Abstract
The intense fishing mortality imposed on Atlantic cod in Icelandic waters during recent decades has resulted in marked changes in stock abundance, as well as in age and size composition. Using a molecular marker known to be under selection (Pan I) along with a suite of six neutral microsatellite loci, we analysed an archived data set and revealed evidence of distinct temporal changes in the frequencies of genotypes at the Pan I locus among spawning Icelandic cod, collected between 1948 and 2002, a period characterized by high fishing pressure. Concurrently, temporal stability in the composition of the microsatellite loci was established within the same data set. The frequency of the Pan IBB genotype decreased over a period of six decades, concomitant with considerable spatial and technical changes in fishing effort that resulted in the disappearance of older individuals from the fishable stock. Consequently, these changes have likely led to a change in the genotype frequencies at this locus in the spawning stock of Icelandic cod. The study highlights the value of molecular genetic approaches that combine functional and neutral markers examined in the same set of individuals for investigations of the selective effects of harvesting and reiterates the need for an evolutionary dimension to fisheries management.
Keywords: cod, fisheries selection, Gadus morhua, genetic composition, Iceland, Pan I locus, temporal trend
Introduction
Worldwide, there have been significant changes in life history traits of fish stocks that are consistent with a response to fishing pressure (Jørgensen et al. 2007). However, in most cases it remains inconclusive whether these changes reflect an evolutionary response to fishing mortality or merely phenotypic plasticity (Marshall and Browman 2007; Kuparinen and Merilä 2008). Evidence in support of contemporary fisheries-induced evolution has accumulated from both experimental (reviewed by Conover and Baumann 2009) and retrospective studies (Walsh et al. 2006; Conover and Munch 2007). The latter include investigations of long-term trends in empirical field data (Edeline et al. 2007; Kendall et al. 2009), which have most frequently utilized observations on the age and size at which individuals mature (Jørgensen et al. 2007). The analysis of such data has been facilitated by the development of the probabilistic maturation reaction norm (PMRN) approach (Heino et al. 2002; Barot et al. 2004), a statistical tool developed to help disentangle genetic variation in maturation from phenotypic plasticity, resulting from variation in growth and survival (Heino and Dieckmann 2008). Indeed, the major difficulty faced by investigations of fisheries-induced evolution in natural populations is that genetic changes and phenotypic plasticity are confounded in the phenotypic trait upon which most previous studies have had to rely. The need for a direct genetic approach has been noted (Kuparinen and Merilä 2007; Allendorf et al. 2008), but unambiguous genetic evidence of contemporary evolution in marine fish populations has proven elusive because of the limited availability of adaptive variation estimates, as well as historical molecular data sets (Conover et al. 2006). Population genetic analyses of historical samples can provide invaluable insights into human-induced changes in the genetic composition of fish populations (Nielsen and Hansen 2008; Hansen et al. 2009). In recent years, attention has thus been drawn to archived collections of otoliths and scales as a source of DNA for long-term temporal genetic studies of natural fish populations (Nielsen and Hansen 2008; Palstra et al. 2009; Palstra and Ruzzante 2010). Although genetic studies have commonly focused on neutral variation, recent approaches have shown that non-neutral or functional markers (loci subject to selection) are more likely than neutral markers to reveal processes leading to changes in allele frequencies over short temporal scales (Conover et al. 2006). Indeed, because of the large effective population sizes of exploited marine fishes, neutral markers are unlikely to exhibit changes in a signal over a relatively short time period, while loci under selection might readily respond to changes in selection regime.
Like most cod (Gadus morhua) stocks in the North Atlantic, Icelandic cod has experienced a drastic reduction in abundance and spawning stock biomass (SSB) during the last few decades. SSB has declined from about 1 million tonnes in the early 1950s to <200 000 tonnes at the beginning of this century (Fig. 1). Since the 1980s, estimates of SSB have remained below the long-term average, rarely exceeding 200 000 tonnes. Concurrently, there has been a long-term increase in fishing mortality (F), with exploitation rates exceeding 0.8 during three different time periods (Fig. 1). These levels of F are considerably higher than the target of 0.3–0.4, which would enable a sustainable fishery that is within biologically safe limits (Marine Research Institute 2007).
Figure 1.
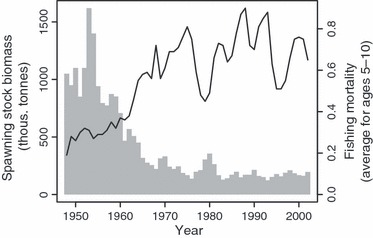
Estimated total spawning stock biomass (histogram bars) and average fishing mortality of age groups 5–10 (line) of cod in Icelandic waters during the period 1948–2002 (Marine Research Institute 2009).
Prior to World War II (WWII), both international trawlers and the Icelandic fleet focused their fishing effort on the relatively shallow inshore areas around Iceland (Þór 2005), with landings of cod reaching a maximum of just short of 52 000 tonnes in 1933 (Marine Research Institute 2009). During the war, the international fleet was absent from Icelandic waters and therefore fishing pressure was considerably lower. Since WWII there have been substantial spatial and seasonal changes in the exploitation pattern of Icelandic cod. Immediately after the war, the inshore cod fishery increased again because of the continued exploitation by the national fleet and return of international vessels, resulting in a peak catch of 548 000 tonnes in 1954 (Marine Research Institute 2009). In 1952, Icelandic authorities banned the use of bottom trawls and Danish seine within four nautical miles of the shoreline. This regulation was followed by a gradual extension of the national fishery jurisdiction, culminating at 200 nautical miles in 1976 (Ministry of Fisheries 2004; Þór 2005). This, combined with the stricter inshore fishing regulations, improvements in fishing technology, and the introduction of larger vessels, resulted in the redirection of fishing effort into deeper offshore waters. Concurrently, trawlers were able to follow the seasonal migration of cod (Jónsson 1996), rather than being restricted to inshore breeding grounds during the spawning season.
Alongside the declines in stock abundance, there have been notable alterations in the life history of Icelandic cod, whereby far fewer fish now survive beyond 7 years of age, resulting in severely truncated age and size distributions (Schopka 1994; Marteinsdóttir and Thorarinsson 1998). Furthermore, Icelandic cod now reach maturity at younger ages and smaller sizes (Marteinsdóttir and Begg 2002). A recent study, which estimated PMRNs for 36 cohorts of Icelandic cod, found evidence that a shift towards maturation at smaller sizes and younger ages has occurred independently of changes in growth, condition and temperature (Pardoe et al. 2009). This change in the maturation schedule of Icelandic cod, along with the loss of older and larger repeat spawners, raises concerns for the stock's reproductive potential, and consequently its ability to withstand harvesting and future environmental change (Law 2007; Árnason et al. 2009). An investigation into whether the fishery has also had a long-term effect on genetic variation in the Icelandic cod stock is thus imperative.
In this study, we used archived otoliths, collected between 1948 and 2002, to jointly examine phenotypic and genetic data. Our goal was to examine historical trends in the genetic composition of the Icelandic cod stock using both selected and neutral markers. Temporal stability in the allele frequency distribution of neutral markers would reflect stability in population structure and minimal drift, as is expected when effective population sizes are large. When such stability at neutral markers is accompanied by temporal changes in allele frequency distributions at functional loci then one can infer the population has been subjected to changes in selection regimes. We examined polymorphism at the Pantophysin (Pan I) locus (Pogson et al. 1995; Pogson 2001), a genetic marker known to be under selection or closely linked to a gene under selection, as well as a suite of six microsatellite loci assumed to be neutral. The two main allele classes within the Pan I locus, Pan IA and Pan IB, are differentiated by a Dra I restriction site (Pogson 2001). The distribution of this diallelic system has been the focus of numerous population genetics studies in cod (Pampoulie et al. 2006, 2008; Nielsen et al. 2007; Skarstein et al. 2007; Árnason et al. 2009 and references within these studies).
We found clear evidence that major changes in genetic variation at the Pan I locus of Icelandic cod have occurred alongside those in fishing patterns and age composition of the stock, whereas no temporal changes were detected with neutral genetic markers. These results strongly demonstrate the importance of long-term genetic data, and more specifically the use of both functional and neutral genes, when studying anthropogenic disturbances in a highly dynamic system such as that characterizing the Icelandic cod stock.
Material and methods
Sampling
A total of 16 samples, comprising 1471 spawning or near-spawning individuals, were collected from three relatively shallow locations known to be major spawning grounds for Icelandic cod (South West = SW, Faxaflói Bay = FAX and the West Coast Bay, Breidafjördur = BRE; see Table 1 for exact details on location). DNA from 1286 of those individuals was successfully amplified and used for genetic analysis. Otoliths collected during the period 1948–1996 were recovered from the archived collections at the Icelandic Marine Research Institute (MRI). Otoliths collected prior to 2000 were stored in dry paper envelopes, while the more recent historical samples (collected in 2000) were stored in specially designed plastic storage wells. A contemporary sample (2002) was obtained from fresh gill tissue. The choice of sample years was based on the availability of individuals (sample size) stratified by years of known high and low stock size. Biological information, i.e. total length, age, sex and maturity status, for all individuals used in the genetic analyses was retrieved directly from the MRI database. Individuals were pooled into 10-year cohort classes based on their year of birth (1931–1940 through to 1991–2000).
Table 1.
Overview of Icelandic cod samples used in this study: year, date and subarea of sampling (see Note for corresponding coordinates), sampling depth (meters; entries with asterisks show depth distributions [10–90% quantiles] within the subarea for those samples where actual depth of the catch was not available), number of individuals (n), amplification success (Asucc), proportion of females (Pfem), mean total length (cm; TLmean), length range (cm; TLrange), mean age (years; Amean), age range (years; Arange). For all samples, DNA was derived from archived otoliths with the exception of 2002 for which DNA was obtained from fresh gills
| Year | Date | Subarea | Depth | n | Asucc | Pfem | TLmean | TLrange | Amean | Arange |
|---|---|---|---|---|---|---|---|---|---|---|
| 1948 | 18/4–22/4 | BRE | 100 | 132 | 68.9 | 83.9 | 87.48 | 67–150 | 9.84 | 5–16 |
| 1957 | 20/3–2/4 | SW | 37 | 111 | 95.6 | 40.7 | 93.95 | 73–136 | 9.38 | 5–19 |
| 1959 | 14/4 | SW | 37–74* | 90 | 83.3 | 21.3 | 96.22 | 78–115 | 9.64 | 6–14 |
| 1966 | 15/4 | SW | 37–74* | 95 | 94.7 | 51.1 | 91.81 | 74–112 | 8.42 | 5–13 |
| 1972 | 6/4–8/4 | FAX | 38–83* | 94 | 88.3 | 44.6 | 87.12 | 75–100 | 8.54 | 4–11 |
| 1973 | 3/5 | FAX | 68 | 95 | 64.2 | 59.0 | 90.37 | 76–124 | 8.84 | 6–12 |
| 1973 | 26/4 | SW | 70 | 86 | 97.7 | 46.4 | 88.26 | 76–103 | 8.84 | 6–11 |
| 1976 | 19/4 | FAX | 55 | 88 | 77.5 | 55.1 | 88.27 | 61–121 | 7.26 | 4–13 |
| 1976 | 10/4 | SW | 45 | 90 | 100.0 | 30.0 | 88.77 | 47–130 | 6.93 | 3–11 |
| 1979 | 26/4 | FAX | 50 | 85 | 97.6 | 34.9 | 85.39 | 69–115 | 6.98 | 6–10 |
| 1985 | 18/4 | SW | 37–74* | 93 | 100.0 | 54.8 | 96.53 | 70–129 | 8.84 | 5–18 |
| 1996 | 16/4–19/4 | BRE | 63 | 87 | 82.2 | 50.0 | 86.28 | 57–128 | 7.26 | 4–11 |
| 1996 | 25/3–9/4 | FAX | 55 | 98 | 68.4 | 56.7 | 97.24 | 79–130 | 7.36 | 5–14 |
| 1996 | 27/3 | SW | 56 | 85 | 100.0 | 43.0 | 101.20 | 82–128 | 7.55 | 5–13 |
| 2000 | 6/4–8/4 | BRE | 150 | 68 | 97.1 | 45.5 | 77.50 | 50–104 | 6.75 | 5–11 |
| 2002 | 5/4 | SW | 54 | 74 | 98.5 | 40.5 | 85.54 | 53–117 | 6.18 | 3–12 |
| 1471 | 87.4 | 47.4 |
Note: Exact coordinates and mean depth of subareas are as follows: subarea South West (SW): 63.5–64°N, 20–21°W; subarea Faxaflói bay (FAX): 64–64.5°N, 22–23°W; subarea West Coast Bay, Breiðafjörður (BRE): 65–65.5°N, 23–24°W.
DNA extraction and analysis
DNA was extracted from the otolith surface using a Chelex/Proteinase K protocol (Estoup et al. 1996). The DNA was preserved at 4°C during this study and subsequently moved to a −80°C freezer for long-term storage. Primers specially designed to amplify a short fragment (142 bp) of the Pan I locus from DNA that has experienced considerable degradation (Nielsen et al. 2007; forward: 5′-GGCAAATGAAACCCAGAAAA, rev: 5′-ATGACACTTGTGGCAAGCAG) were used for the polymerase chain reaction (PCR). PCR was performed in a 17 μL volume containing 3 μL of DNA product, 1.7 μL 10× buffer, 0.51 μL of 1.5 mm MgCl2, 1.7 μL of 2.5 mm DNTP, 0.25 μL of each 10 μm primer solution, 0.25 μL of 20 mg/mL BSA (Fermentas) and 0.5 units of DyNAzyme polymerase (Finnzymes Oy, Espoo, Finland). Cycles were performed on GeneAmp2700 thermal block using ‘Touchdown’ procedures as follows: initial denaturation step of 2 min at 95°C followed by 10 cycles of 30 s at 94°C, 45 s of annealing temperature that decreases in each cycle by 0.5°C until 55°C was reached and 30 s of 70°C. This was followed by 25 cycles of 30 s at 94°C, 50 s at 55°C and 30 s at 70°C. A final elongation step of 5 min at 72°C was performed. Immediately after the PCR, a restriction analysis was carried out on the PCR product (17 μL) using 18 units of the enzyme DraI (Fermentas) and accompanying buffer (2 μL). DraI cuts at the diagnostic restriction sites in the PCR product, discriminating between the Pan IA and the Pan IB alleles. Fragments were visualized on a 3.5% agarose gel stained with ethidium bromide. As expected, the digestion pattern was similar to that described by Nielsen et al. (2007): a single band at 142 bp for the Pan IAA homozygote, two bands of 40 and 102 for the Pan IBB homozygote and all three bands for the heterozygotes.
We also analysed genetic variation at six microsatellite loci assumed to be neutral using a subset of those individuals from the Pan analysis: Gmo2 (Brooker et al. 1994), Gmo8, Gmo19 (Miller et al. 2000), Tch5, Tch14 and Tch 22 (O'Reilly et al. 2000). PCR were performed in 10 μL volumes containing 2 μL of DNA, 1 μL of 10× Buffer, 1 μL of 2.5 mm DNTP, 0.2–0.4 units of DyNAzyme™ DNA polymerase (Finnzymes) and 1–2 μm of each primer. PCR were performed on a GeneAmp®2700 thermal block using the ‘Touchdown’ procedure described earlier. PCR products were multiplexed and detected on an ABI-automatic sequencer (ABI 377; Applied Biosystem) using GeneScan 3.1.2 (Biosystems 2000) and scored using GeneMapper 3.7 (Biosystems 2004). The occurrence of genotypic errors resulting from technical artefacts (null alleles) or DNA quality (large allele dropouts) was assessed using the program MICRO-CHECKER (Van Oosterhout et al. 2004).
Extra care was taken to minimize chances of potential contamination during laboratory procedures. Samples were processed in a designated area that was decontaminated between procedures using a chlorine solution and UV light. Negative controls were run, and comparisons to microsatellite readings were conducted to test for possible contaminations between neighbouring DNA wells.
Statistical analyses
Changes in the age distribution between sampling years were tested using chi-square tests. In situations of low expected values (<5), samples from cod aged 6 years or younger, and eleven years or older, were pooled together. The changes in Pan I genotype frequencies between age groups and cohort classes were also tested using chi-square tests. For these tests, individuals aged 5 years or younger, and aged twelve years or older, were pooled, while all intermediate age classes were kept separate.
To investigate the long-term consistency of genotypic change by age, we pooled the samples in three time periods. The earliest period consisted of samples collected in 1948, 1957, 1959 and 1966 when average fishing mortality is estimated to have been <0.3. The middle period, characterized by an average fishing mortality of 0.6, contained samples from 1972, 1973, 1976 and 1979. The latest period comprised samples collected in 1985, 1996, 2000 and 2002 and featured several peaks of very high fishing mortality (>0.8).
We used a multinomial log-linear model (Venables and Ripley 2002) to explore changes in genotype frequencies with time. The purpose of the model was to describe the long-term changes in genotype frequencies of cohorts 1932–1999, after taking into account that some cohorts were only sampled at a younger age and other cohorts only at an older age. In a single model, we fitted the frequencies of Pan I genotypes Pan IAA, Pan IAB and Pan IBB as a smooth function of both cohort and age. The smooth functions for cohort and age were natural cubic splines, each with 3 degrees of freedom. The multinomial model ensures that the predicted relative frequencies of the three Pan I genotypes sum to one within each cohort-age stratum.
Potential differences in growth between the Pan I genotypes were investigated by analysing length-at-age data, combined for all years. The analyses included cod aged 7–9 years only since those age classes contained sufficient individuals from all three Pan I genotypes in all time periods. To test whether differences in growth of Icelandic cod were related to their Pan I genotype, mean length was modelled using analysis of covariance (ANCOVA) with Pan I genotype as the categorical independent variable and age as continuous covariate.
For the suite of six microsatellite loci, observed (HO) and expected (HE) heterozygosity were calculated in GENETIX 4.03 (Belkhir et al. 1999). Tests for deviations from Hardy–Weinberg equilibrium (HWE) were conducted for both the Pan I locus and the suite of six microsatellite loci using the exact test in GENEPOP 3.1 (Raymond and Rousset 1995). Genetic differentiation between sampling sites and all pairs of populations was estimated with pairwise FST estimates following Weir and Cockerham (1984), and 95% confidence intervals were determined by bootstrapping over loci. The program FSTAT 2.9.2 (Goudet 1995) was used for this analysis. An analysis of molecular variance (AMOVA) was carried out for both the Pan I locus and the suite of six microsatellite loci in ARLEQUIN 3.0 (Excoffier et al. 2005) to assess hierarchical partitioning of genetic variance. The genetic relationships among samples were further analysed using principal component analysis and visualized with a multidimensional scaling (MDS) plot based on a matrix of pairwise FST′s.
Results
A hierarchical AMOVA revealed that overall variation at the Pan I locus was because of temporal (FSC = 0.122, P < 0.001) rather than spatial variation (FCT = −0.025, P = 0.92) (Table 2). Furthermore, no trend was detected in sampling depth (Table 1). Therefore, in the following analyses, samples were grouped into sample years (or cohort classes) independently of sampling location (and depth). Results from the analysis of genetic variation at the Pan I locus are presented in Table 3. Genetic diversity among sample years was highly variable, with observed heterozygosities ranging from 0.153 (2002) to 0.988 (1979), with an average value of 0.617. Genetic diversity within cohort classes was relatively moderate, with observed heterozygosities ranging from 0.395 (1991–2000 cohort class) to 0.761 (1951–1960 cohort class). Significant deviations from HWE were found in 50% of the sample years, whereas five of seven cohort classes contained samples that were not in HWE. Heterozygote excess was the cause of all of the deviations from HWE (Table 3).
Table 2.
Hierarchical partitioning of genetic variance at the Pan I locus, based on an analysis of molecular variance (AMOVA) of samples of Icelandic cod (see Table 1)
| Source of variation | d.f. | Variance components | Percentage variation | Fixation indices | P-values |
|---|---|---|---|---|---|
| Among locations | 2 | −0.006 | −2.49 | CT = −0.025 | 0.917 |
| Among years, within locations | 13 | 0.031 | 12.47 | SC = 0.122 | <0.001 |
| Within years | 2556 | 0.220 | 90.02 | ST = 0.100 | <0.001 |
| Total | 2571 | 0.244 | 100 |
Table 3.
Observed (HO) and expected (HE) heterozygosity and observed frequencies of Pan IB allele in Icelandic cod samples (n: sample size) by cohort class and sample year. Bold values indicate significant deviations of FIS values from Hardy–Weinberg expectations, after correction for multiple tests
| Cohort class | n | HO | HE | FIS | Freq. of Pan IB allele |
|---|---|---|---|---|---|
| 1931–1940 | 62 | 0.629 | 0.490 | −0.278 | 0.573 |
| 1941–1950 | 182 | 0.621 | 0.499 | −0.242 | 0.525 |
| 1951–1960 | 113 | 0.761 | 0.495 | −0.534 | 0.549 |
| 1961–1970 | 383 | 0.640 | 0.496 | −0.290 | 0.453 |
| 1971–1980 | 184 | 0.712 | 0.500 | −0.422 | 0.497 |
| 1981–1990 | 215 | 0.502 | 0.385 | −0.302 | 0.261 |
| 1991–2000 | 147 | 0.395 | 0.377 | −0.044 | 0.252 |
| 1286 |
| Sample year | n | HO | HE | Freq. of Pan IB allele | |
|---|---|---|---|---|---|
| 1948 | 91 | 0.637 | 0.497 | −0.277 | 0.539 |
| 1957 | 106 | 0.670 | 0.499 | −0.338 | 0.476 |
| 1959 | 75 | 0.480 | 0.453 | −0.053 | 0.653 |
| 1966 | 90 | 0.867 | 0.499 | −0.734 | 0.522 |
| 1972 | 83 | 0.723 | 0.484 | −0.490 | 0.590 |
| 1973 | 145 | 0.731 | 0.493 | −0.480 | 0.559 |
| 1976 | 158 | 0.449 | 0.375 | −0.195 | 0.250 |
| 1979 | 83 | 0.988 | 0.500 | −0.976 | 0.506 |
| 1985 | 93 | 0.559 | 0.495 | −0.124 | 0.548 |
| 1996 | 224 | 0.500 | 0.384 | −0.301 | 0.259 |
| 2000 | 66 | 0.652 | 0.486 | −0.333 | 0.417 |
| 2002 | 72 | 0.153 | 0.187 | −0.188 | 0.104 |
| 1286 |
As has been previously observed with an independent data set (Marteinsdóttir and Thorarinsson 1998), the age composition of Icelandic cod changed markedly through the study period. The observed shift towards a narrower age distribution resulted from the loss of older age classes and an overall reduction in mean age (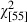 = 851.0, P < 0.001) (Table 1, Fig. 2). Moreover, our analyses revealed that the Pan I locus has been subject to major changes in allele frequencies during the study period, in relation to both age and cohort class. Within cohorts, the frequencies of the homozygous genotypes were age dependent (Figs 3 and 4). The Pan IBB genotype was more common among the older spawning cod, while the Pan IAA genotype was most frequently observed among the younger age classes (
= 851.0, P < 0.001) (Table 1, Fig. 2). Moreover, our analyses revealed that the Pan I locus has been subject to major changes in allele frequencies during the study period, in relation to both age and cohort class. Within cohorts, the frequencies of the homozygous genotypes were age dependent (Figs 3 and 4). The Pan IBB genotype was more common among the older spawning cod, while the Pan IAA genotype was most frequently observed among the younger age classes (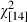 = 187, P < 0.001; Fig. 3). In fact, no spawning cod younger than 6 years of age carried the Pan IBB genotype. These age-dependent trends in homozygote frequency were still present when the data were split into three different time periods (Fig. 4). A high proportion of heterozygote individuals were found in all age classes (Figs 3 and 4). Indeed, as stated earlier, a lack of HWE both within sample years and cohort classes was mainly because of heterozygote excess (Table 3).
= 187, P < 0.001; Fig. 3). In fact, no spawning cod younger than 6 years of age carried the Pan IBB genotype. These age-dependent trends in homozygote frequency were still present when the data were split into three different time periods (Fig. 4). A high proportion of heterozygote individuals were found in all age classes (Figs 3 and 4). Indeed, as stated earlier, a lack of HWE both within sample years and cohort classes was mainly because of heterozygote excess (Table 3).
Figure 2.
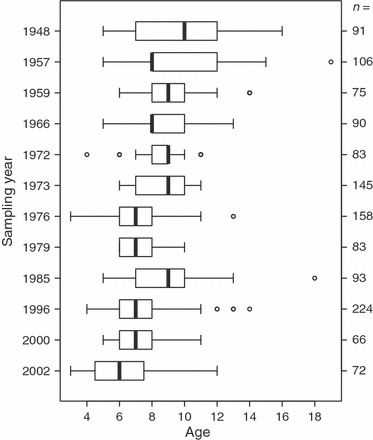
Box and whiskers plot of the age distribution of Icelandic cod within each sampling year. The corresponding sample size is shown on the right axis.
Figure 3.
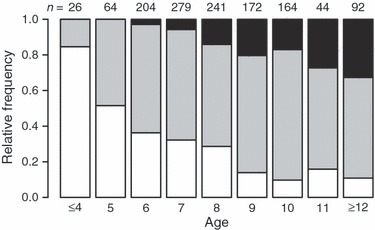
Age-specific distributions of Pan I genotype frequencies (white: Pan IAA, grey: Pan IAB, black: Pan IBB) in samples of Icelandic cod collected between 1948 and 2002. The corresponding sample size is listed above each column.
Figure 4.
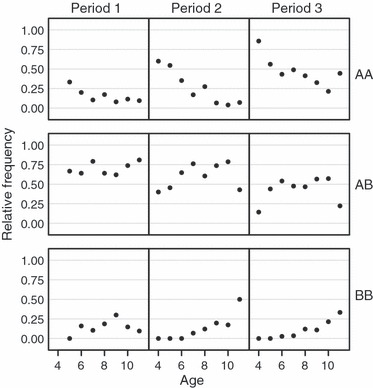
Observed age-specific frequencies of Pan I genotypes (Pan IAA, Pan IAB and Pan IBB) for Icelandic cod sampled in three time periods: Period 1, 1948–1966; Period 2, 1972–1979; Period 3, 1984–2002 (see Table 1 for actual years sampled within each period).
At the inter-cohort level, there was a gradual but strong decline in the frequency of Pan IBB over the study period (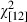 = 199.3, P < 0.001) (Fig. 5). Concomitantly, the Pan IAA genotypes increased in frequency, exceeding 50% in the last 20 years of the study at the expense of the Pan IBB and Pan IAB individuals (
= 199.3, P < 0.001) (Fig. 5). Concomitantly, the Pan IAA genotypes increased in frequency, exceeding 50% in the last 20 years of the study at the expense of the Pan IBB and Pan IAB individuals (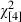 = 85.9, P < 0.0010) (Fig. 5). For those ages represented in our data throughout the study period, predicted genotype frequencies from a multinomial model showed similar temporal trends (Fig. 6). According to the model (likelihood ratio test versus null model
= 85.9, P < 0.0010) (Fig. 5). For those ages represented in our data throughout the study period, predicted genotype frequencies from a multinomial model showed similar temporal trends (Fig. 6). According to the model (likelihood ratio test versus null model 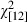 = 276.4, P < 0.001), the gradual increase in Pan IAA genotype frequency started in the 1960s and was accompanied by decreases in the relative proportions of the two other genotypes.
= 276.4, P < 0.001), the gradual increase in Pan IAA genotype frequency started in the 1960s and was accompanied by decreases in the relative proportions of the two other genotypes.
Figure 5.
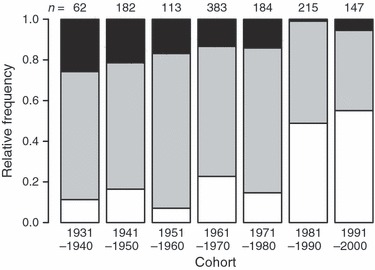
Observed Pan I genotype frequencies (white: Pan IAA, grey: Pan IAB, black: Pan IBB) for Icelandic cod from different (10 year) cohort classes. The corresponding sample size is listed above each column.
Figure 6.
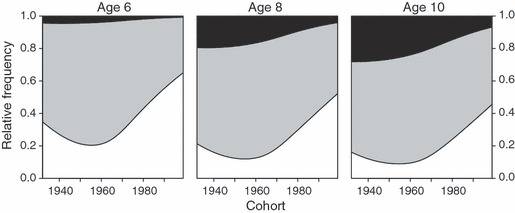
Predicted distributions of cohort-specific Pan I genotype frequencies for Icelandic cod aged 6, 8 and 10 years, from a multinomial model (smoothing allowing for three degrees of freedom).
Distinct differences were observed in length-at-age among the Pan I genotypes, whereby Icelandic cod carrying the Pan IAA genotype had significantly higher length-at-age on average than cod of the Pan IAB and Pan IBB genotypes (P = 0.048) (Fig. 7).
Figure 7.
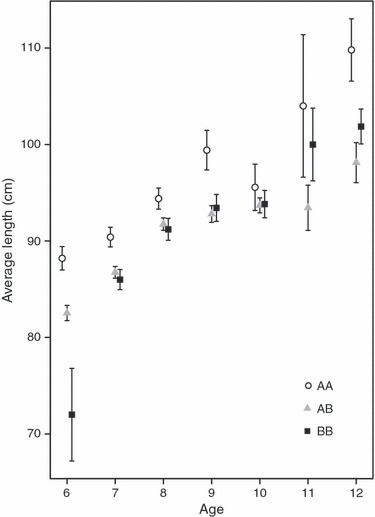
Average length (±1 SE) of Icelandic cod aged 6–12 years, sampled in the period 1948–2002, and carrying either Pan IAA, Pan IAB or Pan IBB genotype.
Multilocus genotype information from six microsatellites was obtained from 892 of the individuals utilized in the Pan I analyses, representing the same sampling years and sites (SW, FAX, BRE; Table 1). Genetic diversity in the samples was moderate to high, with observed heterozygosities ranging from 0.594 (Tch22) to 0.951 (Tch14). The numbers of alleles ranged from 5 (Tch22) to 35 (Tch14). Deviations from HWE were detected in 3 of 105 tests after Bonferroni correction for multiple tests and were not because of any specific loci or sample. The microsatellite analysis revealed that there was no genetic differentiation (FST) among sampling sites (FST = 0.0004, P = 0.100). Of 105 pairwise (FST) comparisons among all samples, none were found to be significantly different from zero and there was an overall FST = 0.0018 (P = 0.538). This pattern was also evident in a hierarchical AMOVA, which showed that only 0.03% of total genetic variance (FCT = 0.0003) could be attributed to variance among sampling locations. The variance between temporal samples was slightly higher (0.13%, FSC = 0.0013). However, both values were nonsignificant (P > 0.05). This lack of differentiation confirms long-term temporal stability under the assumption of neutrality and a MDS analysis (not shown) further confirmed the lack of spatial or temporal pattern in the data.
Discussion
Our study is the first, to our knowledge, to analyse functional and neutral genetic markers in combination with fisheries data, over a multidecadal time period, in an attempt to explain distinct changes in genetic and phenotypic variation in a fish stock that has been heavily exploited since the 1940s and 50s. By isolating DNA from archived cod otoliths, gathered over a 60-year period at spawning sites on the SW coast of Iceland, we were able to detect long-term temporal changes in genotype frequencies at the Pantophysin (Pan I) locus, whereas no changes were observed at six microsatellite loci. The significant decline in the Pan IBB genotype, which represented 26% of the sampled population in the 1930s but only 5% in the 1990s, followed changes in the exploitation pattern, which were characterized by increased effort in deeper waters facilitated by larger ships and technological advancements in fishing gear.
In the current study, the frequencies of Pan I genotypes varied both within and between cohorts. Within cohorts, the frequency of the Pan IBB genotype increased with age (Figs 3 and 4). Currently, the age-specific distribution of the Pan I genotypes is unknown for immature Icelandic cod. However, this result strongly indicates that fish of different genotypes mature at different ages. The absence of the Pan IBB genotype among spawning fish aged 3–5 years is therefore most likely due to cod carrying that genotype not having matured yet and thus not being present on the spawning sites, which provided the data for this study. The gradual increase in the frequency of the Pan IBB genotype from 6 to 9 years of age is thus likely to reflect differences in the rate of maturation. The potential existence of life history variation between cod of different Pan I genotypes was also supported by the observation that cod exhibiting the Pan IAA genotype grew at a significantly higher rate than those carrying the Pan IAB and Pan IBB genotypes (Fig. 7). Furthermore, our findings complement emerging evidence from tagging studies using data storage tags of two groups of Icelandic cod associated with foraging strategy and habitat selection during the feeding season (Pálsson and Thorsteinsson 2003; Pampoulie et al. 2008). The so-called coastal cod inhabit shallow shelf waters characterized by seasonal temperature trends. In contrast, ‘frontal’ cod undertake migrations to cold, deep waters (>250 m) where they forage at thermal fronts, making frequent vertical migrations between temperature extremes (<0 and >7°C). Pampoulie et al. (2008) revealed that individuals carrying the Pan IAA genotype are most likely to be classified as coastal cod based on their behaviour during feeding migrations, while individuals carrying the Pan IBB genotype most frequently exhibit the migratory behaviour of frontal cod. The heterozygotes were found to exhibit mixed feeding migration strategies.
In the present study, we have shown a gradual decline in the frequency of the Pan IBB genotype among spawning cod during the last 60 years. Concurrently, the Pan IAA genotype increased in frequency, exceeding 50% in the last 20 years of the study at the expense of the Pan IBB and Pan IAB individuals (Fig. 5). We cannot be certain of the primary driving force behind these changes in the proportions of the different genotypes. However, four different hypotheses, three of which can be linked to changes in exploitation patterns, warrant our scrutiny.
First, changes in genotypic frequencies may have resulted from changes in stock composition because of the consistent removal of older fish during a period of increased fishing mortality (Marteinsdóttir and Thorarinsson 1998; Fig. 2 in this study). By the end of the last century, relatively few cod survived to 10 years of age in Icelandic waters (Marine Research Institute 2009). Accordingly, the mean age of spawning cod declined from 10 to 6 years during the study period (Table 1, Fig. 2). As previously discussed, the oldest cod in our samples were predominantly of the Pan IBB or Pan IAB genotype, whereas relatively few cod carrying the Pan IAA genotype were found to be aged 9 years or older. Therefore, the truncation of the age distribution of Icelandic cod, a common feature of commercially exploited fish stocks, is likely to have resulted in the removal of the Pan IBB genotype from the spawning stock at a proportionally faster rate than that of the Pan IAA genotype.
Secondly, the gradual decline in the frequency of the Pan IBB genotype may also have resulted from the substantial spatial and seasonal changes in fishing effort on Icelandic cod that have occurred since WWII. Following the termination of fishing by Danish seine and bottom trawl within four nautical miles of the coast, the fishery jurisdiction was gradually extended to 200 nautical miles (see also the Introduction). At the same time, the introduction of larger vessels resulted in the redirection of fishing effort into deeper offshore waters. This also enabled trawlers to follow the seasonal migration of adult cod, rather than being restricted to inshore breeding grounds during the spawning season (Jónsson 1996). The bathymetric distribution of the Pan I genotypes has been recorded for both spawning and migrating cod around Iceland (Pampoulie et al. 2006, 2008; Árnason et al. 2009), while this information for juvenile cod is unfortunately scarce. Spawning cod sampled in 2002–2003 in the SW of Iceland, the area covered by this study, ranged from being predominantly of the Pan IAA genotype at the shallowest stations to predominantly of the Pan IBB and Pan IAB genotypes at the deeper stations (Pampoulie et al. 2006). Similar changes in genotypic frequency with depth, at stations all around Iceland, were observed by Árnason et al. (2009). Therefore, changes in the exploitation pattern of Icelandic cod during the study period are likely to have resulted in relatively greater fishing mortality in deeper water and on the feeding grounds and thus may have led to the disproportionate removal of the Pan IBB genotype from the stock by the commercial fishery.
Thirdly, we may not be able to fully explain the causes of these long-term temporal changes in genotypic frequency in Icelandic cod until our understanding of the population structure further improves. The distinct variation in life history traits, behaviour and Pan I genotype frequencies of Icelandic cod is persuasive evidence that the current stock designation does not capture the actual geographic scale of population differentiation (Pampoulie et al. 2006, 2008; Marteinsdóttir and Pardoe 2008). Traditionally, studies of genetic population structuring have utilized neutral molecular markers. Accordingly, our microsatellite analysis did not indicate any spatial structure and the long-term stability of the population under the assumption of neutrality could not be rejected. However, the absence of heterogeneity using neutral genetic markers does not contradict the hypothesized existence of different life history components in our data. Neutral markers are poorly suited to the detection of changes in genetic diversity because they segregate independently of the selected loci, and in populations with relatively large effective population sizes in particular the level of genetic drift will be low (Conover et al. 2006; Allendorf and Hard 2009). Indeed, the population components might experience differential postsettlement selection, likely influenced by environmental factors, but still exhibit a relatively high level of gene flow as a consequence of larval dispersal or mixing on breeding sites, or both. However, if there is some degree of gene flow restriction between the stock components, as a recent study suggests (T. B. Grabowski, B. McAdam, V. Thorsteinsson and G. Marteinsdóttir, unpublished manuscript), high effective population sizes and recent separation history are two nonexclusive explanations that could mask underlying stock structure (Case et al. 2006). If the Icelandic cod stock indeed comprises populations with limited gene flow and/or different selection schedules, then the changes in exploitation patterns described above will have affected the stock complex by removing the Pan IBB genotype at a faster rate than the Pan IAA genotype. Furthermore, the different populations may also have been targeted by the fishery in an uneven manner. The overall effects are similar, i.e. they result in long-term changes in genotype frequencies.
Finally in our fourth hypothesis, we recognize that the genotype distribution at the Pan I locus may have changed because of selection by other agents such as those of climatic origin. Temperature is the only available long-term proxy for potential environmental changes during this time period. Examination of temporal changes in sea surface temperature (SST) off the northern coast of Iceland (Hanna et al. 2006) did not reveal any significant correlations with changes in frequency of the Pan I genotypes (P > 0.05). This is perhaps not surprising as ocean conditions in this area have fluctuated extensively during the study period. Between 1925 and 1964, there was a high inflow of warm Atlantic water onto the Icelandic shelf resulting in relatively mild temperatures (Malmberg 1986; Astthorsson et al. 2007). In the late 1960s, the climate shifted dramatically because of increased inflow of icy freshwater from the Arctic (Jakobsson 1980; Malmberg 1986; Dickson et al. 1988). During this period, identified locally as the ‘Ice Years’, temperatures dropped far below the long-term average, reaching the lowest recordings in 1968–1969 (Astthorsson et al. 2007). Conditions subsequently remained more stable, until the current situation of gradual warming, which started in 1996 (Astthorsson et al. 2007). These striking fluctuations in temperature and salinity of Icelandic waters thus do not appear responsible for the gradual and consistent changes in the genotype frequencies observed in this study. Other studies have also shown that there does not appear to be a direct link between Pan I genotype frequencies in cod and water temperature (Nielsen et al. 2007). Unfortunately, the quantitative investigation of this potential relationship for Icelandic cod is limited by the quality and quantity of available environmental data. Although the SST time series from northern Icelandic waters (Hanna et al. 2006) is thought to be relatively representative of the average environmental conditions experienced by the stock in general, Icelandic cod are confronted with highly fluctuating environmental conditions throughout their lifespan (Jakobsson and Stefánsson 1998); variability that can probably only be reliably captured through data storage tags. Furthermore, the life history stage(s) at which temperature affects the genotypic distribution of the Pan I locus of cod is unknown. In conclusion, although the observed changes in genotype frequencies at the Pan I locus could be the result of simultaneous selection by both trends in temperature and fishing, the information that does exist indicates that although there have been persistent changes in environmental conditions in Icelandic waters, those changes alone are insufficient to explain the strong selection against the Pan IBB observed during the last 20 years of the study.
A recent study has also attempted to explore the effects of exploitation on genetic diversity at the Pan I locus in Icelandic cod using samples from spawning areas around Iceland over a 3-year period (2005–2007) (Árnason et al. 2009). The authors estimated the fitness across age groups at the inter-cohort level and concluded that, in contrast to our study, the changes in the frequencies of the Pan IAA and Pan IBB genotypes were because of selection against the Pan IAA genotype (the coastal component) by high near-shore fishing pressure. Two potential explanations for the contrasting conclusions from these studies are evident. First, the changes in Pan IBB homozygote frequencies observed in our historical time series appear concomitant with the known changes that took place in the fishery during that period. It is, in principle, possible for an abrupt shift in the selection regime to have taken place in recent years, and this is the conclusion reached by Árnason et al. (2009) in their study using contemporary, but not historical, samples. It is worth noting though that the frequencies of the Pan IAA genotype did not appear to have declined in our contemporary sample (collected in 2002) in comparison with the earlier years. Secondly, our study indicates that the genotypic variation observed within cohorts is because of differences in maturation rates, with cod carrying the Pan IBB genotype more likely to mature at a later age. Árnason et al. (2009) also detected distinct changes in Pan I genotype frequencies at the intra-cohort level, i.e. the Pan IBB genotype increased in frequency with age of the cod. Therefore, the reported change in the frequency of the Pan IAA genotype over the 3-year period in their study may actually have been because of a gradual increase in the number of Pan IBB individuals as they matured and entered the spawning grounds, rather than the intense removal of Pan IAA cod by the fishery. A thorough investigation of maturation rates of cod in relation to Pantophysin genotypes would therefore help resolve the conflicting conclusions from these two studies. What is apparent is that historical or temporally replicated data, such as that used in our study, are of the utmost importance when examining changes in the genetic composition of populations that could have been potentially brought about by anthropogenic effects.
Significant excess of Pan I heterozygotes was observed in five of seven cohort classes and 50% of sampling years. Heterozygote excess at the Pantophysin locus has been reported in other studies of Atlantic cod (Jónsdóttir et al. 1999, 2001; Beacham et al. 2002; Karlsson and Mork 2003; Árnason et al. 2009); however, the magnitudes in the present study are higher than typically reported. The possibility of genotyping errors because of incomplete enzyme digestion, leading to inflated heterozygote scores (this would not affect Pan IAA homozygotes) was thus examined. However, repetitions of samples (ca. 30%) showed consistent results thus rendering the hypothesis of mis-scoring because of partial digestion unlikely. Another explanation for the heterozygote excess observed in this study is that heterozygotes have a selective advantage i.e. in a highly fluctuating environment, such as that inhabited by Icelandic cod, preservation of both alleles could be beneficial both in the short and in the longer term. The fact that polymorphisms have been maintained at the Pan I locus for at least two million years (Pogson and Mesa 2004) indicates there are ongoing natural selection processes influencing the genotypic distribution (Canino et al. 2005). However, heterozygotes do not appear to have an advantage in growth or longevity. Indeed, even if a fluctuating environment can provide a favourable basis for protected polymorphisms, the conditions required for this are often quite restrictive (Hedrick 2006). The strength of selection needed to explain such departures from HWE would probably thus be unrealistically elevated. Significant heterozygote excess at the Pan I locus has also been attributed to the confounding effect of sexes (Karlsson and Mork 2003); however, this was not supported by our data. We therefore propose another explanation: if heterozygotes have maturation rates that are similar to those of Pan IAA homozygotes, i.e. allele A exhibits some dominance (Fig. 3), the recruitment of Pan IAA and Pan IAB cod to the spawning grounds would occur before the Pan IBB homozygotes and would thus generate excesses of heterozygotes. We therefore find it likely that the observed heterozygote excess represents a dynamical system influenced by behavioural differences in the stock (Árnason et al. 2009). Once again, this hypothesis could be examined through future investigations of maturation rates associated with the Pan I genotypes.
In conclusion, decades of high fishing pressure have resulted in evident changes in the age composition of Icelandic cod (Marteinsdóttir and Thorarinsson 1998; this paper) and are also likely to have altered the maturation schedule of this important fish stock (Pardoe et al. 2009). In the present study, we also show that commercial fishing has likely led to loss of adaptive variation at the Pantophysin locus over a period of six decades. The analysis of historical genetic material from archived otolith collections, and more specifically non-neutral markers such as the Pan I locus, has thus proved their worth in the assessment of the long-term effects of exploitation on commercial fish stocks such as Atlantic cod. Our results suggest that fisheries can shape the genetic composition of a fish population over a relatively short time period, thus supporting the calls for an evolutionary dimension to fisheries management (e.g. Conover 2000; Law 2000; Jørgensen et al. 2007; Kuparinen and Merilä 2007), ideally through the establishment of genetic monitoring programs based on an examination of functional as well as neutral markers (Bradbury et al. 2010) and incorporation of genetic perspectives (André et al. 2010) into management objectives that are concerned with the long-term sustainability of harvested populations.
Acknowledgments
We thank anonymous referees for valuable comments and suggestions that led to improvement of the manuscript. We thank Thóra Jörundsdóttir and Kristín Hardardottir for technical work, Ingibjörg G. Jónsdóttir and Dr Gunnar Stefánsson for help with statistical analysis. Further we are indebted to Dr Anna K. Daníelsdóttir for supervision in the laboratory and Dr Pétur H. Petersen for valuable comments and discussions. Financial support was provided by the Marine Research Institute in Iceland (MRI), The Icelandic Centre for Research (RANNIS) and the EU project METACOD (Q5RS-2001-00 953).
Literature cited
- Allendorf FW, Hard JJ. Human-induced evolution caused by unnatural selection through harvest of wild animals. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9987–9994. doi: 10.1073/pnas.0901069106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allendorf FW, England PR, Luikart G, Ritchie PA, Ryman N. Genetic effects of harvest on wild animal populations. Trends in Ecology & Evolution. 2008;23:327–337. doi: 10.1016/j.tree.2008.02.008. [DOI] [PubMed] [Google Scholar]
- André C, Larsson LC, Laikre L, Bekkevold D, Brigham J, Carvalho GR, Dahlgren TG, et al. Detecting population structure in a high gene-flow species, Atlantic herring (Clupea harengus): direct, simultaneous evaluation of neutral vs putatively selected loci. Heredity 1–11. 2010 doi: 10.1038/hdy.2010.71. doi: 10.1038/hdy.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Árnason E, Hernandez UB, Kristinsson K. Intense habitat-specific fisheries-induced selection at the molecular Pan I locus predicts imminent collapse of a major cod fishery. PLoS ONE. 2009;4:e5529. doi: 10.1371/journal.pone.0005529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astthorsson OS, Gislason A, Jonsson S. Climate variability and the Icelandic marine ecosystem. Deep Sea Research Part II: Topical Studies in Oceanography. 2007;54:2456–2477. [Google Scholar]
- Barot S, Heino M, O'Brien L, Dieckmann U. Long-term trend in the maturation reaction norm of two cod stocks. Ecological Applications. 2004;14:1257–1271. [Google Scholar]
- Beacham TD, Brattey J, Miller KM, Le KD, Withler RE. Multiple stock structure of Atlantic cod (Gadus morhua) off Newfoundland and Labrador determined from genetic variation. ICES Journal of Marine Science. 2002;59:650–665. [Google Scholar]
- Belkhir K, Borsa P, Goudet J, Bonhomme F. GENETIX: logiciel sous Windows pour la génétique des populations, Version 4.03. Montpellier, France: Laboratoire Génome & Population, CNRS-UPR, Université de Montpellier II; 1999. [Google Scholar]
- Biosystems. GeneScan, Version 3.1.2. Foster City, CA: Applied Biosystems; 2000. [Google Scholar]
- Biosystems. GeneMapper, Version 3.7. Foster City, CA: Applied Biosystems; 2004. [Google Scholar]
- Bradbury IR, Hubert S, Higgins B, Borza T, Bowman S, Paterson IG, Snelgrove PVR, et al. Parallel adaptive evolution of Atlantic cod on both sides of the Atlantic Ocean in response to temperature. Proceedings of the Royal Society B: Biological Sciences. 2010;277:3725–3734. doi: 10.1098/rspb.2010.0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker AL, Cook D, Bentzen P, Wright JM, Doyle RW. Organization of microsatellites differs between mammals and cold-water teleost fishes. Canadian Journal of Fisheries and Aquatic Sciences. 1994;51:1959–1966. [Google Scholar]
- Canino MF, O'Reilly PT, Hauser L, Bentzen P. Genetic differentiation in walleye pollock (Theragra chalcogramma) in response to selection at the Pantophysin (Pan I) locus. Canadian Journal of Fisheries and Aquatic Sciences. 2005;62:2519–2529. [Google Scholar]
- Case RAJ, Hutchinson WF, Hauser L, Buehler V, Clemmesen C, Dahle G, Kjesbu OS, et al. Association between growth and Pan I genotype within Atlantic cod full-sibling families. Transactions of the American Fisheries Society. 2006;135:241–250. [Google Scholar]
- Conover DO. Darwinian fishery science. Marine Ecology Progress Series. 2000;208:299–313. [Google Scholar]
- Conover DO, Baumann H. The role of experiments in understanding fishery-induced evolution. Evolutionary Applications. 2009;2:276–290. doi: 10.1111/j.1752-4571.2009.00079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover DO, Munch SB. Faith, evolution, and the burden of proof. Fisheries. 2007;32:90–91. [Google Scholar]
- Conover DO, Clarke LM, Munch SB, Wagner GN. Spatial and temporal scales of adaptive divergence in marine fishes and the implications for conservation. Journal of Fish Biology. 2006;69:21–47. [Google Scholar]
- Dickson R, Meincke J, Malmberg SA, Lee A. The ‘‘great salinity anomaly’’ in the northern North Atlantic 1968–1982. Progress in Oceanography. 1988;20:103–151. [Google Scholar]
- Edeline E, Carlson SM, Stige LC, Winfield IJ, Fletcher JM, Ben James J, Haugen TO, et al. Trait changes in a harvested population are driven by a dynamic tug-of-war between natural and harvest selection. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15799–15804. doi: 10.1073/pnas.0705908104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estoup A, Largiader CR, Perrot E, Chourrout D. Rapid one-tube DNA extraction for reliable PCR detection of fish polymorphic markers and transgenes. Molecular Marine Biology and Biotechnology. 1996;5:295–298. [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver.3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Goudet J. FSTAT (Version 1.2): a computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Hanna E, Jónsson T, Ólafsson J, Valdimarsson H. Icelandic coastal sea surface temperature records constructed: putting the pulse on air-sea-climate interactions in the northern north Atlantic. Part I: comparison with hadisst1 open-ocean surface temperatures and preliminary analysis of long-term patterns and anomalies of SSTs around Iceland. Journal of Climate. 2006;19:5652–5666. [Google Scholar]
- Hansen MM, Fraser DJ, Meier K, Mensberg KLD. Sixty years of anthropogenic pressure: a spatio-temporal genetic analysis of brown trout populations subject to stocking and population declines. Molecular Ecology. 2009;18:2549–2562. doi: 10.1111/j.1365-294X.2009.04198.x. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. Genetic polymorphism in heterogeneous environments: the age of genomics. Annual Review of Ecology. Evolution and Systematics. 2006;37:67–93. [Google Scholar]
- Heino M, Dieckmann U. Detecting fisheries-induced life-history evolution: an overview of the reaction-norm approach. Bulletin of Marine Science. 2008;83:69–93. [Google Scholar]
- Heino M, Dieckmann U, Godo OR. Measuring probabilistic reaction norms for age and size at maturation. Evolution. 2002;56:669–678. doi: 10.1111/j.0014-3820.2002.tb01378.x. [DOI] [PubMed] [Google Scholar]
- Jakobsson J. The north Icelandic herring fishery and environmental conditions, 1960–1968. Rapport et Procesverbau Réunion Conseil International Exploration de la Mer. 1980;177:460–465. [Google Scholar]
- Jakobsson J, Stefánsson G. Rational harvesting of the cod-capelin-shrimp complex in the Icelandic marine ecosystem. Fisheries Research. 1998;37:7–21. [Google Scholar]
- Jónsdóttir ÓDB, Imsland AK, Daníelsdóttir AK, Thorsteinsson V, Naedval G. Genetic differentiation among Atlantic cod in south and south-east Icelandic waters: synaptophysin (Syp I) and haemoglobin (HBI) variation. Journal of Fish Biology. 1999;54:1259–1274. [Google Scholar]
- Jónsdóttir ÓDB, Daníelsdóttir AK, Naedval G. Genetic differentiation among Atlantic cod (Gadus morhua L.) in Icelandic waters: temporal stability. ICES Journal of Marine Science. 2001;58:114–122. [Google Scholar]
- Jónsson J. Tagging of cod (Gadus morhua) in Icelandic waters 1948–1986 and Tagging of haddock (Gadus aeglefinus) in Icelandic waters 1953–1965. Reykjavík: Rit Fiskideildar –Journal of the Marine Research Institute; 1996. [Google Scholar]
- Jørgensen C, Enberg K, Dunlop ES, Arlinghaus R, Boukal DS, Brander K, Ernande B, et al. Ecology: managing evolving fish stocks. Science. 2007;318:1247–1248. doi: 10.1126/science.1148089. [DOI] [PubMed] [Google Scholar]
- Karlsson S, Mork J. Selection-induced variation at the Pantophysin locus (Pan I) in a Norwegian fjord population of cod (Gadus morhua L.) Molecular Ecology. 2003;12:3265–3274. doi: 10.1046/j.1365-294x.2003.01993.x. [DOI] [PubMed] [Google Scholar]
- Kendall NW, Hard JJ, Quinn TP. Quantifying six decades of fishery selection for size and age at maturity in sockeye salmon. Evolutionary Applications. 2009;2:523–536. doi: 10.1111/j.1752-4571.2009.00086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuparinen A, Merilä J. Detecting and managing fisheries-induced evolution. Trends in Ecology & Evolution. 2007;22:652–659. doi: 10.1016/j.tree.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Kuparinen A, Merilä J. The role of fisheries-induced evolution. Science. 2008;320:47–48. [PubMed] [Google Scholar]
- Law R. Fishing, selection, and phenotypic evolution. ICES Journal of Marine Science. 2000;57:659–668. [Google Scholar]
- Law R. Fisheries-induced evolution: present status and future directions. Marine Ecology Progress Series. 2007;335:271–277. [Google Scholar]
- Malmberg SA. The ecological impact of the East Greenland Current on the North Icelandic waters. In: Skreslet S, editor. The Role of Freshwater Outflow in Coastal Marine Ecosystems. Berlin: Springer; 1986. pp. 389–404. NATO ASI Series, vol. G7, pp. [Google Scholar]
- Marine Research Institute. State of Marine Stocks in Icelandic Waters 2006/2007. Prospects for the Quota Year 2006/2007. Reykjavik: Marine Research Institute; 2007. Marine Research Technical Report. [Google Scholar]
- Marine Research Institute. State of Marine Stocks in Icelandic Waters 2008/2009. Prospects for the Quota Year 2009/2010. Reykjavik: Marine Research Institute; 2009. Marine Research Technical Report. [Google Scholar]
- Marshall CT, Browman HI. Disentangling the causes of maturation trends in exploited fish populations. Marine Ecology Progress Series. 2007;335:249–251. [Google Scholar]
- Marteinsdóttir G, Begg GA. Essential relationships incorporating the influence of age, size and condition on variables required for estimation of reproductive potential in Atlantic cod Gadus morhua. Marine Ecology Progress Series. 2002;235:235–256. [Google Scholar]
- Marteinsdóttir G, Pardoe H. Effects of fishing on inter- and intra-stock diversity of marine resources. In: Tsukamoto K, Kawamura T, Takeuchi T, Beard DB, Kaiser MJ, editors. Fisheries for Global Welfare and Environment, 5th World Fisheries Congress. Tokyo, Japan: Terrapub; 2008. pp. 27–43. [Google Scholar]
- Marteinsdóttir G, Thorarinsson K. Improving the stock-recruitment relationship in Icelandic cod (Gadus morhua L.) by including age diversity of spawners. Canadian Journal of Fisheries and Aquatic Sciences. 1998;55:1372–1377. [Google Scholar]
- Miller KM, Le KD, Beacham TD. Development of tri- and tetranucleotide repeat microsatellite loci in Atlantic cod (Gadus morhua. Molecular Ecology. 2000;9:238–239. doi: 10.1046/j.1365-294x.2000.00804-2.x. [DOI] [PubMed] [Google Scholar]
- Ministry of Fisheries. 2004. Áfangaskýrsla nefndar um líffræðilega fiskveiðistjórnun til sjávarútvegsráðherra (Status report from the Commission of biological fishery management for the Ministry of Fisheries). Sjávarútvegsráðuneytið (Ministry of Fisheries Iceland). pp. 64.
- Nielsen EE, Hansen MM. Waking the dead: the value of population genetic analyses of historical samples. Fish and Fisheries. 2008;9:450–461. [Google Scholar]
- Nielsen EE, MacKenzie EBR, Magnussen E, Meldrup D. Historical analysis of Pan I in Atlantic cod (Gadus morhua): temporal stability of allele frequencies in the southeastern part of the species distribution. Canadian Journal of Fisheries and Aquatic Sciences. 2007;64:1448–1455. [Google Scholar]
- O'Reilly PT, Canino MF, Bailey KM, Bentzen P. Isolation of twenty low stutter di- and tetranucleotide microsatellites for population analyses of walleye pollock and other gadoids. Journal of Fish Biology. 2000;56:1074–1086. [Google Scholar]
- Pálsson OK, Thorsteinsson V. Migration patterns, ambient temperature, and growth of Icelandic cod (Gadus morhua): evidence from storage tag data. Canadian Journal of Fisheries and Aquatic Sciences. 2003;60:1409–1423. [Google Scholar]
- Palstra FP, Ruzzante DE. A temporal perspective on population structure and gene flow in Atlantic salmon (Salmo salar) in Newfoundland, Canada. Canadian Journal of Fisheries and Aquatic Sciences. 2010;67:225–242. [Google Scholar]
- Palstra FP, O'Connell MF, Ruzzante DE. Age structure, changing demography and effective population size in Atlantic Salmon (Salmo salar. Genetics. 2009;182:1233–1249. doi: 10.1534/genetics.109.101972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampoulie C, Ruzzante DE, Chosson V, Jorundsdottir TD, Taylor L, Thorsteinsson V, Danielsdottir AK, et al. The genetic structure of Atlantic cod (Gadus morhua) around Iceland: insight from microsatellites, the Pan I locus, and tagging experiments. Canadian Journal of Fisheries and Aquatic Sciences. 2006;63:2660–2674. [Google Scholar]
- Pampoulie C, Jakobsdóttir KB, Marteinsdóttir G, Thorsteinsson V. Are vertical behaviour patterns related to the Pantophysin locus in the Atlantic cod (Gadus morhua L.)? Behavior Genetics. 2008;38:76–81. doi: 10.1007/s10519-007-9175-y. [DOI] [PubMed] [Google Scholar]
- Pardoe H, Vainikka A, Thordarson G, Marteinsdóttir G, Heino M. Temporal trends in probabilistic maturation reaction norms and growth of Atlantic cod (Gadus morhua) on the Icelandic shelf. Canadian Journal of Fisheries and Aquatic Sciences. 2009;66:1719–1733. [Google Scholar]
- Pogson GH. Nucleotide polymorphism and natural selection at the Pantophysin (Pan I) locus in the Atlantic Cod, Gadus morhua (L.) Genetics. 2001;157:317–330. doi: 10.1093/genetics/157.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson GH, Mesa KA. Positive darwinian selection at the Pantophysin (Pan I) locus in marine gadid fishes. Molecular Biology and Evolution. 2004;21:65–75. doi: 10.1093/molbev/msg237. [DOI] [PubMed] [Google Scholar]
- Pogson GH, Mesa KA, Boutilier RG. Genetic population structure and gene flow in the Atlantic cod Gadus morhua: a comparison of allozyme and nuclear RFLP loci. Genetics. 1995;139:375–385. doi: 10.1093/genetics/139.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. Genepop (version 1.2): population-genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Schopka SA. Fluctuations in the cod stock off Iceland during the twentieth century in relation to changes in the fisheries and environment. ICES Marine Science Symposia reports. 1994;198:175–193. [Google Scholar]
- Skarstein TH, Westgaard JI, Fevolden SE. Comparing microsatellite variation in north-east Atlantic cod (Gadus morhua L.) to genetic structuring as revealed by the pantophysin (Pan I) locus. Journal of Fish Biology. 2007;70:271–290. [Google Scholar]
- þór Jþ. 2005. Nýsköpunaröld. Saga sjávarútvegs áÍslandi III. Bindi 1939–1973. Saga Sjávarútvegs áÍslandi, Bókaútgáfan Hólar.
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- Venables WN, Ripley BD. Modern applied statistics with S. 4th edn. Springer: New York; 2002. [Google Scholar]
- Walsh MR, Munch SB, Chiba S, Conover DO. Maladaptive changes in multiple traits caused by fishing: impediments to population recovery. Ecology Letters. 2006;9:142–148. doi: 10.1111/j.1461-0248.2005.00858.x. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-Statistics for the analysis of population-structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]


