Abstract
Population genetics plays an increasingly important role in the conservation and management of declining species, particularly for defining taxonomic units. Subspecies are recognized by several conservation organizations and countries and receive legal protection under the US Endangered Species Act (ESA). Two subspecies of spotted owls, northern (Strix occidentalis caurina) and Mexican (S. o. lucida) spotted owls, are ESA-listed as threatened, but the California (S. o. occidentalis) spotted owl is not listed. Thus, determining the boundaries of these subspecies is critical for effective enforcement of the ESA. We tested the validity of previously recognized spotted owl subspecies by analysing 394 spotted owls at 10 microsatellite loci. We also tested whether northern and California spotted owls hybridize as suggested by previous mitochondrial DNA studies. Our results supported current recognition of three subspecies. We also found bi-directional hybridization and dispersal between northern and California spotted owls centered in southern Oregon and northern California. Surprisingly, we also detected introgression of Mexican spotted owls into the range of northern spotted owls, primarily in the northern part of the subspecies’ range in Washington, indicating long-distance dispersal of Mexican spotted owls. We conclude with a discussion of the conservation implications of our study.
Keywords: conservation, introgression, long-distance dispersal, microsatellites, spotted owl, Strix occidentalis, subspecies, US Endangered Species Act
Introduction
Population genetic theory and methods play an important role in the conservation of threatened, endangered and declining species. There are many different applications of genetics in conservation (Frankham et al. 2002a; Allendorf and Luikart 2007), and these applications continue to grow as new techniques and theory are developed. These applications include, but are not limited to, estimation of demographic parameters, such as effective population size, changes in population size, and gene flow (Roman and Palumbi 2003; Schwartz et al. 2005; Goossens et al. 2006); forensic identification of protected species (Baker et al. 1996); characterization of hybridization (Spruell et al. 2001; Funk et al. 2007a); predicting the effects of small population size and inbreeding depression on fitness and population viability (Newman and Pilson 1997; Saccheri et al. 1998); individual identification using noninvasive samples (Mills et al. 2000) and characterizing the effects of habitat fragmentation on population connectivity (Jump and Peñuelas 2006). Genetics does not replace the fundamental role of natural history and ecological studies in conservation. When used and interpreted appropriately, however, genetics can provide critical insights into the biology and management of declining species. One of the most important and common uses of genetics in conservation is to define taxonomic and conservation units, including species, subspecies, evolutionary significant units (ESUs; Ryder 1986; Waples 1991; Moritz 1994), and management units (MUs; Moritz 1994; Palsbøll et al. 2007).
Definition of subspecies has become an increasingly contentious issue in recent years. Subspecies are incipient species (Darwin 1868; Frankham et al. 2002b) and have been defined as ‘a collection of populations occupying a distinct breeding range and diagnosably distinct from other such populations’ (Mayr and Ashlock 1991). Although defining subspecies was originally the concern of evolutionary biologists, this problem has become of interest to a larger audience because many conservation organizations and countries recognize subspecies, including the World Conservation Union (IUCN) Red List of Threatened Species, appendices of the Convention on International Trade in Endangered Species of Wild Flora and Fauna (CITES), TRAFFIC (wildlife trade monitoring network), Brazil’s Lista Nacional das Espécies de Fauna Brasileira Amenaçadas de Extinção, Canada’s Species at Risk Act, Australia’s Environmental Protection and Biodiversity Act, and South Africa’s Biodiversity Act. The US Endangered Species Act (ESA) allows listing (i.e., legal protection) of subspecies and other groupings below the species level as threatened or endangered (US Fish and Wildlife Service 1978). Approximately, one-quarter of all ESA-listed taxa are subspecies, and 43% of all listed birds are subspecies (Haig et al. 2006). However, uncertainty regarding which criteria should be used for defining subspecies has hampered listing and delisting decisions. Subspecies have been defined based on phenotypic (e.g., morphological, ecological, and behavioral) and genetic criteria (Mayr 1942; Amadon 1949; Futuyma 1998; Patten and Unitt 2002; Haig et al. 2004a; Zink 2004). Thus, part of the controversy over subspecies definitions stems from conflicting morphological and genetic results (Zink 1989, 2004; Ball and Avise 1992; Zink et al. 2000; Phillimore and Owens 2006) and disagreement over which genetic markers (mitochondrial versus nuclear DNA) and analyses are most appropriate for defining subspecies (Zink 2004; Phillimore and Owens 2006).
There is a long-standing interest in determining the number and boundaries of spotted owl (Strix) subspecies because of ESA-listing of northern (Strix occidentalis caurina) and Mexican (S. o. lucida) spotted owls as threatened (US Fish and Wildlife Service 1990, 1993), and lack of listing for the California subspecies (S. o. occidentalis). The northern spotted owl is found from southwestern British Columbia to northwestern California; the California spotted owl in the Sierra Nevada Mountains, Coastal Mountains of California, and northern Baja California, Mexico; and the Mexican spotted owl in isolated mountain ranges from southern Utah and Colorado south into the Sierra Occidental and Sierra Oriental of Mexico (Fig. 1). Northern spotted owls were listed as threatened because of declines stemming from harvest of the subspecies’ old forest habitat in the US Pacific Northwest. Despite dramatic reduction of timber harvest under the Northwest Forest Plan in 1994 (Stokstad 2005; Noon and Blakesley 2006), recent field studies indicate that northern spotted owls have continued to decline at an average rate of 3.7% per year and that declines are most severe in Washington state (Anthony et al. 2006). One possible cause for continued decline is competition and hybridization with invasive barred owls (Strix varia) which have rapidly expanded into the range of northern spotted owls from their historic range in eastern North America (Kelly et al. 2003; Haig et al. 2004b; Kelly and Forsman 2004; Olson et al. 2005; Anthony et al. 2006; Funk et al. 2007a). Genetic factors, such as inbreeding depression or loss of adaptive genetic variation, may also play a role in declines (W. C. Funk, E. D. Forsman, T. D. Mullins, and S. M. Haig, unpublished manuscript). Determining the distinctness of spotted owl subspecies and their geographic distributions is essential for enforcing their protection under the ESA.
Figure 1.
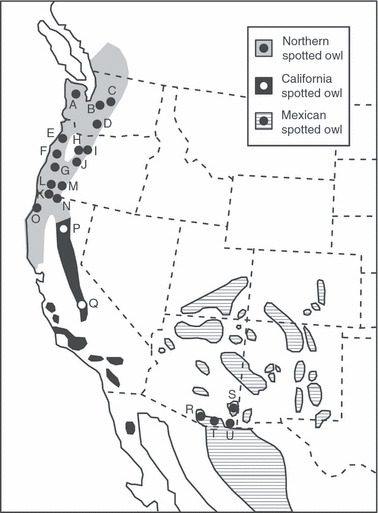
Locations of northern, California, and Mexican spotted owl study areas. Study area locations (and sample sizes) are: A. Olympic Peninsula, WA (n = 22); B. Western Washington Cascades (13); C. Cle Elum (eastern Cascades), WA (51); D. Yakima (eastern Cascades), WA (18); E. Northern Oregon Coast Ranges (12); F. Middle Oregon Coast Ranges (47); G. Southern Oregon Coast Ranges (31); H. Northwestern Oregon Cascades (15); I. Warm Springs (eastern Cascades), OR; J. Western Oregon Cascades (28); K. Siskiyou Mountains, OR and CA (17); L. South Umpqua River area, OR (10); M. Southern Oregon Cascades (32); N. Klamath National Forest, CA (14); O. Humboldt and Del Norte counties, CA (28); P. Lassen National Forest, CA (15); Q. Fresno, CA (8); R. Pima County, AZ (5); S. Graham County, AZ (1); T. Santa Cruz County, AZ (7); U. Cochise County, AZ (6). Subspecies ranges are based on Gutiérrez et al. (1995).
Historically, spotted owl subspecies have been recognized based on body size, plumage coloration, and geographic range (Gutiérrez et al. 1995). Genetic work based on mtDNA suggests that the boundary between northern and California spotted owls is in northern California in a region of low density (Fleischer et al. 2004; Haig et al. 2004a; Barrowclough et al. 2005). These studies have also documented the presence of California spotted owl haplotypes in the range of northern spotted owls, and vice versa, indicating hybridization and/or long-distance dispersal. However, it is not possible to distinguish these alternatives with mtDNA alone because both leave the same genetic signature (Fleischer et al. 2004). Microsatellite loci and other nuclear markers, in contrast, should be able to distinguish hybrid from nonhybrid (immigrant) owls as long as allele frequencies vary significantly among subspecies. Microsatellites have also proven useful for defining several other avian subspecies (Chan and Arcese 2002; Funk et al. 2007b; Draheim and Haig, unpublished manuscript). As of yet, however, there are no published microsatellite studies that have tested the validity of spotted owl subspecies and the extent of introgression between northern and California spotted owls.
The goal of the present study was to define spotted owl subspecies and test for introgression among subspecies using microsatellite loci to allow effective protection of these subspecies under the ESA. More specifically, our three main questions were: (i) Do microsatellite data support currently recognized subspecies? (ii) Is there introgression between northern and California spotted owls? and (iii) If so, what is the geographic extent of introgression?
Materials and methods
Sampling
We collected blood samples from 352 northern spotted owls from 15 study areas from across the subspecies’ range in Washington, Oregon, and California; 23 California spotted owls from two areas in the Sierra Nevada; and 19 Mexican spotted owls from four areas in southeastern Arizona from 1990 to 2006 (Fig. 1). Samples were collected from all regions over multiple years, so there was no systematic spatial pattern of collection over time. Study areas were bounded by landscape features such as mountain ridges, rivers, and nonforested habitat. Our choice of study area boundaries, however, does not affect our conclusions as many of our analyses were individual-based and thus do not require a priori definition of sampling units. No known close relatives (parent-offspring or siblings) were included. Samples were collected following the protocol of the American Ornithologists’ Union (Gaunt and Oring 1997). Blood was stored in cryogenic tubes containing a buffer solution (100 mm Tris–HCl, pH 8.0; 100 mm EDTA, pH 8.0; 10 mm NaCl; 0.5% SDS) and frozen at −80°C until analysis.
Microsatellite genotyping
DNA extraction, PCR, and fragment analysis were performed as described previously (Funk et al. 2007a). All owls were genotyped at 10 variable microsatellite loci developed for Mexican spotted owls (loci: 6H8, 15A6, 13D8, and 4E10.2; Thode et al. 2002), Lanyu scops owls (Otus elegans botelensis; loci: Oe37, Oe53, Oe128, Oe129, and Oe149; Hsu et al. 2003, 2006), and ferruginous pygmy-owls (Glaucidium brasilianum; locus: FEPO5; Proudfoot et al. 2005). One of these loci (Oe128) and an additional microsatellite marker (Bb126; Isaksson and Tegelström 2002) are diagnostic of spotted versus barred owls (Funk et al. 2007a) and were genotyped to assure that no spotted owl-barred owl hybrids were included in the analysis. PCR conditions and annealing temperatures were the same as those described in the original primer notes.
Standard population genetic analyses
We calculated exact probabilities for Hardy–Weinberg proportions, genotypic disequilibrium, and genic differentiation using Genepop 3.4 (Raymond and Rousset 1995). Weir and Cockerham’s (1984)FST values among study areas and subspecies were also calculated in Genepop. Critical α values for pairwise tests of allelic differentiation were determined using a sequential Bonferroni adjustment (Rice 1989). Expected heterozygosities were calculated with Microsatellite Analyzer 2.39 (Dieringer and Schlötterer 2003). Microchecker (van Oosterhout et al. 2004) was used to test for null alleles. California and Mexican spotted owls were each treated as a single population for these analyses to increase sample sizes (which gives n = 17 study areas after pooling). We also tested for isolation-by-distance among individuals using a Mantel test implemented in Alleles in Space (AIS; Miller 2005). Isolation-by-distance was tested using all spotted owl samples, as well as just northern spotted owls.
Subspecies and introgression analyses
We used a Bayesian clustering approach implemented in Structure 2.0 (Pritchard et al. 2000) to test whether subspecies grouped into distinct population clusters and to determine the zone of introgression between northern and California spotted owls. This approach estimates the number of populations (K) in a sample by minimizing deviations from Hardy–Weinberg proportions and linkage equilibrium within populations and then assigns individuals to one or more of these populations (k). The estimation procedure consists of running the program for different trial values of the number of populations, K, and then comparing the estimated log probability of the data under each K, ln[Pr(X|K)], called ln P(D) in Structure. We used the admixture model that assumes gene flow among populations and correlated allele frequencies. The admixture model assigns a proportion of each individual’s genome to each population (qk). We performed 20 runs for each K, from K = 1–10, and calculated the mean ln P(D) across runs for each K (e.g., Waples and Gaggiotti 2006). For each run, we used a burn-in of 30 000 and a total length of 100 000 which gave consistent results across runs in a pilot study. Typically, the correct value of K is taken to be the value with the highest ln P(D) (Pritchard et al. 2000). However, Pritchard and Wen (2003) warned that incremental increases in ln P(D) with increasing K can lead to overestimation of K. Therefore, we chose between the two values of K with the highest values of mean ln P(D) by calculating ΔK, a parameter which takes into account the shape of the log-likelihood curve (Evanno et al. 2005). Once the number of genetic clusters was estimated, we calculated mean membership of each individual to each cluster across runs.
We then used Geneclass2 (Piry et al. 2004) to identify potential migrants, or offspring of migrants, from the range of one subspecies to that of another. This method uses Bayesian techniques to calculate the probability of individual assignment to source and nonsource populations. We chose the partially Bayesian classification method (Rannala and Mountain 1997) paired with a Monte Carlo re-sampling method for computation of assignment probability to each subspecies (Paetkau et al. 2004) using 10 000 simulated individuals. This method is the most accurate of the frequentist assignment methods (Cornuet et al. 1999). Mis-assignments with high probabilities (≥95%) were genotypes unlikely to occur from a random combination of alleles, and thus were interpreted as migration events. This assignment test has been shown to be accurate in a study of individuals of known origin (Berry et al. 2004), as well as in simulation studies (Paetkau et al. 2004).
Finally, we used analysis of molecular variance (AMOVA; Excoffier et al. 1992) to test the proportion of variance explained by: (i) the four clusters identified by Structure (treating northern spotted owls north of the Columbia River as cluster 1 and northern spotted owls south of the Columbia River as cluster 2, as this was the pattern observed in the Structure analysis); (ii) currently recognized subspecies (collapsing the two northern spotted owl clusters identified by Structure into a single northern spotted owl group); and (iii) north versus south of the Columbia River (only including northern spotted owls) to test the validity of the two northern spotted owl clusters. AMOVA was performed in Arlequin 3.01 (Excoffier et al. 2005). We used 10 000 permutations to determine the significance of variance components. Mexican spotted owls from Graham and Cochise counties (study areas S and U, respectively; Fig. 1) were combined for this AMOVA as there was only one individual from Graham County.
Results
Standard population genetic analyses
Genotypic frequencies within study areas generally corresponded to expected Hardy–Weinberg proportions. Only 8 out of 170 tests for deviation from Hardy–Weinberg proportions were significant at the α = 0.05 level. This is less than the value (8.5 = 0.05 × 170) expected to deviate by chance. No loci had a consistent excess of homozygotes. Moreover, Microchecker only found evidence for low frequency null alleles at three loci in three different study areas (locus Oe149 in study area N, 4E10.2 in K, and 13D8 in L). Similarly, 33 out of 745 possible tests for departure from linkage equilibrium were significant, less than the 37.25 tests expected to be significant by chance. No pairs of loci consistently departed from linkage equilibrium across study areas, indicating that loci were independent. Expected heterozygosity (HE) was similar across study areas, ranging from 0.685 to 0.764 (mean = 0.736).
Pairwise FST values among northern spotted owl study areas ranged from 0.001 to 0.061 (Table 1). Allelic differentiation was significant at the α = 0.05 level for 61 out of 105 (58.1%) of these pairwise comparisons after correcting for multiple tests. Pairwise FST values between northern spotted owl study areas and California spotted owls ranged from 0.062 to 0.130. Between northern spotted owl study areas and Mexican spotted owls, pairwise FST values were 0.082–0.131. When all northern spotted owl study areas were lumped into a single group, pairwise FST values between northern and California, northern and Mexican, and California and Mexican spotted owls were 0.084, 0.095, and 0.141, respectively. All pairwise tests for allelic differentiation among subspecies were significant. Correlation coefficients of Mantel tests of isolation-by-distance were r = 0.297 (P < 0.001) and r = 0.089 (P < 0.001) for all spotted owls and just northern spotted owls, respectively.
Table 1.
Pairwise FST values at microsatellite loci among spotted owl study areas and subspecies.
| Site | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Site | A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | CSO |
| A – Olympic Peninsula, WA | ||||||||||||||||
| B – Western Cascades, WA | 0.023 | |||||||||||||||
| C – Cle Elum (E. Cascades), WA | 0.030 | 0.010 | ||||||||||||||
| D – Yakima (E. Cascades), WA | 0.040 | 0.028 | 0.034 | |||||||||||||
| E – Northern Coast Range, OR | 0.043 | 0.021 | 0.020 | 0.042 | ||||||||||||
| F – Middle Coast Range, OR | 0.039 | 0.012 | 0.014 | 0.044 | 0.003 | |||||||||||
| G – Southern Coast Range, OR | 0.019 | 0.001 | 0.016 | 0.045 | 0.011 | 0.007 | ||||||||||
| H – Northwest Cascades, OR | 0.037 | 0.015 | 0.032 | 0.027 | 0.016 | 0.025 | 0.014 | |||||||||
| I – Warm Springs (E. Cascades), OR | 0.051 | 0.001 | 0.038 | 0.059 | 0.022 | 0.033 | 0.015 | 0.013 | ||||||||
| J – Western Cascades, OR | 0.041 | 0.008 | 0.017 | 0.023 | 0.013 | 0.015 | 0.008 | 0.015 | 0.004 | |||||||
| K – Siskiyou Mountains, OR and CA | 0.058 | 0.036 | 0.036 | 0.057 | 0.029 | 0.040 | 0.026 | 0.043 | 0.048 | 0.027 | ||||||
| L – South Umpqua River area, OR | 0.041 | 0.011 | 0.022 | 0.042 | 0.027 | 0.020 | 0.011 | 0.021 | 0.014 | 0.010 | 0.012 | |||||
| M – Southern Cascades, OR | 0.043 | 0.021 | 0.025 | 0.061 | 0.026 | 0.024 | 0.013 | 0.035 | 0.025 | 0.015 | 0.022 | 0.021 | ||||
| N – Klamath National Forest, CA | 0.054 | 0.020 | 0.019 | 0.053 | 0.030 | 0.023 | 0.011 | 0.039 | 0.037 | 0.013 | 0.004 | 0.003 | 0.009 | |||
| O – Humboldt & Del Norte Cos., CA | 0.030 | 0.031 | 0.029 | 0.047 | 0.026 | 0.031 | 0.020 | 0.030 | 0.045 | 0.028 | 0.028 | 0.027 | 0.018 | 0.028 | ||
| CSO – California spotted owls | 0.130 | 0.096 | 0.095 | 0.103 | 0.119 | 0.109 | 0.099 | 0.111 | 0.116 | 0.090 | 0.062 | 0.104 | 0.088 | 0.075 | 0.066 | |
| MSO – Mexican spotted owls | 0.119 | 0.104 | 0.082 | 0.131 | 0.122 | 0.121 | 0.100 | 0.114 | 0.102 | 0.102 | 0.107 | 0.099 | 0.101 | 0.103 | 0.122 | 0.141 |
FST values significant at the α = 0.05 level after sequential Bonferroni correction are bold. WA, Washington; OR, Oregon; CA, California.
Subspecies and introgression
In the Structure analysis, the number of populations (K) with the highest mean ln P(D) was 5 [ln P(D) = −12529.36]. However, mean ln P(D) was only 46.38 lower for K = 4 and ΔK was higher for K = 4 than K = 5 (ΔK = 8.5 and 3.3, respectively). Therefore, we chose K = 4 as the most appropriate value for the number of spotted owl population clusters. Our conclusions regarding subspecies delineations and introgression, however, were qualitatively the same for K = 5 and 4.
The Structure analysis supported current subspecies designations. In 16 out of 20 runs, two clusters (clusters 1 and 2; Fig. 2) were almost completely restricted to northern spotted owls, whereas California spotted owls had high membership in cluster 3 (mean membership of 88.6%) and Mexican spotted owls had high membership in cluster 4 (mean of 95.3%). Individuals from north of the Columbia River in Washington state had higher mean membership in cluster 1 than those from south of the river (47.1% vs 29.9%), while the pattern of membership in cluster 2 north and south of the river was opposite (26.6% vs 44.3%). In the other four runs, Structure lumped California and Mexican spotted owls into a single cluster. Thus there was four-times as much support for current recognition of separate California and Mexican spotted owl subspecies as there was for lumping these subspecies into a single cluster.
Figure 2.
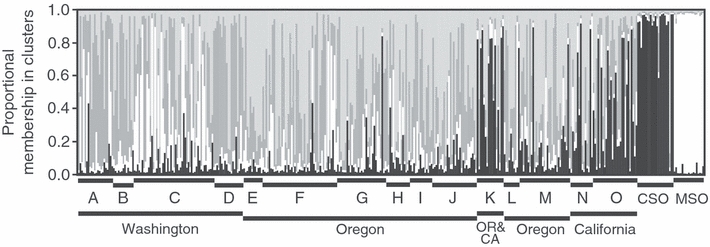
Population structure inferred by Bayesian clustering method implemented in Structure for 394 spotted owls. Four population clusters were identified: cluster 1 (dark grey); cluster 2 (light grey); cluster 3 (black); and cluster 4 (white). The figure shows mean individual membership to each of these four clusters. Letters refer to study areas (see Fig. 1); CSO = California spotted owls, MSO = Mexican spotted owls. California and Mexican spotted owls had high mean membership in clusters 3 and 4, respectively, and clusters 1 and 2 were largely restricted to northern spotted owls. A high level of introgression of California spotted owls into the range of northern spotted owls was observed in the Siskiyou Mountains (study area K), Klamath National Forest (N), and in Humboldt and Del Norte counties (O). Introgression of Mexican spotted owls into the range of northern spotted owls was also observed, especially in Cle Elum, WA (C).
Introgression between northern and California spotted owls was detected and was centered in northern California and southern Oregon, with relatively high membership in the California spotted owl cluster (cluster 3) found in the range of northern spotted owls in the Siskiyou Mountains (study area K; mean membership in cluster 3 = 52.9%; Fig. 2), Klamath National Forest (area N; mean = 28.0%), and Humboldt and Del Norte counties (area O; mean = 38.7%). Two individuals from the range of California spotted owls in Lassen National Forest (study area P) had high membership in northern spotted owl clusters (clusters 1 and 2), suggesting gene flow from northern to California spotted owls as well. Surprisingly, there was also evidence for substantial introgression of Mexican spotted owls into the range of northern spotted owls (Fig. 2). This was most pronounced in Cle Elum (study area C) with a mean membership of 29.1% in the Mexican spotted owl cluster (cluster 4).
The assignment test performed in Geneclass2 identified 10 individuals as migrants from other subspecies. Out of these 10 migrants, seven were identified as California spotted owl migrants in the range of northern spotted owls (one in Cle Elum, three in the Siskiyou Mountains, one in the Klamath National Forest, and two in Humboldt and Del Norte counties; refer to Fig. 1); two were northern spotted owl migrants in the range of California spotted owls (both in Lassen National Forest); and one was a Mexican spotted owl in the range of northern spotted owls (in the Klamath National Forest).
AMOVA also supported current subspecies recognitions. The percent variation explained by the subspecies grouping (8.64%) was highly significant and over four-times greater than the percentage of variation explained by study areas within subspecies (2.09%; Table 2). AMOVA also suggested that northern spotted owls represent a single population, rather than two clusters as estimated by Structure. First, grouping study areas into four clusters (including both northern spotted owl clusters) explained much less variation (3.83%) than grouping by subspecies (in which northern spotted owls were treated as a single group). Second, although clusters 1 and 2 identified by Structure were unevenly distributed on either side of the Columbia River, suggesting possible genetic differentiation, only 0.58% of the variation was explained in the AMOVA by grouping northern spotted owls into two groups found north versus south of the Columbia River.
Table 2.
Results from analysis of molecular variance (AMOVA) for different grouping methods. Study areas were divided by: (i) the four clusters identified in program Structure; (ii) subspecies (lumping the two northern spotted owl clusters into a single group); or (iii) north versus south of the Columbia River (only including individuals from the range of northern spotted owls).
| Grouping | No. of groups | Variance components | Variation (%) | P-value |
|---|---|---|---|---|
| Structure clusters | 4 | Among groups | 3.83 | <0.001 |
| Among study areas | 1.95 | <0.001 | ||
| Within study areas | 94.22 | <0.001 | ||
| Subspecies | 3 | Among groups | 8.64 | <0.001 |
| Among study areas | 2.09 | <0.001 | ||
| Within study areas | 89.27 | <0.001 | ||
| N versus S of Columbia R. | 2 | Among groups | 0.58 | 0.016 |
| Among study areas | 2.19 | <0.001 | ||
| Within study areas | 97.23 | <0.001 |
Discussion
Spotted owl subspecies and introgression
Analysis of our microsatellite data with pairwise FST values (Table 1), Structure (Fig. 2), and AMOVA (Table 2) all supported current recognition of three spotted owl subspecies (Fig. 1). Our results therefore agree with previous mtDNA analyses that support current subspecies designations (Barrowclough et al. 1999, 2005; Haig et al. 2004a), but disagree with a RAPD study which failed to find a distinct boundary between northern and California spotted owls (Haig et al. 2001). Our results also support the currently recognized porous boundary between northern and California spotted owls in northern California (Gutiérrez et al. 1995; Fig. 1). A similar location for this boundary was found using mtDNA (Haig et al. 2004a; Barrowclough et al. 2005). A limitation of our study was the relatively small sample sizes of California (n = 23) and Mexican (n = 19) spotted owls, but this should not affect our conclusions regarding subspecies boundaries and introgression. If anything, additional samples would be expected to increase the power to detect differences among subspecies.
A limitation of Structure is that it sometimes incorrectly identifies more population clusters than are actually present. Because of this, Pritchard and Wen (2003) warned that investigators should be skeptical of population clusters identified by Structure that have no clear biological interpretation, particularly cases in which the proportion of the sample assigned to each cluster is roughly symmetric (i.e., 1/K in each population). In our analysis, individuals from the range of California and Mexican spotted owls were clearly assigned to cluster 3 or 4, respectively, but individuals from the range of northern spotted owls were assigned fairly evenly to clusters 1 and 2. Although there was a somewhat higher mean membership in cluster 1 north of the Columbia River than south, and vice versa for cluster 2, the AMOVA and pairwise FST values indicated that there was little genetic differentiation across the Columbia River. Our data thus suggest that northern spotted owls consist of just one population.
The Structure analysis revealed substantial introgression between northern and California spotted owls centered in northern California and southern Oregon in the Siskiyou Mountains, Klamath National Forest, Humboldt and Del Norte counties, and Lassen National Forest (study areas K, N, O, and P, respectively; Fig. 2). This suggests that at least some previous observations of California spotted owl mtDNA haplotypes in the range of northern spotted owls (and vice versa; Haig et al. 2004a; Barrowclough et al. 2005) represent introgression, not just long-distance dispersal without mating. This matches the previously reported geographic distribution of introgression between these two subspecies. Introgression was observed in both directions, from California spotted owls to the range of northern spotted owls, and vice versa. Moreover, nine first generation northern and California spotted owl migrants were detected using assignment tests in Geneclass2. Seven of these migrants were California spotted owls found in the range of northern spotted owls; the other two were in the opposite direction. There was also limited introgression and dispersal between northern and California spotted owls over larger distances, including one first generation California spotted owl migrant in Cle Elum, WA. These results indicate that there is a limited amount of ongoing dispersal among subspecies. Nonetheless, the main area of introgression is restricted to a relatively narrow zone in northern California and southern Oregon.
Surprisingly, the Structure analysis also revealed introgression of Mexican spotted owls into the range of northern spotted owls (Fig. 2), indicating long-distance dispersal of Mexican spotted owls. The highest level of introgression between these two subspecies was found in Washington, particularly Cle Elum in the northern portion of the range of northern spotted owls. To reach the range of northern spotted owls, Mexican spotted owls would have to fly through the range of California spotted owls up the Sierra Nevada, or through the Rocky Mountains and/or Great Basin. Higher levels of introgression in Washington than in Oregon or California suggest that Mexican spotted owls may primarily use a northern, Rocky Mountain dispersal route. Spotted owls have been reported from the northern Rocky Mountains in northwestern Montana (Weydemeyer 1927; Hoffman et al. 1959), but these observations were dismissed as mis-identified barred owls (Wright 1976; Gutiérrez et al. 1995; Holt et al. 2001). However, the observation of a spotted owl in 1922 by Weydemeyer occurred well before any previous reports of barred owls in the northwestern USA or western Canada (Mazur and James 2000). This fact and our observation of introgression of Mexican spotted owls in Washington suggest that it is possible that these birds were migrant Mexican spotted owls. The spatiotemporal patterns and causes of long-distance dispersal of Mexican spotted owls warrants further study, as among-subspecies dispersal may have important consequences for the ecology and conservation of both subspecies.
Implications for subspecies delineation and characterizing introgression
Analysis of microsatellite data using clustering algorithms and assignment tests as performed here should prove useful for delineating subspecies in other taxa as well. In the past, mtDNA has been the workhorse of genetic studies of subspecies (Fry and Zink 1998; Valliantoes et al. 2002; Benedict et al. 2003; Bhagabati et al. 2004; Eggert et al. 2004; Idaghdour et al. 2004; Pitra et al. 2004; Solorzano et al. 2004), but the limitations of reliance on mtDNA alone have recently come to the forefront (reviewed by Ballard and Whitlock 2004). MtDNA is inherited as a single locus with unique properties; thus, inferences based solely on mtDNA may not accurately reflect the evolutionary and demographic history of populations. In addition, mtDNA cannot distinguish long-distance dispersal from hybridization among subspecies (Fleischer et al. 2004), but these alternatives can be distinguished with microsatellites. In the case of spotted owls, previous observations of California spotted owl haplotypes in the range of northern spotted owls, and vice versa, could be explained by long-distance dispersal or hybridization. Our microsatellite analyses revealed that both are occurring. Thus microsatellite data complement mtDNA results and add additional biological detail regarding the nature of introgression. Additionally, our study demonstrates the importance of using large sample sizes to detect unexpected patterns of hybridization and long-distance dispersal that might be missed using smaller samples sizes.
Conservation implications
Our results have important implications for the conservation of spotted owls, particularly for ESA-listed northern and Mexican spotted owls. First, our data indicate that the three traditionally recognized spotted owl subspecies are valid, distinct subspecies, supporting previous mtDNA studies. These subspecies thus clearly meet the criterion of being distinct from the rest of the species as required for legal protection under the ESA. A practical offshoot of this result is that spotted owls can be identified to subspecies using these microsatellite markers. This will be particularly useful for identifying individual owls as ESA-protected northern or nonlisted California spotted owls in the area of introgression in southern Oregon and northern California. Second, our data show that northern spotted owls extend at least as far south as Klamath National Forest and Humboldt County, California, indicating that spotted owls from these study areas are legally protected under the ESA. Furthermore, the observation of two northern spotted owls in the traditionally recognized range of the California spotted owl in Lassen National Forest, California, means that legally protected northern spotted owls may be going unnoticed and unprotected in the range of California spotted owls. Once again, the microsatellite markers used here should be useful for identifying such immigrant owls.
Hybridization and long-distance dispersal among subspecies may also have important implications for the conservation of spotted owls. On the one hand, naturally occurring hybridization can increase genetic variation in declining species, ameliorating inbreeding depression via genetic rescue (Thrall et al. 1998; Tallmon et al. 2004) and increasing potential for adaptation to environmental variation (Grant et al. 2003). On the other hand, unnatural or increasing rates of hybridization may break down local adaptations (Parris 2004) or cause outbreeding depression (Lynch and Walsh 1998), potentially reducing fitness, causing population declines, and increasing extinction probabilities. It is not possible to determine from existing data from this or previous studies whether the rate or geographic extent of introgression among spotted owl subspecies is stable, increasing, or decreasing, or what the demographic and evolutionary impacts of hybridization are (Fleischer et al. 2004). Moreover, competition and hybridization with invasive barred owls may also affect the hybridization dynamics between northern and California spotted owls, further complicating the problem. Future microsatellite analysis of the northern- California and northern-Mexican spotted owl hybrid zones at different time steps will be important for understanding the dynamics of these hybrid zones.
Conclusions
This study provides another example of the utility of genetic studies in the conservation of declining species, particularly with regards to delineation of taxonomic units for conservation. This is one of the largest genetic studies of any threatened bird species and the first to test subspecies boundaries in spotted owls and dissect the pattern of introgression among subspecies using microsatellite loci. Our results support recognition of current subspecies boundaries used for conservation and management of northern spotted owls under the US Endangered Species Act. Moreover, we confirm previous suggestions based on mtDNA data that northern and California spotted owls hybridize in a narrow zone of introgression in southern Oregon and northern California. Several first generation California spotted owl migrants were also identified in the range of northern spotted owls, and two northern spotted owl migrants were found in the range of California spotted owls, indicating ongoing dispersal between these subspecies. Surprisingly, we also found evidence for substantial levels of introgression between northern and Mexican spotted owls. This is the first evidence for introgression between northern and Mexican spotted owls and reveals unexpectedly long-distance dispersal of Mexican spotted owls. Our study illustrates the utility of microsatellite markers coupled with modern clustering and assignment methods for defining subspecies and characterizing introgression among subspecies, and provides essential data for enforcement of ESA-protection of northern and Mexican spotted owls.
Acknowledgments
We are indebted to many people who helped with sample collection and logistics, particularly P. Loschl, R. Anthony, S. Ackers, R. Claremont, D. Herter, S. Hopkins, J. Reid, and S. Sovern. We also thank R. Anthony, L. Bernatchez, J. Matthews, M. Miller, and two anonymous reviewers for comments on the manuscript. Funding was provided by the USGS Forest and Rangeland Ecosystem Science Center and the USDA Forest Service Pacific Northwest Research Station. Samples were collected under US Fish and Wildlife Service threatened species permit number TE026280-11 and Oregon State University Animal Care and Use permit number 3091.
Literature cited
- Allendorf FW, Luikart G. Conservation and the Genetics of Populations. Oxford: Blackwell Publishing; 2007. [Google Scholar]
- Amadon D. The seventy-five percent rule for subspecies. The Condor. 1949;51:250–258. [Google Scholar]
- Anthony RG, Forsman ED, Franklin AB, Anderson DR, Burnham KP, White GC, Schwarz CJ, et al. Status and trends in demography of northern spotted owls, 1985–2003. Wildlife Monographs. 2006;163:1–47. [Google Scholar]
- Baker CS, Cipriano F, Palumbi SR. Molecular genetic identification of whale and dolphin products from commercial markets in Korea and Japan. Molecular Ecology. 1996;5:671–687. [Google Scholar]
- Ball RM, Avise JC. Mitochondrial DNA phylogeographic differentiation among avian populations and the evolutionary significance of subspecies. The Auk. 1992;109:626–636. [Google Scholar]
- Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Molecular Ecology. 2004;13:729–744. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- Barrowclough GF, Gutiérrez RJ, Groth JG. Phylogeography of spotted owl (Strix occidentalis) populations based on mitochondrial DNA sequences: gene flow, genetic structure, and a novel biogeographic pattern. Evolution. 1999;53:919–931. doi: 10.1111/j.1558-5646.1999.tb05385.x. [DOI] [PubMed] [Google Scholar]
- Barrowclough GF, Groth JG, Mertz LA, Gutiérrez RJ. Genetic structure, introgression, and a narrow hybrid zone between northern and California spotted owls (Strix occidentalis. Molecular Ecology. 2005;14:1109–1120. doi: 10.1111/j.1365-294X.2005.02465.x. [DOI] [PubMed] [Google Scholar]
- Benedict NG, Oyler-McCance SJ, Taylor SE, Braun CE, Quinn TW. Evaluation of the eastern (Centrocercus u. urophasianus) and western (Centrocercus urophasianus phaios) subspecies of sage-grouse using mitochondrial control-region sequence data. Conservation Genetics. 2003;4:301–310. [Google Scholar]
- Berry O, Tocher MD, Sarre SD. Can assignment tests measure dispersal? Molecular Ecology. 2004;13:551–561. doi: 10.1046/j.1365-294x.2004.2081.x. [DOI] [PubMed] [Google Scholar]
- Bhagabati NK, Brown JL, Bowen BS. Geographic variation in Mexican Jays (Aphelocoma ultramarina): local differentiation, polyphyly, or hybridization? Molecular Ecology. 2004;13:2721–2734. doi: 10.1111/j.1365-294X.2004.02290.x. [DOI] [PubMed] [Google Scholar]
- Chan Y, Arcese P. Subspecific differentiation and conservation of song sparrows (Melospiza melodia) in the San Francisco Bay region inferred by microsatellite loci analysis. The Auk. 2002;119:641–657. [Google Scholar]
- Cornuet JM, Piry S, Luikart G, Estoup A, Solignac M. New methods employing multilocus genotypes to select or exclude populations of origin of individuals. Genetics. 1999;153:1989–2000. doi: 10.1093/genetics/153.4.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. The Variation of Animals and Plants under Domestication. London: Murray; 1868. [PMC free article] [PubMed] [Google Scholar]
- Dieringer D, Schlötterer C. Microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Molecular Ecology Notes. 2003;3:167–169. [Google Scholar]
- Eggert LS, Mundy NI, WoodRuff DS. Population structure of loggerhead shrikes in the California Channel Islands. Molecular Ecology. 2004;13:2121–2133. doi: 10.1111/j.1365-294X.2004.02218.x. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software Structure: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fleischer R, Dumbacher J, Moritz C, Monahan W. Assessment of the subspecies and genetics. In: Courtney SP, Blakesley JA, Bigley RE, Cody ML, Dumbacher JP, Fleischer RC, Franklin AB, et al., editors. Scientific Evaluation of the Status of the Northern Spotted Owl. Portland, OR: Sustainable Ecosystem Institute; 2004. pp. 3-1–3-31. [Google Scholar]
- Frankham R, Ballou JD, Briscoe DA. Introduction to Conservation Genetics. Cambridge: Cambridge University Press; 2002a. [Google Scholar]
- Frankham R, Ballou JD, Briscoe DA. Resolving taxonomic uncertainties and defining management units. In: Frankham R, Ballou JD, Briscoe DA, editors. Introduction to Conservation Genetics. Cambridge: Cambridge University Press; 2002b. pp. 365–392. [Google Scholar]
- Fry AJ, Zink RM. Geographic analysis of nucleotide diversity and song sparrow (Aves: Emberizidae) population history. Molecular Ecology. 1998;7:1303–1313. doi: 10.1046/j.1365-294x.1998.00462.x. [DOI] [PubMed] [Google Scholar]
- Funk WC, Mullins TD, Forsman ED, Haig SM. Microsatellite loci for distinguishing spotted owls (Strix occidentalis), barred owls (S. varia), and their hybrids. Molecular Ecology Notes. 2007a;7:284–286. [Google Scholar]
- Funk WC, Mullins TD, Haig SM. Conservation genetics of snowy plovers (Charadrius alexandrinus) in the Western Hemisphere: population genetic structure and delineation of subspecies. Conservation Genetics. 2007b;8:1287–1309. [Google Scholar]
- Futuyma DJ. Evolutionary Biology. 3rd edn. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- Gaunt AS, Oring LW. Guidelines to the Use of Wild Birds in Research. Washington, DC: The Ornithological Council; 1997. [Google Scholar]
- Goossens B, Chikhi L, Ancrenaz M, Lackman-Ancrenaz I, Andau P, Bruford MW. Genetic signature of anthropogenic population collapse in orang-utans. PLoS Biology. 2006;4:285–291. doi: 10.1371/journal.pbio.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PRB, Grant R, Keller LF, Markert JA, Petren K. Inbreeding and interbreeding in Darwin’s finches. Evolution. 2003;57:2911–2916. doi: 10.1111/j.0014-3820.2003.tb01532.x. [DOI] [PubMed] [Google Scholar]
- Gutiérrez RJ, Franklin AB, Lahaye WS. Spotted owl (Strix occidentalis. In: Poole A, editor. The Birds of North America Online. Ithaca, New York: Cornell Lab of Ornithology; 1995. http://0-bna.birds.cornell.edu.oasis.oregonstate.edu:80/bna/species/179(accessed on 10 December 2007) [Google Scholar]
- Haig SM, Wagner RS, Forsman ED, Mullins TD. Geographic variation and genetic structure in spotted owls. Conservation Genetics. 2001;2:25–40. [Google Scholar]
- Haig SM, Mullins TD, Forsman ED. Subspecific relationships and genetic structure in the spotted owl. Conservation Genetics. 2004a;5:683–705. [Google Scholar]
- Haig SM, Mullins TD, Forsman ED, Trail PW, Wennerberg L. Genetic identification of spotted owls, barred owls, and their hybrids: legal implications of hybrid identity. Conservation Biology. 2004b;18:1347–1357. [Google Scholar]
- Haig SM, Beever EA, Chambers SM, Draheim HM, Dugger BD, Dunham S, Elliott-Smith E, et al. Taxonomic considerations in listing subspecies under the US Endangered Species Act. Conservation Biology. 2006;20:1584–1594. doi: 10.1111/j.1523-1739.2006.00530.x. [DOI] [PubMed] [Google Scholar]
- Hoffman RS, Hand RL, Wright PL. Recent bird records from western Montana. The Condor. 1959;61:147–151. [Google Scholar]
- Holt DW, Domenech R, Paulson Anne. Status and distribution of the barred owl in Montana. Northwestern Naturalist. 2001;82:102–110. [Google Scholar]
- Hsu Y-C, Severinghaus LL, Lin Y-S, Li S-H. Isolation and characterization of microsatellite DNA markers from the Lanyu scops owl (Otus elegans botelensis. Molecular Ecology Notes. 2003;3:595–597. [Google Scholar]
- Hsu Y-C, Li S-H, Lin Y-S, Severinghaus LL. Microsatellite loci from Lanyu scops owl (Otus elegans botelensis) and their cross-species application in four species of Strigidae. Conservation Genetics. 2006;7:161–165. [Google Scholar]
- Idaghdour Y, Broderick D, Korrida A, Chbel F. Mitochondrial control region diversity of the Houbara bustard Chlamydotis undulata complex and genetic structure along the Atlantic seaboard of North Africa. Molecular Ecology. 2004;13:43–54. doi: 10.1046/j.1365-294x.2003.02039.x. [DOI] [PubMed] [Google Scholar]
- Isaksson M, Tegelström H. Characterization of polymorphic microsatellite markers in a captive population of the eagle owl (Bubo bubo) used for supportive breeding. Molecular Ecology Notes. 2002;2:91–93. [Google Scholar]
- Jump AS, Peñuelas J. Genetic effects of chronic habitat fragmentation in a wind-pollinated tree. Proceedings of the National Academy of Sciences of the USA. 2006;103:8096–8100. doi: 10.1073/pnas.0510127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly EG, Forsman ED. Recent records of hybridization between barred owls (Strix varia) and northern spotted owls (S. occidentalis caurina. The Auk. 2004;121:806–810. [Google Scholar]
- Kelly EG, Forsman ED, Anthony RG. Are barred owls displacing spotted owls? The Condor. 2003;105:45–53. [Google Scholar]
- Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- Mayr E. Systematics and the Origin of Species. New York: Columbia University Press; 1942. [Google Scholar]
- Mayr E, Ashlock PD. Principles of Systematic Zoology. 2nd edn. New York, NY: McGraw-Hill; 1991. [Google Scholar]
- Mazur KM, James PC. Barred owl (Strix varia. In: Poole A, editor. The Birds of North America Online. Ithaca, New York: Cornell Lab of Ornithology; 2000. http://0-bna.birds.cornell.edu.oasis.oregonstate.edu:80/bna/species/508 (accessed on 10 December 2007) [Google Scholar]
- Miller MP. Alleles in Space (AIS): computer software for the joint analysis of interindividual spatial and genetic information. Journal of Heredity. 2005;96:722–724. doi: 10.1093/jhered/esi119. [DOI] [PubMed] [Google Scholar]
- Mills LS, Citta JJ, Lair KP, Schartz MK, Tallmon DA. Estimating animal abundance using noninvasive DNA sampling: promise and pitfalls. Ecological Applications. 2000;10:283–294. [Google Scholar]
- Moritz C. Defining ‘evolutionarily significant units’ for conservation. Trends in Ecology and Evolution. 1994;9:373–375. doi: 10.1016/0169-5347(94)90057-4. [DOI] [PubMed] [Google Scholar]
- Newman D, Pilson D. Increased probability of extinction due to decreased genetic effective population size: experimental populations of Clarkia pulchella. Evolution. 1997;51:354–362. doi: 10.1111/j.1558-5646.1997.tb02422.x. [DOI] [PubMed] [Google Scholar]
- Noon BR, Blakesley JA. Conservation of the northern spotted owl under the Northwest Forest Plan. Conservation Biology. 2006;20:288–296. doi: 10.1111/j.1523-1739.2006.00387.x. [DOI] [PubMed] [Google Scholar]
- Olson GS, Anthony RG, Forsman ED, Ackers SH, Loschl PJ, Reid JA, Dugger KM, et al. Modeling of site occupancy dynamics for northern spotted owls, with emphasis on the effects of barred owls. The Journal of Wildlife Management. 2005;69:918–932. [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Willis DPM, Shipley P. Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- Paetkau D, Slade R, Burden M, Estoup A. Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Molecular Ecology. 2004;13:55–65. doi: 10.1046/j.1365-294x.2004.02008.x. [DOI] [PubMed] [Google Scholar]
- Palsbøll PJ, Bérubé M, Allendorf FW. Identification of management units using population genetic data. Trends in Ecology and Evolution. 2007;22:11–16. doi: 10.1016/j.tree.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Parris MJ. Hybrid response to pathogen infection in interspecific crosses between two amphibian species (Anura: Ranidae) Evolutionary Ecology Research. 2004;6:457–471. [Google Scholar]
- Patten MA, Unitt P. Diagnosability versus mean differences of sage sparrow subspecies. The Auk. 2002;119:26–35. [Google Scholar]
- Phillimore AB, Owens IPF. Are subspecies useful in evolutionary and conservation biology. Proceedings of the Royal Society B: Biological Sciences. 2006;273:1049–1053. doi: 10.1098/rspb.2005.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piry S, Alapetite A, Cornuet J-M, Paetkau D, Baudouin L, Estoup A. Geneclass2: a software for genetic assignment and first-generation migrant detection. Journal of Heredity. 2004;95:536–539. doi: 10.1093/jhered/esh074. [DOI] [PubMed] [Google Scholar]
- Pitra C, D’Aloia M-A, Lieckfeldt D, Combreau O. Genetic variation across the current range of the Asian Houbara bustard (Chlamydotis undulata macqueenii. Conservation Genetics. 2004;5:205–215. [Google Scholar]
- Pritchard JK, Wen W. Documentation for Structure Software: Version 2. 2003. http://pritch.bsd.uchicago.edu/structure.html (accessed on 10 December 2007) [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot G, Honeycutt R, Slack RD. Development and characterization of microsatellite DNA primers for ferruginous pygmy-owls (Glaucidium brasilianum. Molecular Ecology Notes. 2005;5:90–92. [Google Scholar]
- Rannala B, Mountain JL. Detecting immigration by using multilocus genotypes. Proceedings of the National Academy of Sciences of the USA. 1997;94:9197–9201. doi: 10.1073/pnas.94.17.9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. Genepop (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;83:248–249. [Google Scholar]
- Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Roman J, Palumbi SR. Whales before whaling in the North Atlantic. Science. 2003;301:508–510. doi: 10.1126/science.1084524. [DOI] [PubMed] [Google Scholar]
- Ryder OA. Species conservation and systematics: the dilemma of subspecies. Trends in Ecology and Evolution. 1986;1:9–10. [Google Scholar]
- Saccheri I, Kuussaari M, Kankare M, Vikman P, Fortelius W, Hanski I. Inbreeding depression and extinction in a butterfly metapopulation. Nature. 1998;392:491–494. [Google Scholar]
- Schwartz MK, Ralls K, Williams DF, Cypher BL, Pilgrim KL, Fleischer RC. Gene flow among San Joaquin kit fox populations in a severely changed ecosystem. Conservation Genetics. 2005;6:25–37. [Google Scholar]
- Solorzano S, Baker AJ, Oyama K. Conservation priorities for resplendent quetzals based on analysis of mitochondrial DNA control region sequences. The Condor. 2004;106:449–456. [Google Scholar]
- Spruell P, Bartron ML, Kanda N, Allendorf FW. Detection of hybrids between bull trout (Salvelinus confluentus) and brook trout (Salvelinus fontinalis) using PCR primers complementary to interspersed nuclear elements. Copeia. 2001;2001:1093–1099. [Google Scholar]
- Stokstad E. Learning to adapt. Science. 2005;309:688–690. doi: 10.1126/science.309.5735.688. [DOI] [PubMed] [Google Scholar]
- Tallmon DavidA, Luikart Gordon, Waples RobinS. The alluring simplicity and complex reality of genetic rescue. Trends in Ecology and Evolution. 2004;19:489–496. doi: 10.1016/j.tree.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Thode AB, Maltbie M, Hansen LA, Green LD, Longmire JL. Microsatellite markers for the Mexican spotted owl (Strix occidentalis lucida. Molecular Ecology Notes. 2002;2:446–448. [Google Scholar]
- Thrall PH, Richards CM, McCauley DE, Antonovics J. Metapopulation collapse: the consequences of limited gene-flow in spatially structured populations. In: Bascompte J, Sole RV, editors. Modeling Spatiotemporal Dynamics in Ecology. Berlin: Springer-Verlag; 1998. pp. 83–104. [Google Scholar]
- US Fish and Wildlife Service. Determination that 11 plant taxa are endangered species and two plant taxa are threatened species. Federal Register. 1978;43:17910–17916. [Google Scholar]
- US Fish and Wildlife Service. Determination of threatened status for the northern spotted owl. Federal Register. 1990;55:26114–26194. [Google Scholar]
- US Fish and Wildlife Service. Final rule to list the Mexican spotted owl as a threatened species. Federal Register. 1993;58:14248–14271. [Google Scholar]
- Valliantoes M, Lougheed SC, Boag PT. Conservation genetic of the loggerhead shrike (Lanius ludovicianus) in central and eastern North America. Conservation Genetics. 2002;3:1–13. [Google Scholar]
- Waples RS. Pacific salmon, Oncorhynchus spp., and the definition of ‘species’ under the Endangered Species Act. Marine Fisheries Review. 1991;53:11–22. [Google Scholar]
- Waples RS, Gaggiotti O. What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Molecular Ecology. 2006;15:1419–1439. doi: 10.1111/j.1365-294X.2006.02890.x. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Weydemeyer W. Some new birds from western Montana. The Condor. 1927;39:159. [Google Scholar]
- Wright PL. Further bird records from western Montana. The Condor. 1976;78:418–420. [Google Scholar]
- Zink RM. The study of geographic variation. The Auk. 1989;106:157–160. [Google Scholar]
- Zink RM. The role of subspecies in obscuring avian biological diversity and misleading conservation policy. Proceedings of the Royal Society B: Biological Sciences. 2004;271:561–564. doi: 10.1098/rspb.2003.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink RM, Barrowclough GF, Atwood JL, Blackwell-Rago RC. Genetics, taxonomy, and conservation of the threatened California gnatcatcher. Conservation Biology. 2000;14:1394–1405. [Google Scholar]


