Cells are enclosed by a membrane that is readily penetrated by water but does not allow free diffusion of most solutes. Consequently, changes in external osmolality result in osmotic stress because of unequal rates of movement of water and solutes across the cell membrane. Cells respond to osmotic stress by osmoregulation, i.e., regulatory compensation of changes in cell volume, water content, and intracellular solute concentration. Osmoregulation minimizes changes in the concentration of intracellular inorganic ions—in particular Na+ and K+, macromolecules, and metabolites—and is essential for cell metabolism to operate properly. This is achieved by adjusting the levels of compatible osmolytes. Compatible osmolytes are small organic solutes, such as glycine betaine or myo-inositol, that are generally accumulated by transporters or enzymes, many of which are transcriptionally regulated. In this issue of the Proceedings, Miyakawa et al. (1) report the cloning and characterization of the first animal transcription factor responsible for regulating osmolyte transporter genes during osmotic stress. They name this transcription factor TonE binding protein (TonEBP) because it specifically binds to and activates the tonicity-responsive enhancer element (TonE) of osmoprotective genes, also known as osmotic response element. This important discovery should open up new avenues for addressing fundamental questions of cellular osmoregulation. In particular, it greatly contributes to understanding the molecular basis of osmosensory signal transduction.
Most fundamental aspects of cell metabolism have been optimized to a conserved ionic milieu found in the majority of extant cells. Of particular importance is the homeostasis of intracellular inorganic ion concentrations, cell volume, and macromolecular/metabolite concentration. Most, if not all, cells maintain lower intracellular Na+ and higher K+ concentrations than those present in the external milieu. The resulting Na+ gradient is maintained at an energetic cost but has the benefit of being the driving force for energy-dependent processes such as nutrient uptake, motility, or neuronal activity. Osmotic stress disturbs the conserved intracellular ionic milieu and interferes with cell function. Primordial cells were probably very sensitive to osmotic stress and had only a limited capacity to withstand fluctuations in cell volume and intracellular Na+ and K+ concentration because of, for example, evaporation or rain. To meet osmotic stress, life has evolved the so-called osmoregulatory responses, which enable cells to maintain a constant volume and intracellular ionic milieu in face of variations in the external osmolality. For instance, increases in external osmolality lead to H2O efflux and consequently a decrease in cell volume and an increase in the concentration of all cellular constituents. The change in cell volume and water content is initially compensated by osmoregulatory responses that result in volume increase because of uptake of inorganic solutes. However, this still leaves cells with disturbed Na+ and K+ concentrations, which are restored to normal by other osmoregulatory responses, including the accumulation of compatible osmolytes. Because of their central importance in cell physiology, such responses presumably arose very early in evolution and show remarkable conservation across all biological kingdoms. In cells in which the cytoplasmic membrane is surrounded by a stress-bearing cell wall, such as most bacteria, some fungi, and plant cells, the osmolality generated by compatible osmolytes can exceed the external osmolality. These cells can maintain a positive difference between the internal and external osmolality, which promotes the influx of water and generates an outwardly directed pressure supported by the cell wall. This pressure, termed turgor, has been suggested to be required for the expansion of bacterial and plant cells (2) and as an osmoregulatory signal (3). In contrast to these walled cells, animal cells, some algae, yeast, and Mollicute bacteria do not possess cell walls and cannot maintain an osmotic pressure difference between the internal and external milieu. Both types of cells, those with and without walls, accumulate a conserved set of organic compounds, i.e., glycine betaine, myo-inositol, other methylamines, polyols, and amino acids or their derivatives, during hyperosmotic stress to down-regulate intracellular Na+ and K+ levels (4–6).
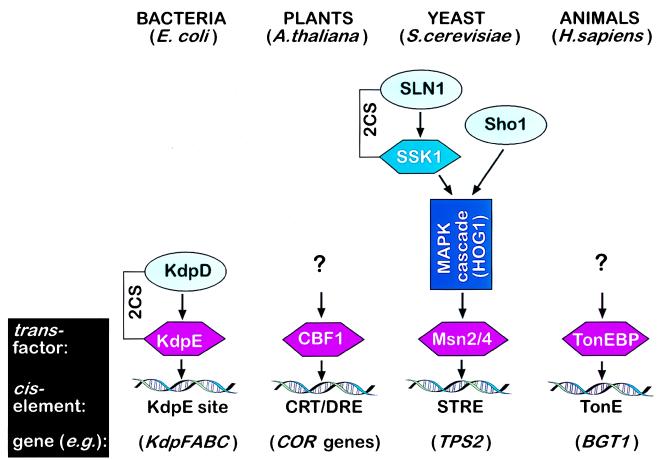
TonEBP, the protein discovered by Miyakawa et al. (1), is responsible for transcriptional activation of the genes encoding the glycine betaine transporter, BGT1, and the myo-inositol transporter, SMIT, during hyperosmotic stress in mammalian cells. Of interest, this protein bears no similarity to transcription factors that regulate osmoprotective genes in prokaryotes, plant cells, or yeast (Fig. 1). In prokaryotes, a variety of mechanisms is responsible for osmosensory signal transduction (7). In the bacteria Escherichia coli and Salmonella typhimurium, the prominent response to hyperosmotic stress is uptake of K+ and compatible osmolytes. These organisms have three transport systems for the uptake of K+—Trk, Kup, and Kdp—and two transporters with overlapping specificities for proline and glycine betaine—ProP and ProU (8). The activity of these transporters is enhanced by high osmolality, although by different mechanisms. Hyperosmolality regulates Trk, Kup, and ProP primarily by posttranslational activation, and it controls Kdp and ProU by >100-fold transcriptional induction (8). The osmotic control of transcription of the kdpFABC operon (encoding the elements of the Kdp system) is mediated by KdpD and KdpE, which constitute a two-component system. KdpD is a membrane-bound kinase, which phosphorylates KdpE in response to two environmental signals: K+ insufficiency and high osmolality. Phosphorylated KdpE then binds to a specific 22-bp region upstream of the kdpFABC promoter and activates its transcription (9). However, an important unresolved issue concerns what constitutes the high osmolality signal and how it is perceived by the KdpD protein (7).
Figure 1.
Transcriptional regulation of osmoprotective genes in different biological kingdoms. Transcription factors induced by osmotic stress are shown in purple, and osmosensor proteins are shown in light blue. 2CS, two-component system; CRT/DRE, C-repeat/dehydration responsive element.
Genes induced by osmotic stress or drought, some of which are also involved in adaptation to cold stress, have been identified in plants (10). In Arabidopsis thaliana, these genes are regulated by CBF1, the C-repeat/dehydration responsive element binding factor (11). Like TonEBP in animal cells, CBF1 is specifically up-regulated during hyperosmotic stress in plant cells and targets osmoprotective genes. CBF1 encodes a protein with a molecular mass of 24 kDa and belongs to a small family of closely related proteins that includes CBF2 and CBF3 (12). The C-repeat/dehydration responsive element consensus sequence for CBF1 is the pentanucleotide motif CCGAC (11). A different pentanucleotide motif, CCCCT or AGGGG, constitutes the stress-responsive enhancer element (STRE) in the yeast Saccharomyces cerevisiae (13). STRE is present in several genes, notably in TPS2, the yeast gene encoding trehalose phosphate phosphatase, which catalyzes the accumulation of the compatible osmolyte trehalose during hyperosmotic stress (14). The transcription factors that induce genes via STRE are the two zinc finger proteins Msn2 and Msn4 (15, 16). Because the high-osmolarity-glycerol response (HOG1) pathway has been shown to mediate hyperosmotic induction of genes via STRE (17), it is likely that Msn2 and Msn4 are activated by the HOG1 mitogen-activated protein kinase cascade (Fig. 1). Signal transduction via the HOG1 cascade depends on hyperosmolality and is directly controlled by two primary osmosensor proteins, SLN1 and Sho1 (18). Of interest, SLN1 is a sensor histidine kinase and part of a two-component system similar to the one that is responsible for osmosensory signal transduction in bacteria (Fig. 1). However, two-component systems have not been found in animals, and genes for sensor histidine kinases and response regulators are absent from the Caenorhabditis elegans genome (19). Thus, TonEBP may be regulated by different osmosensory signaling pathways than those found in bacteria and yeast. This notion is supported by the fact that TonEBP has very different properties compared with the transcription factors induced by hyperosmolality in other eukaryotes such as yeast (Msn2, Msn4) and plants (CBF1). This difference is apparent in the lack of sequence conservation and size. In contrast to proteins that mediate osmotic regulation of gene expression in bacteria, yeast, and plants, TonEBP is a very large protein consisting of 1,455 amino acids with a calculated molecular mass of 160 kDa (1). In addition, the consensus sequence of TonE is TGGAAANN(C/T)N(C/T) (1), which does not resemble the binding site for KdpE (TTTATACTTTTTTTACACCCCG), the yeast STRE consensus sequence (CCCCT or AGGGG), or the plant C-repeat/dehydration responsive element consensus sequence (CCGAC). Thus, TonEBP is a novel osmotically regulated transcription factor that is activated in animal cells exposed to hyperosmotic stress.
The cloning and characterization of TonEBP (1) represents a significant step in our search for understanding the molecular basis of cellular osmosensory signal transduction. TonEBP is regulated by means of its abundance, but the identification of a region of homology to the transcription factor rel, the rel homology region (RHR), in the N-terminal part of TonEBP also points to the possibility of posttranslational regulation by interaction with other proteins (1). An RHR is also present in nuclear factor of activated T cells (NFAT) but the degree of homology between rel/nuclear factor κB and NFAT in the RHR is low. The RHR in TonEBP also shows only a low degree of homology to the RHR in rel/nuclear factor κB. On the contrary, ≈150 amino acids of the RHR are highly conserved between TonEBP and NFAT. The strong similarity between the RHR of TonEBP and NFAT implies that they have similar functions. An important function of the NFAT RHR is DNA binding. In addition, the NFAT RHR also contains binding sites for the transcription factors Jun and Fos. Thus, NFAT is bound to DNA as part of a multiprotein complex, a scenario that is likely for TonEBP (1). Interaction of TonEBP with other transcription factors and cooperativity of a multiprotein complex during gene induction may explain the modulation of BGT1 gene expression via the stress-activated protein kinase 2 (p38) pathway (20), even though stress-activated protein kinase pathways do not seem to target TonE directly (21). Consistent with this notion, the transactivation capacity of the TonE consensus element decreases considerably when isolated from its sequence context, which may be necessary for all factors of a multiprotein complex to bind properly (22, 23).
Based on these considerations, we may speculate that TonEBP acts as a targeting transcription factor that recruits activating transcription factors such as Jun and Fos to particular genes. The unusually slow time course of TonEBP accumulation (10 h) reported by Miyakawa et al. (1) may reflect a more immediate need to use activating transcription factors for genes other than those encoding transporters and enzymes for compatible osmolytes. Such genes may encode heat shock proteins, cellular chaperones, and other proteins that are induced much more rapidly and whose rapid induction is more critical compared with BGT1 or SMIT. Activating transcription factors could be targeted to such genes by proteins other than TonEBP, and the maintenance of low initial levels of TonEBP would reduce competition for ubiquitous activating transcription factors such as Fos or Jun. Later, activating transcription factors may be redistributed to compatible osmolyte transporter genes by interaction with TonEBP to ensure long-term cell survival. Such a targeting mechanism for activating transcription factors, if existent, would explain gene- and stressor-specific transcriptional regulation by ubiquitous activating transcription factors, including Fos and Jun, and would provide a great target for manipulating cellular responses to stress by genetic engineering of targeting transcription factors such as TonEBP.
Surprisingly, the amino acids involved in the interaction of NFAT with DNA are extremely well conserved in TonEBP (1, 24). Twelve of fourteen amino acids involved in NFAT binding to DNA are identical in TonEBP, even though NFAT does not bind to TonE. Perhaps, this indicates that the site within the TonEBP RHR that binds to DNA is different from the site that recognizes TonE. In contrast to the strong conservation of the DNA binding site, only 3 of 11 amino acids involved in the interaction with Jun are identical between NFAT and TonEBP (1, 24). The same is true for those amino acids interacting with Fos (1, 24). Because the sequence conservation between NFAT and TonEBP of the DNA binding sites is greater than, and of the Jun and Fos binding sites considerably less than, that of the RHR on average, selection against Jun and Fos binding in TonEBP has to be assumed. Other activating transcription factors may be part of the TonEBP multiprotein complex. The study by Miyakawa et al. (1) lays the groundwork for the identification of proteins that interact with TonEBP, which constitutes a very important link in the chain of osmosensory signal transduction in animal cells. The investigation of signaling pathways that target TonEBP should enable us to better understand the molecular basis of cellular osmoregulation and the molecular contingencies involved in cellular stress responses.
Footnotes
A commentary on this article begins on page 2538.
References
- 1.Miyakawa H, Woo S K, Dahl S C, Handler J S, Kwon H M. Proc Natl Acad Sci USA. 1999;96:2538–2542. doi: 10.1073/pnas.96.5.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koch A. ASM News. 1991;57:633–637. [Google Scholar]
- 3.Epstein W. FEMS Microbiol Rev. 1986;39:73–78. [Google Scholar]
- 4.Brown A D. Microbial Water Stress Physiology: Principles and Perspectives. New York: Wiley; 1990. [Google Scholar]
- 5.Burg M B, Kwon E D, Kültz D. FASEB J. 1996;10:1598–1606. doi: 10.1096/fasebj.10.14.9002551. [DOI] [PubMed] [Google Scholar]
- 6.Somero G N, Yancey P. In: Handbook of Physiology. Hoffmann J F, Jamieson J D, editors. Vol. 14. Oxford: Oxford Univ. Press; 1997. pp. 441–484. [Google Scholar]
- 7.Csonka L N, Hanson A D. Annu Rev Microbiol. 1991;45:569–606. doi: 10.1146/annurev.mi.45.100191.003033. [DOI] [PubMed] [Google Scholar]
- 8.Csonka L N, Epstein W. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, Curtiss I I, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1210–1223. [Google Scholar]
- 9.Sugiura A, Nakashima K, Tanaka K, Mizuno T. Mol Microbiol. 1992;6:1769–1776. doi: 10.1111/j.1365-2958.1992.tb01349.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhu J-K, Hasegawa P M, Bressan R A. Crit Rev Plant Sci. 1997;16:253–277. [Google Scholar]
- 11.Stockinger E J, Gilmour S J, Thomashow M F. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilmour S J, Zarka D G, Stockinger E J, Salazar M P, Houghton J M, Thomashow M F. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- 13.Ruis H, Schüller C. BioEssays. 1995;17:959–965. doi: 10.1002/bies.950171109. [DOI] [PubMed] [Google Scholar]
- 14.Gounalaki N, Thireos G. EMBO J. 1994;13:4036–4041. doi: 10.1002/j.1460-2075.1994.tb06720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Pastor M T, Marchler G, Schüller C, Marchler-Bauer A, Ruis H, Estruch F. EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitt A P, McEntee K. Proc Natl Acad Sci USA. 1996;93:5777–5782. doi: 10.1073/pnas.93.12.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schüller C, Brewster J L, Alexander M R, Gustin M C, Ruis H. EMBO J. 1994;13:4382–4389. doi: 10.1002/j.1460-2075.1994.tb06758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wurgler-Murphy S M, Saito H. Trends Biochem Sci. 1997;22:172–176. doi: 10.1016/s0968-0004(97)01036-0. [DOI] [PubMed] [Google Scholar]
- 19.Chervitz S A, Aravind L, Sherlock G, Ball C A, Koonin E V, Dwight S S, Harris M A, Dolinski K, Mohr S, Smith T, et al. Science. 1998;282:2022–2028. doi: 10.1126/science.282.5396.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheikh-Hamad D, Di Mari J, Suki W N, Safirstein R, Watts B A, Rouse D. J Biol Chem. 1997;273:1832–1837. doi: 10.1074/jbc.273.3.1832. [DOI] [PubMed] [Google Scholar]
- 21.Kültz D, Garcia-Perez A, Ferraris J D, Burg M B. J Biol Chem. 1997;272:13165–13170. doi: 10.1074/jbc.272.20.13165. [DOI] [PubMed] [Google Scholar]
- 22.Takenaka M, Preston A S, Kwon H M, Handler J S. J Biol Chem. 1994;269:29379–29381. [PubMed] [Google Scholar]
- 23.Ferraris J D, Williams C K, Jung K-Y, Bedford J J, Burg M B, Garcia-Perez A. J Biol Chem. 1996;271:18318–18321. doi: 10.1074/jbc.271.31.18318. [DOI] [PubMed] [Google Scholar]
- 24.Chen L, Glover J N, Hogan P G, Rao A, Harrison S C. Nature (London) 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]