Abstract
Although continental populations of avian influenza viruses are genetically distinct, transcontinental reassortment in low pathogenic avian influenza (LPAI) viruses has been detected in migratory birds. Thus, genomic analyses of LPAI viruses could serve as an approach to prioritize species and regions targeted by North American surveillance activities for foreign origin highly pathogenic avian influenza (HPAI). To assess the applicability of this approach, we conducted a phylogenetic and population genetic analysis of 68 viral genomes isolated from the northern pintail (Anas acuta) at opposite ends of the Pacific migratory flyway in North America. We found limited evidence for Asian LPAI lineages on wintering areas used by northern pintails in California in contrast to a higher frequency on breeding locales of Alaska. Our results indicate that the number of Asian LPAI lineages observed in Alaskan northern pintails, and the nucleotide composition of LPAI lineages, is not maintained through fall migration. Accordingly, our data indicate that surveillance of Pacific Flyway northern pintails to detect foreign avian influenza viruses would be most effective in Alaska. North American surveillance plans could be optimized through an analysis of LPAI genomics from species that demonstrate evolutionary linkages with European or Asian lineages and in regions that have overlapping migratory flyways with areas of HPAI outbreaks.
Keywords: avian influenza, genetic, low pathogenic, migration, northern pintail, virus, waterfowl
Introduction
Current surveillance programs for detection of highly pathogenic avian influenza (HPAI) viruses focus on monitoring avian populations for mortality events and the sampling of live wild and domestic birds (Munster et al. 2007; Komar and Olsen 2008). Viruses isolated from avian samples can then be characterized genetically to determine their subtype and pathogenicity. Previous molecular surveys have revealed substantial levels of sequence divergence among continental populations of low pathogenic avian influenza (LPAI) viruses in Europe, Asia, and North America (Ito et al. 1995; Widjaja et al. 2004) and, although less studied, South America (Pereda et al. 2008). This broad-scale genetic differentiation among continents yields a methodology based on phylogenetic assignment to identify intercontinental virus exchange (Webster et al. 1992; Widjaja et al. 2004) and also indicates that intercontinental dispersal of LPAI viruses is relatively rare (Webster et al. 2007).
However, there is growing evidence of genetic exchange between North American and Eurasian strains of LPAI avian influenza via reassortment in northern pintails (Anas acuta) and dunlin (Calidris alpina) in Alaska (Koehler et al. 2008; Wahlgren et al. 2008), ruddy turnstones (Arenaria interpres) and herring gulls (Larus argentatus) along the Atlantic Coast of North America (Marakova et al. 1999), in mallards (Anas platyrhynchos) in Minnesota (Jackwood and Stallnecht 2007) and waterfowl in Alberta, Canada (Krauss et al. 2007). Furthermore, intercontinental genetic exchange appears to be bi-directional. RNA segments from North American avian influenza viruses have been observed in guillemots (Uria aalge) in Europe (Wallensten et al. 2005) and waterfowl in Asia (Bean et al. 1992; Liu et al. 2004), North American RNA segments were found in a South American influenza virus isolated from cinnamon teal (Anas cyanoptera; Spackman et al. 2007), and some Asian lineages of the N8 RNA segment were more similar to North American virus isolates from northern pintails (Anas acuta) than to other Asian reference samples (Koehler et al. 2008).
On the basis of these studies, it appears that the relative frequency of occurrence of Asian origin RNA segments in North American LPAI isolates could be used to identify species and regions where the relative risk of introduction of a foreign HPAI virus is highest. For example, nearly half of avian influenza viruses isolated from northern pintails in Alaska contained at least one Asian RNA segment (Koehler et al. 2008). Conversely, Krauss et al. (2007) found little evidence of Eurasian origin RNA in Alberta, Canada, and Delaware Bay (New Jersey), United States, and concluded that foreign virus introduction was likely to be rare. The higher frequency of Asian lineage RNA segments in virus isolates from northern pintails probably results from the proximity of Alaska to Asian sources of LPAI viruses and the movement of this species between Asia and Alaska.
Genomic analyses of LPAI viruses could serve as a powerful approach to evaluate whether North American surveillance for HPAI has targeted bird species and regions most likely to exhibit transcontinental virus exchange. To assess the applicability of such an approach, we conducted a phylogenetic and population genetic analysis of LPAI viral genomes isolated from the northern pintail at opposite ends of a migratory flyway in North America (Fig. 1). The connectivity of the Pacific migratory flyway between Alaska and California is well documented for the northern pintail through banding and satellite telemetry data (Bellrose 1980; Miller et al. 2005; Nicolai et al. 2005). We used genomic sequencing and phylogenetic analysis of northern pintail LPAI viruses collected from wintering areas in California to determine the frequency of Asian lineage viruses present and to examine the similarity of RNA segments between Alaska and California isolates.
Figure 1.
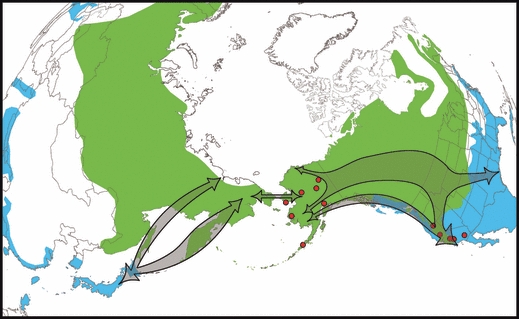
Breeding (green) and wintering (blue) distributions of the northern pintail. General migration routes (arrows) are shown for northern pintails that winter in Eastern Asia and Western North America based on banding and satellite telemetry studies (Bellrose 1980; Miyabayashi and Mundkur 1999; Miller et al. 2005; Nicolai et al. 2005). The geographic location of virus isolates (red dots) used in this study from Alaska and California are also shown.
Materials and methods
Sampling, virus isolation and sequencing
A total of 3045 samples, collected from live and hunter-killed wild northern pintail ducks, was subjected to virus isolation in embryonated eggs as previously described (Ip et al. 2008). Samples were collected during fall and winter months of 2006 and 2007 from across California in Siskiyou, Butte, Fresno, Kern, and Imperial counties (Fig. 1). A total of 30 LPAI viruses was isolated from California samples and compared to 38 viruses isolated from samples collected across Alaska (Fig. 1) by Koehler et al. (2008).
Viral RNA was extracted from allantoic fluid with the MagMAX AI/NDV RNA extraction kit (Ambion Inc., Austin, TX, USA). All eight RNA segments were amplified with the one-step RT PCR kit (Qiagen, Inc., Valencia, CA, USA) using a combination of previously published primers (Zou 1997; Hoffmann et al. 2001; Phipps et al. 2004; Bragstad et al. 2005; Chan et al. 2006; Obenauer et al. 2006; Li et al. 2007; Koehler et al. 2008) or primers specifically designed for this study which are available from the authors upon request. PCR products were gel purified and extracted using the QIAquick gel extraction kit (Qiagen, Inc.) or treated with ExoSap-IT (USB Inc., Cleveland, OH, USA) without additional purification before sequencing. Cycle sequencing was performed with identical primers used for PCR along with BigDye Terminator version 3.1 mix (Applied Biosystems, Foster City, CA, USA). Samples were analyzed on an Applied Biosystems 3730xl automated DNA sequencer (Applied Biosystems).
We sequenced a total of 236 (out of 240) individual segments from the 30 California virus isolates. The total number of nucleotides sequenced for each RNA segment was: M (737), NP (1406), NS (671), PA (2142), PB1 (2257), PB2 (2243), HA (1650–1704), and NA (1334–1394). Sequences were assembled and edited with Sequencher version 4.7 (Gene Codes Corp., Ann Arbor, MI, USA). Sample chromatograms that contained multiple peaks for all or some nucleotides were considered co-infections and excluded from analysis. GenBank accession numbers for the California isolates are as follows: FJ519990–FJ520225.
Phylogenetic relationships
To address our first objective of examining the placement of all RNA segments from each California isolate into either Asian or North American clades, we compared sequences obtained from California isolates to reference sample sequences (see Table S1) from the GenBank database at the National Center for Biotechnology Information (Bao et al. 2008). Five reference samples isolated from dabbling ducks from across the United States were selected for each RNA segment to represent North American lineages. A total of 4–35 reference samples isolated from water birds from Southeast Asia (China, Japan, and South Korea) was selected for each RNA segment to represent Asian lineages. All sequences were aligned using Sequencher version 4.7.
We used paup* version 4.0b (Swofford 2003) to generate neighbor-joining trees using the branch-and-bound method with 10 000 bootstrap replicates. We incorporated the best approximating model of nucleotide evolution as determined by Akaike's Information Criterion in modeltest version 3.06 (Posada and Crandall 1998) for each RNA segment analysis. Estimates of the proportion of invariable sites (I) and the gamma distribution parameter (G), calculated in modeltest, were also incorporated. For nearly all genes, the best approximating model was the General Time Reversible model (Lanave et al. 1984) plus I and G. For N6 and H7, better fit to the data was achieved with the Kimura (1981) 3-parameter model plus I. For Bayesian analysis, we used MrBayes version 3.1.2 (Ronquist and Huelsenbeck 2003) to construct posterior probabilities of clade support. Each analysis was run for ∼1 × 106 generations or until the average standard deviation of split frequencies was ≤1.00. We also verified that the potential scale reduction factor was ∼1.00 as another indicator of convergence (see Ronquist and Huelsenbeck 2003). Average posterior probabilities of the 50% majority rule consensus tree topologies were estimated by sampling likelihood parameters every 100 generations. Trees were visualized with TreeView (Page 1996). Following the construction of phylograms, we determined that a viral reassortment event had occurred between Asian and North American viruses when a lineage from a northern pintail in North America was most closely related to Asian reference sequences.
Genetic diversity
To characterize genetic diversity and differentiation of North American lineages of LPAI viruses between Alaska and California northern pintails, we first collapsed all sequences into individual alleles or haplotypes and computed their frequencies in each sampling area. We focused these analyses on the six non-surface glycoprotein RNA segments (M, NP, NS, PA, PB1, and PB2) as not all HA and NA subtypes were represented in both locations. We tabulated general measures of diversity, including percentage of unique sequences, sequence diversity (h), and the mean number of pairwise differences (p) within each sampling area using arlequin version 3.0.1 (Excoffier et al. 2005). We evaluated the level of population differentiation between Alaska and California by computing the effective number of distinct subpopulations, ΔST, and the true level of differentiation, D (Jost 2008). Traditional measures of population differentiation, such as F-statistics, typically demonstrate incorrectly low levels when comparing groups composed of numerous rare alleles that occur when using highly polymorphic loci (Jost 2008) such as virus sequence data. However, because estimators such as ΔST may fail to reveal other patterns in allelic data, we included calculations of FST after incorporating a model of nucleotide evolution (ΦST) from arlequin for comparative purposes. In these assessments of population diversity and differentiation we restricted our analysis to North American lineages and excluded those lineages from Alaska identified by Koehler et al. (2008) which are related to Asian clades. To visually examine the diversity of North American lineages, we constructed phylograms for the six internal gene segments using the neighbor-joining and Bayesian methods described above.
Results
Subtype variation
Among the 30 isolates from California sampled in 2006 (n = 24) and 2007 (n = 6), we found eight HA and six NA subtypes (Fig. 2). The most frequent subtype combination among northern pintail viruses isolated in California was H11N9 (23%), followed by H5N2 (20%). Twelve additional subtypes were observed in <7% of isolates (H1N2, H3N6, H3N8, H4N2, H4N6, H5N9, H6N1, H6N2, H7N3, H10N3, H10N8, and H11N2). These subtype combinations differed substantially from virus isolates from Alaska northern pintails analyzed by Koehler et al. (2008), where H3N8 was the most frequent subtype (Fig. 2).
Figure 2.
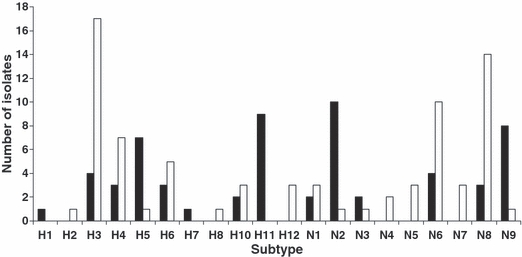
Distribution of hemagglutinin (HA) and neuraminidase (NA) subtypes observed among 30 northern pintail avian influenza isolates from California sampled in 2006–2007 (black bars) in comparison to those observed among 38 northern pintail isolates from Alaska sampled in 2006 (white bars) by Koehler et al. (2008).
Phylogenetic relationships
We found no evidence of Asian lineage genes within California isolates for the 177 RNA segments of the M, NP, NS, PA, PB1, and PB2 genes. In all cases, sequences from California isolates for these genes clustered more closely with North American reference samples and were distinct and well supported, with neighbor-joining and Bayesian methods, from Asian reference samples (data not shown). Similarly, we observed a clear separation between Asian reference samples and California northern pintail virus sequences for all HA and NA subtypes in Fig. 2, with the exception of four (H6, N8, N1 and N2). In phylogenetic trees for the H6 (Fig. 3) and N8 (Fig. 4) subtypes, there was evidence for multiple and differentiated clades. In the H6 tree, we observed four groups (Fig. 3), with group 1 composed of a mixture of North American and Asian lineages, including three California and four Alaska northern pintail isolates that were identified as derived from reassortment events (Koehler et al. 2008). Group 3 of the H6 tree contained only Asian reference samples and group 4 was composed of two North American reference samples. Similarly, group 1 lineages of the N8 tree (Fig. 4) contained sequences from North American and Asian isolates, including three isolates from California northern pintails and 15 from Alaska, whereas group 3 contained only Asian lineages. A similar pattern was observed in the N2 tree (not shown), in which all sequences clustered into one large group of North American and Asian lineages, including 10 California segments that were distinct from an outgroup of swine sequences. Thus, a total of 16 (6.6%) California segments were observed in mixed lineage clades, containing sequences from Asia and North America, for the H6, N8, and N2 genes. Lastly, we observed three Asian reference samples (AJ410565/A/duck/Hong Kong, AB292405/A/duck/Hong Kong and AB298280/A/duck/Hokkaido) within the North American clade of lineages for the N1 gene (not shown) that included a California isolate from this study (FJ520158/A/pintail/California).
Figure 3.
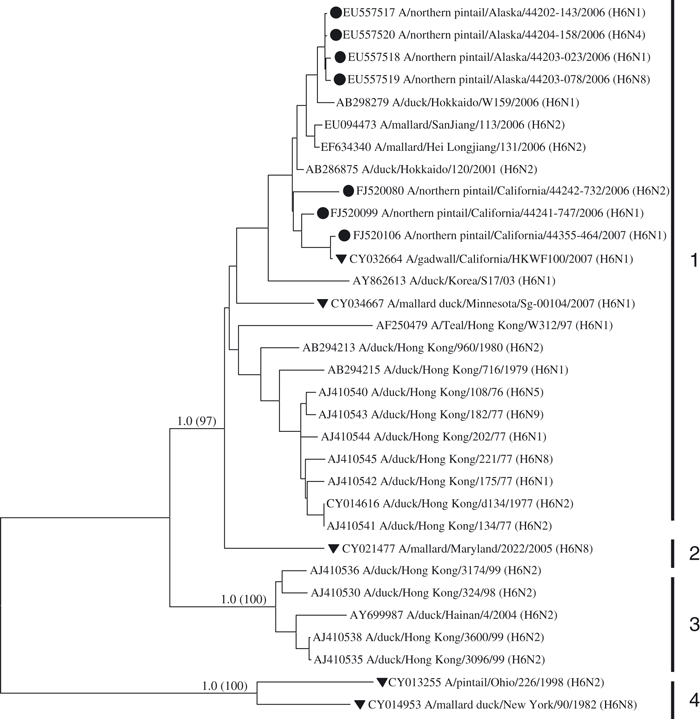
Phylogenetic relationship of sequences from the H6 subtype. North American isolates of northern pintails from Alaska (Koehler et al. 2008) and California (this study) are indicated with black circles. North American reference samples taken from GenBank are shown with a black triangle. Two North American waterfowl reference samples that exhibited substantial divergence from all other sequences serve as an outgroup. Bayesian posterior probabilities and levels of neighbor-joining bootstrap support >70% between major clades are shown on branches. Numbered vertical bars delineate major clades.
Figure 4.
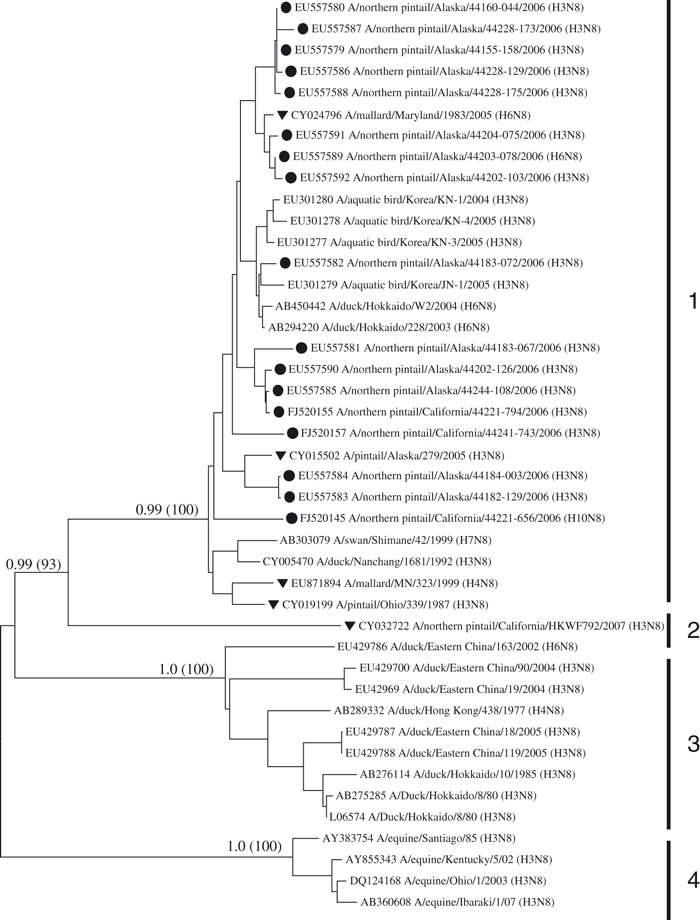
Phylogenetic relationship of sequences from the N8 subtype. North American isolates of northern pintails from Alaska (Koehler et al. 2008) and California (this study) are indicated with black circles. North American reference samples taken from GenBank are shown with a black triangle. Four equine reference samples serve as an outgroup. Bayesian posterior probabilities and levels of neighbor-joining bootstrap support >70% between major clades are shown on branches. Numbered vertical bars delineate major clades.
Genetic diversity
In comparing genetic diversity of LPAI isolates between Alaska and California, we observed a substantial number and diversity of RNA segments for the M, NP, NS, PA, PB1, and PB2 genes (Table 1). These six RNA segments exhibited a great deal of sequence diversity and no homologous sequences were observed between Alaska and California (Table 1). Sequence diversity (h) was similar between areas, but pairwise differences (p) among sequences within each area were not consistent. However, the indices for these six RNA segments are underestimated for Alaska as Asian reassortment events identified by Koehler et al. (2008) were excluded from diversity summaries.
Table 11.
Percentage of unique sequences, diversity (h), and pairwise differences (P) among samples from Alaska and California low pathogenic avian influenza virus isolates of northern pintails
| Alaska | California | ||||||
|---|---|---|---|---|---|---|---|
| RNA segment | % Unique* | h† | P | % Unique | h | P | ΦST |
| M | 74 | 0.988 | 23.1 | 88 | 0.987 | 20.3 | 0.014 |
| NP | 91 | 0.994 | 75.9 | 88 | 0.987 | 98.7 | 0.029 |
| NS | 76 | 0.989 | 101.7 | 81 | 0.989 | 103.0 | 0.093 |
| PA | 97 | 0.998 | 170.2 | 97 | 0.997 | 160.1 | 0.156 |
| PB1 | 97 | 0.986 | 95.4 | 84 | 0.987 | 94.8 | 0.017 |
| PB2 | 100 | 0.999 | 139.5 | 93 | 0.995 | 140.8 | 0.060 |
Levels of population differentiation (ΦST) after incorporating a model of nucleotide evolution are also shown. Asian lineage segments identified in Alaska samples by Koehler et al. (2008) are not included.
Percentage of alleles (sequences) that are not shared by other virus isolates in the sampling area. No sequences were shared between Alaska and California.
Probability that any two randomly chosen sequences will differ.
The lack of shared LPAI virus sequences for the six internal genes between Alaska and California yielded maximum values for both the number of effective subpopulations (ΔST = 2.0) and true differentiation (D = 1.0) as defined in Jost (2008). However, these metrics are potentially misleading because they do not account for the small number of mutations that occur among some lineages of each RNA segment. Although the mean number of pairwise differences among sequences for each of the six RNA segments analyzed was large (Table 1), the range of those differences also included very low values (≤4). Phylograms of the six non-surface glycoprotein RNA segments demonstrate the similar levels of genetic diversity within Alaska and California virus isolates (Fig. 5). As a result, levels of population differentiation using traditional F-statistics (ΦST) were low to moderate with an estimated 1.5–15% of the total genetic variation occurring between Alaska and California (Table 1). The greatest level of differentiation was observed for the PA segment (ΦST = 0.156) and a phylogram of this segment revealed a greater degree of region-specific clustering of lineages than other RNA segments (Fig. 5). One clade of the PA segment was composed entirely of Alaskan isolates, as well as three Asian reference samples (Fig. 6), and similar lineages were not observed among California wintering samples. Moderate levels of ΦST in the NS and PB2 RNA segments were also associated with small clusters of Alaska- and California-specific lineages (Fig. 5).
Figure 5.
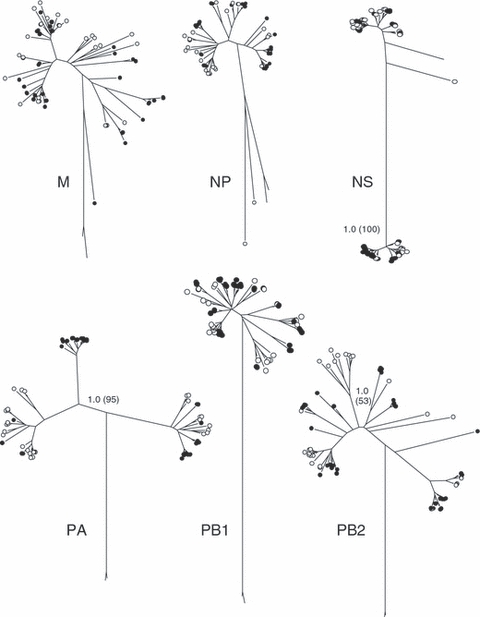
Phylograms depicting genetic diversity among sequences from the six non-surface glycoprotein RNA segments isolated from northern pintails in Alaska (black circles) and California (white circles). Unlabelled roots of each tree are equine reference samples included as outgroups. Bayesian posterior probabilities and levels of neighbor-joining bootstrap support >70% between major clades are shown on branches for three segments (NS, PA, and PB2) that exhibit patterns of Alaska- or California-specific lineages (see text).
Figure 6.
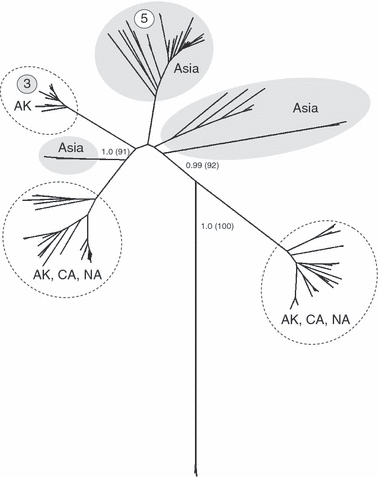
Phylogenic relationship of sequences from the PA segment of avian influenza viruses isolated from northern pintails in Alaska (AK; Koehler et al. 2008) and California (CA; this study) in comparison to 36 reference samples from Asia (shaded ovals) and five from North America (NA). Circled numbers indicate the frequency of outsider events in each clade, including the five reassortment events identified by Koehler et al. (2008) for this RNA segment. The tree is rooted using two equine influenza isolates from North America. Bayesian posterior probabilities and levels of neighbor-joining bootstrap support >70% between major clades are shown on branches.
Discussion
Host migration ecology and viral population genomics
To our knowledge, this is the first survey of LPAI variation in a single species at opposite ends of a migratory flyway. The northern pintail is known to migrate between North America and Eurasia (Miller et al. 2005; Nicolai et al. 2005) and LPAI viruses isolated from northern pintails in Alaska, an area where migratory birds from both North American and Asian flyways intersect (Fig. 1), contain a higher frequency of Asian lineage genes than observed elsewhere in North America (Koehler et al. 2008). Genetic evidence suggests that even though migratory connectivity between Alaska and California is well documented for northern pintails (Bellrose 1980; Miller et al. 2005; Nicolai et al. 2005), the number of Asian LPAI lineages and exact nucleotide composition of North American LPAI segments is not maintained through fall migration. Thus, LPAI genetic data indicate that Asian lineage viruses have been introduced to northern pintails in Alaska, but Asian-related lineages were difficult to detect at greater distances from Alaska along an established migratory flyway. We speculate that the most likely mechanisms that contribute to these findings are the rapid nucleotide mutation rate and reassortment among RNA segments that are common features of influenza A viruses (Macken et al. 2006; Dugan et al. 2008). As a result, the genomic characteristics of viruses isolated from birds in Alaska and California appear similarly diverse and differ by only a small number of mutations. In a study with wild mallards (Anas platyrhynchos), (Latorre-Margalef et al. 2008) reported that the mean value for the maximum duration of avian influenza virus infection was 8.3 days. Therefore, birds likely acquire entirely new LPAI infections during the fall migratory period and once they arrive on wintering grounds. However, Latorre-Margalef et al. (2008) also noted that the temporal range of virus shedding was large (2–34 days).
We observed some region-specific lineages in Alaska (NS and PA segments) and California (PB2), suggesting that population structures of host and viruses may be similar on breeding and wintering areas. That is, the sample of LPAI virus isolates from Alaska may come from northern pintails that winter in both Asia and North America. Conversely, the sample of LPAI isolates from California may also come from northern pintails that breed in Alaska and other locales. However, we acknowledge that independent data from satellite telemetry and banding studies are needed to assess this possibility, especially since not all virus RNA segments showed similar evidence for population genetic differentiation, as did the NS, PA, and PB2 segments. The mechanisms of mutation and resulting differences in population structure are not, however, as applicable to our results for the HA and NA RNA subtypes. Some evidence for transcontinental exchange was noted among four segments of HA and NA, but such polymorphism is characteristic of these segments (Clark and Hall 2006) and likely results from selection to adapt to a wide variety of host species (Dugan et al. 2008).
Frequency of Asian lineages and LPAI subtype diversity
We observed no Asian lineages among the 177 sequences of the M, NP, NS, PA, PB1, and PB2 genes isolated from viruses of northern pintails wintering in California in contrast to 12 (5.4%) Asian lineages among the 222 segments for these same RNA segments in LPAI viruses isolated from northern pintails in Alaska (Koehler et al. 2008). Additionally, 11 of the 14 HA and NA subtype segments showed no evidence for Asian lineages among California samples as North American and Asian sequences were well differentiated with high support. Four surface glycoprotein subtypes in California isolates (H6, N8, N1, and N2), representing 16 (26%) segments, yielded phylogenetic trees that suggest reassortment between continental gene pools of avian influenza viruses, followed by persistence and evolution of these mixed lineage clades in Asia and North America. The tree for the HA H6 yields a complex pattern in which North American and Asian lineages do not cluster as distinct clades. Similarly, mixed lineage topologies have been observed previously for the H6 subtype (see Webby et al. 2003; Spackman et al. 2005; Dugan et al. 2008). For the H6 tree, we included a slightly different suite of North American and Asian reference samples than either Spackman et al. (2005) or Koehler et al. (2008). Thus, one possible conclusion is that the northern pintail isolates from Alaska (n = 4) and California (n = 3) can be considered outsider events, along with at least three North American reference samples. However, these results may also demonstrate the persistence of Asian-like lineages in North American water birds following initial introduction events.
The tree for the N8 subtype is similar to that of Koehler et al. (2008), in which most Asian reference samples cluster as a single group separate from a clade that is a mixture of North American (including Alaska and California samples) and Asian lineages. The NA N2 segment tree is similarly a heterogeneous composition of North American and Asian lineages, with all California samples appearing as derived from Asian and North American reference samples (not shown). These results did not change following the inclusion of two completely Asian genotype LPAI viruses of the N2 subtype (A/Chicken/Nanchang/4-301/2001 and A/Wild Duck/Nanchang/2-0480/2000; Obenauer et al. 2006). Thus, this phylogram does not appear to result from the use of Asian reference samples that may have arisen from reassortment events. These results may also arise from the persistence of Asian-like lineages in North American water birds following initial introduction events or vice versa (e.g., our N1 results) and suggest selective pressures for maintaining genetic diversity in the NA surface glycoprotein to evade host immunity (Dugan et al. 2008).
In addition to sequence diversity, we also observed substantial variation in the subtype distribution of the transmembrane glycoproteins HA and NA between northern pintail isolates from Alaska and California. These frequency distributions also differ substantially from those observed among northern pintail LPAI viruses collected and isolated in Japan during the same time period as in our analysis (Jahangir et al. 2008). Categorizing virus isolates by subtype is imperative for surveillance programs to identify possible pathogenic strains, such as H2, H5, and H7, but may be of limited use for assessments of reassortment and virus exchange among continents due to the lack of complete geographic separation of these lineages (e.g., Figs 3 and 4). Indeed, the HA and NA segments were excluded from a recent methodological review of how to best determine rates of reassortment and virus turnover (Macken et al. 2006).
North American LPAI genomic diversity
Low pathogenic avian influenza genetic diversity was high and the lack of shared sequences between Alaska and California suggests initially that they could be considered as differentiated subpopulations of LPAI viruses. Such results are expected for a sample drawn from a large, diverse population characterized with a highly polymorphic molecular marker (see Jost 2008). However, the Pacific migratory flyway between Alaska and California is well documented for the northern pintail. During 4 years of marking wintering northern pintails in the Central Valley of California, Miller et al. (2005) noted that approximately 50–70% of adult females were present in Alaska during the breeding season. Additionally, 64–85% of northern pintails banded in Alaska during the breeding season were recovered in California winering areas (Bellrose 1980; Nicolai et al. 2005). Thus, both reassortment and rapid nucleotide evolution, which are hallmarks of avian influenza A viruses (Clark and Hall 2006), probably contribute substantially to our findings of some differentiation between breeding and wintering areas. Summarizing LPAI genome data across several additional years of sampling is needed to determine if avian influenza viruses in northern pintails exhibit an evolutionary pattern similar to the migration model proposed by Nelson et al. (2007) for the H3N2 human influenza strain. We observed a small number of distinct phylogenetic clades in the PA, NS, and PB2 segments. Similar patterns for the PA segment were observed in wild ducks in Canada (Hatchette et al. 2004) and Japan (Liu et al. 2004). For the PA and NS segments, clades of mostly Alaskan isolates may suggest an admixture in our sample of breeding birds from Asian and North American wintering locales.
Viral genomics as an aid to optimize surveillance plans
Our results demonstrate that genome sequencing of LPAI viruses can be used as a methodology to determine levels of connectivity for continental virus populations in migratory birds and to optimize surveillance efforts. Persistence of the H5N1 HPAI virus in Eurasia and Africa, and concerns that the virus might be transported among continents by migratory birds have resulted in wide-spread surveillance programs. From 2005 to 2008, state and federal agencies tested >326 000 wild bird samples from across the United States for the HPAI H5N1 virus (Highly Pathogenic Avian Influenza Early Detection Data System 2008). While these samples have not yielded detection of HPAI H5N1, numerous LPAI strains have been isolated. Genetic evidence from this study suggests that analyses of virus isolates from these surveillance efforts would provide a wealth of information on intercontinental movement of LPAI viruses in other species and other regions of North America that could greatly increase the efficiency of national HPAI early detection programs. Such an approach could address outstanding questions, such as: Do priority species targeted for surveillance in North America routinely carry LPAI viruses with Eurasian lineage RNA segments? Does the frequency of these RNA segments vary in time and space? Are there geographic ‘hot spots’ and carrier species in which Eurasian lineages are more common? Currently, 26 priority species have been selected for surveillance sampling in Alaska (Ip et al. 2008), but further optimization of this list, and timing of sampling, could be based on analysis of LPAI viral genetics as summarized in this study. A list of priority surveillance species will probably differ across regions of North America, but can be validated by a similar genetic analysis. Thus, a molecular prioritization is not only needed for Alaska, but also for species along the North Atlantic coast of North America, especially for those that exhibit initial evidence for transcontinental exchange (e.g., ruddy turnstones and herring gulls; Marakova et al. 1999).
With few exceptions, genetic evidence for transcontinental virus exchange in studies of North American LPAI has come from samples collected in coastal regions that are closest to Europe or Asia (i.e., Alaska and North Atlantic). These probably represent the first or primary areas of contact for foreign viruses, yet only 28% of birds tested for HPAI H5N1 in the United States to date have been from Alaska and the North Atlantic coast (Highly Pathogenic Avian Influenza Early Detection Data System 2008). The lower frequency of occurrence of transcontinental lineages from the areas of the continent that are more distant from these areas, such as California for northern pintails, is probably caused by functional dilution by this distance, virus mutation rates, reassortment and the short temporal nature of virus infections. Thus, based on genetic evidence, we conclude that one strategy would be to target surveillance efforts on species of coastal regions in the areas of geographic proximity to current sources of the highly pathogenic H5N1 virus.
Conclusions
We found limited evidence for the persistence of Asian lineage gene segments from breeding areas to a major wintering ground for the northern pintail, a species that is known to migrate between Asia and North America and that has shown a high frequency of Asian lineages in Alaska (Koehler et al. 2008). Thus, phylogenetic analysis of LPAI genomic variation can be used as an indicator of species and regions that should be targeted for HPAI surveillance. For example, if there is no evidence of transcontinental LPAI exchange for a certain species or regional pathway, then the introduction of HPAI via that route seems unlikely. However, species and or regions that do show genetic evidence of exchange with European or Asian LPAI lineages would be more likely pathways for the introduction of HPAI via wild migratory birds. Because of the pronounced diversity of HA and NA genes, determination of reassortment events with these segments is difficult. Therefore, we suggest that whole genome sequencing of LPAI be used as a methodology to determine levels of connectivity for continental virus populations in migratory birds, thereby informing future surveillance programs.
Acknowledgments
This research was funded by the U.S. Geological Survey (USGS), and the U.S. Fish and Wildlife Service (USFWS). California Department of Fish and Game (CDFG) provided support for collection of additional California samples. We thank other members of the northern pintail avian influenza research team including J. Hupp, S. Talbot and M. Petersen. We are grateful to S. Haseltine, R. Kearney, and P. Bright (USGS); D. Rocque, K. Trust, B. Bortner, and J. Hoskins (USFWS); and D. Yparraguirre (CDFG) for their support of field collections and laboratory analyses. Y. Gillies (USGS Alaska Science Center), D. Goldberg, Z. Najacht and R. Zane (USGS National Wildlife Health Center; NWHC) coordinated distribution of sampling materials, receipt of samples, and data verification. We thank the numerous state, federal, and California Waterfowl Association (CWA) field biologists who participated in sampling wild birds, including D. Loughman, J. Laughlin (CWA), and C. Tierney and G. Yarris (USGS). We also thank past and current members of the Diagnostic Virology Laboratory at the USGS NWHC, including T. Egstad, K. Griffin, A. Hauser, K. Kooiman, R. Long, A. Miyamoto, J. Montez, Z. Najacht, S. Nashold, J. TeSlaa, J. Tuscher and A. Ray. J. Terenzi created the map. C. Ely, J. Peters, C. Brand and K. Trust provided comments on an earlier version of this manuscript. None of the authors have any financial interests or conflict of interest with this article. Any use of trade names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Supporting information
Additional Supporting Information may be found in the online version of this article:
Table S1. Genbank accession numbers for Asian and North American reference samples used in phylogenetic anlyses of northern pintail viruses from California.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Literature cited
- Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavasky L, Tatusova T, Ostell J, et al. The influenza virus resource at the National Center for Biotechnology Information. Journal of Virology. 2008;82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean WJ, Schell M, Katz J, Kawaoka Y, Naeve C, Gorman O, Webster RG. Evolution of the H3 influenza virus hemagglutinin from human and nonhuman hosts. Journal of Virology. 1992;66:1129–1138. doi: 10.1128/jvi.66.2.1129-1138.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellrose FC. Ducks, Geese and Swans of North America. 3rd edn. Harrisburg, Pennsylvania: Stackpole Books; 1980. [Google Scholar]
- Bragstad K, Jørgensen PH, Handberg KJ, Mellergaard S, Corbet S, Fomsgaard A. New avian influenza A virus subtype combination H5N7 identified in Danish mallard ducks. Virus Research. 2005;109:181–190. doi: 10.1016/j.virusres.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Chan C, Lin K, Chan Y, Wang Y, Chi Y, Tu H, Shieh HK, et al. Amplification of the entire genome of influenza A virus H1N1 and H3N2 subtypes by reverse-transcription polymerase chain reaction. Journal of Virology Methods. 2006;136:38–43. doi: 10.1016/j.jviromet.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Clark L, Hall J. Avian influenza in wild birds: status as reservoirs, and risks to humans and agriculture. Ornithological Monographs. 2006;60:3–29. [Google Scholar]
- Dugan VG, Chen R, Spiro DJ, Sengamalay N, Zaborsky J, Ghedin E. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathogens. 2008;4:e1000076. doi: 10.1371/journal.ppat.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. arlequin, version 3.1: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Hatchette TF, Walker D, Johnson C, Baker A, Pryor SP, Webster RG. Influenza A viruses in feral Canadian ducks: extensive reassortment in nature. Journal of General Virology. 2004;85:2327–2337. doi: 10.1099/vir.0.79878-0. [DOI] [PubMed] [Google Scholar]
- Highly Pathogenic Avian Influenza Early Detection Data System. 2008. National avian influenza surveillance information. http://wildlifedisease.nbii.gov/ai/index.jsp (last accessed on 8 April 2009)
- Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Archives of Virology. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- Ip HS, Flint PL, Franson JC, Dusek RJ, Derksen DV, Gill RE, Ely CR, et al. Prevalence of influenza A viruses in wild migratory birds in Alaska: patterns of variation in detection at a crossroads of intercontinental flyways. Virology Journal. 2008;5:71. doi: 10.1186/1743-422X-5-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Okazaki K, Kawaoka Y, Takada A, Webster RG, Kida H. Perpetuation of influenza A viruses in Alaskan waterfowl reservoirs. Archives of Virology. 1995;140:1163–1172. doi: 10.1007/BF01322743. [DOI] [PubMed] [Google Scholar]
- Jackwood MW, Stallnecht DE. Molecular epidemiologic studies on North American H9 avian influenza virus isolates from waterfowl and shorebirds. Avian Diseases. 2007;51:448–450. doi: 10.1637/7536-032706R.1. [DOI] [PubMed] [Google Scholar]
- Jahangir A, Watanabe Y, Chinen O, Yamazaki S, Sakai K, Okamura M, Nakamura M, et al. Surveillance of avian influenza viruses in northern pintails (Anas acuta) in Tohoku District, Japan. Avian Diseases. 2008;52:49–53. doi: 10.1637/8035-062507-Reg. [DOI] [PubMed] [Google Scholar]
- Jost L. GST and its relatives do not measure differentiation. Molecular Ecology. 2008;17:4015–4026. doi: 10.1111/j.1365-294x.2008.03887.x. [DOI] [PubMed] [Google Scholar]
- Kimura M. Estimation of evolutionary distances between homologous nucleotide sequences. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:454–458. doi: 10.1073/pnas.78.1.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler AV, Pearce JM, Flint PL, Franson JC, Ip H. Genetic evidence of intercontinental movement of avian influenza in a migratory bird: the northern pintail (Anas acuta. Molecular Ecology. 2008;17:4754–4762. doi: 10.1111/j.1365-294X.2008.03953.x. [DOI] [PubMed] [Google Scholar]
- Komar N, Olsen B. Avian influenza virus (H5N1) mortality surveillance. Emerging Infectious Diseases. 2008;14:1176–1178. doi: 10.3201/eid1407.080161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S, Obert CA, Franks J, Walker D, Jones K, Seiler P, Niles PL, et al. Influenza in migratory birds and evidence of limited intercontinental virus exchange. PloS Pathogens. 2007;3:1684–1693. doi: 10.1371/journal.ppat.0030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanave C, Preparata G, Saccone C, Serio G. A new method for calculating evolutionary substitution rates. Journal of Molecular Evolution. 1984;20:86–93. doi: 10.1007/BF02101990. [DOI] [PubMed] [Google Scholar]
- Latorre-Margalef N, Gunnarsson G, Munster VJ, Fouchier RAM, Osterhaus ADME, Elmberg J, Olsen B, et al. Effects of influenza A virus infection on migrating mallard ducks. Proceedings of the Royal Society of London. Series B: Biological Sciences. 2008;276:1029–1036. doi: 10.1098/rspb.2008.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li OTW, Barr I, Leung CYH, Honglin C, Yi G, Malik Peiris JS, Poon LLM. Reliable universal RT-PCR assays for studying influenza polymerase subunit gene sequences from all 16 hemagglutinin subtypes. Journal of Virology Methods. 2007;142:218–222. doi: 10.1016/j.jviromet.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Liu JH, Okazaki K, Bai GR, Shi WM, Mweene A, Kida H. Interregional transmission of the internal protein genes of H2 influenza virus in migratory ducks from North America to Eurasia. Virus Genes. 2004;29:81–86. doi: 10.1023/B:VIRU.0000032791.26573.f1. [DOI] [PubMed] [Google Scholar]
- Macken CA, Webby RJ, Bruno WJ. Genotype turnover by reassortment of replication complex genes from avian influenza A virus. Journal of General Virology. 2006;87:2803–2815. doi: 10.1099/vir.0.81454-0. [DOI] [PubMed] [Google Scholar]
- Marakova NV, Kaverin NV, Krauss S, Senne D, Webster RG. Transmission of Eurasian avian H2 influenza virus to shorebirds in North America. Journal of General Virology. 1999;80:3167–3171. doi: 10.1099/0022-1317-80-12-3167. [DOI] [PubMed] [Google Scholar]
- Miller MR, Takekawa JY, Fleskes JP, Orthmeyer DL, Cassaza ML, Perry WM. Spring migration of northern pintails from California's Central Valley wintering area tracked with satellite telemetry: routes, timing and destinations. Canadian Journal of Zoology. 2005;83:314–332. [Google Scholar]
- Miyabayashi Y, Mundkur T. Atlas of Key Sites for Anatidae in the East Asian Flyway. Tokyo: Wetlands International; 1999. [Google Scholar]
- Munster VJ, Baas C, Lexmond P, Waldenstrom J, Wallensten A, Fransson T, Rimmelzwaan GF, et al. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathogens. 2007;3:e61. doi: 10.1371/journal.ppat.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Simonsen L, Viboud C, Miller MA, Holmes EC. Phylogenetic analysis reveals the global migration of seasonal influenza A viruses. PLoS Pathogens. 2007;3:e131. doi: 10.1371/journal.ppat.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolai CA, Flint PL, Wege ML. Annual survival and site fidelity of northern pintails banded on the Yukon-Kuskokwim Delta, Alaska. Journal of Wildlife Management. 2005;69:1202–1210. [Google Scholar]
- Obenauer JC, Denson J, Mehta PK, Xiaping S, Mukatira S, Finkelstein DB, Xu X, et al. Large-scale sequence analysis of avian influenza isolates. Science. 2006;311:1562–1563. doi: 10.1126/science.1121586. [DOI] [PubMed] [Google Scholar]
- Page RDM. TreeView: an application to display phylogenetic trees on personal computers. Bioinformatics. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Pereda AJ, Uhart M, Perez AA, Zaccagnini ME, La Sala L, Decarre J, Goijman A, et al. Avian influenza virus isolated in wild waterfowl in Argentina: evidence of a potentially unique phylogenetic lineage in South America. Virology. 2008;37:363–370. doi: 10.1016/j.virol.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps LP, Essen SC, Brown IH. Genetic subtyping of influenza A viruses using RT-PCR with a single set of primers based on conserved sequences within the HA2 coding region. Journal of Virology Methods. 2004;122:119–122. doi: 10.1016/j.jviromet.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Spackman E, Stallknecht DE, Slemons RD, Winker K, Suarez DL, Scott M, Swayne DE. Phylogenetic analyses of type A influenza genes in natural reservoir species in North America reveals genetic variation. Virus Research. 2005;114:89–100. doi: 10.1016/j.virusres.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Spackman E, McCracken KG, Winker K, Swayne DE. An avian influenza virus from waterfowl in South America contains genes from North American avian and equine lineages. Avian Diseases. 2007;51:273–274. doi: 10.1637/7529-032106R.1. [DOI] [PubMed] [Google Scholar]
- Swofford DL. paup*. Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, Massachusetts: Sinauer Associates, Inc; 2003. Version 4. [Google Scholar]
- Wahlgren J, Waldenstrom J, Sahlin S, Haemig PD, Fouchier RAM, Osterhaus ADME, Pinhassi J, et al. Gene segment reassortment between American and Asian lineages of avian influenza virus from waterfowl in the Beringia area. Vector Borne and Zoonotic Diseases. 2008;8:783–790. doi: 10.1089/vbz.2007.0274. [DOI] [PubMed] [Google Scholar]
- Wallensten A, Munster VJ, Elmberg J, Osterhaus ADME, Foucheir RAM, Olsen B. Multiple gene segment reassortment between Eurasian and American lineages of influenza A virus (H6N2) in guillemot (Uria aalge. Archives of Virology. 2005;150:1685–1692. doi: 10.1007/s00705-005-0543-8. [DOI] [PubMed] [Google Scholar]
- Webby RJ, Woolcock PR, Krauss SL, Walker DB, Chin PS, Shortridge KF, Webster RG. Multiple genotypes of nonpathogenic H6N2 influenza viruses isolated from chickens in California. Avian Diseases. 2003;47:905–910. doi: 10.1637/0005-2086-47.s3.905. [DOI] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiology Review. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Krauss S, Hulse-Post D, Sturm-Ramirez K. Evolution of influenza A viruses in wild birds. Journal of Wildlife Diseases. 2007;43(Suppl):S1–S6. [Google Scholar]
- Widjaja L, Krauss SL, Webby RJ, Xie T, Webster RG. Matrix gene of influenza A viruses from wild aquatic birds: ecology and emergence of influenza A viruses. Journal of Virology. 2004;78:8771–8779. doi: 10.1128/JVI.78.16.8771-8779.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S. A practical approach to genetic screening for influenza virus variants. Journal of Clinical Microbiology. 1997;35:2623–2627. doi: 10.1128/jcm.35.10.2623-2627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


