundance of Chondrilla juncea in south-eastern Australia. Journal of Applied Ecology. 1981a;18:957–966. [Google Scholar]
- Burdon JJ, Marshall DR, Luig NH. Isozyme analysis indicates a virulent cereal rust is a somatic hybrid. Nature. 1981b;293:565–566. [Google Scholar]
- Burdon JJ, Marshall DR, Luig NH, Gow DJS. Isozyme studies on the origin and evolution of Puccinia graminis f.sp. tritici in Australia. Australian Journal of Biological Sciences. 1982;35:231–238. [Google Scholar]
- Burdon JJ, Oates JD, Marshall DR. Interactions between Avena and Puccinia species. I. The wild hosts: Avena barbata Pott ex Link, A. fatua L. and A. ludoviciana Durieu. Journal of Applied Ecology. 1983;20:571–585. [Google Scholar]
- Burdon JJ, Marshall DR, Oates JD. Interactions between wild and cultivated oats in Australia. In: Barr AR, Medd RW, editors. Proceedings of 4th International Oat Conference. Vol. 2. Adelaide: Intern. Oat Conf. Comm; 1992. pp. 82–87. [Google Scholar]
- Carlsson-Granér U, Thrall PH. The spatial distribution of plant populations, disease dynamics and evolution of resistance. Oikos. 2002;97:97–110. [Google Scholar]
- Chin KM, Wolfe MS. Selection on Erysiphe graminis in pure and mixed stands of barley. Plant Pathology. 1984;33:535–546. [Google Scholar]
- Dinoor A. Sources of oat crown rust resistance in hexaploid and tetraploid wild oats in Israel. Canadian Journal of Botany. 1970;48:153–161. [Google Scholar]
- Espiau C, Riviere D, Burdon JJ, Gartner S, Daclinat B, Hasan S, Chaboudez P. Host-pathogen diversity in a wild system: Chondrilla juncea — Puccinia chondrillina. Oecologia. 1998;113:133–139. doi: 10.1007/s004420050361. [DOI] [PubMed] [Google Scholar]
- Ewald P. Evolution of Infectious Disease. Oxford: Oxford University Press; 1994. [Google Scholar]
- Frenkel O, Shtienberg D, Abbo S, Sherman A. Sympatric ascochyta complex on wild Cicer judaicum and domesticated chickpea. Plant Pathology. 2007;56:464–471. [Google Scholar]
- Friesen TL, Stukenbrock EH, Liu Z, Meinhardt S, Ling H, Faris JD, Rasmussen JB, et al. Emergence of a new disease as a result of interspecific virulence gene transfer. Nature Genetics. 2006;38:953–956. doi: 10.1038/ng1839. [DOI] [PubMed] [Google Scholar]
- Gandon S. Evolution of multihost parasites. Evolution. 2004;58:455–469. [PubMed] [Google Scholar]
- Gandon S, Capowiez Y, Dubois Y, Michalakis Y, Olivieri I. Local adaptation and gene-for-gene coevolution in a metapopulation model. Proceedings of the Royal Society of London, Series B. 1996;263:1003–1009. [Google Scholar]
- Glass NL, Jacobson DJ, Shiu PKT. The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycete fungi. Annual Review of Genetics. 2000;34:165–186. doi: 10.1146/annurev.genet.34.1.165. [DOI] [PubMed] [Google Scholar]
- Hanson BD, Mallory-Smith CA, Price WJ, Bahman S, Thill DC, Zemetra RS. Interspecific hybridization: potential for movement of herbicide resistance from wheat to jointed goatgrass (Aegilops cylindrical. Weed Technology. 2005;19:674–682. [Google Scholar]
- Hennessy C, Walduck G, Daly A, Padovan A. Weed hosts of Fusarium oxysporum f.sp. cubense tropical race 4 in northern Australia. Australasian Plant Pathology. 2005;34:115–117. [Google Scholar]
- Jarosz AM, Burdon JJ. Resistance to barley scald (Rhynchosporium secalis) in wild barley grass (Hordeum glaucum and H. leporinum) populations in south-eastern Australia. Australian Journal of Agricultural Research. 1996;47:413–425. [Google Scholar]
- Jenkinson P, Parry DW. Isolation of Fusarium species from common broad-leaved weeds and their pathogenicity to winter wheat. Mycological Research. 1994;98:776–780. [Google Scholar]
- Johnson T. Man-guided evolution in plant rusts. Science. 1961;133:357–362. doi: 10.1126/science.133.3450.357. [DOI] [PubMed] [Google Scholar]
- Lawrence GJ, Boelen MG, Pryor A. Transmission of double-stranded RNAs in flax rust, Melampsora lini. Canadian Journal of Botany. 1988;66:61–66. [Google Scholar]
- Lebeda A, Boukema IW. Further investigation of the specificity of interactions between wild Lactuca spp. and Bremia lactucae isolates from Lactuca serriola. Journal of Phytopathology. 1991;133:57–64. [Google Scholar]
- Lebeda A, Zinkernagel V. Characterization of new highly virulent German isolates of Bremia lactucae and efficiency of resistance in wild Lactuca spp. germplasm. Journal of Phytopathology. 2003;151:274–282. [Google Scholar]
- Little R, Manners JG. Somatic recombination in yellow rust of wheat (Puccinia striiformis). 2. Germ tube fusions, nuclear number and nuclear size. Transactions of the British Mycological Society. 1969;53:259–267. [Google Scholar]
- Luig NH, Watson IA. The role of barley, rye and grasses in the 1973-74 wheat stem rust epiphytotic in southern and eastern Australia. Proceedings of the Linnean Society of New South Wales. 1977;101:65–76. [Google Scholar]
- Manners JM, Masel A, Braithwaite KS, Irwin JAG. Molecular analysis of Colletotrichum gloeosporioides pathogenic on the tropical pasture legume Stylosanthes spp. In: Bailey JA, Jeger MJ, editors. Colletotrichum: Biology, Pathology and Control. Wallingford, UK: CABI; 1992. pp. 250–268. [Google Scholar]
- Mundt CC, Brophy LS, Kolar SC. Effect of genotype unit number and spatial arrangement on severity of yellow rust in wheat cultivar mixtures. Plant Pathology. 1996;45:215–222. [Google Scholar]
- Newcombe G, Stirling B, McDonald S, Bradshaw Jr HD. Melampsora×Columbiana, a natural hybrid of M. medusae and M. occidentalis. Mycological Research. 2000;104:261–274. [Google Scholar]
- Oates JD, Burdon JJ, Brouwer JB. Interactions between Avena and Puccinia species. II. The pathogens: Puccinia coronata Cda and P. graminis Pers. f.sp. avenae Eriks. and Henn. Journal of Applied Ecology. 1983;20:585–596. [Google Scholar]
- Roelfs AP, Groth JV. A comparison of virulence phenotypes in wheat stem rust populations reproducing sexually and asexually. Phytopathology. 1980;70:855–862. [Google Scholar]
- Spiers AG, Hopcroft DH. Comparative studies of the poplar rusts Melampsora medusaeM. larisi-populina and their interspecific hybrid M. medusae-populina. Mycological Research. 1994;98:889–903. [Google Scholar]
- Thompson JN. The Geographic Mosaic of Coevolution. Chicago: University of Chicago Press; 2005. [Google Scholar]
- Thrall PH, Burdon JJ. The spatial scale of pathogen dispersal: consequences for disease dynamics and persistence. Evolutionary Ecology Research. 1999;1:681–701. [Google Scholar]
- Thrall PH, Burdon JJ. Evolution of gene-for-gene systems in metapopulations: the effect of spatial scale of host and pathogen dispersal. Plant Pathology. 2002;51:169–184. [Google Scholar]
- Thrall PH, Burdon JJ. Evolution of virulence in a plant host-pathogen metapopulation. Science. 2003;299:1735–1737. doi: 10.1126/science.1080070. [DOI] [PubMed] [Google Scholar]
- Thrall PH, Burdon JJ. Host-pathogen life-history interactions affect the success of biological control. Weed Technology. 2004;18:S1269–S1274. [Google Scholar]
- Thrall PH, Hochberg ME, Burdon JJ, Beaver JD. Coevolution of symbiotic mutualists and parasites in a community context. Trends in Ecology & Evolution. 2007;22:120–126. doi: 10.1016/j.tree.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Wallwork H, Preece P, Cotterill PJ. Puccinia hordei on barley and Ornithogalum umbellatum in South Australia. Australasian Plant Pathology. 1992;21:95–97. [Google Scholar]
- Wang B, Brubaker CL, Burdon JJ. Fusarium species and Fusarium wilt pathogens associated with native Gossypium populations in Australia. Mycological Research. 2004;108:35–44. doi: 10.1017/s0953756203008803. [DOI] [PubMed] [Google Scholar]
- Warren J, James P. The ecological effects of exotic disease resistance genes introgressed into British gooseberries. Oecologia. 2006;147:69–75. doi: 10.1007/s00442-005-0257-3. [DOI] [PubMed] [Google Scholar]
- Wellings CR. Puccinia striiformis in Australia: a review of the incursion, evolution, and adaptation of stripe rust in the period 1979-2006. Australian Journal of Agricultural Research. 2007;58:567–575. [Google Scholar]
- Wisler GC, Norris RF. Interactions between weeds and cultivated plants as related to management of plant pathogens. Weed Science. 2005;53:914–917. [Google Scholar]
- Woolhouse MEJ, Haydon DT, Antia R. Emerging pathogens: the epidemiology and evolution of species jumps. Trends in Ecology & Evolution. 2005;20:238–244. doi: 10.1016/j.tree.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]


