Abstract
The incidence of hybridization between coastal cutthroat (Oncorhynchus clarki clarki) and rainbow trout (Oncorhynchus mykiss) varies widely among populations. The breakdown of reproductive isolation is of concern to managers, and raises the question: how have the two species retained their genetic and morphological divergence? Using a combination of mitochondrial DNA and nuclear DNA markers coupled with watershed attribute and disturbance data, we determined the distribution and frequency of trout hybridization on Vancouver Island, BC and the environmental factors associated with the hybridization. We found 284 hybrids (among 1004 fish) in 29 of 36 sampled populations. High variation in levels of hybridization was observed among populations, and no single environmental factor was found to dominate in determining hybridization levels. However, logging activity, urban infrastructure development, and stocking of hatchery rainbow trout played significant roles in determining hybridization levels, and populations in small watersheds are more at risk of reproductive barrier breakdown. This study illustrates that cutthroat–rainbow trout reproductive barrier breakdown is widespread on Vancouver Island and that anthropogenic disturbance plays a role in the process. As similar environmental disturbance is common in much of coastal trout habitat, large-scale hybridization may be occurring elsewhere and thus may represent a critical management issue for Pacific trout species.
Keywords: anthropogenic, forest harvesting, GIS, hybrid, reproductive isolation, trout
Introduction
Many forms of reproductive barriers have been postulated to contribute to maintaining species integrity. The best documented reproductive isolating mechanisms are physical barriers; however, other reproductive isolating mechanisms (i.e., temporal, behavioral, ecological, and/or genetic) are also known to have evolved to maintain species boundaries (e.g., Mallet 2005; Butlin et al. 2008). For example, species that have the potential to inter-breed (i.e., are sympatric) may exhibit prezygotic reproductive barriers, due to the effects of reinforcement (see Coyne and Orr 2004; Mallet 2005). The nature and strength of the various reproductive isolating mechanisms in nature have been shown to vary widely among taxa (Coyne and Orr 2004; Mallet 2005). Thus, systems where reproductive barriers have failed, and hybridization results, provide natural experiments that allow a better understanding of the evolution of reproductive isolation and the conservation consequences of its erosion.
Areas of hybridization are usually spatially limited (hybrid zones), and the underlying causes of the variation in the magnitude and distribution of reproductive barrier breakdown are typically not well understood (e.g., Nolte et al. 2006). Although generalizations are difficult to make, anthropogenic change, or disturbance, appear as common themes in studies examining the causes of hybridization (e.g., Docker et al. 2003; Lamont et al. 2003; Schwarz and McPheron 2007; Keller et al. 2008). However, there are other published studies where anthropogenic factors were considered, yet no significant effects were found (e.g., Mallet 2005; Williams et al. 2007). The form of the environmental disturbance varies; in plants, physical disturbance such as roadways or building sites may foster hybridization (e.g., Lamont et al. 2003), while in animals the introduction of non-native species is often implicated (Grosholz 2002; Rubidge and Taylor 2005; Schwarz and McPheron 2007). Few studies have attempted to partition the relative contribution of various factors that contribute to the erosion of reproductive isolation between sympatric species.
Hybridization occurs frequently among fish taxa, perhaps more often than in any other vertebrate group (Allendorf and Waples 1996). Several factors have been hypothesized as contributing to the high incidence of hybridization in fish including: (i) weak behavioral isolating mechanisms; (ii) external fertilization; (iii) unequal species abundance among parental taxa; (iv) competition for limited spawning habitat; and (v) loss of habitat complexity (Hubbs 1955; Campton 1987; Scribner et al. 2001). Hybridization is common in most major lineages of salmonids (Taylor 2004), and has been observed in all genera (Salmo, Verspoor 1988; Coregonus, Lu and Bernatchez 1998; Salvelinus, Baxter et al. 1997; Redenbach and Taylor 2004; Oncorhynchus, Dowling and Child 1992; Rosenfield et al. 2000; Rubidge et al. 2001; Docker et al. 2003), although many species in the genus Oncorhynchus are not reported to hybridize. In some cases, salmonid species have been shown to maintain their genetic integrity in the face of hybridization. For example, mating between naturally sympatric bull trout (Salvelinus confluentus) and Dolly Varden (Salvelinus malma) has been documented (and evidence exists for ancient hybridization), yet the two taxa have maintained species status (Baxter et al. 1997). Similarly, hybridization has been reported between bull trout and introduced brook trout (Salvelinus fontinalis); however, reduced survival and fertility in hybrids has limited levels of introgression (Kanda et al. 2002).
Cutthroat (Oncorhynchus clarki spp.) and rainbow trout (Oncorhynchus mykiss spp.) diverged from a common ancestor approximately 2 million years ago (Behnke 1992) allowing for considerable genetic (Leary et al. 1987), chromosomal (Gold 1977), and morphological (Behnke 1992) differences to accumulate. Western North American trout species of the genus Oncorhynchus have since evolved into several subspecies within the cutthroat and rainbow trout. Most of those subspecies of trout evolved in allopatry (Young et al. 2001), and thus stocking of non-native rainbow trout has resulted in extensive hybridization between cutthroat and rainbow trout (e.g., Rubidge et al. 2001; Campbell et al. 2002; Boyer et al. 2008; Metcalf et al. 2008). In some instances, hybrid swarms have been documented (Forbes and Allendorf 1991; Bettles et al. 2005) and hybridization has been specifically recognized as the driving force for the extinction of one subspecies of cutthroat trout (Gyllensten et al. 1985; Bartley and Gall 1991). Unlike many of the inland trout subspecies, the distribution of coastal cutthroat (O. clarki clarki) and coastal rainbow/steelhead trout (O. mykiss irideus) has a long history of sympatry, with over 10 000 years of co-occurrence (i.e., since the last glaciation; Behnke 1992). The long-standing reproductive isolation is thought to be due to spatial and temporal differences in spawning behavior (Young et al. 2001; Williams et al. 2007). Campton and Utter (1985) first reported genetic evidence of hybridization between coastal cutthroat and rainbow trout in two streams in Washington State, USA. Since then, coastal cutthroat and rainbow/steelhead trout have been shown to hybridize across their sympatric range and, in some cases, at very high levels (Baker et al. 2002; Docker et al. 2003; Bettles et al. 2005; Williams et al. 2007).
Hybridization between sympatric coastal cutthroat and rainbow/steelhead trout is widespread (Williams et al. 2007); however, neither the magnitude of the introgression, nor the factors contributing to the loss of reproductive isolation are well characterized. Thus, there are two principal goals of this study. The first is to investigate the distribution and frequency of hybridization between sympatric coastal cutthroat and coastal rainbow trout on Vancouver Island, British Columbia. A broad range of hybridization is expected (Docker et al. 2003; Bettles et al. 2005), both in incidence and geographic extent. The second objective is to test quantitatively for anthropogenic disturbance and watershed-level/ecological factor effects on the incidence of hybridization. Based on previous work, we expect that anthropogenic disturbance through urbanization, recreational access (roads), fishery management actions, or logging activity will contribute to the incidence and distribution of trout hybridization on Vancouver Island. Natural attributes of the river/stream systems are also expected to play a role in the breakdown of reproductive isolation in the coastal rainbow and cutthroat trout, and are also included in our models. Our multivariate stepwise regression models showed that primarily anthropogenic factors contribute to hybridization between naturally sympatric trout species in the more than 30 streams sampled. Our analyses provide new insight into the relative roles of disturbance versus natural factors driving reproductive barrier breakdown between two closely related trout species. Our analyses emphasize the need for conservation, management and ecological efforts in systems with sympatric sibling species subject to elevated levels of disturbance.
Materials and methods
Study location and species
Streams on Vancouver Island generally flow out from interior lakes and snowpacks to the ocean. Stream flow commonly peaks during winter months, with low flows during the summer and fall. Approximately half of the forest cover on Vancouver Island is reported as old growth forest (>140 years old), found primarily in higher elevation and more remote western and northern locations. Resident freshwater and anadromous fish populations in Vancouver Island streams are extensive, and are particularly dependent on the forest ecosystems for survival at all life-history stages (Porter et al. 2000). Past and present human activities have resulted in substantial impacts on salmonid spawning and rearing habitats, and the decline of several native salmonid populations has been attributed to anthropogenic effects (Slaney et al. 1996; Porter et al. 2000).
Coastal rainbow and coastal cutthroat trout are both native to the Pacific coast drainages of North America. The native range of coastal rainbow trout ranges from Baja California to southwest Alaska, while coastal cutthroat's native range is somewhat more limited extending from northern California to southeastern Alaska (Behnke 1992; Trotter 1997). Both species have anadromous and resident freshwater life histories; anadromous coastal rainbow trout are specifically referred to as steelhead while anadromous cutthroat trout are referred to as sea-run cutthroat trout. Steelhead trout generally spawn in late winter to early spring (February–April; Pearcy et al. 1990) using primarily deep, fast water of larger rivers. Resident freshwater coastal rainbow trout generally spawn during a similar timeframe as steelhead (February–May) in small to moderately large (but shallow) streams and rivers. Sea-run coastal cutthroat trout return to freshwater in late fall to early winter (i.e., October–December), feed over the winter, and spawn mid/late winter to early spring (January–May; Trotter 1989) depending on locale. Mature resident freshwater cutthroat trout spawn during the same time period as their anadromous counterpart, and both life-history types prefer to utilize smaller headwater streams for spawning (Trotter 1989). Hartman and Gill (1968) reported that where cutthroat and coastal rainbow/steelhead were sympatric, juvenile cutthroat were predominant in headwater tributaries and rainbow/steelhead juveniles in larger river reaches. It has been postulated, however, that habitat preferences for cutthroat and coastal rainbow/steelhead trout may overlap (Campton and Utter 1985).
Sample collection
Samples were collected from 36 streams thought to harbor sympatric populations of coastal cutthroat and rainbow/steelhead trout on Vancouver Island (Fig. 1). All fish were collected during early/mid summer 2002 (22 June–30 July) and 2003 (20 June–7 July) using a 2-pass backpack electroshocking technique. Captured fish were anaesthetized using a mixture of clove oil and stream water (10–15 ppm), fin clips were collected and stored in 95% ethanol (28–44 individuals per site), and fish were released back to sites from which they were collected. All sample locations were recorded in the field using a global positioning system (GPS) to locate accurately sampling sites within specific Vancouver Island watersheds for eventual use in a geographic information system (GIS).
Figure 1.
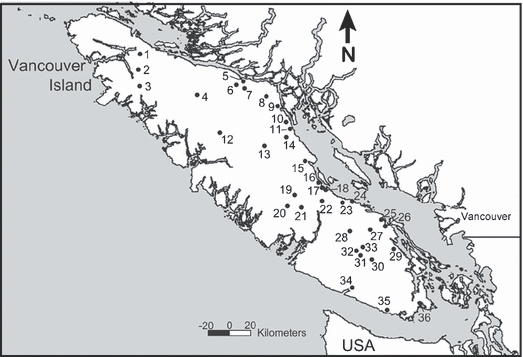
Map of Vancouver Island, British Columbia showing the locations of all streams sampled for coastal rainbow and coastal cutthroat trout and their hybrids. Stream identification numbers correspond to Map ID values in Table 3.
Species markers
Seven polymerase chain reaction (PCR)-based nuclear and one mitochondrial DNA (mtDNA) markers diagnostic for coastal cutthroat and rainbow trout were used in this study. Five of these nuclear loci (GH2D; GTH II-B; IGF-2; Ikaros; RAG) were developed and validated as diagnostic for coastal cutthroat and rainbow trout by Baker et al. (2002). Two additional nuclear species markers based on restriction fragment length polymorphisms (RFLPs; GH1D and Tfex3–5) were developed and validated in Bettles et al. (2005). The mtDNA marker (ND3) was developed and validated as diagnostic by Docker et al. (2003). These species-specific RFLPs (nuclear and mitochondrial) and size polymorphism (GH2D) were further validated as diagnostic using 30 allopatric rainbow and 30 allopatric coastal cutthroat trout taken from several coastal British Columbia populations. These validation runs were in addition to the tests performed by the original authors for the published species markers (see Table 1).
Table 1.
Species identification genetic markers used in this study to characterize the hybridization status of trout collected in streams on Vancouver Island. All markers are nuclear, except ND3 (mitochondrial), and all but GH2D are restriction fragment length polymorphisms. Diagnostic fragment size refers to the band size variation used to identify rainbow trout (RBT) and cutthroat trout (CTT)-specific alleles
| Fragment size (bp) | |||||
|---|---|---|---|---|---|
| Locus name | Annealing Temp. (°C) | Restriction enzyme | PCR fragment size (bp) | RBT | CTT |
| GH2D* | 55 | N/A | 1305/1100 | 1305 | 1100 |
| GTH II-B* | 55 | BglII | 1619 | 1619 | 1050/569 |
| IGF-2* | 62 | BstNI | 922 | 922 | 600/322 |
| Ikaros* | 49 | HinfI | 813 | 813 | 608/205 |
| RAG* | 57 | DdeI | 1013 | 600/240/173 | 600/413 |
| TFex 3-5† | 63 | NciI | 1634 | 917/717 | 717/487/430 |
| GH1D† | 58 | MboI | 1375 | 985/390 | 1375 |
| ND3‡ | 53 | HaeIII | 320 | 320 | 270/50 |
Molecular analysis
Extraction of DNA from fin clips was conducted using the Wizard DNA Purification Kit (Promega Corp., Madison, WI, USA) following manufacturer's instructions. PCR conditions for each genetic marker were in standard 25 μL reactions consisting of 10 mm Tris–HCl (pH-8.4) 50 mm KCl, 2.5 mm MgCl2, 200 μm dNTPs, 0.05 μg of each primer, 0.5 units of DNA Taq polymerase, and approximately 100 ηg of genomic DNA template. PCR conditions consisted of a ‘hot-start’ with a 2-min denaturation (94°C), followed by 35–40 cycles of 1-min denaturation (94°C), 1-min annealing, 1.5-min extension (72°C), and ending with a final 5-min extension cycle (72°C). Five microliters of PCR product was digested for 6 h in a 10-μL reaction mix containing ddH2O (3.5 μL), enzyme optimizing buffer (1 μL), restriction enzyme (0.25 μL), and BSE (0.25 μL). One marker was based on a size polymorphism (GH2D), which was visualized directly after PCR. For specific annealing temperatures and restriction enzymes used, refer to Table 1. Species-specific polymorphisms were visualized and scored on agarose gels, and banding patterns that were ambiguous were repeated to confirm their genotype.
Hybrid distribution and frequency
All fish were genotyped as homozygous rainbow trout, homozygous cutthroat trout, or heterozygous, at each of the seven nuclear loci. We tested for departures from Hardy–Weinberg equilibrium (binomial distribution) using Haldane's (1954) exact test for randomness of mating. mtDNA haplotypes were identified as cutthroat or rainbow trout for all fish. Fish that agreed at all seven nuclear and the mitochondrial markers were classed as ‘pure’ types, while all other fish were identified as various levels of introgression. Backcross (F1 × pure-type cross) and subsequent higher-order hybrid categories have been combined in our analyses as the chances of misidentifying backcross versus higher-order hybrid genotypes, even with seven co-dominant loci, are high (Boecklen and Howard 1997). It is likely that some higher-order hybrids have been misidentified as pure-type. Specifically, Boecklen and Howard (1997) estimated an approximate 12% error rate in identifying the second backcross generation individuals with seven co-dominant species markers. However, our error rate is likely lower than that estimate, as we applied a mtDNA species marker. Additionally, Boecklen and Howard's (1997) model only permitted unidirectional backcross events with pure-type parental fish – an assumption almost certainly incorrect in our study.
Population hybridization levels were quantified in each sample population using two statistics: (i) ‘Hybridization Index’ (HI), which is the percent frequency of introgressed fish in a population (regardless of the nature of the introgression in each fish), and (ii) ‘Genome Mixing Index’ (GMI), which is a measure of the level of mixing of the two species’ genomes in individual fish and was calculated as;
| (1) |
where ARare is the total number of rare nuclear species alleles scored in the fish (i.e., <7 alleles) and AT is the total number of alleles in the fish (AT is constant for our data; 14 alleles). The GMI index thus ranges from zero to one, with pure-species individuals at GMI = 0%, and F1 hybrids at GMI = 100%. The GMI metric reflects not only the proportion of mixed ancestry individuals, but also the level of mixing (i.e., maximum mixing is an F1 hybrid, with 50% of each genome). We then calculated mean GMI for each population. This approach differs from other indices used in hybridization studies of cutthroat and rainbow trout (e.g., Hitt et al. 2003) where emphasis was placed on quantifying the introgression of introduced species alleles (e.g., introduced rainbow trout) into native species genomes (e.g., inland cutthroat trout). As both coastal cutthroat and rainbow/steelhead trout are native to our sample locations, our GMI index reflects bi-directional introgression into either species. To assess the possibility that the direction of introgression may affect the level of genome mixing (i.e., is there a bias in the direction of hybridization?), we calculated the percent frequency of RBT alleles in all hybridized fish, and included the mean RBT allele frequency for each population as a variable in the analysis of environmental effects.
Environmental effects
Environmental data were collected on the watershed scale, as watershed-level assessments have been shown to have predictive capability for evaluating relative environmental and anthropogenic effects on resident fish populations (e.g., Roth et al. 1996; Feist et al. 2003; Regetz 2003).
Watershed data for British Columbia are in a province-wide GIS database, which holds extensive baseline information, particularly for variables pertaining to the effects of logging (BC Watershed Statistics data dictionary, http://ilmbwww.gov.bc.ca/risc/). The study watershed data were extracted from a provincial database in ArcMap (ArcGIS Version 8.1; ESRI, Redlands, CA, USA). GPS coordinates obtained for all sample locations in the field allowed precise identification of stream sample locations within their respective watersheds.
We chose environmental variables to be assessed for correlation with hybrid levels measures based on current understanding of habitat factors deemed important for western North American trout. The environmental variables included fall into two broad categories: (i) stream or ecological attributes and (ii) anthropogenic disturbance. A total of 15 variables were selected for inclusion in the analyses, with six relating to landscape or ecological attributes (see Table 2). The remaining nine are related to various forms of anthropological disturbance, including; logging (four variables; Table 2), urban and road development (four variables; Table 2), and rainbow trout stocking (one variable; Table 2). Our choice of specific variables to include in the analysis was justified based on literature (e.g., Hartman et al. 1996) that identifies logging impacts on streams based on recent logging (i.e., 3–20 years) and long-term logging (i.e., 20–140 years) effects. Data were extracted for each watershed from the GIS watershed database for Vancouver Island, and from the BC government FishWizard website (http://pisces.env.gov.bc.ca). Some variables were transformed or were calculated based on available data; Table 2 provides information on their derivation, and Appendix S1 gives mean values by stream.
Table 2.
List of 15 environmental variables with abbreviation (Abrv) and description included in the analysis of reproductive barrier breakdown in Vancouver Island trout populations. The first six variables relate to stream or ecological attributes, while the remaining nine relate to anthropologic disturbance. Mean values (with range) are across all sampled streams and watersheds
| Environmental variable | Abrv | Mean (range) | Variable description |
|---|---|---|---|
| Watershed area | WA | 85.1 km2 (6.5–390) | Total surface area of watershed containing sample site |
| Stream order | SO | 2.68 (1–5) | Measure of relative stream size [smallest (1) to largest (12)] |
| Mean stream gradient | SG | 2.54% (1.0–11.0) | Total stream elevation rise, divided by total stream length |
| Mean stream discharge | SD | 0.43 m3/s (0.030–1.32) | Mean year-round stream water discharge at mouth |
| Anadromous CTT | aCTT | 0.55 | Presence/absence of sea-run cutthroat trout life history (0 or 1) |
| Anadromous RBT | aRBT | 0.71 | Presence/absence of steelhead trout life history (0 or 1) |
| Total forested area | TFA | 87.9% (30.5–100) | Proportion of watershed surface area forested |
| Young forested area | YFA | 56.1% (18.4–90.4) | Proportion of watershed logged 40–140 years ago |
| Recently logged area | RL | 14.2% (0.0–51.6) | Proportion of watershed logged within the last 20 years |
| Stream length logged | SLL | 14.4% (0.0–56.4) | Proportion of the total stream length logged |
| Road density | RD | 2.04 km/km2 (0.60–4.1) | Total length roads, divided by watershed surface area |
| Urban development | UD | 6.7% (0.0–31.6) | Proportion of watershed area classified as urban |
| Stream crossings | SC | 1.42 #/km2 (0.20–4.3) | Number of stream road crossings, divided by watershed surface area |
| Stream availability | SAv | 73.7% (13.7–100) | Stream length before first impassable barrier, divided by total stream length |
| Trout stocking | TS | 0.42 | History of trout stocking (0 or 1) |
Statistical analysis
All proportional data were arc-sine √ transformed and watershed area was logarithm transformed to correct for normality. We tested for a correlation between our two measures of reproductive barrier breakdown (HI and GMI) to determine the level of overlap between the information contained in the two metrics. We also performed a correlation analysis among all of our environmental variables.
We analyzed our data to identify which variables explained the most variance in the level of hybridization among sampled streams. Our analysis consisted of two approaches: (i) we used forward, backwards and bothways stepwise regression coupled with an analysis of information criteria to identify the environmental variables that significantly contribute to the model explaining variation in hybridization incidence, and (ii) we use individual-variable analyses to examine the nature of specific functional relationships:
Multifactor models
Backwards and bothways stepwise regression gave the same models and thus we refer to the resulting model simply as the bothways model. We compared the forwards and bothways model using the Akaike's information criterion (AIC). Because this metric provides only a qualitative comparison, we also compared the two models using cross-validation (Roff 2006). The approach was as follows: (i) randomly select 20% of the data set to use as the test set; (ii) fit the two best forwards and backwards models using the remaining data (the training set); (iii) for each model, calculate the predicted values for the test set and compute the residual sums of squares,
where 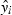 is the predicted value of the ith observation in the test set, yi, using the appropriate model (forwards or bothways); (iv) repeat the foregoing steps 1000 times; and (v) because the paired residual sums of squares come from the same training and test sets, the set of paired residual sums of squares can be compared using a paired t-test.
is the predicted value of the ith observation in the test set, yi, using the appropriate model (forwards or bothways); (iv) repeat the foregoing steps 1000 times; and (v) because the paired residual sums of squares come from the same training and test sets, the set of paired residual sums of squares can be compared using a paired t-test.
Individual factor models
We selected two environmental variables for further investigation of their relationship with the hybrid and introgression levels using a general linear model (SYSTAT® Version 7.01, Chicago, Illinois), and scatterplots. We selected watershed area (WA) as an important landscape variable, and proportion of young forest area within each watershed (YFA) as a good indicator of recent logging activity. We log-transformed WA and arc-sine √ transformed YFA to better approximate normal distributions.
Results
Hybrid distribution and frequency
Six of 36 populations consisted of 100% pure genotypes of only one trout species (i.e., either cutthroat or rainbow/steelhead with no presence of hybrids; Table 3). Consequently, those six populations were excluded from all further analyses because field identification and genetic analyses did not identify a sympatric relationship between the trout species, hence no gene flow between the species was possible.
Table 3.
List of sampled Vancouver Island streams with watershed area (Area), sample size (n) and species (rainbow–cutthroat trout) genotype proportion summary. Map identification (ID) corresponds to those provided in Fig. 1. Total proportions of pure-type fish are given as ‘Pure CTT’– pure cutthroat trout, and ‘Pure RBT’– pure rainbow/steelhead trout. ‘F1 hybrids’, ‘Backcross genotypes’ and ‘HI’ (hybridization index) are proportions of the various hybrid genotypes. The Genome Mixing Index (GMI) is calculated as described in the text. The number of loci that departed from Hardy–Weinberg equilibrium at P < 0.01 [HWE (# loci)] is given for each stream. Streams indicated with an asterisk (*) had only one species present, and were not included in the analyses
| Map ID | Stream name | Area (km2) | n | Pure CTT | Pure RBT | F1 hybrids | Backcross genotypes | HI | GMI | HWE (# loci) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Waukwaas Cr | 29.6 | 37 | 0.00 | 0.97 | 0.03 | 0.00 | 0.03 | 0.03 | 0 |
| 2 | Howlal Cr | 7.9 | 29 | 0.48 | 0.04 | 0.10 | 0.38 | 0.48 | 0.23 | 0 |
| 3 | Marble R trib. | 144.8 | 28 | 0.86 | 0.07 | 0.00 | 0.07 | 0.07 | 0.03 | 5 |
| 4 | Lukwa Cr | 33.0 | 31 | 0.48 | 0.13 | 0.16 | 0.23 | 0.39 | 0.31 | 0 |
| 5 | Bear Bight Cr* | – | 44 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | – |
| 6 | Elk Cr | 58.2 | 33 | 0.94 | 0.00 | 0.03 | 0.03 | 0.06 | 0.03 | 0 |
| 7 | Stowe Cr | 252.3 | 30 | 0.10 | 0.67 | 0.00 | 0.23 | 0.23 | 0.09 | 6 |
| 8 | Roberts Cr | 46.6 | 34 | 0.91 | 0.00 | 0.00 | 0.09 | 0.09 | 0.04 | 0 |
| 9 | Menzies Cr | 21.0 | 30 | 0.43 | 0.00 | 0.00 | 0.57 | 0.57 | 0.30 | 0 |
| 10 | Cold Cr | 6.5 | 30 | 0.27 | 0.37 | 0.36 | 0.00 | 0.36 | 0.37 | 0 |
| 11 | Black Cr* | – | 35 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | – |
| 12 | Nameless Cr | 65.4 | 32 | 0.25 | 0.63 | 0.06 | 0.06 | 0.12 | 0.08 | 7 |
| 13 | Miller Cr* | – | 33 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | – |
| 14 | Woodhus Cr | 34.9 | 30 | 0.00 | 0.90 | 0.00 | 0.10 | 0.10 | 0.04 | 4 |
| 15 | Morrison Cr | 10.3 | 33 | 0.42 | 0.03 | 0.03 | 0.52 | 0.55 | 0.25 | 0 |
| 16 | Cowie CS Cr | 18.9 | 32 | 0.03 | 0.09 | 0.04 | 0.84 | 0.88 | 0.48 | 0 |
| 17 | Rosewall Cr | 35.9 | 27 | 0.00 | 0.96 | 0.00 | 0.04 | 0.04 | 0.02 | 0 |
| 18 | Cook Cr | 27.0 | 32 | 0.13 | 0.47 | 0.22 | 0.18 | 0.40 | 0.26 | 0 |
| 19 | Atluck L. trib.* | – | 37 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | – |
| 20 | Taylor R trib. | 95.5 | 30 | 0.87 | 0.00 | 0.00 | 0.13 | 0.13 | 0.05 | 0 |
| 21 | Friesen Cr | 200.9 | 33 | 0.30 | 0.21 | 0.22 | 0.27 | 0.49 | 0.34 | 1 |
| 22 | Esary Cr | 147.6 | 37 | 0.92 | 0.00 | 0.00 | 0.08 | 0.08 | 0.03 | 0 |
| 23 | Whisky Cr | 96.3 | 36 | 0.81 | 0.08 | 0.03 | 0.08 | 0.11 | 0.11 | 7 |
| 24 | French Cr | 68.1 | 28 | 0.00 | 0.89 | 0.04 | 0.07 | 0.11 | 0.06 | 0 |
| 25 | Millstone R | 99.7 | 35 | 0.37 | 0.26 | 0.31 | 0.06 | 0.37 | 0.35 | 0 |
| 26 | Chase R ‘02 | 37.1 | 35 | 0.00 | 0.14 | 0.06 | 0.80 | 0.86 | 0.54 | 0 |
| 26 | Chase R ‘03 | 37.1 | 37 | 0.00 | 0.19 | 0.02 | 0.79 | 0.81 | 0.41 | 0 |
| 27 | N Nanaimo R | 62.8 | 38 | 0.05 | 0.74 | 0.03 | 0.18 | 0.21 | 0.16 | 0 |
| 28 | Rockyrun Cr | 9.0 | 37 | 0.00 | 0.70 | 0.05 | 0.25 | 0.30 | 0.13 | 0 |
| 29 | Stocking Cr | 37.6 | 32 | 0.00 | 0.81 | 0.00 | 0.19 | 0.19 | 0.04 | 0 |
| 30 | Meade Cr | 42.2 | 30 | 0.47 | 0.03 | .017 | 0.33 | 0.50 | 0.30 | 7 |
| 31 | Misery Cr | 389.7 | 32 | 0.97 | 0.03 | 0.00 | 0.00 | 0.00 | 0.00 | 0 |
| 32 | Wardroper Cr | 389.7 | 34 | 0.97 | 0.00 | 0.00 | 0.03 | 0.03 | 0.02 | 0 |
| 33 | Croft Cr* | – | 35 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | – |
| 34 | Fairy Cr | 13.9 | 31 | 0.06 | 0.87 | 0.00 | 0.07 | 0.07 | 0.05 | 3 |
| 35 | Kirby Cr | 18.7 | 31 | 0.16 | 0.74 | 0.00 | 0.10 | 0.10 | 0.06 | 0 |
| 36 | Colquitz R* | – | 30 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | – |
Across all sample locations, 284 fish (29%) had some level of hybridization (Table 3). First generation (F1) hybrids were least abundant, making up 7% (n = 62; Table 3) of the fish genotyped in this study. Backcross hybrids made up 22% (n = 222; Table 3) of the total number of genotyped fish. Pure coastal cutthroat and rainbow/steelhead consisted of 36% (n = 365) and 35% (n = 355) of the sample, respectively. The Hybrid and Genome Mixing indices were highly correlated (R2 = 0.88; P < 0.0001), with a slope of 0.58. Only seven populations showed evidence for departures from Hardy–Weinberg equilibrium (at P < 0.01; Table 3). Those seven populations departed from Hardy–Weinberg equilibrium at three to seven of the marker loci (Table 3), and in all cases the deviations were due to deficiencies in heterozygote (hybrid) genotypes.
Only one stream (Misery Creek) had no evidence of hybrids despite the presence of both trout species (Table 3). Five populations (Menzies Creek, Morrison Creek, Cowie Cougar-Smith Creek, Chase River, and Meade Creek) demonstrated hybridization levels of 50% or higher, with Cowie Cougar-Smith Creek and Chase River (2002 and 2003) displaying the highest levels (all above 80%; Table 3). Only seven populations (Waukwaas Creek, Marble River tributary, Elk Creek, Roberts Creek, Rosewall Creek, Wardroper Creek, and Fairy Creek) demonstrated hybridization <10% (Table 3). Genome Mixing Index (GMI) values indicate highly variable levels of genetic mixing between the two trout species throughout Vancouver Island (Table 3). Chase River and Cowie Cougar-Smith Creek showed very high mixing levels (Table 3), and the high incidence of mixing coupled with a rarity of pure trout in both species, indicates they may be approaching hybrid swarm status. Since the frequency of pure and hybrid genotypes at all seven loci conform to Hardy–Weinberg equilibrium in those populations (Table 3), the identified pure-type fish are likely valid, and hence the populations do not represent true hybrid swarms.
Environmental factor analysis
Environmental variable correlations
Generally, the selected variables were not highly correlated; five out of 61 pairwise correlations were significant based on post hoc Bonferroni significance, with 9/61 without Bonferroni correction (Table 4). The significant environmental variable correlations are consistent with expectation, for example total forested area is negatively correlated with urban development and road density (Table 4). There are also surprising nonsignificant results, for example stream availability (SAv) was not correlated with any of the other environmental variables (Table 4).
Table 4.
Pearson correlation matrix of pairwise correlation coefficients for all continuous environmental factors used in analyzing the incidence of hybridization between rainbow and cutthroat trout on Vancouver Island. Asterisks indicate significant correlations, while boldface type indicates significance after post hoc Bonferroni correction
| WA | SO | TFA | YFA | RL | SLL | UD | RD | SC | SAv | SG | SD | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Watershed area (WA) | 1.00 | |||||||||||
| Stream order (SO) | −0.55* | 1.00 | ||||||||||
| Total forested area (TFA) | −0.04 | 0.18 | 1.00 | |||||||||
| Young forested area (YFA) | −0.01 | 0.04 | 0.04 | 1.00 | ||||||||
| Recently logged area (RL) | −0.02 | 0.21 | 0.14 | −0.47* | 1.00 | |||||||
| Stream length logged (SLL) | −0.04 | 0.19 | 0.04 | −0.54* | 0.97* | 1.00 | ||||||
| Urban development (UD) | 0.01 | −0.12 | −0.89* | 0.03 | −0.26 | −0.17 | 1.00 | |||||
| Road density (RD) | −0.05 | 0.00 | −0.60* | 0.22 | −0.18 | −0.13 | 0.83* | 1.00 | ||||
| Stream crossings (SC) | −0.10 | 0.15 | 0.14 | −0.22 | 0.39* | 0.36* | −0.14 | 0.00 | 1.00 | |||
| Stream availability (SAv) | −0.24 | 0.12 | 0.25 | −0.17 | 0.20 | 0.23 | −0.28 | −0.23 | 0.24 | 1.00 | ||
| Mean stream gradient (SG) | 0.07 | 0.12 | 0.26 | 0.06 | −0.03 | −0.08 | −0.29 | −0.23 | 0.29 | 0.13 | 1.00 | |
| Mean stream discharge (SD) | 0.08 | 0.02 | 0.28 | 0.06 | −0.09 | −0.15 | −0.30 | −0.30 | −0.09 | −0.05 | 0.09 | 1.00 |
Stepwise regression
The forward stepwise regression model for Genome Mixing (GMI) included all environmental variables, and resulted in a three factor model (SLL, WA and SAv), while the bothways stepwise regression model resulted in an eight factor model (Table 5). The AIC values for the forward and bothways models were −19.5 and −25.9, respectively, and thus the bothways model is preferred using this criterion. Cross-validation also showed that the fit of the bothways model was significantly better than that of the forwards model (t = 7.11, df = 999, P < 0.0001). The bothways stepwise regression model explained 72% of the observed variance in GMI.
Table 5.
Stepwise (bothways) regression results for analysis of the environmental factors contributing to the breakdown of reproductive barriers in Vancouver Island trout populations
| Dependant variable | Factor | Slope | t-Value | P-value | Univariate R |
|---|---|---|---|---|---|
| Hybrid Index (HI) | WA | −0.0018 | −4.44 | 0.0002 | −0.37 |
| TFA | −0.46 | −2.54 | 0.017 | −0.25 | |
| YFA | 0.49 | 2.82 | 0.0092 | 0.42 | |
| SAv | −0.38 | −2.97 | 0.0065 | −0.39 | |
| TS | −0.21 | 2.21 | 0.036 | − | |
| Genome Mixing Index (GMI) | WA | −0.001 | −3.38 | 0.0027 | −0.34 |
| TFA | −0.64 | −2.68 | 0.014 | −0.22 | |
| RL | −0.39 | −2.36 | 0.028 | −0.50 | |
| UD | 1.05 | 2.71 | 0.013 | 0.21 | |
| RD | 0.17 | 3.00 | 0.0066 | 0.35 | |
| SAv | −0.25 | −2.89 | 0.0085 | −0.44 | |
| aRBT | 0.13 | 2.06 | 0.051 | – | |
| TS | −0.12 | 1.78 | 0.088 | – |
‘Factors’ refer to environmental variables with the abbreviations defined in Table 2. Student's t-values and two-tailed probability (P) are given. Factors highlighted in bold were retained in the model for both Hybrid Index (HI) and Genome Mixing Index (GMI). Transformation used: logarithm (WA); arc-sine √ (TFA, YFA, SAv, RL, UD, RD).
The forward stepwise regression model for the Hybrid Index (HI) included all environmental variables and resulted in a three factor model (SLL, WA and SAv), while the bothways stepwise regression model resulted in a five factor model (Table 5). As with the GMI model, the bothways model was selected using both the AIC (5.8 vs 0.1) and cross-validation (t = 2.65, df = 999, P = 0.0081). The bothways stepwise regression model explained 60% of the observed variance in HI.
The inclusion of the mean RBT allele frequency in the multivariate regression model did not change the outcome: the RBT allele frequency variable was not retained in any of the stepwise models, and the AIC did not differ between the model with and without the RBT allele frequency variable included. Examination of the residuals from the multivariate regressions and separate regressions on the dependant variables show no evidence of outliers that could be affecting our results.
Linear regression
We found significant negative log-linear relationships between watershed area (WA) and both the Hybrid and Genome Mixing indices (Fig. 2A,B). The relationships explained 9% (HI) and 14% (GMI) of the observed variation (Fig. 2A,B). We also found a significant positive linear relationship between the arc-sine √ proportion of new forest cover (YFA) and both the Hybrid and Genome Mixing indices (Fig. 2C,D). The relationships explained a somewhat larger component of the observed variation [15% (HI) and 19% (GMI); Fig. 2C,D].
Figure 2.
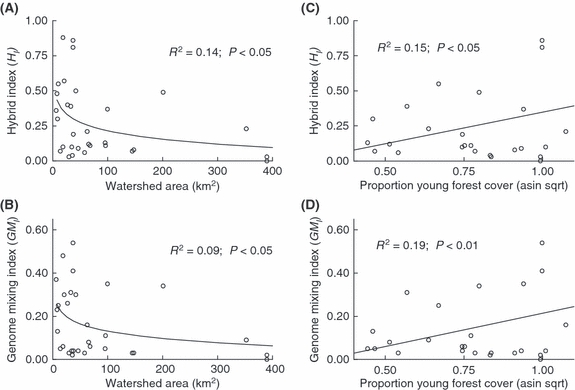
Regression plots of selected environmental variables with Hybrid Index (HI) and Genome Mixing Index (GMI) for 31 sympatric populations of rainbow and cutthroat trout on Vancouver Island. Panels (A) and (B): log-transformed watershed area versus HI and GMI. Panels (C) and (D): arc-sine √ transformed proportion of young forested area per watershed versus HI and GMI.
Discussion
Although many studies have been published examining hybridization between introduced rainbow trout and a variety of cutthroat trout subspecies (e.g., Boyer et al. 2008; Metcalf et al. 2008), substantially less is known of the geographic extent and magnitude of hybridization (and introgression) between sympatric coastal cutthroat and coastal rainbow/steelhead in their native range. This study adds to the growing body of literature that shows that hybridization between the sympatric coastal trout species is widespread and can reach very high levels (e.g., Docker et al. 2003; Ostberg et al. 2004; Williams et al. 2007). Previous studies have documented hybridization between coastal cutthroat trout and native rainbow/steelhead trout in the lower Columbia River (Spruell et al. 1998), in British Columbia, Canada (Docker et al. 2003; Bettles et al. 2005), in Washington State (Ostberg et al. 2004), and in Alaska (Williams et al. 2007). In most cases, the frequency of hybridization or level of introgression among populations experiencing reproductive barrier breakdown was highly variable. Although considerable interest exists in identifying the factors that drive the observed variation in hybrid frequency, so far only a few broad categories of factors have been explored. The most widely reported cause of intraspecific introgression is the introduction of non-native rainbow trout (e.g., Boyer et al. 2008; Metcalf et al. 2008); however, in such cases the native species has had limited opportunity to strengthen reproductive barriers through reinforcement, and thus hybridization is perhaps not surprising. In cases where cutthroat and rainbow trout exist sympatrically, the introduction of rainbow trout of a different origin (usually hatchery-bred and reared) has been shown to accelerate the breakdown of reproductive isolation (Docker et al. 2003). Ecological disturbance, either anthropogenic or natural, has been shown (or speculated) to contribute to loss of reproductive isolation between sympatric species (Lamont et al. 2003; Taylor et al. 2006; Schwarz and McPheron 2007; Keller et al. 2008). However, studies of sympatric trout populations experiencing no obvious disturbance, have shown evidence of substantial levels of hybridization (e.g., Williams et al. 2007). Our study is the first designed to test for the effect of a broad range of environmental and disturbance factors on the observed variation in the magnitude of hybridization in multiple sympatric cutthroat and rainbow trout populations.
Despite obvious associations between habitat disturbance and threatened or endangered species, the relationship between environmental variables and population viability is often difficult to quantify (Feist et al. 2003). For example, identifying relationships between habitat conditions and salmonid demography has proven extremely difficult (e.g., Regetz 2003). However, it appears that reproductive isolation between sympatric sibling species may be particularly sensitive to habitat disturbance (Hubbs 1955). In this study, no single environmental factor dominates as the driving mechanisms of reproductive barrier breakdown, despite a range of environmental variables assayed for over 30 sampled sympatric trout populations. Perhaps as might be expected, our analyses indicate that the loss of reproductive isolation in the Vancouver Island coastal cutthroat and rainbow trout results from the interaction of multiple stressors and ecological processes. Nevertheless, some generalizations can be made that are of relevance to other impacted systems.
Total WA was a consistently significant factor contributing to our models predicting hybridization and introgression: WA was negatively correlated with elevated levels of hybridization and genome mixing. The frequency of hybridization between coastal cutthroat and coastal rainbow/steelhead trout was higher in smaller watersheds irrespective of the effects of the other environmental factors included in the models. As watershed size, by itself, is unlikely to affect the reproductive isolation directly, it probably reflects some other, not measured, property of the environment that does influence hybridization. The relationship does not appear to be due to a watershed location bias, as the small watersheds examined in this study were distributed uniformly throughout the sampled area. Furthermore, the only environmental factor significantly correlated with WA was stream order (SO), thus the contribution of WA to the incidence of hybridization is not due to autocorrelational effects with other measured environmental variables. It may be that smaller watersheds, in general, experience greater cumulative environmental effects, due to their relative paucity of buffering capacity (e.g., Walling 1999) when disturbed. Such variability could be related to stronger terrestrial linkages and hydrological instability associated with smaller streams. Furthermore, Rosenfeld et al. (2002) pointed out that small watersheds have been viewed by planners and resource managers as having poor fisheries value, and thus may have been excluded from specific protection during resource extraction. Finally, it could simply be that small watersheds have less habitat and smaller trout populations; therefore hybridization would be more likely due to reduced mate choice. Independent of the mechanism behind the correlation, smaller watersheds should be treated with caution since they appear to magnify disturbance effects.
In general, anthropogenic disturbance appears to be the dominant factor in our models predicting introgression and hybridization. We found that variables reflecting logging (e.g., proportion of total forested area, recent logging activity, and newly regenerated forest area), urban development and road density, and fishery management practices (rainbow trout stocking) significantly contributed to our models. Other studies of reproductive barrier breakdown have implicated anthropogenic disturbance in the process (e.g., Allendorf et al. 2001); however, few studies have used empirical approaches to address the specific role of disturbance in hybridization.
Logging practices clearly play a role in the breakdown of reproductive barriers between coastal cutthroat and coastal rainbow/steelhead trout on Vancouver Island. The slope of the relationships between genome mixing and hybridization and total forested area (TFA) was negative, indicating that watersheds with more forested area generally have lower incidence of hybridization and lower genome mixing levels. This is consistent with the positive slope between YFA and hybridization, and is indicative that even after substantial recovery time, the watersheds are affected by the change in forest type. This result is perhaps not surprising given the fact that logging activities have previously been correlated with population reduction in other Pacific salmonids (e.g., Slaney et al. 1996; Porter et al. 2000; Deschênes et al. 2007). Interestingly, recent logging activity (RL) was negatively correlated with genome mixing, indicating that the increased light and sediment load associated with logging activity is associated with lower levels of GMI (although it was not a significant factor for overall hybridization levels). Thus, our analyses indicate that the long-term effects of logging, more so than recent logging effects, erode reproductive isolation between the sympatric rainbow and cutthroat trout on Vancouver Island. The persistent long-term effects of erosion and transport of sediment over several decades may be driving the pattern of hybridization, as medium-term increased sediment load into streams has been shown to reduce critical spawning habitat for salmonids. Alternatively, recovering forests may provide different temperature and nutrient environments (Holtby 1988; Hartman et al. 1996). Deschênes et al. (2007) showed that the effects of forest-related variables on the density and abundance of juvenile Atlantic salmon (Salmo salar), were critically dependant on spatial scale. Our analyses are all at the watershed scale, and the effects of active logging might be quite localized.
Bettles et al. (2005) reported that sympatric populations of coastal rainbow and cutthroat trout in the Chase River constituted a ‘hybrid swarm’ (Allendorf et al. 2001). As the Chase River runs through the city of Nanaimo on Vancouver Island, it would be tempting to conclude that the complete loss of reproductive isolation in those populations was a result of the multifarious habitat disturbances associated with urbanization. However, to our knowledge, no previous study has empirically examined the contribution of urban development to the incidence of hybridization in native sympatric species. We found that road density (RD) and urban development (UD) were positively correlated with genome mixing. Reproductive isolation breakdown may be facilitated in high road density urban areas due to culvert and other stream barriers associated with road crossings; however, we found no significant effect of the actual number of stream crossings associated with the study streams. Urbanization and road density may also serve as a proxy for contaminant runoff that may affect reproductive behaviors (Jones and Reynolds 1997), and ultimately conspecific recognition and hybridization avoidance (Fisher et al. 2006). Finally, although we did not have data for the relative exploitation levels (fishing pressure) in the sampled streams, it would seem likely that elevated urbanization and road access (i.e., road density) would correlate with fishing pressure. Hence, fishing pressure may indirectly contribute to the loss of reproductive isolation in the Vancouver Island rainbow and cutthroat trout, especially in streams where hatchery fish are stocked (e.g., Evans and Willox 1991).
Docker et al. (2003) found that the frequency of hybridization and introgression was significantly higher in systems where hatchery rainbow trout were introduced. A similar effect is seen when rainbow trout are introduced into allopatric populations of cutthroat trout (e.g., Rubidge et al. 2001; Boyer et al. 2008; Metcalf et al. 2008). We found that rainbow trout stocking was associated with elevated hybridization and genome mixing in our study streams; however, the effect size was small. We do not know if the elevated level of hybridization observed in the hatchery-stocked populations is due to mating between the stocked rainbow trout and native cutthroat trout, or if the presence of the non-native rainbow trout facilitates the breakdown of reproductive barriers between the native fish species. Given the strong published evidence that the introduction of rainbow trout into either sympatric rainbow/cutthroat trout populations or allopatric cutthroat trout populations leads to hybridization, it may be that the introduced trout are directly involved in the intraspecific breeding and subsequent reproductive barrier breakdown on Vancouver Island.
This study provides additional evidence for the widespread and substantial loss of reproductive isolation between sympatric rainbow and cutthroat trout on the west coast of North America. Although surveying levels of hybridization between sibling species is important for management and conservation (Allendorf et al. 2001), our analysis of variation in the incidence of hybridization also provides a powerful tool to detect and characterize factors affecting evolutionary processes (Dowling and Secor 1997). Although sympatric species may experience a loss of reproductive barriers in situations with no identified disturbance or stressor (e.g., Williams et al. 2007), it begs the question of how did the coastal cutthroat and rainbow trout develop and maintain their genetic and morphological divergence? In our study, we identify primarily anthropogenic disturbance as contributing to the loss of reproductive isolation; however, factors not directly associated with human activities (i.e., watershed area and the presence of anadromous life histories) were also significant. This study highlights the value of examining evolutionary processes and patterns as a bellwether for population or ecosystem changes of conservation concern.
Acknowledgments
We thank B. Dufour for his help in the laboratory and field, and D. Minty for providing GIS data for Vancouver Island watersheds. G. Reid, L. Carswell, and B. Hooton provided valuable field advice and guidance. P. Graniero provided valuable GIS support and advice. This work was supported by BC Habitat Conservation Trust Fund, the Natural Sciences and Engineering Research Council (NSERC) of Canada, and Yellow Island Aquaculture Ltd.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Appendix S1. List of sampled Vancouver Island stream variables used in the analysis of factors contributing to hybridization between native coastal cutthroat and coastal rainbow trout.
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Literature cited
- Allendorf FW, Waples RS. Conservation genetics of salmonid fishes. In: Avise JC, Hamrick JL, editors. Conservation Genetics: Case Histories from Nature. New York, NY, USA: Chapman and Hall; 1996. pp. 238–280. [Google Scholar]
- Allendorf FW, Leary RF, Spruell P, Wenburg JK. The problems with hybrids: setting conservation guidelines. Trends in Ecology and Evolution. 2001;16:613–622. [Google Scholar]
- Baker J, Bentzen P, Moran P. Molecular markers distinguish coastal cutthroat trout from coastal rainbow trout/steelhead and their hybrids. Transactions of the American Fisheries Society. 2002;131:404–417. [Google Scholar]
- Bartley DM, Gall GAE. Genetic identification of native cutthroat (Oncorhynchus clarki) and introgressive hybridization with introduced rainbow trout (O. mykiss) in streams associated with the Alvord basin, Oregon and Nevada. Copeia. 1991;1991:854–859. [Google Scholar]
- Baxter JS, Taylor EB, Devlin RH, Hagen J, McPhail JD. Evidence for natural hybridization between Dolly Varden (Salvelinus malma) and bull trout (S. confluentus) in a northcentral British Columbia watershed. Canadian Journal of Fisheries and Aquatic Science. 1997;54:421–429. [Google Scholar]
- Behnke RJ. Native Trout of Western North America. Bethesda, MD: American Fisheries Society; 1992. Monograph 6. [Google Scholar]
- Bettles CM, Docker MF, Dufour B, Heath DD. Hybridization dynamics between sympatric species of trout: loss of reproductive isolation. Journal of Evolutionary Biology. 2005;18:1220–1233. doi: 10.1111/j.1420-9101.2005.00935.x. [DOI] [PubMed] [Google Scholar]
- Boecklen WJ, Howard DJ. Genetic analysis of hybrid zones: numbers of markers and power of resolution. Ecology. 1997;78:2611–2616. [Google Scholar]
- Boyer MC, Muhlfeld CC, Allendorf FW. Rainbow trout (Oncorhynchus mykiss) invasion and the spread of hybridization with native westslope cutthroat trout (Oncorhynchus clarki lewisi. Canadian Journal of Fisheries and Aquatic Science. 2008;65:658–669. [Google Scholar]
- Butlin RK, Galindo J, Grahame JW. Sympatric, parapatric or allopatric: the most important way to classify speciation? Philosophical Transactions of the Royal Society B: Biological Sciences. 2008;363:2997–3007. doi: 10.1098/rstb.2008.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MR, Dillon J, Powell MS. Hybridization and introgression in a managed, native population of Yellowstone cutthroat trout: genetic detection and management implications. Transactions of the American Fisheries Society. 2002;131:364–375. [Google Scholar]
- Campton DE. Natural hybridization and introgression in fishes: methods of detection and genetic interpretations. In: Ryman N, Utter FM, editors. Population Genetics and Fisheries Management. Seattle, WA: University of Washington Press; 1987. pp. 161–192. [Google Scholar]
- Campton DE, Utter FM. Natural hybridization between steelhead trout (Salmo gairdneri) and coastal cutthroat trout (Salmo clarki clarki) in two Puget Sound streams. Canadian Journal of Fisheries and Aquatic Science. 1985;42:110–119. [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer Associates, Inc; 2004. [Google Scholar]
- Deschênes J, Rodríguez M, Bérubé P. Context-dependent responses of juvenile Atlantic salmon (Salmo salar) to forestry activities at multiple spatial scales within a river basin. Canadian Journal of Fisheries and Aquatic Science. 2007;64:1069–1079. [Google Scholar]
- Docker MF, Dale A, Heath DD. Erosion of interspecific reproductive barriers resulting from hatchery supplementation of rainbow trout sympatric with cutthroat trout. Molecular Ecology. 2003;12:3515–3521. doi: 10.1046/j.1365-294x.2003.02000.x. [DOI] [PubMed] [Google Scholar]
- Dowling TE, Child MR. Impact of hybridization on a threatened trout of the Southwestern United States. Conservation Biology. 1992;6:355–364. [Google Scholar]
- Dowling TE, Secor CL. The role of hybridization and introgression in the diversification of animals. Annual Review of Ecology and Systematics. 1997;28:593–619. [Google Scholar]
- Evans DO, Willox CC. Loss of exploited, indigenous populations of Lake Trout, Salvelinus namaycush, by stocking of non-native stocks. Canadian Journal of Fisheries and Aquatic Sciences. 1991;48(S1):134–147. [Google Scholar]
- Feist BE, Steel EA, Pess GR, Bilby RE. The influence of scale on salmon habitat restoration priorities. Animal Conservation. 2003;6:271–282. [Google Scholar]
- Fisher HS, Wong BBM, Resenthal GG. Alteration of the chemical environment disrupts communication in a freshwater fish. Proceedings of the Royal Society B: Biological Sciences. 2006;273:1187–1193. doi: 10.1098/rspb.2005.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SH, Allendorf FW. Associations between mitochondrial and nuclear genotypes in cutthroat hybrid swarms. Evolution. 1991;45:1332–1349. doi: 10.1111/j.1558-5646.1991.tb02639.x. [DOI] [PubMed] [Google Scholar]
- Gold JR. Systematics of western North American trout with notes on the redband trout of Sheepheaven Creek, California. Canadian Journal of Zoology. 1977;55:1858–1873. [Google Scholar]
- Grosholz E. Ecological and evolutionary consequences of coastal invasions. Trends in Ecology and Evolution. 2002;17:22–27. [Google Scholar]
- Gyllensten U, Leary RF, Allendorf FW, Wilson AC. Introgression between two cutthroat subspecies with substantial karyotypic nuclear and mitochondrial genomic divergence. Genetics. 1985;111:905–915. doi: 10.1093/genetics/111.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS. An exact test for randomness of mating. Journal of Genetics. 1954;52:631–635. [Google Scholar]
- Hartman GF, Gill CA. Distributions of juvenile steelhead and cutthroat trout (Salmo Gairdneri and S. clarki clarki) within streams in southwestern British Columbia. Journal of the Fisheries Research Board of Canada. 1968;25:33–48. [Google Scholar]
- Hartman GF, Scrivener JC, Miles MJ. Impacts of logging in Carnation Creek, a high-energy coastal stream in British Columbia, and their implication for restoring fish habitat. Canadian Journal of Fisheries and Aquatic Science. 1996;53:237–251. [Google Scholar]
- Hitt NP, Frissell CA, Muhlfeld CC, Allendorf FW. Spread of hybridization between native westslope cutthroat trout, Oncorhynchus clarki lewisi, and nonnative rainbow trout, Oncorhynchus mykiss. Canadian Journal of Fisheries and Aquatic Science. 2003;60:1440–1451. [Google Scholar]
- Holtby LB. Effects of logging on stream temperatures in Carnation Creek, British Columbia, and associated impacts on the coho salmon (Oncorhynchus kisutch. Canadian Journal of Fisheries and Aquatic Science. 1988;45:502–515. [Google Scholar]
- Hubbs CL. Hybridization between fish species in nature. Systematic Zoology. 1955;4:1–20. [Google Scholar]
- Jones JC, Reynolds JD. Effects of pollution on reproductive behaviour of fishes. Reviews in Fish Biology and Fisheries. 1997;7:463–491. [Google Scholar]
- Kanda N, Leary RF, Spruell P, Allendorf FW. Molecular genetic markers identifying hybridization between Colorado River – Greenback cutthroat trout complex and Yellowstone cutthroat trout or rainbow trout. Transactions of the American Fisheries Society. 2002;131:312–319. [Google Scholar]
- Keller B, Wolinska J, Manca M, Spaak P. Spatial, environmental and anthropogenic effects on the taxon composition of hybridizing Daphnia. Philosophical Transactions of the Royal Society B: Biological Sciences. 2008;363:2943–2952. doi: 10.1098/rstb.2008.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont BB, He T, Enright NJ, Krauss SL, Miller BP. Anthropogenic disturbance promotes hybridization between Banksia species by altering their biology. Journal of Evolutionary Biology. 2003;16:551–557. doi: 10.1046/j.1420-9101.2003.00548.x. [DOI] [PubMed] [Google Scholar]
- Leary RF, Allendorf FW, Phelps SR, Knudsen KL. Genetic divergence and identification of seven cutthroat trout subspecies and rainbow trout. Transactions of the American Fisheries Society. 1987;116:580–587. [Google Scholar]
- Lu GQ, Bernatchez L. Experimental evidence for reduced hybrid viability between dwarf and normal ecotypes of lake whitefish (Coregonus clupeaformis Mitchill) Proceedings of the Royal Society B: Biological Sciences. 1998;265:1025–1030. [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. Trends in Ecology and Evolution. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Metcalf JL, Siegle MR, Martin AP. Hybridization dynamics between Colorado's native cutthroat trout and introduced rainbow trout. Journal of Heredity. 2008;99:149–156. doi: 10.1093/jhered/esm118. [DOI] [PubMed] [Google Scholar]
- Nolte AW, Freyhof J, Tautz D. When invaders meet locally adapted types: rapid molding of hybrid zones between sculpins (Cottus, Pisces) in the Rhine system. Molecular Ecology. 2006;15:1983–1993. doi: 10.1111/j.1365-294X.2006.02906.x. [DOI] [PubMed] [Google Scholar]
- Ostberg CO, Slatton SL, Rodriguez RJ. Spatial partitioning and asymmetric hybridization among sympatric coastal steelhead trout (Oncorhynchus mykiss irideus), coastal cutthroat trout (O. clarki clarki) and interspecific hybrids. Molecular Ecology. 2004;13:2773–2788. doi: 10.1111/j.1365-294X.2004.02268.x. [DOI] [PubMed] [Google Scholar]
- Pearcy WG, Brodeur RD, Fisher JP. Distribution and biology of juvenile cutthroat trout Oncorhynchus clarki clarki and steelhead O. mykiss in coastal waters off Oregon and Washington. Fisheries Bulletin. 1990;88:697–711. [Google Scholar]
- Porter M, Haas G, Parkinson E. Fisheries Management Report 114. Victoria, British Columbia: British Columbia Ministry of Agriculture, Food and Fisheries, Fisheries Research Section; 2000. Sensitivity of British Columbia's freshwater fish to timber harvest: using species traits as predictors of species risk. [Google Scholar]
- Regetz J. Landscape-level constraints on recruitment of chinook salmon (Oncorhynchus tshawytscha) in the Columbia River basin, USA. Aquatic Conservation: Marine and Freshwater Ecosystems. 2003;13:35–49. [Google Scholar]
- Roff DA. Introduction to Computer-Intensive Methods of Data Analysis in Biology. Cambridge: Cambridge University Press; 2006. [Google Scholar]
- Rosenfeld JS, MacDonald S, Foster D, Amrhein S, Bales B, Williams T, Race F, et al. Importance of small streams as rearing habitat for coastal cutthroat trout. North American Journal of Fisheries Management. 2002;22:177–187. [Google Scholar]
- Rosenfield JA, Todd T, Greil R. Asymmetric hybridization and introgression between pink salmon and chinook salmon in the Laurentian Great Lakes. Transactions of the American Fisheries Society. 2000;129:670–679. [Google Scholar]
- Roth NE, Allan JD, Erickson DL. Landscape influences on stream biotic integrity assessed at multiple spatial scales. Landscape Ecology. 1996;11:141–156. [Google Scholar]
- Rubidge EM, Taylor EB. An analysis of spatial and environmental factors influencing hybridization between native westslope cutthroat trout (Oncorhynchus clarkia lewisi) and introduced rainbow trout (O. mykiss) in the upper Kootenay River drainage, British Columbia. Conservation Genetics. 2005;6:369–384. [Google Scholar]
- Rubidge E, Corbett P, Taylor EB. A molecular analysis of hybridization between native cutthroat trout and introduced rainbow trout in southeastern British Columbia, Canada. Journal of Fish Biology. 2001;59:42–54. [Google Scholar]
- Schwarz D, McPheron BA. When ecological isolation breaks down: sexual isolation is an incomplete barrier to hybridization between Rhagoletis species. Evolutionary Ecology Research. 2007;9:829–841. [Google Scholar]
- Scribner KT, Page KS, Barton ML. Hybridization in freshwater fishes: a review of case studies and cytonuclear methods of biological inference. Reviews in Fish Biology and Fisheries. 2001;10:293–323. [Google Scholar]
- Slaney TL, Hyatt KD, Northcote TG, Fielden RJ. Status of anadromous salmon and trout in British Columbia and Yukon. Fisheries. 1996;21:20–35. [Google Scholar]
- Spruell P, Pearce Smithwick JW, Knudsen KL, Allendorf FW. Progress Report to Oregon Department of Fish and Wildlife. Oregon, USA: Oregon Department of Fish and Wildlife; 1998. Genetic analysis of rainbow and cutthroat trout from the lower Columbia River. WTSGL98-103. [Google Scholar]
- Taylor EB. Evolution in mixed company – evolutionary inferences from studies of natural hybridzation in Salmonidae. In: Hendry AP, Stearns SC, editors. Evolution Illuminated: Salmon and Their Relatives. New York, NY: Oxford University Press; 2004. pp. 231–263. [Google Scholar]
- Taylor EB, Boughman JW, Groenenboom M, Sniatynski M, Schluter D, Gow JL. Speciation in reverse: morphological and genetic evidence of the collapse of a three-spined stickleback (Gasterosteus aculeatus) species pair. Molecular Ecology. 2006;15:343–355. doi: 10.1111/j.1365-294X.2005.02794.x. [DOI] [PubMed] [Google Scholar]
- Trotter PC. Coastal cutthroat trout: a life-history compendium. Transactions of the American Fisheries Society. 1989;118:463–473. [Google Scholar]
- Trotter PC. Sea-run cutthroat trout: life history profile. In: Hall D, Bisson PA, Gresswell RE, editors. Sea-run Cutthroat Trout: Biology, Management, and Future Conservation. Corvallis, OR: American Fisheries Society, Oregon Chapter; 1997. pp. 7–15. [Google Scholar]
- Verspoor E. Widespread hybridization between native Atlantic salmon, Salmo salar, and introduced brown trout, Salmo trutta, in eastern Newfoundland. Journal of Fish Biology. 1988;32:327–334. [Google Scholar]
- Walling DE. Linking land use, erosion and sediment yields in river basins. Hydrobiologia. 1999;410:223–240. [Google Scholar]
- Williams I, Reeves GH, Graziano SL, Nielsen JL. Genetic investigation of natural hybridization between rainbow and coastal cutthroat trout in the Copper River delta, Alaska. Transactions of the American Fisheries Society. 2007;136:926–942. [Google Scholar]
- Young WP, Ostberg CO, Keim P, Thorgaard GH. Genetic characterization of hybridization and introgression between anadromous rainbow trout (Oncorhynchus mykiss irideus) and coastal cutthroat trout (O. clarki clarki. Molecular Ecology. 2001;10:921–930. doi: 10.1046/j.1365-294x.2001.01247.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


