Abstract
Hybridization has been hypothesized to influence invasion through the generation of novel phenotypes and/or increased levels of genetic variance. Based on morphology, hybrids between diffuse knapweed and spotted knapweed, two invasive plants in North America, are present in the invaded range. Some individuals within most diffuse knapweed sites in North America exhibit intermediate diffuse × spotted floral morphology. We examined hybridization at the molecular level, using amplified fragment length polymorphisms. Approximately a quarter of the assayed North American diffuse knapweed individuals exhibited evidence of introgression from spotted knapweed. However, plants with intermediate morphology did not show evidence of mixed ancestry more often than the plants with typical diffuse knapweed morphology. The high proportion of hybrid individuals in North American diffuse knapweed sites found here, combined with evidence from recent studies, suggests that diffuse knapweed was likely introduced with admixed individuals, and the hybrids are not newly created postintroduction. A century of backcrossing with diffuse knapweed has likely decoupled the relationship between morphology and admixture at the molecular level. In contrast to the scenario encountered in North America, in the native range where diploid diffuse and spotted knapweed overlap, hybrid swarms are common. In such sites, the floral phenotype aligns more closely with the genotype.
Keywords: amplified fragment length polymorphism, biological invasion, diffuse knapweed, hybridization, spotted knapweed, STRUCTURE
Hybridization can have profound evolutionary consequences (Stebbins 1959; Arnold 1992; Rieseberg et al. 2003; Gompert et al. 2006). Recently, attention has focused on the role that hybridization may play in successful biological invasions (Ellstrand and Schierenbeck 2000;Rieseberg et al. 2007). Hybridization may result in evolutionary novelty and/or increased genetic variation, either of which may provide the genetic material for rapid adaptation to new abiotic and biotic conditions (Ellstrand and Schierenbeck 2000; Rieseberg et al. 2007). Additionally, hybridization can cause increased heterozygosity, which may increase fitness (Reed and Frankham 2003). The outcomes of hybridization, however, are not always positive; hybridization can result in outbreeding depression, as two disparate genomes are brought together (Price and Waser 1979). Yet, even if low fitness is the rule for most early generation hybrid individuals, gene flow and the creation of new evolutionary lineages is still possible (Arnold et al. 1999).
In a review of plant hybridization and invasion, 28 confirmed examples were found where invasiveness resulted after interspecific hybridization, and approximately 24 additional examples were found but molecular evidence was not available to confirm the hybrid status of the taxa (Ellstrand and Schierenbeck 2000). For example, Gaskin and Schaal (2002) discovered that the invasive Tamarix in North America is a hybrid undetected in the native range. The authors posit that multiple introductions brought together historically isolated genotypes from the native range. Another example is Spartina anglica, an allopolyploid hybrid capable of invading salt marshes and becoming a dominant species across a variety of such habitats (Thompson 1991). This hybrid differs significantly from its parent species, which do not demonstrate this aggressive, dominating capability (Thompson 1991). Thus, it appears hybridization may play an important role in some invasions. Presently, hybridizing non-native species warrant intense scrutiny and should be ‘guilty until proven innocent,’ as enough is not yet known about the importance of this mechanism in invasion.
This paper focuses on spotted knapweed (Centaurea stoebe L.) and diffuse knapweed (C. diffusa Lam.), two of the most ecologically and economically devastating introduced plants in western North America (Watson and Renney 1974; Roché and Roché 1991;Sheley et al. 1999). The diploid variants of these species are capable of hybridization (Ochsmann 1998, 1999), and the spotted × diffuse hybrid is called Centaurea xpsammogena (Gáyer 1909). We refer here and elsewhere (Blair et al. 2008; Blair and Hufbauer 2009) to individuals matching Gáyer's description of C. xpsammogena as hybrid-like, as the designation of ‘hybrid’ is based on morphological, and not molecular data. In a recent study conducted across western North America, such hybrid-like plants were found in 38 out of 39 diffuse knapweed sites, but in none of the 22 spotted knapweed sites (Blair and Hufbauer 2009). While the plants with intermediate morphology in North America are suggestive of hybridization between the two knapweeds, their presence has been interpreted in a variety of ways. For example, Watson and Renney (1974) suggested that ‘the degree of variation within the diffuse knapweed populations is possibly due to more than one introduction of the species into the area, and that the variable genotypes expressed by flower color in diffuse knapweed populations may be due to loose multiple gene control’ rather than hybridization. Moore and Frankton (1954) reached a similar conclusion that putative hybrids are simply morphological variants of diffuse knapweed in North America. Contrary to these conclusions, Ochsmann (2001a) argued that C. xpsammogena is present in North America based on herbaria records from seven different states in the USA. Blair and Hufbauer's (2009) recently conducted field surveys of spotted and diffuse knapweed also extended to the native range. In Europe they visited 19 diffuse knapweed sites in three countries (Romania, Ukraine, and Turkey) and 12 spotted knapweed sites in five countries (Austria, Hungary, Romania, Switzerland, and Ukraine). By conducting these cross-continent surveys, they found evidence suggesting that the hybrid-like plants in the diffuse knapweed sites in North America are likely of hybrid origin; regions exist in the native range where typical diffuse knapweed is found in the absence of hybrid-like plants. This is different from the nearly ubiquitous presence of hybrid-like plants in North American diffuse knapweed sites and suggests something more than morphological variation. Additionally, F1 and Back-Cross 1 hybrids created for a greenhouse common garden experiment exhibited similar intermediate traits as those seen in hybrid-like plants in the field (Blair, unpublished data).
The goal of this study was to examine if hybrid-like individuals are indicative of hybridization at the molecular level. Even if hybrids were created prior to introduction, hybridization may have contributed to the imminent success of diffuse knapweed as this plant encountered a novel selection regime (see above). We also wanted to gain a deeper understanding of morphological patterns encountered in the native and introduced ranges. Thus, we used amplified fragment length polymorphisms (AFLPs) (Vos et al. 1995) to examine hybridization between spotted and diffuse knapweed at the genome level.
Materials and methods
Study species
The genus Centaurea L. (Asteraceae) contains approximately 300 species (Garcia-Jacas et al. 2006), a number of which have been introduced globally and become invasive. In North America, at least 34 Centaurea species have been introduced, 14 of which are defined as noxious weeds in one or more states in the United States at the time of this writing (http://plants.usda.gov/). The taxonomy of the genus is complicated: sections within the genus are still being revised, and relationships within sections are not well resolved (Garcia-Jacas et al. 2006).
Our research focused on members of the Centaurea genus within the section Acrolophus-Phaelolepsis (Garcia-Jacas et al. 2006). More specifically, we focused on two members, C. stoebe (sensu stricto) and C. diffusa and their hybrids, of the C. stoebe (sensu latto) species group. This group encompasses approximately 33 named taxa (Ochsmann 2000). It is reported that both species have diploid (2n = 18) and tetraploid (4n = 36) cytotypes (Ochsmann 2000). Both cytotypes of diffuse knapweed are referred to simply as C. diffusa Lam. The tetraploid has only been reported twice in the literature from one specimen in Bulgaria (Löve 1979) and one in the former Yugoslavia (Löve 1978). The diploid is thus much more common and likely the only cytotype in North America (A.C. Blair, unpublished data; Marrs et al. 2008a). In its native range, diffuse knapweed grows in the Eastern Mediterranean area, western Asia, and from the southern part of the former U.S.S.R to western Germany (Rees et al. 1996). It is common in Bulgaria, Romania, former Yugoslavia, northern Italy, Greece, Ukraine, Turkey, and Syria (Watson and Renney 1974). The two cytotypes of spotted knapweed are both under C. stoebe L., a name that takes precedence over the commonly used C. maculosa (Ochsmann 2000). The monocarpic diploid is designated C. stoebe subsp. stoebe L, and the polycarpic tetraploid is designated C. stoebe subsp. micranthos (Gugler) Hayek (for which C. biebersteinii DC. is a synonym). Ploidy number is the only way to unambiguously distinguish these subspecies (Ochsmann 2001b). Centaurea stoebe subsp. stoebe occurs across western, central, and eastern Europe, spanning a broad west to east distribution from France to Ukraine and western Russia, and as far north as Belarus and Lithuania and as far south as Romania (see distribution map in Ochsmann 2001b). Centaurea stoebe subsp. micranthos originally occurred from south central to south-eastern Europe and northwest Asia, but it has been introduced to almost all parts of Europe (see distribution map in Ochsmann 2001b). All of the North American spotted knapweed plants that have been assayed for chromosome number are tetraploids (i.e., C. stoebe micranthos) (Moore and Frankton 1954; Müller 1989; Treier et al. 2009; H. Müller-Schärer, personal communication). Both spotted and diffuse knapweed are self-incompatible (A.C. Blair, personal observation; Harrod and Taylor 1995).
Floral traits are often used to distinguish species in the Centaurea genus. Both the diploid and tetraploid variants of spotted knapweed have larger flowering heads than diffuse knapweed, purple (rarely white) flowers, and a pronounced dark spot on each bract (Watson and Renney 1974; Ochsmann 2000). Diffuse knapweed has a terminal spine on each bract with no pigmentation and white (occasionally pink) flowers (Watson and Renney 1974; Ochsmann 2000). The hybrid C. xpsammogena, is characterized by distinct spotted bracts in addition to a terminal spine (Ochsmann 2000). Individual inflorescences often have both purple ray flowers and white disc flowers.
Tetraploid spotted knapweed and diffuse knapweed were accidentally introduced into North America from Eurasia in the late 1800s or early 1900s (Watson and Renney 1974; Roché and Roché 1991); both species were likely introduced several times (Hufbauer and Sforza 2008; Marrs et al. 2008a,b). They have become a major threat to rangeland productivity and quality across western North America (Watson and Renney 1974; Roché and Roché 1991; Sheley et al. 1999). These plants increase soil erosion (Lacey et al. 1989; Sheley et al. 1997), can alter plant community composition (Tyser and Key 1988), negatively impact biodiversity (Ortega et al. 2006), and are thought to have allelopathic effects on other plants (Fletcher and Renney 1963; Callaway and Aschehoug 2000; but see Locken and Kelsey 1987; Blair et al. 2005, 2006; Duke et al. 2009).
Collection sites and specimens
Tissue for molecular analysis was collected across Europe and North America (Table 1), following the sampling approach successfully employed to study interspecific hybridization by O'Hanlon et al. (1999), Kronforst et al. (2006), and Gompert et al. (2006) wherein both parental species, and a positive control and negative control are included in analyses of putative hybrids. Spotted × diffuse hybrids are diploid (A.C. Blair, unpublished data; Ochsmann 1998, 1999), so morphologically typical diploid diffuse knapweed and diploid spotted knapweed from the native range were included in the molecular analysis as the parental species (Fig. 1, Table 1). Hereafter, ‘spotted knapweed’ and ‘diffuse knapweed’ refer to the diploid parental variants unless otherwise stated. Ploidy of the parent was confirmed with flow cytometry (see Treier et al. 2009 for methods). Because the diffuse knapweed sites surveyed in North America almost always contained some percentage of hybrid-like plants (n = 38 out of 39 sites, Blair and Hufbauer 2009) and therefore might not be pure, and diploid spotted knapweed appears to be absent from North America, we could not include samples of the parent species from the introduced range. As positive controls, to determine if the AFLP technique could reliably detect recent hybridization, individuals from apparent hybrid swarms of the diploid parent species in the Ukraine were included; such sites were never encountered in North America (Blair and Hufbauer 2009). Plants from these Ukrainian sites were morphologically classified as hybrid-like, diffuse-like, or spotted-like based on a hierarchical cluster analysis of a suite of floral traits as part of a larger data set (n = 419 plants) in a previous study (Blair and Hufbauer 2009). Briefly, the following floral traits were included in that analysis: bract pigmentation ranked 0 (no pigmentation, golden) to 3 (deeply pigmented), capitula width, capitula length, and spine length. Data were standardized prior to analysis by the variable mean and standard deviation, and we used ‘Ward's minimum variance’ clustering method. As a negative control, we included an outgroup species, meadow knapweed (C. pratensis Thuill), to test the ability of our markers to distinguish between the closely related spotted and diffuse knapweed. If we could not clearly distinguish the two species with AFLP markers, inclusion of this outgroup would help discern between inherent technique and analysis problems versus actual difficulty in distinguishing between the species because of close relatedness. To determine if interspecific hybrids are present in North American diffuse knapweed sites, we analyzed individuals from sites that contained both morphologically typical diffuse knapweed and hybrid-like plants (Fig. 2, Table 1). We included approximately equal numbers of typical diffuse knapweed and hybrid-like individuals from each site, objectively classified by a hierarchical cluster analysis of floral traits, as described above, except flower color was included in the analysis [ranked 1 (white) to 5 (solid purple)] (Blair and Hufbauer 2009). For all samples, tissue was either collected in the field and dried by temporary storage in Drierite (W.A. Hammond Drierite Co., Xenia, OH) prior to transfer to a −80 freezer, or collected fresh from plants grown from seed in the greenhouse.
Table 1.
Sites of European diffuse knapweed (verified diploid), spotted knapweed (verified diploid), and spotted × diffuse knapweed hybrid swarms, and North American diffuse knapweed + hybrid-like plants and meadow knapweed used in the AFLP analyses
| Site ID | State/Country | Species | GPS | Number of introgressed individuals*/total |
|---|---|---|---|---|
| Europe | ||||
| Ro 6 | Romania | Diffuse knapweed | N 45°11′8.8″ E 28°47′8.3″ | 0/4 |
| Ro 5 | Romania | Diffuse knapweed | N 44°94′34.3″ E 28°91′4.5″ | 0/4 |
| Ro 4 | Romania | Diffuse knapweed | N 44°23′22.8″ E 28°31′35.9″ | 0/4 |
| Crimea 21 | Ukraine | Diffuse knapweed | N 44°33′0.0″ E 34°16′0.0″ | 0/4 |
| Crimea 20 | Ukraine | Diffuse knapweed | N 44°36′0.0″ E 34°10′0.0″ | 0/3 |
| Rus 1119 | Russia | Diffuse knapweed | N 44°3′0.0″ E 43°3′36″ | 0/4 |
| Rus 1142 | Russia | Diffuse knapweed | N 51°22′48″ E 56°48′0.0″ | 0/4 |
| UA 2-2n-SK | Ukraine | Spotted knapweed | N 49°55′48.5″ E 24°50.1′8.9″ | 0/5 |
| UA 5-2n-SK | Ukraine | Spotted knapweed | N 49°46′19.2″ E 27°17.5′27.6″ | 0/5 |
| SUAC-2n-SK | Ukraine | Spotted knapweed | N 49°13′13.4″ E 24°42.3′17.6″ | 0/6 |
| UA 6 | Ukraine | Active hybrid zone | N 48°34′51.9″ E 37°54′36.9″ | 4/4 |
| UA 4 | Ukraine | Active hybrid zone | N 48°53′31.2″ E 30°40′33.2″ | 1/4 |
| UA 14 | Ukraine | Active hybrid zone | N 50°28′50.7″ E 30°29′10.7″ | 4/4 |
| North America | ||||
| 1 W.USA | CO, USA | Diffuse knapweed + hybrid-like | N 39°40′17.0″ W102°33′01.3″ | 1/4 |
| 6 W.USA | WA, USA | Diffuse knapweed + hybrid-like | N 46°35′06.7″ W120°27′33.0″ | 0/4 |
| 11 W.USA | WA, USA | Diffuse knapweed + hybrid-like | N 47°33′40.4″ W120°16′11.3″ | 0/4 |
| 13 W.USA | WA, USA | Diffuse knapweed + hybrid-like | N 47°28′14.4″ W120°20′11.5″ | 1/4 |
| 20 W.USA | WA, USA | Diffuse knapweed + hybrid-like | N 46°43′16.9″ W117°9′50.5″ | 0/4 |
| 21 W.USA | OR, USA | Diffuse knapweed + hybrid-like | N 45°54′58.8″ W119°33′31.8″ | 0/4† |
| 25 W.USA | OR, USA | Diffuse knapweed + hybrid-like | N 45°36′17.1″ W121°11′02.3″ | 0/4† |
| 41 W.USA | WY, USA | Diffuse knapweed + hybrid-like | N 43°23′07.9″ W107°03′45.6″ | 4/4† |
| 43 W.USA | CO, USA | Diffuse knapweed + hybrid-like | N 39°42′10.2″ W106°40′32.8″ | 4/4† |
| Cow Creek | OR, USA | Meadow knapweed | N 42°52′47.2″ W123°31.6′36.8″ | 0/2 |
| Wyeth | OR, USA | Meadow knapweed | N 45°41′20.0″ W121°47′56.6″ | 0/2 |
Significant introgression is assumed if the 95% posterior probability interval around the individual's admixture proportions did not include 1.
Only diploid individuals detected in site.
Figure 1.
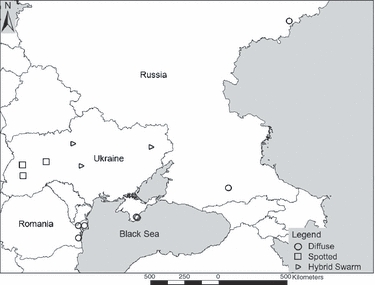
European site locations for diffuse knapweed (2n), spotted knapweed (2n), and active hybrid swarms in Ukraine (i.e. both parent species and a morphological gradient of hybrids) used in AFLP analyses.
Figure 2.
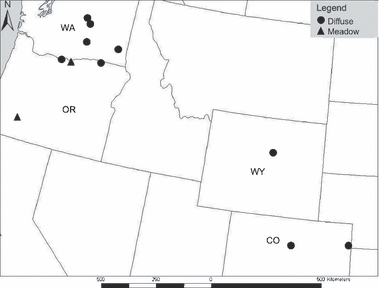
North American site locations for diffuse knapweed + hybrid-like plants and meadow knapweed used in AFLP analyses. All diffuse knapweed sites contained hybrid-like plants at varying frequencies.
DNA extraction
Total DNA was extracted from 95 individuals from fresh (100 mg) or dry (25 mg) leaf tissue with QIAGEN Mini Plant Extraction kits (QIAGEN Inc., Valencia, CA). Leaf tissue from individual plants was ground under liquid nitrogen in mortar and pestle, and then the Qiagen protocol was followed.
AFLP protocol
Because we anticipated that we would need a large number of markers to be able to clearly discern the closely related spotted and diffuse knapweeds, we used AFLPs. AFLPs provide a powerful and frequently used approach for the reliable generation of numerous markers (Vos et al. 1995). AFLP markers are mainly made up of noncoding DNA and are distributed throughout the genome (for a review of the technique and its limitations, see Meudt and Clarke 2007).
The AFLP method followed Vos et al. (1995) but included the following changes: restriction and ligation were performed during a single step in an 11-μL reaction containing genomic DNA, 1 U MseI, 5 U EcoRI, 1X T4 DNA ligase buffer, 60 U T4 DNA ligase, 0.05 m NaCl, 0.5X BSA, 4.5 μm MseI adaptor, 0.45 μm EcoRI adapter, and water. This mixture was incubated at room temperature overnight. The next day, 5.5 μL of the reaction was diluted to 100 μL in TE (15 mm Tris and 0.1 mm EDTA). A preselective polymerase chain reaction (PCR) was performed in a 20-μL reaction containing the following: 4 μL of the diluted restriction-ligation product, 1X PCR buffer, 1.5 mm MgCl2, 0.2 mm each dNTP, 0.2 μm of each preselective amplification primer (MseI + C and EcoRI + A), 0.5 U Taq polymerase, and water. The preselective PCR cycles were as follows: 20 cycles of 30 s at 94°C, 60 s at 56°C, and 60 s at 72°C. Ten microliters of the preselective amplification product was diluted to 200 μL in TE (15 mm Tris and 0.1 mm EDTA). The selective amplification was performed in a 20 μL reaction with 3 μL of the diluted preselective amplification product. The following reagents were included in the reaction: 1X PCR buffer, 1.5 mm MgCl2, 0.2 mm each dNTP, 0.1 μmMseI selective primer, 0.05 μmEcoRI selective primer dye-tagged with D4 (blue), 0.5 U of Taq polymerase, and water. The selective PCR cycles were as follows: 120 s at 94°C, 10 cycles of 20 s at 94°C and 30 s at 66°C (decreasing by 1°C each cycle), 120 s of 72°C; 25 cycles of 20 s at 94°C, 30 s at 56°C, and 120 s at 72°C, and a final 30 min at 60°C. One microliter of each selective PCR product was combined with 0.3 μL of 600 bp size standard and 28.7 μL of deionized formamide. All selective primer combinations of MseI + CAA, CAC, CAT, CTA, or CTC and EcoRI + AAG, ACC, or ACT were prescreened with five individuals, and the three most polymorphic primer pairs were chosen (MseI + CAC/EcoRI + AAG; MseI + CAT/EcoRI + AAG; and MseI + CTA/EcoRI + AAG). Samples were analyzed on a Beckman Coulter (Fullerton, CA) CEQ 8000 fragment analyzer.
AFLP data analyses
Amplified fragment length polymorphism fragments between 100 and 600 bp were scored using the fragment analysis software Genemarker® (Softgenetics®, State College, PA). Initially, we set the program to call only peaks above 200 reflectance units; thus, bins for markers were created that had at least one peak >200 reflectance units. We then ran these data with the new bin set and lowered the threshold to 100 reflectance units. This approach was used to minimize ambiguity in subjectively defining ‘real’ peaks. After these two passes of the data, we went through each electropherogram trace by hand to ensure that peaks were correctly called and placed in the appropriate bins. If a peak was a borderline call (i.e. around 100 reflectance units), we compared it to other traces to see if the shape and position matched other individuals for that marker.
To test the repeatability of the method, 10 individuals (≍ 10%) were selected at random, and AFLP fragments were generated starting from the restriction/ligation step for each of the three primer pairs. Repeat runs were scored blindly and compared to original runs to calculate error rate.
Statistical analysis
We used a Bayesian clustering method (STRUCTURE v. 2.2; Pritchard et al. 2000; Falush et al. 2007) to determine if the AFLP markers could (i) distinguish amongst the three species (spotted knapweed, diffuse knapweed, and meadow knapweed) and (ii) detect interspecific hybridization between spotted and diffuse knapweed. Briefly, STRUCTURE works as follows: a model is used which assumes there are K populations (either known or unknown), and each of these K populations is defined by a unique set of allele frequencies at each locus. STRUCTURE then assigns individuals to these populations based on their allele frequencies, while at the same time, estimating population allele frequencies. The most recent version of STRUCTURE (v. 2.2) can analyze dominant markers, like AFLPs, by defining a null allele at each locus (Falush et al. 2007). For each analysis, we had a burn-in length of 100 000 iterations; an appropriate burn-in length is critical to minimize the effect of the starting configuration. This was followed by 1 000 000 iterations of data collection; an appropriate number of iterations is necessary to get accurate parameter estimates. These two values produced highly consistent results across runs, and the summary statistics were stable before the end of the burn-in. For all runs, we provided only genetic data to the model with no prior information about the location of collection or morphological species status. For all analyses, we used the admixture model in STRUCTURE to estimate the proportion of each individual's genotype (q) from the K populations. Using the ANCESTDIST option in STRUCTURE, we computed the 95% posterior probability interval around each individual's admixture proportion. If an individual's proportion did not include one, introgression was likely to have occurred.
We analyzed our data with a two-step approach. In the first STRUCTURE analysis, we included the AFLP data from parental European diffuse knapweed, parental European spotted knapweed, Ukrainian hybrid sites (positive control), and North American meadow knapweed (negative control). This first analysis was conducted to verify that the individuals from the parental species were pure and to test if the analysis would correctly detect admixture in the European hybrid swarms. For this analysis, we assumed that three genetic clusters would likely best explain the data, as among species differences would presumably be greater than within species differences. We validated that K = 3 clusters yielded the highest log likelihood value by running replicated runs (K = 2–10 with four iterations) (Fig. 3). Upon confirming that parental species were pure and that the technique could detect admixture (see Results), we then conducted a STRUCTURE analysis with the parental European diffuse knapweed, parental European spotted knapweed, and the North American diffuse knapweed + hybrid-like individuals to examine if plants with detectable admixture are present in North America. For this analysis, as before, we validated that K = 2 clusters best explains the data (data not shown).
Figure 3.
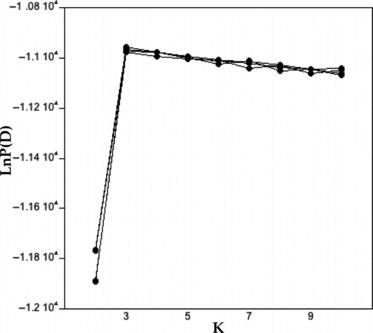
Log-likelihood probabilities of the number of clusters (K) for four independent series of K = 2 through 10 estimated using STRUCTURE v. 2.2 with admixture (Pritchard et al. 2000; Falush et al. 2007). The value ln P(D) is the probability that K is the correct number of clusters given the data. The larger, or less negative, the log likelihood value, the better the K fits the data.
All other statistical analyses were conducted using JMP v. 6.0 (SAS Institute Inc., Cary, NC). Based on both the hierarchical cluster analysis of floral morphology and the molecular data (see below), hybridization was detected within the North American diffuse knapweed sites. We therefore wanted to know if the phenotype matched the genotype (i.e., did morphological hybrids demonstrate significant admixture at the molecular level). To determine if North American individuals in the morphological hybrid cluster (see above) exhibited greater admixture from spotted knapweed than plants that morphologically clustered as typical diffuse knapweed, ANOVA was used to compare the posterior mean proportion of ancestry associated with the diffuse knapweed cluster between the two plant types. We then used this same approach to compare the posterior mean proportion of ancestry associated with the diffuse knapweed cluster among all of the plant groups included in the study (except for North American meadow knapweed), and we used a Tukey-Kramer post-hoc test to determine which plant types had different posterior mean proportions of ancestry associated with the diffuse knapweed cluster. Thus, six plant groups were included in this final analysis: European diffuse knapweed, North American diffuse knapweed, Ukrainian diffuse knapweed, Ukrainian hybrid, Ukrainian spotted, and European spotted knapweed. These six plant groups span the range of morphologies from pure European diffuse knapweed to pure European spotted knapweed found across the introduced and native range.
Results
AFLP analyses
For the first and second STRUCTURE analyses, we used a total of 375 and 374 AFLP bands, respectively, after removing uninformative bands that were either present in all individuals surveyed or found in only one individual. In the duplicated runs to examine the consistency of this technique, ≍ 94% of the bands were scored similarly across the three primer pairs.
In the first STRUCTURE (v 2.2) analysis (admixture model; K = 3), all European spotted and diffuse knapweed and North American meadow knapweed individuals had a population of origin genome probability interval that included one, indicating that admixture was unlikely within those groups (Fig. 4A). As predicted based on morphology, those species groups seem genetically isolated, and the admixture model performed well at distinguishing at the species level. Within the actively hybridizing sites in the Ukraine, nine out of 12 individuals had population of origin genome probability intervals that did not include one, and all individuals had pure ancestry proportions <0.9 (Fig. 4A).
Figure 4.
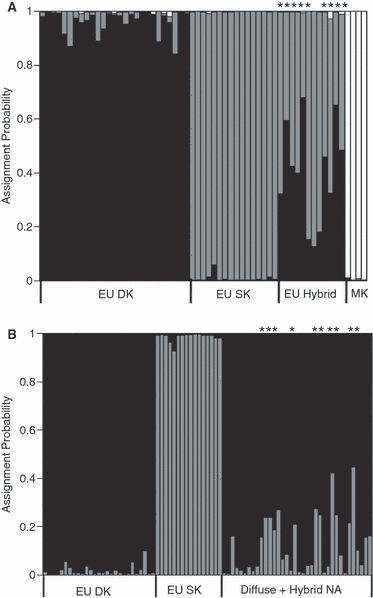
Bayesian assignment probabilities with admixture and (A) K = 3 and (B) K = 2 [STRUCTURE v. 2.2; Pritchard et al. (2000); Falush et al. (2007)]. Each vertical bar represents one individual. The black, grey, and white coloring represents the posterior mean proportion of ancestry from diffuse knapweed (2n), spotted knapweed (2n), and in (A) meadow knapweed, respectively. *population of origin genome probability does not include one, indicating hybridization. EU, Europe; NA, North America; EU hybrid, individuals from spotted × diffuse hybrid swarms in the Ukraine; MK, meadow knapweed (out-group). Based on morphology, diffuse and hybrid-like plants were included in approximately equal numbers from North America sites (see text).
In the second STRUCTURE analysis (admixture; K = 2), a number of individuals were identified with mixed ancestry in the North American diffuse knapweed sites (Fig. 4B). Ten out of 36 individuals had population of origin genome probability intervals that did not include one (Table 1), and 15 out of the same 36 individuals had pure ancestry proportions <0.9, further confirming the presence of admixture.
Based on the hierarchical cluster analysis of the floral characters from the plants within the North American diffuse knapweed sites, two clusters were identified with 17 and 14 members. The first cluster was dominated by plants identified visually as diffuse knapweed in the field (15 out of 17), while the second cluster contained only plants identified as hybrid-like in the field (14 out of 14) (Fig. 5).
Figure 5.
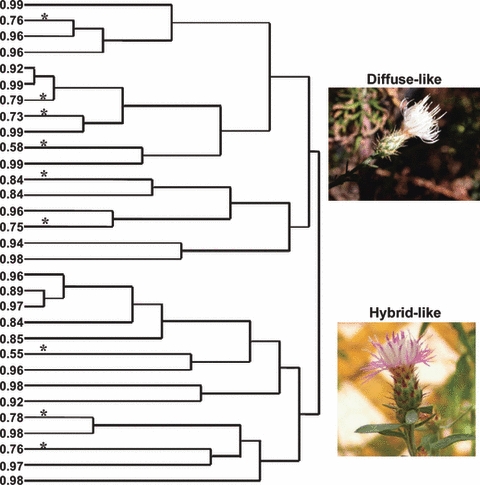
A dendrogram from hierarchical cluster analysis (Ward's method) of diffuse-like and hybrid-like plants surveyed across nine diffuse knapweed sites in western North America in 2005. Five morphological floral characters were analyzed. The top cluster includes plants with typical diffuse knapweed morphology, while the bottom cluster includes plants with hybrid morphology. The numbers to the left of the branches are the genome proportions associated with the diffuse knapweed group [(K = 2 with admixture, STRUCTURE v. 2.2; Pritchard et al. (2000); Falush et al. (2007)]. *population of origin genome probability interval does not include one, indicating interspecific hybridization. See text for details.
While the presence of individuals with intermediate floral morphology encountered in North American diffuse knapweed sites correctly suggested interspecific hybridization, the floral morphology did not correctly predict the genetic classification within the North American sites (Fig. 5). Out of the 31 individuals included in the hierarchical cluster analysis, STRUCTURE classified slightly less than half (14/31) as predicted by the phenotype (i.e. a plant that looked like typical diffuse knapweed had a population of origin genome probability interval that included one). In fact, counter to expectation, six out of 17 individuals in the morphological diffuse cluster demonstrated admixture from spotted knapweed, while 11 of the 14 individuals in the morphological hybrid cluster did not exhibit evidence of mixed ancestry. The two morphological clusters did not differ in the proportion of the genome associated with the diffuse knapweed cluster (F1,29 = 0.01, P = 0.92; morphological diffuse cluster = 0.88, morphological hybrid cluster = 0.88).
Interestingly, when we ran the second STRUCTURE analysis with K = 3 clusters (data not shown), we consistently found that two individuals from a diffuse knapweed site near Yakima, WA, USA had a significant portion of their genome that grouped with no other individuals in the analysis. This suggests that some of the diffuse knapweed individuals from this site may have hybridized with a presently unidentified species. As Centaurea species are known to hybridize frequently in their native range (Ochsmann 2000), it is possible that we have detected a separate instance of diffuse knapweed either introduced as an inter-specific hybrid or currently undergoing hybridization with a different introduced Centaurea species.
Of the 12 individuals included from the actively hybridizing Ukraine sites, based on floral morphology, three plants were classified as diffuse knapweed, six as hybrid-like, and three as spotted knapweed (from Blair and Hufbauer 2009). Contrary to the North American data, plants that appeared more like typical diffuse knapweed had a significantly greater posterior mean proportion of ancestry associated with the diffuse knapweed cluster than those that appeared visually as typical spotted knapweed (Fig. 6). As predicted by morphology, hybrids were intermediate between the two. Interestingly, the plants that appeared as typical diffuse knapweed in the hybrid swarms exhibited greater levels of admixture than either the pure European diffuse knapweed or the North American + hybrid-like plants (Fig. 6).
Figure 6.
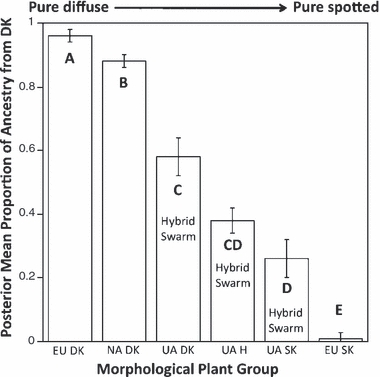
The posterior mean proportion of ancestry associated with the diffuse knapweed cluster, as calculated by STRUCTURE v. 2.2 [Pritchard et al. (2000); Falush et al. (2007)]. EU, Europe; NA, North America; UA, Ukraine; DK, diffuse knapweed; H, hybrid; SK, spotted knapweed. Based on morphology, the European diffuse and spotted knapweed sites did not contain any hybrid-like plants. Diffuse and hybrid-like plants from North America were combined into one group for this analysis because they had identical posterior mean proportion of ancestry values from the diffuse knapweed cluster (see text). Individuals from Ukraine came from apparently active hybrid swarms, and the morphological grouping of an individual plant from these sites as spotted, diffuse, or hybrid was done by hierarchical cluster analysis (Blair and Hufbauer 2009). Values represent mean ± 1 SE. Different letters denote significantly different means (Tukey's test P < 0.05).
Discussion
We have shown at the molecular level that some individuals of the North American noxious weed, diffuse knapweed, contain detectable admixture from a closely related species, spotted knapweed. STRUCTURE identified evidence of mixed ancestry in 28% of the assayed plants in the North American diffuse knapweed sites. Thus, we have likely identified a new example of an invasive organism that has undergone interspecific hybridization. Neither parental species from the native range or the outgroup meadow knapweed demonstrated such admixture, while 75% of the plants in the hybrid swarms in the Ukraine were of hybrid origin.
While the field surveys (Blair and Hufbauer 2009), hand pollinations (Blair, unpublished data), and molecular work presented here suggest interspecific hybridization, we must point out that a history of hybridization can never be fully proved. It is possible that lineage sorting from a common ancestor would leave a misleading imprint of past hybridization. As Gottlieb (1972) eloquently stated when attempting to deal with the problems of past events, ‘Depending on the available evidence…, the best we can do is establish levels of confidence for the inferences we make.’ We feel reasonably confident that our multiple lines of evidence allow us to infer hybridization, but we can never be certain.
Assuming a history of hybridization, recent evidence most parsimoniously suggests that the hybrid-like plants encountered in North American diffuse knapweed sites are not from recent spotted × diffuse knapweed hybridization events in the introduced range (Blair and Hufbauer 2009). Two geographically comprehensive surveys of spotted knapweed in the introduced range support that only tetraploid spotted knapweed is likely present in North America (Marrs et al. 2008b; Treier et al. 2009), while the diffuse knapweed is diploid (A.C. Blair, unpublished data; Marrs et al. 2008a). Triploids have never been found in North America (A.C. Blair, unpublished data; H. Müller-Schärer, unpublished data; Moore and Frankton 1954), and multiple attempts to create F1 hybrids between North American tetraploid spotted and diploid diffuse knapweed from various populations failed, in spite of the successful production of other crosses (i.e. European diploid spotted knapweed × North American diffuse knapweed). Apparently, genetic incompatibilities between North American tetraploid spotted and diploid diffuse knapweed largely prevent successful mating. Therefore, plants with hybrid ancestry were most likely introduced with diffuse knapweed. That plants with hybrid morphology were found in nearly all diffuse knapweed sites sampled in North America suggests that they were introduced early in the invasion of North America rather than recently (Blair and Hufbauer 2009). While the specific location(s) of where diffuse knapweed was introduced from are unknown, it seems conceivable that this plant was introduced one or multiple times from the regions where diploid spotted and diffuse knapweed overlap and hybridize in certain parts of Romania or Ukraine (U. Schaffner, personal communication). It is also possible that the hybrids in the introduced range stem from previous crosses between diploid diffuse and diploid spotted knapweed in North America, the latter which could have theoretically been present early in the invasion of these species but later went extinct. This scenario is less parsimonious, though, as it requires the additional assumption that diploid spotted knapweed was at one point present in North America. Treier et al. (2009) conducted extensive surveys of spotted knapweed ploidy in its introduced and native range, and they have yet to find diploid spotted knapweed in North America (H. Müller-Schärer, personal communication).
Floral traits correctly suggested the presence of hybridization in both the Ukraine and North America; however, individuals in North America with intermediate floral traits were no more likely to show evidence of mixed ancestry than those that appeared as typical diffuse knapweed (Fig. 5). It is likely that the diagnostic floral traits are controlled by a small number of genes, and the randomly distributed AFLP markers were probably not located within those genes. This is similar to the situation that Kronforst et al. (2006) encountered; a butterfly they morphologically identified as a hybrid had a population of origin genome proportion that included one when admixture clustering was implemented with AFLP data in STRUCTURE, indicating that individual was not of hybrid origin. They used wing pattern to diagnose hybridization between butterflies, and concluded that ‘within a few generations of initial hybridization, many individuals with hybrid ancestry are unlikely to be distinguishable based on phenotype alone.’ Wing patterning provides few loci for determining ancestry of a butterfly (Kronforst et al. 2006), perhaps similar to floral morphology in the knapweeds.
Additionally, in the introduced range diffuse knapweed has been isolated from diploid spotted knapweed for approximately 100 years. Diffuse knapweed is an annual to short-lived perennial that flowers in one to three years (Watson and Renney 1974). Assuming a mid-point time to flower (i.e., two years), there have been approximately 50 generations since introduction. Reproductive barriers do not exist between hybrid-like individuals and those with typical diffuse knapweed morphology within a site; seeds from hybrid-like plants often result in plants with typical diffuse knapweed morphology and vice versa (A.C. Blair, unpublished data). There has been ample time for genetic shuffling, and the floral hybrid traits may no longer be strongly associated with hybridization, per se. It is interesting that after a century, portions of the spotted knapweed genome have been retained in some diffuse knapweed individuals. This long period of time might also explain why we did not detect plants with hybrid ancestry in some of the diffuse knapweed sites that contained morphological hybrids. Extensive back-crossing and drift have possibly erased the signature of hybridization in some locations.
In the Ukraine, we encountered several sites where there were both morphologically typical parental species and a gradient of intermediate plants. Individuals within those sites that grouped with the diffuse or spotted knapweed clusters based on floral characters still demonstrated admixture. Different than within North America, however, plants that appeared more similar to diffuse knapweed had a greater posterior mean proportion of ancestry associated with the diffuse knapweed genetic cluster, while plants that appeared more similar to spotted knapweed had a greater posterior mean proportion of ancestry associated with the spotted knapweed cluster. In locations of recent and/or on-going hybridization, it seems that the floral morphology is associated with the predicted species at the genetic level. These data further support that the hybrid-like plants in North America are not newly created.
In conclusion, some diffuse knapweed plants in North America contain detectable admixture from diploid spotted knapweed, and we found one instance that suggests diffuse knapweed may contain introgression from a presently unidentified species. While morphological floral traits in the field correctly suggested the presence of plants with hybrid ancestry in North America, the individual phenotype did not align with the genotype; individuals with diffuse knapweed morphology often showed evidence of mixed ancestry, while hybrid-like plants often did not. These discrepancies likely result from the long time period since the hybridization event prior to introduction. In sites where hybridization is ongoing in Ukraine, the genotype and phenotype were more closely aligned. Further research is exploring whether the inclusion of plants with hybrid ancestry in the introduction of diffuse knapweed influenced the invasion process through, for example, increased genetic variation and/or evolutionary novelty (Ellstrand and Schierenbeck 2000).
Acknowledgments
We gratefully acknowledge J. Gaskin and K. Mann for the use of their equipment and sharing their expertise in generating the AFLPs; S. Keller generously shared his opinions on how to score AFLPs objectively; S. Knudson, S. Rauth, and M. Bruce assisted with trouble shooting; P. Häfliger, O. Kornyenko, S. Mosyakin, H. Müller-Schärer, and U. Schaffner helped with the European tissue collection and V. Blair and E. Coombs helped in the field in North America; D. Baker kindly help to make the site maps; and M. Antolin and two anonymous reviewers provided helpful comments on an earlier version of this manuscript. Funding for this research was provided by the Colorado Agricultural Experiment Station, an EPA STAR fellowship FP-91676001-0, CIPM G257-05-W0094, NSF DEB #0508922, NSF DDIG #0607974, and USDA #2002-35320-12137.
Literature cited
- Arnold ML. Natural hybridization as an evolutionary process. Annual Review in Ecology and Systematics. 1992;23:237–261. [Google Scholar]
- Arnold ML, Bulger MR, Burke JM, Hempel AL, Williams JH. Natural hybridization: how low can you go and still be important? Ecology. 1999;80:371–381. [Google Scholar]
- Blair AC, Hufbauer RA. Geographic patterns of interspecific hybridization between spotted knapweed (Centaurea stoebe) and diffuse knapweed (C. diffusa. Invasive Plant Science and Management. 2009;2:55–69. [Google Scholar]
- Blair AC, Hanson BD, Brunk GR, Marrs RA, Westra P, Nissen SJ, Hufbauer RA. New techniques and findings in the study of a candidate allelochemical implicated in invasion success. Ecology Letters. 2005;8:1039–1047. [Google Scholar]
- Blair AC, Nissen SJ, Brunk GR, Hufbauer RA. A lack of evidence for a role of the putative allelochemical (±)-catechin in spotted knapweed invasion success. Journal of Chemical Ecology. 2006;32:2327–2331. doi: 10.1007/s10886-006-9168-y. [DOI] [PubMed] [Google Scholar]
- Blair AC, Schaffner U, Häfliger P, Meyer SK, Hufbauer RA. How do biological control and hybridization affect enemy escape? Biological Control. 2008;46:358–370. [Google Scholar]
- Callaway RM, Aschehoug ET. Invasive plants versus their new and old neighbors: a mechanism for exotic invasion. Science. 2000;290:521–523. doi: 10.1126/science.290.5491.521. [DOI] [PubMed] [Google Scholar]
- Duke SO, Blair AC, Dayan FE, Johnson RD, Meepagala KM, Cook D, Bajsa J. Is (−)-catechin a novel weapon of spotted knapweed (Centaurea stoebe)? Journal of Chemical Ecology. 2009;35:141–153. doi: 10.1007/s10886-008-9587-z. [DOI] [PubMed] [Google Scholar]
- Ellstrand NC, Schierenbeck KA. Hybridization as a stimulus for the evolution of invasiveness in plants? Proceedings of the National Academy of Sciences of the USA. 2000;97:7043–7050. doi: 10.1073/pnas.97.13.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher RA, Renney AJ. A growth inhibitor found in Centaurea spp. Canadian Journal of Plant Science. 1963;43:475–481. [Google Scholar]
- Garcia-Jacas J, Uysal T, Romashchenko K, Suarez-Santiago VN, Ertugrul K, Susanna A. Centaurea revisited: a molecular survey of the Jacea group. Annals of Botany. 2006;98:741–753. doi: 10.1093/aob/mcl157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskin JF, Schaal BA. Hybrid Tamarix widespread in U.S. invasion and undetected in native Asian range. Proceedings of the National Academy of Sciences of the USA. 2002;99:11256–11259. doi: 10.1073/pnas.132403299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gáyer G. Vier neue Centaureen der Flora Botanikai von Ungarn. Magyar Botanikai Lapok. 1909;8:58–61. [Google Scholar]
- Gompert Z, Fordyce JA, Forister ML, Shapiro AM, Nice CC. Homoploid hybrid speciation in an extreme habitat. Science. 2006;314:1923–1925. doi: 10.1126/science.1135875. [DOI] [PubMed] [Google Scholar]
- Gottlieb LD. Levels of confidence in the analysis of hybridization in plants. Annals of the Missouri Botanical Garden. 1972;59:435–446. [Google Scholar]
- Harrod RJ, Taylor RJ. Reproduction and pollination biology of Centaurea and Acroptilon species, with emphasis on C. diffusa. Northwest Science. 1995;69:97–105. [Google Scholar]
- Hufbauer RA, Sforza R. Multiple introductions of two invasive Centaurea taxa inferred from cpDNA haplotypes. Diversity and Distributions. 2008;14:252–261. [Google Scholar]
- Kronforst MR, Young LG, Blume LM, Gilbert LE. Multilocus analyses of admixture and introgression among hybridizing Heliconius butterflies. Evolution. 2006;60:1254–1268. [PubMed] [Google Scholar]
- Lacey JR, Marlow CB, Lane JR. Influence of spotted knapweed (Centaurea maculosa) on surface water runoff and sediment yield. Weed Technology. 1989;3:627–631. [Google Scholar]
- Locken LJ, Kelsey RG. Cnicin concentrations in Centaurea maculosa, spotted knapweed. Biochemical Systematics and Ecology. 1987;15:313–320. [Google Scholar]
- Löve A. IOPB Chromosome Number Reports LIX. Taxon. 1978;27:53–61. [Google Scholar]
- Löve A. IOPB Chromosome Number Reports LXIV. Taxon. 1979;28:391–408. [Google Scholar]
- Marrs RA, Sforza R, Hufbauer RA. When invasion increases population genetic structure: a study with Centaurea diffusa. Biological Invasions. 2008a;10:561–572. [Google Scholar]
- Marrs RA, Sforza R, Hufbauer RA. Evidence for multiple introductions of Centaurea stoebe micranthos (spotted knapweed, Asteraceae) to North America. Molecular Ecology. 2008b;17:4197–4208. doi: 10.1111/j.1365-294x.2008.03903.x. [DOI] [PubMed] [Google Scholar]
- Meudt RM, Clarke AC. Almost forgotten or latest practice? AFLP applications, analyses, and advances. Trends in Plant Science. 2007;12:106–117. doi: 10.1016/j.tplants.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Moore RJ, Frankton C. Cytotaxonomy of three species of Centaurea adventive in Canada. Canadian Journal of Botany. 1954;32:182–186. [Google Scholar]
- Müller H. Growth pattern of diploid and tetraploid spotted knapweed, Centaurea maculosa Lam (Compositae), and effects of the root-mining moth Agapeta zoegana (L.) (Lep.:Cochylidae) Weed Research. 1989;29:103–111. [Google Scholar]
- O'Hanlon PC, Peakall R, Briese DT. Amplified fragment length polymorphism (AFLP) reveals introgression in weedy Onopordum thistles: hybridization and invasion. Molecular Ecology. 1999;8:1239–1246. doi: 10.1046/j.1365-294x.1999.00646.x. [DOI] [PubMed] [Google Scholar]
- Ochsmann J. Ein bestand von Cenaurea xpsammogena Gáyer (Centaurea diffusa Lam. x Centaurea stoebe L.) am NSG Sonnenstein (Thüringen) Florist. Rundbr. 1998;31:118–125. [Google Scholar]
- Ochsmann J. Chromosomenzahlen einiger europäischer Centaurea-Sippen. Haussknechtia. 1999;7:59–65. [Google Scholar]
- Ochsmann J. Morphologische und molekularsystematische Untersuchungen an der Centaurea stoebe L.-Gruppe (Asteraceae-Cardueae) in Europa. Diss. Bot. 2000;324:242. (Ph.D. Dissertation) [Google Scholar]
- Ochsmann J. An overlooked hybrid in North America: Centaurea x psammogena Gáyer (diffuse knapweed x spotted knapweed) In: Smith L, editor. The First International Symposium of the Twenty-First Century. Idaho: Coeur d'Alene; 2001a. p. 76. (oral presentation abstract) [Google Scholar]
- Ochsmann J. On the taxonomy of spotted knapweed (Centaurea stoebe L.) In: Smith L, editor. The First International Knapweed Symposium of the Twenty-First Century. ID: Couer d'Alene; 2001b. pp. 33–41. [Google Scholar]
- Ortega YK, McKelvey KS, Six DL. Invasion of an exotic forb impacts reproductive success and site fidelity of a migratory songbird. Oecologia. 2006;149:340–351. doi: 10.1007/s00442-006-0438-8. [DOI] [PubMed] [Google Scholar]
- Price MV, Waser NM. Pollen dispersal and optimum outcrossing in Delphinium nelsoni. Nature. 1979;277:294–297. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DH, Frankham R. Correlation between fitness and genetic diversity. Conservation Biology. 2003;17:230–237. [Google Scholar]
- Rees NE, Quimby PC, Jr, Piper GL, Coombs EM, Turner CE, Spencer NR, Knutson LV. Biological Control of Weeds in the West. Western Society of Weed Sceince, USDA-ARS. Bozeman, MT: Montana State University; 1996. [Google Scholar]
- Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, Nakazato T, Durphy JL, et al. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;301:1211–1216. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Kim SC, Randell RA, et al. Hybridization and the colonization of novel habitats by annual sunflowers. Genetica. 2007;129:149–165. doi: 10.1007/s10709-006-9011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roché BF, Roché CT. Identification, introduction, distribution, and economics of Centaurea species. In: James LF, Evans JO, Ralphs MH, Child RD, editors. Noxious Range Weeds. Boulder, CO: Westview Press; 1991. pp. 274–291. [Google Scholar]
- Sheley RL, Olson BE, Larson LL. Effect of weed seed rate and grass defoliation level on diffuse knapweed seedlings. Journal of Range Management. 1997;50:39–43. [Google Scholar]
- Sheley RL, Jacobs JS, Carpinelli ML. Spotted knapweed. In: Sheley RL, Petroff JK, editors. Biology and Management of Noxious Rangeland Weeds. Corvallis, OR: Oregon State University Press; 1999. pp. 350–361. [Google Scholar]
- Stebbins GL. The role of hybridization in evolution. Proceeding of the American Philosophical Society. 1959;103:231–251. [Google Scholar]
- Thompson JD. The biology of an invasive plant. BioScience. 1991;41:393–401. [Google Scholar]
- Treier UA, Broennimann O, Normand S, Guisan A, Schaffner U, Steinger T, Müller-Schärer H. Shift in cytotype frequency and niche space in the invasive plant Centaurea maculosa. Ecology. 2009;90:1366–1377. doi: 10.1890/08-0420.1. [DOI] [PubMed] [Google Scholar]
- Tyser RW, Key CH. Spotted knapweed in natural areas fescue grasslands – an ecological assessment. Northwest Science. 1988;62:151–160. [Google Scholar]
- Vos P, Hogers R, Bleeker M, et al. A new technique for DNA-fingerprinting. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson AK, Renney AJ. The biology of Canadian weeds: Centaurea diffusa and C. maculosa. Canadian Journal of Plant Science. 1974;54:687–701. [Google Scholar]


