Abstract
Conservation genetics can be seen as the effort to influence the evolutionary process in ways that enhance the persistence of populations. Much published research in the field applies genetic sampling techniques to infer population parameters from the patterns of variation in threatened populations. The limited resolution of these inferences seems to yield limited confidence which results in conservative policy recommendations. As an alternative, I suggest that conservation genetics focus on the relationships between those variables conservationists can control, and the probability of desirable evolutionary outcomes. This research would involve three phases – a greater use of existing evolutionary theory; testing management options using experimental evolution; and ‘field trials’ under an adaptive management framework. It would take a probabilistic approach that recognizes the stochasticity inherent in evolutionary change. This would allow a more nuanced approach to conservation policy than rule of thumb guidelines. Moreover, it would capitalize on the fact that evolution is a unifying theory in biology and draw on the substantial body of evolutionary knowledge that has been built up over the last half a century.
Keywords: adaptation, adaptive management, effective population size, experimental evolution, genetic inference, population structure, theoretical modelling
Introduction
Human influence on the evolutionary process has been evident ever since we domesticated plants and animals. The diversity of domestic species (e.g. Canis domesticus, Wayne 2001; Brassica oleracea, Tsunoda et al. 1980), and the divergence from their closest wild relatives (e.g. Zea mays, Doebley 1992) are a powerful testament to the ability of humans to harness the evolutionary process to our own ends. What has become more recently apparent is the speed with which evolutionary change can occur under direct artificial selection (e.g. Dudley and Lambert 1992), as a by-product of domestication (Heath et al. 2003), and under human altered natural selection (Stockwell et al. 2003). Evolutionary adaptation is not a slow process observable only on geological time scales, but an active process that occurs within human lifespans (Grant and Grant 1995; Reznick et al. 1997; Cook 2003; Soltis et al. 2004).
Two ironies must be recognized at the outset, which challenge our ability to control and mitigate our impact on evolution. First, humans tend to select against the traits that they find most useful, especially in harvested or managed populations. Fish that are easy to catch are removed from the population leaving smaller less desirable fish behind (Bell et al. 1977; Lande et al. 1997). Pests or pathogens that are susceptible to pesticides and antibiotics are killed off, leaving resistant forms in their wake (Gould 1998). Even trophy hunting selects against the large spectacular traits that are prized by hunters (Coltman et al. 2003). The second irony is that those populations that we would most hope would evolve tolerance to human activities are typically the very populations least likely to be capable of doing so, while those which we would prefer to remain static are most likely to evolve. It is much easier for an invasive or otherwise troublesome species to adapt to our control measures simply because such species tend have large populations and high reproductive rates (Sakai et al. 2001; Lee et al. 2007). Populations of conservation concern are typically much smaller and less reproductive, and consequently more likely to go extinct before they can adapt to new environmental challenges (Lynch and Lande 1992).
Evolutionary processes have been prominent in conservation biology almost from the outset (Frankel 1974; Soule and Wilcox 1980; Frankel and Soule 1981), featuring both in debates over what taxonomic groupings deserve conservation protections, and in the desire to preserve the evolutionary potential or process in addition to the species themselves (Moritz 2002). Genetic threats to population persistence are a standard topic for conservation biology textbooks, and several textbooks (e.g. Avise and Hamrick 1996; Frankham et al. 2002; Allendorf and Luikhart 2006) and a journal are now devoted to the subfield of conservation genetics. Broadly speaking, these threats include: inbreeding depression and mutation accumulation in small populations; assimilation through hybridization with more common species; outbreeding depression through the mating of distantly related lineages; maladaptation through environmental change that exceeds the evolutionary potential of the population; and translocation into environments to which the populations are not locally adapted.
Addressing these threats involves answering a series of basic practical questions for biological managers: how large and how genetically variable a population should we maintain in order for it to remain viable? When should we maintain barriers to reproductive exchange between related species, or between populations of the same species? How much environmental change can we allow and expect a population to adapt in response? When is stocking or re-stocking a beneficial practice and where should we obtain the colonists to restore degraded populations?
Conservation genetics in the broad sense can be seen as an attempt to manage human influence on the evolutionary process so as to minimize the harmful effects of human activities, and to maintain as much as possible the adaptive potential of natural populations (both large and small). I am going to suggest that as complicated as this challenge is, we have the advantage of applying one of the most elegant and powerful unifying theories in all of science – evolution by natural selection. At the same time, I will argue that much of conservation genetics is so narrowly focused on genetic inferences about individual taxa, that it fails to take advantage of the generality that should be its greatest asset. My perspective is that of a ‘pure’ researcher, and what I will offer will in some degree be a plea (or an apologia) for pure research that can be applied broadly, rather than applied research into more narrowly focused (if nonetheless pressing) questions. I will take many of my examples from the fields with which I am most familiar – that of gene flow and local adaptation, but the criticisms I offer and the approach I am suggesting are not restricted to these topics.
Illustrating the problem – population structure and genetic inference
The study of population structure is one of the major themes within conservation genetics. Electrophoretic surveys of DNA variation are relatively quick and affordable, and can be applied to almost any species from which tissue can be sampled. Statistical methods to infer parameters of interest from DNA data have become legion. As a result of this burgeoning field of enquiry, population structure has come to be recognized on finer and finer scales. A decade ago, Steinberg and Jordan (1998) lamented that molecular techniques had ‘spawned an industry of papers reporting the genetic structure of natural populations’, and this trend has continued. A casual survey of the papers published in 2006 in the journal Conservation Genetics revealed that out of 96 papers, just over half (51) used DNA surveys to explicitly address issues of population subdivision. A further 12 addressed the loss of variation and/or inbreeding as evidenced by DNA markers, and 29 described new markers, laboratory protocols, or analysis techniques for DNA markers. Thus, the research in Conservation Genetics is dominated by DNA surveys with a strong emphasis on patterns of population subdivision. (This is not to criticize either the journal or its editors, because this seems like a reasonable cross-section of activity in the field of conservation genetics).
Given the ubiquity of population structure, local adaptation of populations to their local environmental conditions has become a paramount concern of conservation genetics. In parallel with concerns over invasive species, conservation geneticists have become concerned over the potential spread of genes into novel habitats in applications ranging from restoration (e.g. McKay et al. 2005) and reforestation to fisheries and agriculture (e.g. Ellstrand 2003). Discussions of Evolutionarily Significant Units and Management Units (reviews in Crandall et al. 2000; Fraser and Bernatchez 2001) have also fuelled the concern that mixing genetic material from different environments will have negative consequences for the populations being managed. One of the major questions being debated in this field is whether DNA surveys adequately capture the pattern of adaptive as opposed to neutral variation (Reed and Frankham 2001; van Tienderen et al. 2004). More recent work has come to appreciate the importance of genetic variation in quantitative traits in the adaptation of populations to their habitats, and their evolution in response to habitat change, and methods attempting to address this variation have been developed (Lynch 1996; Storfer 1996; Merila and Crnokrak 2001; van Tienderen et al. 2004).
Identifying population structure is easy. Identifying when population structure represents divergent adaptation is much more difficult, and may often reflect assumption rather than evidence. In an often cited review, Crandall et al. (2000) reviewed 84 studies examining the genetic or ecological ‘exchangeability’ of populations in a broad range of taxa. By ‘exchangeability’ the authors hoped to explicitly address the question of whether one population could be used to replace the other, for example in a restoration effort. Almost in passing, they make a remarkable observation:
Our survey shows that the overwhelming majority of analyses fall within Case 8, either rejecting just recent genetic exchangeability or failing to reject any exchangeability. Interestingly, the authors assigned the term ESU to every category …. even when there was no evidence against exchangeability. (Crandall et al. 2000, Italics added)
This is a striking accusation, implying that conservation geneticists tend to draw the same conclusion regardless of what their research findings suggest. This view is epitomized by one prominent textbook in the field, which observes that when populations are diverged at DNA markers, adaptive divergence is likely given the isolation of the populations that creates the marker divergence. This suggests that marker divergence is indicative of adaptive divergence, but the same paragraph warns that the reverse is not true – that low marker divergence should not be taken as evidence of exchangeability, as adaptive divergence can take place in the face of high gene flow. It would seem from this argument that local adaptation is to be assumed no matter what the pattern of marker divergence. Yet the textbook observes on the previous page that marker divergence is a good predictor of adaptive divergence (a claim that has been debated at length elsewhere – e.g. Reed and Frankham 2001; Merila and Crnokrak 2001; McKay and Latta 2002).
If we accept the reasoning described above, then must we not conclude that research programs into population divergence are at best redundant? We have known for decades that no species exists as a single panmictic unit (Ehrlich and Raven 1969 being the classic statement of this point, albeit in the context of species concepts). If we are to assume that populations are always locally adapted to their particular habitats, and that immigration from other habitats will always result in maladaptive gene flow, then the conservation method is clear, and no further research into genetic exchangeability need take place. The resources that currently go into research on population structure could instead be directed to actual conservation efforts.
Arguably, in the absence of better information, a prudent conservation strategy is to maintain population divergence generally until the specific targets of selection can be identified through further research. This is much more difficult than one might think. Methods have been proposed to infer the action of selection from geographic patterns of electrophoretic variation, using either the Fst-outlier approach for single loci (Lewontin and Krakauer 1973; Beaumont and Balding 2004), or the Fst–Qst comparison for quantitative traits (Merila and Crnokrak 2001; McKay and Latta 2002). Additional methods address diversity rather than divergence (e.g. Kauer et al. 2003). These approaches hinge on the assumption that loci under selection will be recognizably different from neutral loci by having a distinct pattern of divergence among populations. This has intuitive appeal. Broad reviews of trait divergence show that quantitative traits tend to be more often under selection than are putatively neutral molecular markers (Merila and Crnokrak 2001; McKay and Latta 2002). Traits under divergent selection do in fact diverge more than do neutral loci (Endler 1973; Morgan et al. 2005; Porcher et al. 2006). However, what has typically been shown is that selection causes populations to diverge – one can not infer from this that all divergence implies selection.
Thus, genetic survey methods offer a remarkably weak predictor of adaptive divergence in individual populations or traits, because of the stochastic variation inherent in neutral processes. In essence, the ‘signature’ of selection is easily ‘forged’ by neutral processes (and vice versa – many loci under selection may not be recognizably different from neutral markers). It is in fact the expectation for many population models that a long tail of Fst values will be observed at neutral loci. Figure 1 shows the cumulative frequency distribution of Fst for data simulated using Simcoal (Excoffier et al. 2000) under two contrasting neutral models, and analysed using FDist2 (Beaumont and Balding 2004). When the data were simulated with the island model, the distribution is well matched by FDist2, which assumes and island model in its analysis. However, the modest variability in Fst for an island model becomes extreme when populations are grouped into hierarchies of phylogentic relatedness (Robertson 1975). In fairness, the manual distributed with FDIST2 acknowledges this point, but the perception in the literature is that FDist is robust. My point is that many real world populations will likely depart from the island model in ways that seriously undermine the inferences drawn (note that this criticism applies to the Fst-outlier or Fst–Qst approaches in general, and is not restricted to FDist). For example when populations emerge from separate glacial refugia – a common finding of phylogeographic studies (Avise 2004) – then regardless of the number of populations sampled, there will be only as many demes as there are refugia. In this situation, a long tail is present in the Fst of neutral markers (Fig. 1), and comparing against an island model means that the nominal 5% significant threshold gives a false positive rate for adaptive divergence of some 15–20%. It is questionable whether this level of resolution provides meaningful guidance for conservation.
Figure 1.
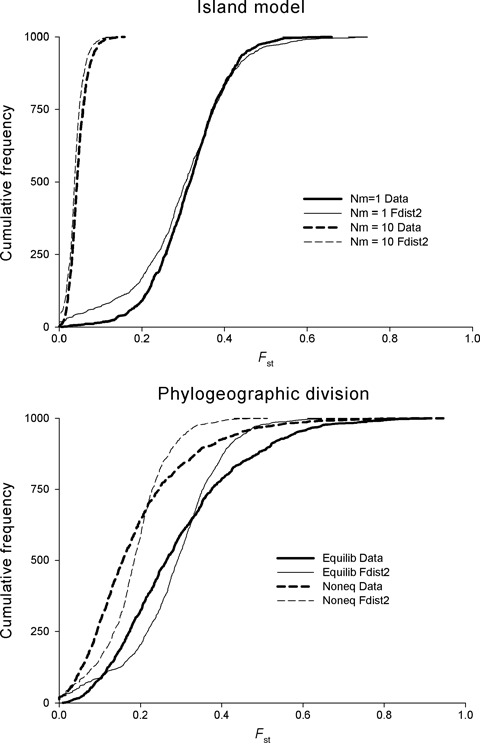
An example of the difficulty in genetic inference. Cumulative frequency distribution of Fst for data simulated using Simcoal (Excoffier et al. 2000– heavy lines) and analysed using FDist2 (Beaumont and Balding 2004– thin lines). Top panel: Island model with 1 and 10 migrants per generation (solid and dashed lines respectively). Because the data were simulated with the island model, the distribution is well matched by FDist2. Bottom panel: Populations phylogeneticaly related within two lineages at migration/drift equilibrium (solid lines) and recently diverged (dashed lines) 5Ne generations ago. Departure from the Island model greatly increases the stochastic variability of the data relative to assumptions of the analysis. Data were simulated assuming 10 populations each with Ne = 10 000, and the infinite alleles model with μ = 0.5 × 10−6. The island model assumed equal migration among all populations, whereas the phylogenetically structured model assumed five populations within each lineage, with Nm = 1 among populations within each lineage and restricted (Nm = 0.001) migration between lineages.
Three inter-related rebuttals are commonly offered to this criticism. Some argue that analyses such as the Fst-outlier technique are merely heuristics, which suggest candidate loci for further analysis. In the conservation context, however, such further analysis is frequently not possible, and assessment methods based on Fst-outliers have been offered as a decision-making tool (Bonin et al. 2007). Another argument posits that ‘more recent techniques’ offer greater promise. Naturally, I have not addressed each of the many available methods in detail. However, I submit that such optimism should be tempered until the reliability of these techniques is established a priori, rather embracing unproven methods. A third rebuttal suggests that with further information, more reliable estimates can be made. Yet, methods that measure population parameters with greater precision typically require large sample sizes, large numbers of markers with detailed marker information (often involving a genetic map), repeated sampling and/or long-term data, detailed field observations, sophisticated analysis and the combinations of multiple approaches (see e.g. Vasemagi and Primmer 2005 for a review). Such effort is only possible only for a few model organisms and the most iconic of endangered species. For example, Kauer et al. (2003) used over 100 loci per chromosome(!) to assess selective sweeps in Drosophila. This level of effort was possible only because of complete sequence data for Drosophila. Yet still only about 5% of those loci departed from neutrality at the 5% significance level. In fairness, Kauer et al. (2003) do not present their method as a conservation tool, but theirs is the type of analysis to which optimistic conservation geneticists often point. This level of return on investment offers little hope of reliable and economical techniques appropriate to conservation issues in the many threatened taxa which are experimentally intractable, of low economic value or have limited charismatic resonance with the public.
The problem I have tried to illustrate then, is that attempting to refine conservation efforts by genetic inference appears to be inefficient. Conceptually simple approaches are in practice complex and often unreliable, and the intensive effort needed to make precise and reliable inferences is often beyond the resources of conservation agencies. Such effort might be even be counterproductive, if the effort and resources necessary to make a solid inference detract from the conservation efforts themselves. Indeed, simpler methods are often seen as more reliable. It seems more economical to assess the environmental differences directly (McKay et al. 2005), as such information is often readily available. When genetic surveys identify divergence between populations from contrasting environments, it is common to infer that the method has worked (cf Bonin et al. 2007). When genetic methods fail to identify such populations as adaptively diverged the tendency is to hedge and assume adaptive divergence anyway (Crandall et al. 2000; Mace and Purvis 2007).
A shift in research emphasis
The alternative approach I am suggesting here is to harness the fact that evolution is a unifying theory that applies to all species, and identify the control points that managers, policymakers and conservationists might have over the process of evolution. I began by referring to cases where humans have harnessed evolution to their own ends and deliberately chose examples of domestication that predate our modern understanding of genetics (or evolution for that matter). This is to highlight the point that humans can exert productive control over evolution without detailed research into the genetics of particular taxa or traits. In the modern conservation crisis, the problems are considerably more challenging, and the ‘levers of control’ are less obvious than they were when harnessing evolution was simply a matter of selecting the individuals with traits we desired and propagating them. We are dealing with wild rather than domestic species, and we are trying to maintain adaptive potential in small slow breeding populations of endangered species, and curtail it in large rapidly reproducing populations of invasive or problematic species. But our understanding of evolution is greater now, and we have the advantage that it is broadly applicable.
Theory presents the obvious starting point. There already exists a large body of evolutionary theory ready to be harnessed to conservation genetic problems. Theory identifies the factors affecting the evolutionary process from which we might choose those factors susceptible of human control or influence. For example, Lynch and Lande (1992) presented a model of adaptative response to changing environmental conditions (see also Burger and Lynch 1995). This model was formulated to predict the realized rate of population increase (r) under the selective load imposed by adapting to a changing environment:
| 1 |
where rm is the maximum rate of population increase (in the optimal environment), σ2z and σ2g are the phenotypic and genetic variances, σ2w is the width of the stabilizing selection fitness function in any given environment, k is the rate of environmental change in the trait optimum and σ2 is the degree of stochastic variation in the environmental conditions.
I have deliberately chosen a model that I suspect few conservation biologists would routinely use, due to its seeming complexity, and because of the difficulty of measuring the terms on the right hand side. This is to emphasize that the utility of such a model is not to substitute values into the equation and predict or infer the rate of growth. Rather it is that the model identifies the relationship between evolutionary processes (drift, selection, effective population size, rate of environmental change) and population persistence. It allows us to evaluate the likely outcome of different impacts on the population and identify actions that are most likely to favour (or threaten) population persistence.
For example, consider two hypothetical (and for illustrative purposes, oversimplified) conservation proposals to help a threatened population adapt to changing environmental conditions. Proposal A would halve the rate of environmental change by changing land use patterns, while Proposal B would double population size. However, proposal B is more expensive, because more land must be purchased for an expanded reserve. Figure 2 plots Eqn 1 for varying values of Ne and k. For small populations, doubling population size has a greater impact on r than halving the rate of environmental change. In general, halving the rate of change is roughly 30% less effective for populations under 200 individuals (Ne). Conservationists can see immediately from Fig. 2 that proposal B is preferred, but if the necessary land is unavailable, that proposal A can have a significant benefit. Beyond 200 individuals, conserving more individuals has less benefit, and more productive action must address the rate of environmental change. This is useful information that can inform practical decision making. It costs nothing, is immediately available to conservationists, and broadly applicable. It is considerably more flexible than a ‘rule’, such as the 50/500 rule (Franklin 1980; Franklin and Frankham 1998), and can be extended (for example to include immigration as a source of variation) as research proceeds, or tailored to specific conservation questions.
Figure 2.
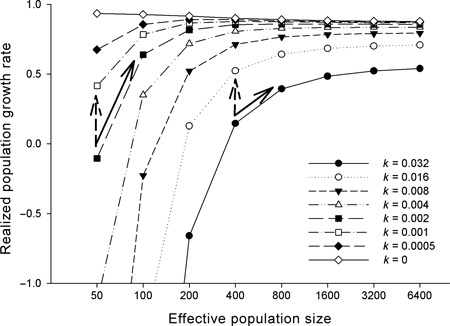
Response of population growth rate (r) to changes in the effective population size (Ne) and rate of environmental change (k) using the model of Lynch and Lande (1992) (Eqn 1). For small populations, doubling Ne (solid arrows) has a greater effect than halving k (dashed arrows). For larger populations, reducing k has the greater effect. Parameters used are: rmax = 1; σ2w = 10; σ2 = 1; σ2z and σ2g were calculated following Lynch and Lande (1992) assuming that the trait under selection is controlled by 25 loci, with mutational variance of 0.001, and heritability of 0.5; k is expressed as the rate of change in the trait optimum in phenotypic standard deviations per generation (i.e. in Haldanes).
Theory is of course only as good as its underlying assumptions, so the second stage of this research approach is experimental. Theories generate predictions which can and must be tested before the theory is put into widespread application. It is rarely considered in the context of conservation, but experimental evolution seems like a hugely powerful tool, allowing us to test outcomes of evolutionary changes over many generations in a short time. With laboratory populations of Chlamydomonas, Brassica, Arabidopsis, Caenorhabditis, Daphnia or Drosophila, among other species, it is possible to manipulate the evolutionary inputs and confirm (or refute) the predicted outcomes. Using the example above, the relative influence of Ne and environmental change on population growth rate might well be examined with vials of Drosophila at different densities, with temperature increments at different rates per generation. Such experiments have been used to investigate genetic rescue (Ball et al. 2000; Swindell and Bouzat 2006); local adaptation in the face of gene flow (Endler 1973) or to novel conditions (Reznick et al. 1997), the fitness consequences of genetic variation (Wise et al. 2002; Swindell and Bouzat 2005), and adaptation to novel CO2 environments (Collins and Bell 2006), among other topics. This approach offers considerable potential to test the effects of human manipulations before costly management efforts are initiated. Moreover, the diversity of available model organisms would allow a science of conservation genetics to distinguish repeatable patterns from the idiosyncrasies of particular species.
The third (and riskiest) component of this research approach is to test the models under actual field conditions (analogous to the ‘clinical trials’ of new medical procedures). Such trials might proceed under an adaptive management framework (Holling 1978; Walters and Hilborn 1978). From the modelling and experimental work, several feasible approaches to a particular problem might be identified, and decision makers would select from these the one(s) that best fit(s) the conditions (both biological and political) of a particular case. Ideally, several modified courses of action would be identified a priori to invoke if monitoring efforts indicate that the original fails to have the desired effect. The key feature of the adaptive management approach (as I see it being applied here) is that criteria for determining whether the proposed actions are working as planned are established before the management strategy is put into effect. Thus as the management program is monitored, the in-course corrections are ‘on deck’, ready to be implemented quickly should the need arise. At the same time, monitoring of management actions would provide valuable tests of the conclusions drawn in the theoretical and experimental stages of the research. An adaptive management framework would thus provide the critical link between model organisms in microcosm experiments (above) and the (often larger and longer-lived) organisms in real, heterogeneous and human-altered habitats that are the focus of conservation concern.
This process may seem excessively abstract for the pressing concerns of conservation, and its implementation has met with considerable resistance (Schreiber et al. 2004; Allan and Curtis 2005; Walters 2007). A common observation in conservation is the need to make decisions now with limited data (Allendorf and Luikhart 2007). However, this approach does not seek to postpone management decisions until experimentation is complete, but permits ongoing decision making based on the best evidence available at any time. We might wish for experimental confirmation, yet theoretical predictions give considerable guidance. If we cannot wait for complex modelling, simple models yield useful insights. Moreover, as such research proceeds, an increasingly solid framework can be built within which decisions can be tailored to particular conservation situations.
The Florida panther
The ‘genetic rescue’ of the Florida panther (Pimm et al. 2006) provides a concrete example of how the approaches I have outlined may be starting to contribute to genetic conservation. The case has been highlighted in several reviews and is likely well known. Briefly, the Florida panther (Puma concolor coryi) occurs as a single isolated population in south Florida. In the early 1990’s its numbers had declined to perhaps as few as 30 individuals (Pimm et al. 2006), and several lines of evidence strongly suggested that the small population was suffering the genetic effects of inbreeding depression. One possible management action was to introduce immigrant cats from the geographically closest related subspecies, and so introduce additional genetic variation to mitigate the inbreeding effects (see also, e.g. Westemeier et al. 1998; Madsen et al. 2004; Hogg et al. 2006; Hedrick and Fredrickson 2008). Eight immigrant females were introduced in 1995, and the population has been closely monitored since. Both before and after the introduction, the decision was controversial (Maehr and Lacy 2002; Beier et al. 2006; Maehr et al. 2006). However, whether due to the rescue (Pimm et al. 2006) or other factors (Maehr et al. 2006), the population has since more than doubled to approximately 87 individuals in 2003.
Whether referred to as genetic ‘rescue’, ‘restoration’ or ‘intervention’ (by authors who are enthusiastic, cautious and skeptical respectively) the introduction of immigrant individuals will influence a series of processes with both positive and negative consequences. The immediate goal is the reduction of inbreeding depression through hybrid vigour (Hedrick 1995; Ball et al. 2000) – more heterozygous outcrossed individuals express fewer deleterious recessive alleles. The addition of novel variation can also boost the evolutionary potential of a population providing the genetic variation on which selection can act to adapt the population to novel environmental conditions (Lynch and Lande 1992; Swindell and Bouzat 2005). On the other hand, if the population is specialized to the local environment, then immigration may introduce maladapted alleles suited to different environmental conditions. Also working against the success of restoration efforts is the fact that recombination between diverged gene pools can disrupt co-adapted gene complexes, causing reduced fitness (termed outbreeding depression, Templeton 1986). This last is particularly problematical in that it often does not manifest itself until later generations. Tallmon et al. (2004) and Hufford and Mazer (2003) provide lucid reviews of these issues, all of which are likely to be occurring simultaneously (Johansen-Morris and Latta 2006).
The first point I would like to highlight about the Florida panther example is that the complexity of the issue was acknowledged and addressed, even if it was controversial, during the planning phase. It is tempting for researchers focused on a single genetic process (inbreeding depression, local adaptation, adaptive potential, coadapted gene complexes, purging of genetic load and no doubt many others) to see a conservation issue through the prism of their own specialty. However, threats to populations do not happen separately like chapters in a textbook, but concurrently, and interactively, so any management action is likely to have both risks and benefits. Both positive and negative effects of the introducing immigrant panthers were considered in detail (Seal 1992, 1994; cited in Hedrick 1995).
Second, the Florida panther illustrates how theoretical models have been successfully used to provide a framework within which to evaluate different management options. Hedrick (1995) used straightforward population genetic models to assess both the positive effects of the introduction. By modelling the effects of introduction on maladaptive, neutral and adaptive variation, practical guidance was obtained on the optimal number of immigrants – that is, the level of immigration that gave the greatest chance of relieving inbreeding depression with the lowest risk of diluting adaptive variation.
Third, while I know of no case to date where experimental evolution has been directly employed in conservation, the case of genetic rescue illustrates the role that evolutionary experiments could play in understanding the mechanisms by which management decisions might affect population health. The work of Ball et al. (2000), and more recently of Swindell and Bouzat (2006) with immigrants into vials of Drosophila comes closest to being ‘experimental evolution’, but experimental studies have also been carried out in native systems (e.g. Ebert et al. 2002– see review in Tallmon et al. 2004). These studies are beginning to define the circumstances under which genetic rescue might work versus when the negative effects of immigration might outweigh the benefits.
Fourth, however, managers in Florida were able to take action based upon the theoretical models without being constrained to wait for these experimental results to confirm or refute the model. Thus the process-oriented approach allows practical conservation to proceed in parallel with research. While further experiments and refined models will doubtless further our understanding, everyone concerned with conservation recognizes the need to make decisions in the present based upon limited data.
Discussion
At the recent summit on Evolutionary Change in Human-Altered Environments (Smith and Bernatchez 2008), it seemed that conservation genetics is dominated by an increasing sense of urgency on the one hand (Mace and Purvis 2008), and by the promise of unlimited genomic information on the other (e.g. Kohn et al. 2006). At the same time that environmental degradation is becoming ever more alarming, the explosion of genetic information seems to promise that we will soon be able to identify all of the adaptively important variation in endangered species, map its distribution (both genetically and geographically), and conserve it. However, as Lewontin (1991) lamented for allozymes, and Hedrick (1996) for DNA markers, new technologies allow us to genetically study organisms that were previously inaccessible, but have often diverted researchers from understanding the basis, or process of evolution. Recently, a third perception may be entering conservation genetics – the appreciation that evolution is not simply a matter of history, but of current events (Palumbi 2001; Stockwell et al. 2003; Hendry et al. 2008) in which humans play a role, and over which we have considerable influence.
I suspect that much of the appeal of molecular genetics in conservation is driven by the culture of research agencies, universities among them. Allan and Curtis (2005) identified seven unspoken ‘imperatives’ that propel the behaviour of both research and management. These are the imperative to act (to ‘move forward’); the imperative to evince certitude; the imperative to maintain control; the imperative convince (to ‘sell’ one’s ideas); the imperative to compete (for funding, status, etc.); the imperative to maintain existing institutions; and the imperative to remain within a comfortable paradigm. Molecular genetic approaches in conservation appeal to most of these imperatives. New technologies and facilities can be established and data generated directly from the species of concern, satisfying the urge to act. The high tech nature of DNA technology and sophisticated genetic analysis give an air of certainty and control. This promise is also easily ‘sold’ to administrators, funding agencies and policy makers where it helps to compete for funding. Most genetic surveys can be published in peer-reviewed journals, allowing the researcher to compete for status. The work can be conducted within existing university departments and research institutions, and the results can be interpreted within a familiar paradigm. But in Hedrick’s (1996) words ‘The fact that sophisticated techniques are available does not mean that the answer is always apparent in the DNA’. He goes on to warn that the uncritical application of genetic tools may not be in the best interests of ‘either endangered species, (or) the future acceptance of molecular genetics research in conservation’.
I have argued for a shift of emphasis away from studying genetic variation per se, to a focus on the evolutionary process (see also Carroll 2008). I suggest that conservation genetics should seek to identify positive control points that humans can use to enhance the chances of population persistence. Genomic tools and electrophoretic screening undoubtedly have a role to play in conservation and I am in no way arguing that they be abandoned. Intensive genomic study of model systems has been and will continue to be a key component of research into evolutionary processes. They will also serve a valuable role to calibrate the application of models to particular conservation situations (e.g. Hedrick 1995). But by themselves, genetic markers and genomic information are insufficient to inform conservation practice.
Striving for precise inference on a case by case basis is an inordinately inefficient way to proceed for two reasons. First, the extremely stochastic nature of evolution renders such precision immensely difficult (Fig. 1). There are simply too many different processes that can lead to the same genetic outcome for inferences to have the level of certitude that many researchers and managers desire. The second is that a thorough understanding of the genetic architecture of ecologically important traits is likely only possible for model organisms (and their close relatives) and organisms of economic importance. Moreover, the quest for greater precision – for the ‘further study’ that will ‘give a clear answer’– can often be used as a political delaying tactic to avoid taking unpopular action.
The approach I have suggested will require that conservation genetics as a field relinquish some of its comfort and certainty, but that this may not be a great price to pay for a valuable payoff in terms of generality. And the potential for greater generality in evolutionary research is very high, simply because the same principles apply everywhere. It should be eminently possible to offer policy-makers guidelines by which the potential risks and benefits associated with particular management actions can be assessed. The panther example illustrates that we can tell decision makers how much immigration is optimal to enhance evolutionary potential while minimizing the genetic load. Such guides will inevitably be probabilistic, simply because the nature of evolution is inherently stochastic (as indeed is demography), and effective management must work with this uncertainty (Steinberg and Jordan 1998; Walters 2007). Yet this should perhaps be seen as a strength, rather than a weakness, because it allows management decisions to consider the extremes of the range of possible outcomes (e.g. unlikely, but catastrophic events) which may be more important than the median expectation.
The value of a process-oriented approach to conservation is that it allows a more nuanced understanding of the consequences of management actions, in place of such dichotomous thinking as the ‘50/500 rule’ (Franklin 1980). Where the 50/500 rule, for example, may appeal to policy makers because it is ‘clear and simple’ (cf Tseng 2007), it has also seen 25 years of caveats and debate (Franklin and Frankham 1998; Lynch and Lande 1998). It has fostered a rigidity of thinking that in its worst classifies populations into ‘doomed’ and ‘safe’ categories based upon whether the population size exceeds the threshold prescribed in the rule, and excusing inaction in either case (Allendorf and Luikhart 2007, p. 359). By instead viewing effective population size or migration rate as variables affecting the evolutionary process, we can help managers choose between suboptimal but feasible plans, and to recognize those modifications to a plan that are benign from those that will undercut its effectiveness. This allows genetic considerations to be balanced with other concerns (ecological, economic, social and political) that fall outside the expertise of geneticists, in formulating overall management policies. In return decision makers must (by definition) be willing to make decisions, using the tools conservation geneticists can provide, and embracing the uncertainty that the evolutionary process entails.
Acknowledgments
These ideas were developed while on sabbatical at the University of California Davis, and were shaped by conversations with a great many colleagues at and following the Summit on Evolution in Human Altered Environments at UCLA in February 2007. However, any errors of omission or commission remain the author’s own. This work was funded by a Discovery Grant from the Natural Science and Engineering Council of Canada.
Literature cited
- Allan C, Curtis A. Nipped in the bud: why regional scale adaptive management is not blooming. Environmental Management. 2005;36:414–425. doi: 10.1007/s00267-004-0244-1. [DOI] [PubMed] [Google Scholar]
- Allendorf FW, Luikhart G. Conservation and the Genetics of Populations. Malden, MA, USA: Blackwell Publishing; 2006. [Google Scholar]
- Avise JC. Genetic Markers, Natural History, and Evolution. 2nd edn. New York: Chapman and Hall; 2004. [Google Scholar]
- Avise JC, Hamrick JL. Conservation Genetics: Case Histories from Nature. New York: Chapman and Hall; 1996. [Google Scholar]
- Ball SJ, Adams M, Possingham HP, Keller MA. The genetic contribution of single male immigrants to small, inbred populations: a laboratory study using Drosophila melanogaster. Heredity. 2000;84:677–684. doi: 10.1046/j.1365-2540.2000.00721.x. [DOI] [PubMed] [Google Scholar]
- Beaumont MA, Balding DJ. Identifying adaptive genetic divergence among populations from genome scans. Molecular Ecology. 2004;13:969–980. doi: 10.1111/j.1365-294x.2004.02125.x. [DOI] [PubMed] [Google Scholar]
- Beier P, Vaughan MR, Conroy MJ, Quigley H. Evaluating scientific inferences about the Florida panther. Journal of Wildlife Management. 2006;70:236–245. [Google Scholar]
- Bell G, Handford P, Dietz C. Dynamics of an exploited population of Lake Whitefish (Coregonus-Clupeaformis. Journal of the Fisheries Research Board of Canada. 1977;34:942–953. [Google Scholar]
- Bonin A, Nicole F, Pompanon F, Miaud C, Taberlet P. Population Adaptive Index: a new method to help measure intraspecific genetic diversity and prioritize populations for conservation. Conservation Biology. 2007;21:697–708. doi: 10.1111/j.1523-1739.2007.00685.x. [DOI] [PubMed] [Google Scholar]
- Burger R, Lynch M. Evolution and extinction in a changing environment – a quantitative-genetic analysis. Evolution. 1995;49:151–163. doi: 10.1111/j.1558-5646.1995.tb05967.x. [DOI] [PubMed] [Google Scholar]
- Carroll SP. Facing change: forms and foundations of contemporary adaptation to biotic invasions. Molecular Ecology. 2008;17:361–372. doi: 10.1111/j.1365-294X.2007.03484.x. [DOI] [PubMed] [Google Scholar]
- Collins S, Bell G. Evolution of natural algal populations at elevated CO2. Ecology Letters. 2006;9:129–135. doi: 10.1111/j.1461-0248.2005.00854.x. [DOI] [PubMed] [Google Scholar]
- Coltman DW, O’Donoghue P, Jorgenson JT, Hogg JT, Strobeck C, Festa-Bianchet M. Undesirable evolutionary consequences of trophy hunting. Nature. 2003;426:655–658. doi: 10.1038/nature02177. [DOI] [PubMed] [Google Scholar]
- Cook LM. The rise and fall of the Carbonaria form of the peppered moth. Quarterly Review of Biology. 2003;78:399–417. doi: 10.1086/378925. [DOI] [PubMed] [Google Scholar]
- Crandall KA, Bininda-Emonds ORP, Mace GM, Wayne RK. Considering evolutionary processes in conservation biology. Trends in Ecology & Evolution. 2000;15:290–295. doi: 10.1016/s0169-5347(00)01876-0. [DOI] [PubMed] [Google Scholar]
- Doebley J. Mapping the genes that made maize. Trends in Genetics. 1992;8:203–207. doi: 10.1016/0168-9525(92)90261-2. [DOI] [PubMed] [Google Scholar]
- Dudley JW, Lambert RJ. 90-Generations of selection for oil and protein in maize. Maydica. 1992;37:81–87. [Google Scholar]
- Ebert D, Haag C, Kirkpatrick M, Riek M, Hottinger JW, Pajunen VI. A selective advantage to immigrant genes in a Daphnia metapopulation. Science. 2002;295:485–488. doi: 10.1126/science.1067485. [DOI] [PubMed] [Google Scholar]
- Ehrlich PR, Raven PH. Differentiation of populations. Science. 1969;165:1228–1232. doi: 10.1126/science.165.3899.1228. [DOI] [PubMed] [Google Scholar]
- Ellstrand NC. Dangerous Liaisons?: When Cultivated Plants Mate with Their Wild Relatives. Baltimore, MD: The Johns Hopkins University Press; 2003. [Google Scholar]
- Endler JA. Gene flow and population differentiation. Science. 1973;179:243–250. doi: 10.1126/science.179.4070.243. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Novembre J, Schneider S. SIMCOAL: a general coalescent program for the simulation of molecular data in interconnected populations with arbitrary demography. Journal of Heredity. 2000;91:506–509. doi: 10.1093/jhered/91.6.506. [DOI] [PubMed] [Google Scholar]
- Frankel OH. Genetic conservation – our evolutionary responsibility. Genetics. 1974;78:53–65. doi: 10.1093/genetics/78.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel OH, Soule ME. Conservation and Evolution. Cambridge, UK: Cambride University Press; 1981. [Google Scholar]
- Frankham R, Ballou JD, Briscoe DA. Introduction to Conservation Genetics. Cambridge, UK: Cambride University Press; 2002. [Google Scholar]
- Franklin IR. Evolutionary changes in small populations. In: Soule ME, Wilcox BA, editors. Conservation Biology: An Evolutionary-Ecological Perspective. Sunderland, MA: Sinauer; 1980. pp. 135–149. [Google Scholar]
- Franklin IR, Frankham R. How large must populations be to retain evolutionary potential? Animal Conservation. 1998;1:69–70. [Google Scholar]
- Fraser DJ, Bernatchez L. Adaptive evolutionary conservation: towards a unified concept for defining conservation units. Molecular Ecology. 2001;10:2741–2752. [PubMed] [Google Scholar]
- Gould F. Sustainability of transgenic insecticidal cultivars: integrating pest genetics and ecology. Annual Review of Entomology. 1998;43:701–726. doi: 10.1146/annurev.ento.43.1.701. [DOI] [PubMed] [Google Scholar]
- Grant PR, Grant BR. Predicting microevolutionary responses to directional selection on heritable variation. Evolution. 1995;49:241–251. doi: 10.1111/j.1558-5646.1995.tb02236.x. [DOI] [PubMed] [Google Scholar]
- Heath DD, Heath JW, Bryden CA, Johnson RM, Fox CW. Rapid evolution of egg size in captive salmon. Science. 2003;299:1738–1740. doi: 10.1126/science.1079707. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. Gene flow and genetic restoration – the Florida panther as a case-study. Conservation Biology. 1995;9:996–1007. doi: 10.1046/j.1523-1739.1995.9050988.x-i1. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. Conservation genetics and molecular techniques: a perspective. In: Smith TB, Wayne RK, editors. Molecular Genetic Approaches in Conservation. New York: Oxford University Press; 1996. pp. 459–477. [Google Scholar]
- Hedrick PW, Fredrickson RJ. Captive breeding and the reintroduction of Mexican and red wolves. Molecular Ecology. 2008;17:344–350. doi: 10.1111/j.1365-294X.2007.03400.x. [DOI] [PubMed] [Google Scholar]
- Hendry AP, Farrugia TJ, Kinnison MT. Human influences on rates of phenotypic change in wild animal populations. Molecular Ecology. 2007 doi: 10.1111/j.1365-294X.2007.03428.x. (online early articles). doi: 10.1111/j.1365-294X.2007.03428.x. [DOI] [PubMed] [Google Scholar]
- Hogg JT, Forbes SH, Steele BM, Luikart G. Genetic rescue of an insular population of large mammals. Proceedings of the Royal Society B – Biological Sciences. 2006;273:1491–1499. doi: 10.1098/rspb.2006.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holling CS. Adaptive Environmental Assessment and Management. New York: John Wiley; 1978. [Google Scholar]
- Hufford KM, Mazer SJ. Plant ecotypes: genetic differentiation in the age of ecological restoration. Trends in Ecology & Evolution. 2003;18:147–155. [Google Scholar]
- Johansen-Morris AD, Latta RG. Fitness consequences of hybridization between ecotypes of Avena barbata: hybrid breakdown, hybrid vigour and transgressive segregation. Evolution. 2006;60:1585–1595. [PubMed] [Google Scholar]
- Kauer MO, Dieringer D, Schlotterer C. A microsatellite variability screen for positive selection associated with the “Out of Africa” habitat expansion of Drosophila melanogaster. Genetics. 2003;165:1137–1148. doi: 10.1093/genetics/165.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn MH, Murphy WJ, Ostrander EA, Wayne RK. Genomics and conservation genetics. Trends in Ecology & Evolution. 2006;21:629–637. doi: 10.1016/j.tree.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Lande R, Saether BE, Engen S. Threshold harvesting for sustainability of fluctuating resources. Ecology. 1997;78:1341–1350. [Google Scholar]
- Lee CE, Remfert JL, Chang YM. Response to selection and evolvability of invasive populations. Genetica. 2007;129:179–192. doi: 10.1007/s10709-006-9013-9. [DOI] [PubMed] [Google Scholar]
- Lewontin RC. 25 Years ago in genetics - electrophoresis in the development of evolutionary genetics - milestone or millstone? Genetics. 1991;128:657–662. doi: 10.1093/genetics/128.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC, Krakauer J. Distribution of gene frequency as a test of theory of selective neutrality of polymorphisms. Genetics. 1973;74:175–195. doi: 10.1093/genetics/74.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. A quantitative genetic perspective on conservation issues. In: Avise JC, Hamrick JL, editors. Conservation Genetics: Case Studies from Nature. New York: Chapman and Hall; 1996. pp. 471–501. [Google Scholar]
- Lynch M, Lande R. Evolution and extinction in response to environmental change. In: Kareive PM, Kingsolver JG, Huey RB, editors. Biotic Interactions and Global Change. Sunderland, MA: Sinauer; 1992. pp. 234–250. [Google Scholar]
- Lynch M, Lande R. The critical effective size for a genetically secure population. Animal Conservation. 1998;1:70–72. [Google Scholar]
- Mace GM, Purvis A. Evolutionary biology and practical conservation: bridging a widening gap. Molecular Ecology. 2008;17:9–19. doi: 10.1111/j.1365-294X.2007.03455.x. [DOI] [PubMed] [Google Scholar]
- Madsen T, Ujvari B, Olsson M. Novel genes continue to enhance population growth in adders (Vipera berus. Biological Conservation. 2004;120:145–147. [Google Scholar]
- Maehr DS, Lacy RC. Avoiding the lurking pitfalls in Florida panther recovery. Wildlife Society Bulletin. 2002;30:971–978. [Google Scholar]
- Maehr DS, Crowley P, Cox JJ, Lacki MJ, Larkin JL, Hoctor TS, Harris LD, Hall PM. Of cats and Haruspices: genetic intervention in the Florida panther. Animal Conservation. 2006;9:127–132. Response to Pimm et al. [Google Scholar]
- McKay JK, Latta RG. Adaptive population divergence: markers, QTL and traits. Trends in Ecology & Evolution. 2002;17:285–291. [Google Scholar]
- McKay JK, Christian CE, Harrison S, Rice KJ. How local is local? – A review of practical and conceptual issues in the genetics of restoration. Restoration Ecology. 2005;13:432–440. [Google Scholar]
- Merila J, Crnokrak P. Comparison of genetic differentiation at marker loci and quantitative traits. Journal of Evolutionary Biology. 2001;14:892–903. [Google Scholar]
- Morgan TJ, Evans MA, Garland T, Swallow JG, Carter PA. Molecular and quantitative genetic divergence among populations of house mice with known evolutionary histories. Heredity. 2005;94:518–525. doi: 10.1038/sj.hdy.6800652. [DOI] [PubMed] [Google Scholar]
- Moritz C. Strategies to protect biological diversity and the evolutionary processes that sustain it. Systematic Biology. 2002;51:238–254. doi: 10.1080/10635150252899752. [DOI] [PubMed] [Google Scholar]
- Palumbi SR. The Evolution Explosion: How Humans Cause Rapid Evolutionary Change. New York: Norton and Company; 2001. [Google Scholar]
- Pimm SL, Dollar L, Bass OL. The genetic rescue of the Florida panther. Animal Conservation. 2006;9:115–122. [Google Scholar]
- Porcher E, Giraud T, Lavigne C. Genetic differentiation of neutral markers and quantitative traits in predominantly selfing metapopulations: confronting theory and experiments with Arabidopsis thaliana. Genetical Research. 2006;87:1–12. doi: 10.1017/S0016672306007920. [DOI] [PubMed] [Google Scholar]
- Reed DH, Frankham R. How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evolution. 2001;55:1095–1103. doi: 10.1111/j.0014-3820.2001.tb00629.x. [DOI] [PubMed] [Google Scholar]
- Reznick DN, Shaw FH, Rodd FH, Shaw RG. Evaluation of the rate of evolution in natural populations of guppies (Poecilia reticulata. Science. 1997;275:1934–1937. doi: 10.1126/science.275.5308.1934. [DOI] [PubMed] [Google Scholar]
- Robertson A. Gene frequency distributions as a test of selective neutrality. Genetics. 1975;81:775–785. doi: 10.1093/genetics/81.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, Baughman S, et al. The population biology of invasive species. Annual Review of Ecology and Systematics. 2001;32:305–332. [Google Scholar]
- Schreiber ESG, Bearlin AR, Nicol SJ, Todd CR. Adaptive management: a synthesis of current understanding and effective application. Ecological Management & Restoration. 2004;5:177–182. [Google Scholar]
- Seal US. Genetic Conservation and Management of the Florida Panther (Felis concolor coryi) Apple Valley, MN: Captive Breeding Specialist Group; 1992. Report to the US Fish and Wildlife Service. [Google Scholar]
- Seal US. A Plan for Genetic Restoration and Management of the Florida Panther (Felis concolor coryi) Apple Valley, MN: Captive Breeding Specialist Group; 1994. Report to the US Fish and Wildlife Service. [Google Scholar]
- Smith TB, Bernatchez L. Evolutionary change in human-altered environments. Molecular Ecology. 2008;7:1–8. doi: 10.1111/j.1365-294X.2007.03607.x. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Pires JC, Kovarik A, Tate JA, Mavrodiev E. Recent and recurrent polyploidy in Tragopogon (Asteraceae): cytogenetic, genomic and genetic comparisons. Biological Journal of the Linnean Society. 2004;82:485–501. [Google Scholar]
- Soule ME, Wilcox BM. Conservation Biology: An Evolutionary-Ecological Perspective. Sunderland, MA: Sinauer; 1980. [Google Scholar]
- Steinberg EK, Jordan CE. Using molecular genetics to learn about the ecology of threatened species: the allure and the illusion of measuring genetic structure in natural populations. In: Fiedler PL, Karieva PM, editors. Conservation Biology. 2nd edn. New York: Chapman and Hall; 1998. pp. 440–460. For the coming decade. [Google Scholar]
- Stockwell CA, Hendry AP, Kinnison MT. Contemporary evolution meets conservation biology. Trends in Ecology & Evolution. 2003;18:94–101. [Google Scholar]
- Storfer A. Quantitative genetics: a promising approach for the assessment of genetic variation in endangered species. Trends in Ecology & Evolution. 1996;11:343–348. doi: 10.1016/0169-5347(96)20051-5. [DOI] [PubMed] [Google Scholar]
- Swindell WR, Bouzat JL. Modeling the adaptive potential of isolated populations: experimental simulations using Drosophila. Evolution. 2005;59:2159–2169. [PubMed] [Google Scholar]
- Swindell WR, Bouzat JL. Gene flow and adaptive potential in Drosophila melanogaster. Conservation Genetics. 2006;7:79–89. [Google Scholar]
- Tallmon DA, Luikart G, Waples RS. The alluring simplicity and complex reality of genetic rescue. Trends in Ecology & Evolution. 2004;19:489–496. doi: 10.1016/j.tree.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Templeton AR. Coadaptation and outbreeding depression. In: Soule ME, editor. Conservation Biology: The Science of Scarcity and Diversity. Sunderland, MA: Sinauer; 1986. pp. 105–116. [Google Scholar]
- Van Tienderen PH, De Hahn AA, Van Der Linden CG, Vosman B. Biodiversity assessment using markers for ecologically important traits. Trends in Ecology & Evolution. 2004;17:577–582. [Google Scholar]
- Tseng M. Evolution in human-altered environments: a summit to turn science into policy. Molecular Ecology. 2007;16:3287–3288. doi: 10.1111/j.1365-294X.2007.03393.x. [DOI] [PubMed] [Google Scholar]
- Tsunoda S, Hirata K, Gomez-Campo C. Brassica Crops and Wild Allies. Tokyo: Japan Scientific Societies Press; 1980. [Google Scholar]
- Vasemagi A, Primmer CR. Challenges for identifying functionally important genetic variation: the promise of combining complementary research strategies. Molecular Ecology. 2005;14:3623–3642. doi: 10.1111/j.1365-294X.2005.02690.x. [DOI] [PubMed] [Google Scholar]
- Walters CJ. Is adaptive management helping to solve fisheries problems? Ambio. 2007;36:304–307. doi: 10.1579/0044-7447(2007)36[304:iamhts]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Walters CJ, Hilborn R. Ecological optimization and adaptive management. Annual Review of Ecology and Systematics. 1978;9:157–188. [Google Scholar]
- Wayne RK. Consequences of domestication: morphological diversity of the dog. In: Ruvinsky A, Sampson J, editors. The Genetics of the Dog. Oxon, UK: CABI Publishing; 2001. pp. 43–60. [Google Scholar]
- Westemeier RL, Brawn JD, Simpson SA, Esker TL, Jansen RW, Walk JW, Kershner EL, et al. Tracking the long-term decline and recovery of an isolated population. Science. 1998;282:1695–1698. doi: 10.1126/science.282.5394.1695. [DOI] [PubMed] [Google Scholar]
- Wise CA, Ranker TA, Linhart YB. Modeling problems in conservation genetics with Brassica rapa: genetic variation and fitness in plants under mild, stable conditions. Conservation Biology. 2002;16:1542–1554. [Google Scholar]


