Abstract
As pest species may evolve resistance to chemical controls, they may also evolve resistance to cultural control methods. Yearly rotation of corn (Zea mays) with another crop interrupts the life cycle of the western corn rootworm beetle (Diabrotica virgifera virgifera, Coleoptera: Chrysomelidae), but behavioral resistance to crop rotation is now a major problem in the Midwest of the USA. Resistant adult females exhibit reduced fidelity to corn as a host and lay their eggs in the soil of both corn and soybean (Glycine max) fields. Behavioral assays suggest that the adaptation is related to increased locomotor activity, but finding molecular markers has been difficult. We used microarray analysis to search for gene expression differences between resistant and wild-type beetles. Candidates validated with real-time polymerase chain reaction exhibit predicted patterns from the microarray in independent samples across time and space. Many genes more highly expressed in the rotation-resistant females have no matches to known proteins, and most genes that were more lowly expressed are involved in antimicrobial defense.
Keywords: behavior, expressed sequence tag, gene expression, microarray, resistance
Introduction
Resistance of pest species to control methods allows us to study evolution on an ecological time scale. Evolution of resistance to synthetic toxins may present information relating to how organisms respond to naturally occurring chemicals in the environment (Scott et al. 1998). Behavioral resistance, however, could provide insight into how animals interact with the environment in activities such as host location or ovipositional choice. Molecular changes associated with resistance to a toxin have been frequently investigated, but no study to date has demonstrated a link between gene expression and behavioral resistance.
In the agriculturally intensive landscape of the Midwest of the USA, the western corn rootworm beetle, Diabrotica virgifera virgifera LeConte (Coleoptera: Chrysomelidae), typically requires fields planted with corn (Zea mays L.) in consecutive years to complete its life cycle. Eggs hatch in late spring, and the larvae feed on corn roots. Adults emerge midsummer, and females lay eggs in the soil of cornfields, after which the eggs overwinter in obligatory diapause. The only crop plant on which larvae can survive is corn, so yearly rotation of crops (i.e., annual alternation of corn with another crop in the same field) has been a major control method. However, D. v. virgifera has evolved behavioral resistance to crop rotation in a large portion of the midwestern USA. Rotation-resistant females also lay eggs in noncorn fields, namely soybean [Glycine max (L.) Merrill], that will be planted with corn the following spring (Levine and Oloumi-Sadeghi 1996). Even though females in regions where resistance is present may lay eggs indiscriminately in a variety of crops, including corn (Rondon and Gray 2004; Schroeder et al. 2005), and soybean fields can also act as a strong selective force. Virtually, all soybean fields are rotated to corn the following spring in areas with rotation resistance (Onstad et al. 2001, 2003), and, indeed, soybean fields may receive more D. v. virgifera oviposition than cornfields (Pierce and Gray 2006). The proportion of the landscape devoted to annual corn:soybean rotation is a major predictor in whether rotation resistance will spread to a given area (Onstad et al. 2003).
Our current hypothesis concerning the rotation-resistant behavioral phenotype is that resistant females show reduced fidelity to corn as a host (Levine et al. 2002; Spencer and Levine 2008). Rotation-resistant females are not necessarily attracted to soybeans (Spencer et al. 1999), nor can they survive on a diet composed strictly of soybean foliage (Mabry and Spencer 2003), but they may benefit more from soybean herbivory than wild-type (WT) females (Mabry et al. 2004). Adult beetles commonly leave corn fields as the plants senesce (Darnell et al. 2000) and are more likely to feed on soybean foliage as corn becomes an undesirable food source (O'Neal et al. 2002). Therefore, O'Neal et al. (2004) proposed a behavioral plasticity model for rotation resistance, in which early senescence of corn because of early planting repels adult beetles from corn fields, and no genetic change has occurred in beetle populations. However, resistant females are found in soybean fields even when corn tissues are optimal for adult feeding (Rondon and Gray 2003; Pierce and Gray 2007; LMK personal observation). Rotation resistance is likely to be a genetic trait, based on characteristics of the geographical spread (Onstad et al. 1999). The spread did not follow latitudinal isolines as would be expected under a scenario of early planting, even though an analysis of planting date has not been conducted. Behavioral studies also suggest that a genetic change has occurred because rotation-resistant females exhibit increased locomotor activity and tendency to take flight, which would increase the chances that they will leave their natal cornfield and lay eggs (Knolhoff et al. 2006). A simple model of the evolution of rotation resistance suggests that this behavioral change is likely because of one major allele (Onstad et al. 2001).
However, many questions remain. A significant piece of the puzzle lies in finding a molecular mechanism or marker for rotation resistance. Mapping approaches to find a marker have been difficult because the genome is rather large (Sappington et al. 2006). None of the eight microsatellites (Kim and Sappington 2005) tested so far are associated with resistance (Miller et al. 2006), but a few newly characterized loci have yet to be tested in this regard (Kim et al. 2008). Miller et al. (2007) conducted an analysis using amplified fragment length polymorphisms, but, of the 253 polymorphic loci they found, only one seems to be weakly associated with resistance. Garabagi et al. (2008) report small expression differences in the D. v. virgifera ortholog of the foraging gene in relation to rotation resistance. The foraging gene codes for a cGMP-dependent protein kinase involved in behaviors relating to host finding and acceptance in Drosophila (Pereira and Sokolowski 1993) and other insects (for a recent review, see Kaun and Sokolowski 2009). However, the two populations used for comparison were laboratory colonies that are no longer subjected to selection for desirable behavioral traits in their given native landscape.
Genetic markers are often derived using mapping approaches, but one desirable feature of microsatellites, for example, might present a problem. One of the reasons microsatellite markers are chosen is because they are selectively neutral among populations. The rotation resistance trait, however, does have a selective advantage in landscapes with a high proportion of yearly corn:soybean rotation (Onstad et al. 2001, 2003), so perhaps it would be easier and more useful to take a more functional approach. One option is to use microarrays to find changes in gene expression relating to the rotation resistance trait. Two results could arise: (i) the proposed mutation for the trait (Onstad et al. 2001) could be present in a sequence of interest, which would be either expressed or not or (ii) observed differences in gene expression could be the result of a mutation that is upstream in a given pathway. The latter result will not directly provide the polymorphism responsible for the rotation resistance trait, so further work would be necessary to determine the responsible mutation. A molecular mechanism would be easier to ascertain in the first scenario, but a marker for the trait could be found with either result, i.e., a marker does not need a mechanism to be useful. A molecular marker for rotation resistance has many applications in studying population dynamics and in sampling for emergent resistance in a given area. Another feature is that colony maintenance of behaviorally resistant insects can be verified with a molecular marker. Finally, a marker could provide insight into novel targets for management of rotation resistance.
Reasoning that genes controlling differences in dispersal and host-selection behavior are likely to be expressed in the brain, we therefore searched for gene expression differences in heads of female beetles using a cDNA-based microarray. Candidate genes from the microarray were validated with real-time polymerase chain reaction (PCR) for possible use as markers and to postulate a potential mechanism for behavioral resistance to crop rotation. This study presents a link between behavioral resistance to crop rotation and an associated molecular trait.
Materials and methods
Female head EST project
An expressed sequence tag (EST) project to study rotation resistance in D. v. virgifera was carried out by the W.M. Keck Center for Biotechnology at the University of Illinois at Urbana-Champaign. The cDNA library was made from heads of gravid female D. v. virgifera. The rationale for selecting heads was the expectation that genes relating to behavior would be most highly expressed in the brain, but the small size of rootworm beetle heads precluded brain dissection. Populations selected for RNA extraction consisted of two WT and two rotation-resistant populations in Illinois (at time of sampling). After ligating a 5-bp linker that was specific for each population, the cDNA was cloned using the pGEM-11Zf vector (Promega, Madison, WI). After transformation into bacteria, individual random colonies were selected for sequencing. To increase the number of unique transcripts, the library was normalized for relative abundance, and furthermore, a subtracted library was later created to obtain extra or rare transcripts that were not included in the original library following the protocol from Whitfield et al. (2002). A total of 16 172 high quality sequences resulted after trimming and filtering steps, and these have been submitted into the dbEST database at GenBank (http://www.ncbi.nlm.nih.gov/dbEST/index.html), with accession nos: EW761110–EW777362. The EST were assembled into contigs as in Whitfield et al. (2002).
Microarray construction
A printed cDNA microarray was constructed from 7947 unique transcribed sequences from the library (4643 singletons and 3304 contigs after assembly). As part of a collaboration, 383 additional sequences derived from a cDNA library from the larval midgut (Siegfried et al. 2005) were added to the array, as well as one cDNA sequence from a D. v. virgifera foraging ortholog (Garabagi et al. 2008), yielding 8331 probes. Microarray construction was similar to that described in Whitfield et al. (2002). Amplified cDNA fragments from representative clones were spotted in duplicate on the array, and these were grouped into 48 blocks consisting of the EST of interest and positive and negative controls. Positive controls included cyclophilin, glyceraldehyde-3-phosphate dehydrogenase, elongation factor-1 alpha (EF1a), beta-actin, and beta-tubulin. Negative controls consisted of sequences from soybean (Glycine max): ribulose biphosphate carboxylase, major latex protein homologue, and chlorophyll a/b-binding protein. Negative controls also included blank spots and spots printed with buffer.
Samples
To test for differences in gene expression in adult females between the two behavioral types, three populations of each type were collected in Illinois in July 2006. Samples of a rotation-resistant and a WT population were collected as pairs on the same day when growing degree-days approximated 750–800 (accumulated since beginning of calendar year, base 11°C, depth 10 cm). This was carried out to minimize any beetle phenology effects resulting from differences in latitude; females were post-teneral and most likely preovipositional. Rotation-resistant adults (Urbana, Pontiac, and Grand Ridge) were collected from first-year cornfields, and WT adults (Perry, Monmouth, and Morrison) were collected from continuous cornfields (Fig. 1A), defined as a field planted with corn two or more consecutive years. Because the sampled populations in this experiment are widely separated (>45 km), even within a given type, observed expression differences are not likely to be an artifact of local adaptation in one population.
Figure 1.
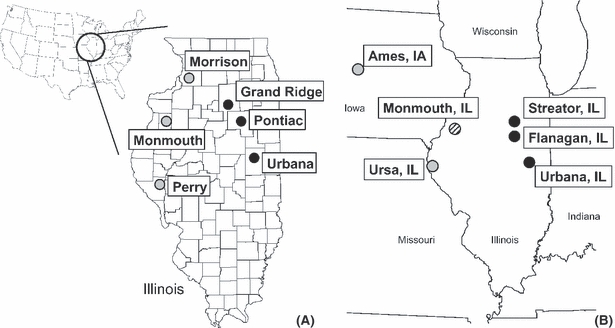
Locations of collections conducted in (A) 2006 and (B) 2007. Rotation-resistant populations are denoted with black circles; wild-type populations are denoted with open circles.
Status of rotation resistance in a population was confirmed by multiple lines of evidence. Most important was whether a given area had experienced consistent damage by D. v. virgifera larvae in first-year corn (defined as corn planted the year after another crop) over recent years. In addition, three sweep net samples of 100 sweeps were taken from soybean fields adjacent to cornfields where D. v. virgifera beetles were collected. Beetles present in sweep net samples from soybean fields indicate the presence of rotation resistance; a range of 10–20 beetles per 100 sweeps is a good predictor of rotation resistance (Onstad et al. 1999, 2003). Finally, behavioral tests on individual beetles [n = 30 (approximately) per population] were conducted in an Urbana cornfield 1–2 days after collection. Rotation-resistant females exhibit increased locomotor behavior and are more likely to take flight in a behavioral bioassay (Knolhoff et al. 2006).
Microarray analysis
Samples of 50 heads of female beetles were used per RNA extraction with Trizol reagent (Invitrogen, Carlsbad, CA); two sets of extractions (2 × 50 heads) were performed from each population. Total RNA was separated into 15 μg aliquots for array hybridization. The mRNA was reverse-transcribed, and cDNA was purified using a PCR purification kit (Qiagen, Valancia, CA). The appropriate cyanine (Cy3 or Cy5) label (General Electric Healthcare, Piscataway, NJ) was incorporated in samples of cDNA; labeled cDNA was also purified. Pairs of labeled samples were applied to the arrays and were left to hybridize for 48 h. After this time, arrays were scanned using a GenePix 4000B scanner and a GenePix Pro v5 software platform (previously Axon Instruments, now Molecular Devices, Sunnyvale, CA). Scanning parameters were set at autoscale values for intensity, and spots were manually checked for contamination before analysis.
The experimental design for array pairing was as follows (Fig. 2). Every array received both a WT and a rotation-resistant sample, and each population was compared with each population of the other behavioral type. Analysis was conducted as a dual-mixed model analysis of variance (Gibson and Wolfinger 2004) using sas software v. 9.1 (SAS Institute 2004). Data were prepared for analysis by (i) local background subtraction and (ii) log2 transformation. These data were then analyzed using a global-mixed model that accounted for the fixed effect of dye and the random effects of array and array–dye interaction. The residuals from this model were used in the second step, where an analysis of variance was conducted for each individual EST. Residuals were modeled for each EST as the fixed effects of type and population nested in type and the random effects of variation at biological and technical levels. Because of the large number of simultaneous tests and to narrow the list of genes for further examination, a stringent significance cutoff (P < 0.00001) was used to control for experiment-wise error rate when determining EST that were differentially expressed by behavioral type. EST of interest were further examined for matches in a search of nonredundant protein sequences in GenBank (BLASTx: http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) and possible matches in Tribolium castaneum, a beetle with a draft genome sequence (Tribolium Genome Sequencing Consortium 2008).
Figure 2.
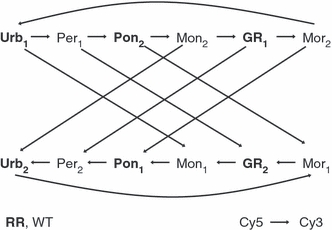
Interwoven loop design of microarray experiment. Each arrow represents one array, where the base of the arrow represents the sample with Cy5 label and the head of the arrow represents the sample with the Cy3 label. Biological (and extraction) replication is represented by subscripts; rotation-resistant populations are in bold. Population are as follows: Urb, Urbana; Per, Perry; Pon, Pontiac; Mon, Monmouth; GR, Grand Ridge; Mor, Morrison.
Candidate list from synthesis of microarray experiments
Three other microarray experiments were previously conducted, and their conditions are briefly described below. Results from these three preliminary experiments are not reported, and their role in this presented work was to assist in selection of candidates for independent verification. Experiments were conducted with sample preparation as described and with pools of 50 female heads representing biological replicates within a population. Two experiments compared gene expression between Urbana and Monmouth (Fig. 1), as representatives of the rotation-resistant and WT behavioral phenotypes, respectively. Both of these experiments included comparisons between beetles collected out of corn and beetles collected out of soybean, but no significant differences in gene expression were detected at a threshold of P < 0.001 between resistant beetles collected out of corn versus those collected out of soybean. A third experiment compared expression between Urbana as representative of the resistant type to expression in Ames, Iowa, and Arlington, Wisconsin, representing WT insects.
Candidates from the microarray experiment with large and significant differences in expression were selected for validation with quantitative real-time RT-PCR (hereafter referred to as real-time PCR). To synthesize results from preliminary work with results from current work, a variable for each EST was created to account for both difference in expression and significance of each particular analysis of variance. This method was expected to enhance the chances of success in finding genes with consistent expression patterns. For each EST's statistical analysis, the difference in expression (log2) was multiplied by the significance (−log10). These values were summed across all four microarray experiments such that extremes on either end of the distribution would be biased toward EST with large, significant differences. Candidates were selected by both rank of score and consensus among experiments.
Validation of expression differences in candidates
Independent collections of beetles from July 2007 were used to validate expression differences for candidates. As described, collections were timed to minimize any effects of beetle phenology because of latitudinal differences. Rotation-resistant adult females were collected from first-year cornfields in Urbana, Flanagan, and Streator, Illinois; WT females were collected from continuous cornfields in Ursa, Illinois, and Ames, Iowa (Fig. 1B). Beetles were also collected from a continuous cornfield in Monmouth, Illinois, but this population may be evolving resistance to crop rotation (Schroeder et al. 2005). A behavioral assay measuring locomotor activity as described in Knolhoff et al. (2006) was repeated to examine whether expression differences in individuals are related to observed behavior in the field. Locomotor activity was assessed by recording the time for a female to exit a cylindrical screen arena. A subset (n = 15) of the females for which gene expression was measured were also previously assessed for behavior; populations were each represented by 1–3 individuals.
Expression differences were independently evaluated in individual whole female adults using real-time PCR. Total RNA was extracted with Trizol reagent (Invitrogen) and subsequently treated with DNase (Turbo DNA-free; Ambion, Austin, TX) to minimize possible genomic DNA contamination. Reverse transcription (Arrayscript; Ambion) was performed with 200 ng of total RNA; cDNA was diluted 10× for real-time PCR. Expression of candidate genes was quantified using SYBR Green as a fluorescence reporter (SYBR Green PCR Master Mix; Applied Biosystems, Foster City, CA). Real-time PCR reactions were performed in triplicate for each combination of beetle and cDNA. Fluorescence was measured and critical threshold was automatically determined using an ABI Prism 7900 Sequence Detection System (Applied Biosystems). Negative controls consisted of reactions without cDNA template and reactions with template that received no reverse transcriptase. Expression of candidates was normalized to expression of EF1a, which was selected because of its low variability among treatments and individuals.
Expression data for candidates were analyzed as the difference in critical threshold between the genes of interest and EF1a (ΔCt) using sas software (SAS Institute 2004). Separate analyses of variance were conducted on each candidate measured in the same females (n = 23); ΔCt values were modeled as effects of behavioral type and extraction date. As noted above, the population of Monmouth may be evolving rotation resistance. Analyses were conducted without this population, which was subsequently added to the dataset to confirm or reject the suggestion that Monmouth has resistance.
Results
Transcriptomic differences
Using a strict threshold of significance, there are 51 EST showing a difference in expression by behavioral type (d.f. 1,>36; logP < −5); 23 of these show a greater than twofold difference (Fig. 3). Note that many genes more highly expressed in rotation-resistant females are either highly significant with a small difference or less significant but with a bigger difference in expression. Analysis of the foraging ortholog (Garabagi et al. 2008) could not be conducted because measurements did not meet the quality and detection standards applied to the dataset.
Figure 3.
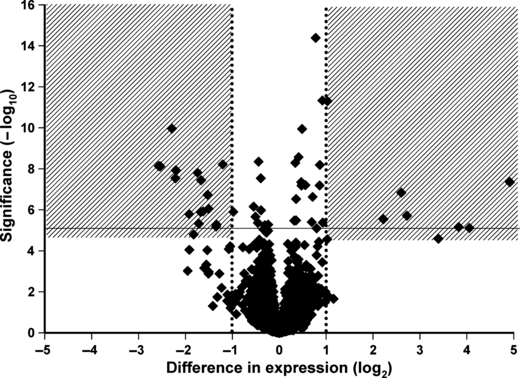
Volcano plot of results from microarray experiment. Each point represents the output from the analysis of variance conducted on each expressed sequence tag (EST). The x-axis is the estimated difference in expression measured in log2; vertical dotted lines refer to a twofold difference in expression between the two behavioral types. Genes more highly expressed in rotation-resistant populations are on the right; genes more highly expressed in wild-type populations are on the left. The y-axis is the significance of the difference measured in −log10 of the P-value; the dotted horizontal line represents our cutoff for significance at P < 0.00001. The shaded boxes encompass EST characterized in Table 1 that shows large, significant differences in expression between behavioral types.
Of interest are those EST with large and highly significant differences in expression between behavioral types, highlighted in Table 1 with sequence information and statistical results from the microarray. Many of the genes that are more highly expressed in rotation-resistant females have no significant matches to known proteins. Many of the genes with higher expression in WT females are similar in sequence and have the same single match in a search for known proteins: an antibacterial peptide from the dung beetle, Copris tripartitus. Interestingly, there is no annotated ortholog in Tribolium. Three others more highly expressed in WT females are defensins, which are involved in immune response to gram-positive bacteria.
Table 1.
Highlights of microarray results consisting of candidates with greater than twofold expression differences at P < 0.00001 (shaded boxes in Fig. 3)
| Type with higher expression | Accession no. | L | A | C | Diff. | P | Top match | Function | Tribolium castaneum match |
|---|---|---|---|---|---|---|---|---|---|
| RR | EW774177 | 452 | − | S | 2.04 | 11.30 | NP_000178 (S = 215, E = 9e-55) | Homogentisate 1,2-dioxygenase | XP_973513: PREDICTED: similar to CG4779-PA (S = 211, E = 1e-53) |
| CN497936 | 827 | − | C | 30.27 | 7.35 | None | |||
| EW761417 | 511 | − | C | 6.07 | 6.83 | None (no open reading frame) | |||
| CN497825 | 742 | − | C | 6.60 | 5.71 | XP_967924 (S = 125, E = 2e-27) | UDP-glucoronosyl and UDP-glucosyl transferase | XP_967924: PREDICTED: similar to CG18578-PA (S = 125, E = 2e-27) | |
| Contig1160* | 889 | − | C | 4.66 | 5.54 | ACI32832 (S = 230, E = 3e-73) | Beta-1,3-glucanase | XP_970010: PREDICTED: similar to beta-1,3-glucanase (S = 256, E = 2e-71) | |
| EW775372 | 333 | − | S | 14.18 | 5.16 | None | |||
| EW762256 | 434 | + | S | 16.61 | 5.11 | None | |||
| EW774489 | 547 | − | C | 10.51 | 4.59 | None | |||
| WT | EW762768 | 453 | − | S | 4.88 | 9.96 | ABP97089 (S = 57.8, E = 3e-07) | Antibacterial peptide | None |
| EW769719 | 633 | + | C | 2.30 | 8.22 | ABP97089 (S = 55.5, E = 1e-06), note only 200b match | Antibacterial peptide | None | |
| EW771833 | 573 | + | C | 5.92 | 8.13 | ABP97089 (S = 48.9, E = 3e-11; S = 42.0, E = 3e-11) | Antibacterial peptide | None | |
| EW768123 | 628 | + | S | 5.80 | 8.12 | ABP97089 (S = 76.3, E = 7e-13) | Antibacterial peptide | None | |
| EW763389 | 453 | + | C | 4.61 | 7.93 | None | |||
| EW770378 | 605 | − | S | 3.34 | 7.81 | ABP97089 (S = 91.3, E = 2e-17) | Antibacterial peptide | None | |
| EW765267 | 603 | + | C | 4.64 | 7.54 | ABP97089 (S = 73.9, E = 3e-12) | Antibacterial peptide | None | |
| EW772775 | 398 | + | C | 3.17 | 7.44 | AAK35160 (S = 77.0, E = 4e-13) | Defensin | XP_967194: PREDICTED: similar to CG1385-PA (S = 72.0, E = 1e-11) | |
| EW770481 | 211 | − | C | 2.88 | 6.73 | AAK35160 (S = 64.7, E = 2e-09) | Defensin | XP_967194: PREDICTED: similar to CG1385-PA (S = 54.3, E = 3e-06) | |
| EW767301 | 547 | + | S | 2.84 | 6.04 | ABP97089: (S = 59.3, E = 9e-08) | Antibacterial peptide | None | |
| EW761497 | 463 | + | S | 3.07 | 5.92 | None | |||
| EW769816 | 627 | + | C | 3.19 | 5.89 | ABP97089: (S = 91.3, E = 2e-17) | Antibacterial peptide | None | |
| EW764671 | 546 | + | S | 3.79 | 5.79 | None | |||
| EW766935 | 416 | + | C | 3.27 | 5.32 | AAW57774: S = 44.7, E = 0.002 | Parcxpwnx03 | None | |
| EW765002 | 343 | − | S | 2.53 | 5.28 | AAK35160 (S = 81.3, E = 2e-14) | Defensin | XP_967194: PREDICTED: similar to CG1385-PA (S = 54.3, E = 3e-06) | |
| EW767372 | 650 | + | C | 2.54 | 5.18 | ABP97089 (S = 56.2, E = 7e-07) | Antibacterial peptide | None | |
| EW774852 | 316 | − | S | 3.54 | 4.79 | NP_650064: CG5214 (S = 102, E = 7e-21) | Dihydrolipoyllysine-residue succinyltransferase activity | XP_971313: PREDICTED: similar to CG5214-PA (S = 97.8, E = 9e-19) |
RR, rotation-resistant; WT, wild-type; L, length of EST; A, presence of polyA tail; C, nontig (C) versus singleton (S); Diff., difference in expression between types as a fold change relative to the other behavioral type; P, significance of difference as −log10 of P-value.
Top match: top match in BLASTx search for known proteins in nonredundant database in GenBank. Cutoff was set at E < 10−4; scores (S) and expected (E) values are reported. Function: putative function ascertained from top match described above, species names of accession nos are as follows: ABP97089, Copris tripartitus (Coleoptera: Scarabeidae); AAK35160, Acalolepta luxuriosa (Coleoptera: Cerambycidae); ACI32832, Anthocharis cardamines (Lepidoptera: Pieridae); NP_000178, Homo sapiens; NP_650064, Drosophila melanogaster.
Contig1160 is composed of at least eight expressed sequence tag (EST), of which most of the consensus sequence in contained in CN497302.
Candidate gene expression
Preliminary results indicated that candidates selected from microarray experiments conducted only on Urbana and Monmouth populations yielded expression patterns consistent with local adaptation. Expression differences are indeed large between Urbana and Monmouth, but differences are not consistent across behavioral types, i.e., differences are not observed in replicates at the population level (results not shown).
Schroeder et al. (2005) have suggested that evolution to rotation resistance is occurring in the Monmouth area, thus we explored the dependence of the results on the classification of that population. Four candidates show significant differences in expression by behavioral type (n = 20, P < 0.05; Table 2). Analyses conducted with Monmouth as either a WT or rotation-resistant population show that, for three candidates (E, H, and I), it does not matter which classification Monmouth receives. These statistical results are highly similar for both analyses because data from Monmouth are variable. For one candidate (A), however, beetles tested from Monmouth cluster with rotation-resistant beetles. Expression differences from analyses conducted without Monmouth data (Fig. 4) yield four candidates with large enough differences to be used as a diagnostic trait for resistance. All expression patterns follow the predicted trends from the microarray, but not all differences are significant at P < 0.05.
Table 2.
Comparisons of statistical results of real-time polymerase chain reaction data for nine candidates
| Statistical results | |||||
|---|---|---|---|---|---|
| Predicted over-expression | ID | Accession no. | Without Monmouth (d.f. 1,16) | Monmouth as WT (d.f. 1,19) | Monmouth as RR (d.f. 1,19) |
| RR | A | EW762256 | F =6.59; P = 0.021 | F = 0.73; P = 0.402 | F = 9.23; P = 0.007 |
| B | Contig1160* | F = 0.01; P = 0.921 | F = 1.13; P = 0.301 | F = 0.19; P = 0.665 | |
| C | CN497825 | Nonparametric, P > 0.10 | F = 0.19; P = 0.671 | Nonparametric, P > 0.10 | |
| D | EW770254 | F = 1.32; P = 0.269 (d.f. 1,15) | F = 1.59; P = 0.225 (d.f. 1,17) | F = 1.14; P = 0.302 (d.f. 1,17) | |
| E | EW773352 | Nonparametric, χ2 = 5.22, P = 0.022 | Nonparametric, χ2 = 3.34,P = 0.068 | Nonparametric, χ2 = 5.54, P = 0.019 | |
| WT | F | EW768123 | Nonparametric, P > 0.10 | Nonparametric, P > 0.10 | Nonparametric, P > 0.10 |
| G | EW765267 | Nonparametric, P > 0.10 | Nonparametric, P > 0.10 | Nonparametric, P > 0.10 | |
| H | EW770481 | F = 4.82; P = 0.043 | F = 5.72; P = 0.027 | F = 3.76; P = 0.067 | |
| I | EW772775 | F = 6.91; P = 0.018 | F = 7.26; P = 0.014 | F = 5.34; P = 0.032 | |
Analyses were conducted as characteristics of residuals would dictate; nonparametric tests were chi-square median tests. Differences at P < 0.10 are presented in bold. RR, rotation-resistant; WT, wild-type.
Contig 1160 is composed of at least eight expressed sequence tag (EST), of which most of the consensus sequence in contained in CN49730.
Figure 4.
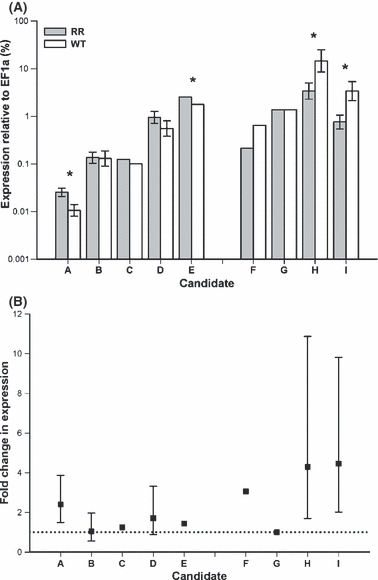
Expression of candidates validated with real-time polymerase chain reaction (PCR). (A) Bars depict means in the statistical analysis conducted without Monmouth data (Table 2). However, candidates C, E, F, and G represent median values because nonparametric analyses were used and therefore error bars could not be calculated. *Significant differences in expression at P < 0.05. (B) Calculated fold change difference in expression relative to the other behavioral type. The dotted line represents no difference in expression. A–E are predicted rotation-resistant candidates; F-I are predicted wild-type candidates. For analyses and accession numbers, see Table 2.
Expression of only one candidate seems to be related to observed locomotor activity, measured in seconds to exit a behavioral arena. In a correlation analysis of expression data for which behavior was recorded in the field the previous summer, there is a positive association between expression of candidate D in ΔCt to locomotor activity in seconds (n = 15, r = 0.74, P = 0.009), meaning beetles that are more active over-express this candidate.
Discussion
Results from microarrays are only as good as the experimental design and sample input. This project demonstrates that replication both within and between populations is important in seeking markers for rotation resistance. It is well recognized that sample size should always be maximized or optimized, but this mantra is often only applied to individuals within populations or strains. The term biological replication is often understood to represent the number of individual organisms, not the number of individual genotypes. Treatment differences found when comparing only two populations (even with robust within-population replication) could possibly turn out to be effects of local adaptation, i.e., not the traits of interest under selection. These artifacts were found in candidates from preliminary microarray experiments conducted on only two populations; differences could not be replicated across populations.
Expression differences relating to rotation resistance in this study are consistent across different populations and multiple years. Therefore, it is expected that the four candidates with significant differences in expression are appropriate for use as markers for this behavioral resistance trait. Real-time PCR results for the Monmouth population give a measure of applicability for the potential markers: three markers suggest the population is in transition, and one suggests it is resistant.
A mechanism for rotation resistance, however, is much harder to extract from the results. A marker does not require a known function to be useful for diagnostics, but a function would facilitate to answer questions about the behavioral causes. Behavioral transcriptomics is an emerging field and has been used in Drosophila melanogaster to study behaviors relating to aggressiveness (Edwards et al. 2006) and locomotor activity (Jordan et al. 2007), as examples. Microarrays have also been used to study temporal division of labor in honey bees (Whitfield et al. 2003) and behavioral phase changes in migratory locusts (Kang et al. 2004). While most of the genes on the microarray were derived from the head library, interestingly, some genes originating from only the larval midgut library were expressed in the head (Table 1). Small size prevented brain dissection of these insects, but perhaps genes originating from the midgut library are expressed in other tissues in the head, such as the salivary gland or the crop.
Three putative defensin genes (two of which are validated candidates) were more highly expressed in WT beetles. Furthermore, nine EST more highly expressed in WT beetles share sequence similarity to a single antibacterial peptide (Table 1) and are putative attacins, as inferred from amino acid sequence comparison with Tribolium castaneum (Zou et al. 2007). Rotation-resistant females exhibit increased expression of a beta-1,3-glucanase gene, which play a role in digestion of bacteria (Pauchet et al. 2009). Increased expression of a glucanase could mean that the glucans eliciting an immune response would be degraded.
Some studies with Drosophila have identified differentially expressed immunity genes in relation to certain behaviors. Carney (2007) found that many of the genes down-expressed in courting males (as compared with noncourting males) were related to innate immunity. Notably, Jordan et al. (2007) found a large number of immune and defense genes that were differentially expressed after 25 generations of selection for high- and low-locomotor activity. Flies in these studies were not challenged with bacterial infection, so perhaps there is a correlated response or developmental cascade associated with locomotor behavior. Domanitskaya et al. (2007) suggest that induction of immune genes by male sex peptide occurs by molecular mimicry of the bacterial cell wall, so there may be other regulatory mechanisms of innate immune response. The reason that expression of immunity genes seems to be related to behavior deserves more study.
Interactions between the immune and nervous systems in insects can occur as immune-induced behavioral changes, for example behavioral fever, where the insect migrates to warmer temperatures as a result of immune response (Adamo 2008). Conversely, changes in immune function can occur through behavioral influences of stress response and biogenic amines (Adamo 2008). Octopamine plays a major role in insect metabolism and behaviors (Roeder 2005) and can also affect immune response. Its broad effects have led Fahrbach and Mesce (2005) to conclude ‘octopamine regulation often links insect behaviors rarely considered to be related.’ This idea is exemplified in crickets (Gryllus spp.), in which octopamine levels in the hemolymph increase after flight activity (Adamo et al. 1995). Similarly, increased physical activity (both running and flying) and octopamine increases susceptibility to bacterial infection in crickets (Adamo and Parsons 2006). We propose that decreased expression of antibacterial genes in rotation-resistant beetles is not a causal mechanism for rotation resistance, but rather a pleiotropic effect of increased locomotor activity. Still consistent with the overall hypothesis is the alternative possibility, that decreased induction of immune-related genes is a side effect of less exposure to bacteria because of increased locomotor activity.
The problem of rotation resistance has been difficult to quantify and understand because it does not seem to be a simple case of altered attraction or food preference. In fact, the trait may even be as a result of some loss of function in the responsible gene product. This behavioral adaptation has presented a special and frustrating challenge to researchers to find markers or mechanisms. Indeed, it is among the four areas of focus of the Diabrotica Genetics Consortium: insecticide resistance, rotation resistance, the recent invasion into Europe, and resistance management for transgenic crops (Sappington et al. 2006). With the examination of differentially transcribed genes, this study provides evidence of a molecular link to behavioral adaptation to crop rotation. The markers proposed here will help in studies of population dynamics of this insect, as well as suggest novel targets for interfering with rotation resistance.
Acknowledgments
The EST project at the Keck Center was conducted by J. Pardinas, A. Hernandez, L. Liu, and R. Schwartz, with beetle collections made by C. Pierce, R. Wright, and J. Schroeder. Construction of microarrays and technical assistance with them was provided by M. Band, A. Bari, and A. Cash. For the microarray, B. Siegfried supplied sequences derived from the larval midgut, and K.P. Pauls provided the D. v. virgifera foraging sequence. The authors thank H. Lewin, M. Gray, E. Levine, J. Spencer, and C. Whitfield for advice and guidance during this project. Assistance in location of beetle populations used in this study was provided by E. Adee, E. Cullen, D. Feltes, M. Rabe, J. Oleson, M. Shier, and M. Vose. This work was funded by a grant from the cFAR grant program of the State of Illinois and a Clark summer research grant from the School of Integrative Biology at UIUC. The authors also thank two anonymous reviewers for thoughtful comments on the manuscript.
Literature cited
- Adamo SA. Bidirectional connections between the immune system and the nervous system in insects. In: Beckage N, editor. Insect Immunology. San Diego, CA: Academic Press; 2008. pp. 129–149. [Google Scholar]
- Adamo SA, Parsons NM. The emergency life-history stage and immunity in the cricket, Gryllus texensis. Animal Behavior. 2006;72:235–244. [Google Scholar]
- Adamo SA, Linn CE, Hoy RR. The role of neurohormonal octopamine during fight or flight behavior in the field cricket Gryllus bimaculatus. Journal of Experimental Biology. 1995;198:1691–1700. doi: 10.1242/jeb.198.8.1691. [DOI] [PubMed] [Google Scholar]
- Carney GE. A rapid genome-wide response to Drosophila melanogaster social interactions. BMC Genomics. 2007;288 doi: 10.1186/1471-2164-8-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell SJ, Meinke LJ, Young LJ. Influence of corn phenology on adult western corn rootworm (Coleoptera: Chrysomelidae) distribution. Environmental Entomology. 2000;29:587–595. [Google Scholar]
- Domanitskaya EV, Liu HF, Chen SJ, Kubli E. The hydroxyproline motif of male sex peptide elicits the innate immune response in Drosophila females. FEBS Journal. 2007;274:5659–5668. doi: 10.1111/j.1742-4658.2007.06088.x. [DOI] [PubMed] [Google Scholar]
- Edwards AC, Rollmann SM, Morgan TJ, Mackay TFC. Quantitative genomics of aggressive behavior in Drosophila melanogaster. PLOS Genetics. 2006;2:1386–1395. doi: 10.1371/journal.pgen.0020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrbach SE, Mesce KA. “Neuroethoendocrinology”: integration of field and laboratory studies in insect neuroendocrinology. Hormones and Behavior. 2005;48:352–359. doi: 10.1016/j.yhbeh.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Garabagi F, French BW, Schaafsma AW, Pauls KP. Increased expression of a cGMP-dependent protein kinase in rotation-adapted western corn rootworm (Diabrotica virgifera virgifera L.) Insect Biochemistry and Molecular Biology. 2008;38:697–704. doi: 10.1016/j.ibmb.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Gibson G, Wolfinger RD. Gene expression profiling using mixed models. In: Saxton AM, editor. Genetic Analysis of Complex Traits Using SAS. Cary, NC: SAS Users Press; 2004. pp. 251–278. [Google Scholar]
- Jordan KW, Carbone MA, Yamamoto A, Morgan TJ, Mackay TF. Quantitative genomics of locomotor behavior in Drosophila melanogaster. Genome Biology. 2007;8:8. doi: 10.1186/gb-2007-8-8-r172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L, Chen XY, Zhou Y, Liu BW, Zheng W, Li RQ, Wang J, et al. The analysis of large-scale gene expression correlated to the phase changes of the migratory locust. Proceedings of the National Academy of Science of the United States of America. 2004;101:17611–17615. doi: 10.1073/pnas.0407753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaun KR, Sokolowski MB. cGMP-dependent kinase: linking foraging to energy homeostasis. Genome. 2009;52:1–7. doi: 10.1139/G08-090. [DOI] [PubMed] [Google Scholar]
- Kim KS, Sappington TW. Genetic structuring of western corn rootworm (Coleoptera: Chrysomelidae) populations in the United States based on microsatellite loci analysis. Environmental Entomology. 2005;34:494–503. [Google Scholar]
- Kim KS, Stolz U, Miller NJ, Waits ER, Guillemaud T, Sumerford DV, Sappington TW. A core set of microsatellite markers for western corn rootworm (Coleoptera: Chrysomelidae) population genetics studies. Environmental Entomology. 2008;37:293–300. doi: 10.1603/0046-225x(2008)37[293:acsomm]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Knolhoff LM, Onstad DW, Spencer JL, Levine E. Behavioral differences between rotation-resistant and wild-type Diabrotica virgifera virgifera (Coleoptera: Chrysomelidae) Environmental Entomology. 2006;35:1049–1057. [Google Scholar]
- Levine E, Oloumi-Sadeghi H. Western corn rootworm (Coleoptera: Chrysomelidae) larval injury to corn grown for seed production following soybeans grown for seed production. Journal of Economic Entomology. 1996;89:1010–1016. [Google Scholar]
- Levine E, Spencer JL, Isard SA, Onstad DW, Gray ME. Adaptation of the western corn rootworm to crop rotation: evolution of a new strain in response to a management practice. American Entomologist. 2002;48:94–107. [Google Scholar]
- Mabry TR, Spencer JL. Survival and oviposition of a western corn rootworm variant feeding on soybean. Entomologia Experimentalis et Applicata. 2003;109:113–121. [Google Scholar]
- Mabry TR, Spencer JL, Levine E, Isard SA. Western corn rootworm (Coleoptera: Chrysomelidae) behavior is affected by alternating diets of corn and soybean. Environmental Entomology. 2004;33:860–871. [Google Scholar]
- Miller NJ, Kim KS, Ratcliffe ST, Estoup A, Bourguet D, Guillemaud T. Absence of genetic divergence between western corn rootworms (Coleoptera: Chrysomelidae) resistant and susceptible to control by crop rotation. Journal of Economic Entomology. 2006;99:685–690. doi: 10.1603/0022-0493-99.3.685. [DOI] [PubMed] [Google Scholar]
- Miller NJ, Ciosi M, Sappington TW, Ratcliffe ST, Spencer JL, Guillemaud T. Genome scan of Diabrotica virgifera virgifera for genetic variation associated with crop rotation tolerance. Journal of Applied Entomology. 2007;131:378–385. [Google Scholar]
- O'Neal ME, Di Fonzo CD, Landis DA. Western corn rootworm (Coleoptera: Chrysomelidae) feeding on corn and soybean leaves affected by corn phenology. Environmental Entomology. 2002;31:285–292. [Google Scholar]
- O'Neal ME, Landis DA, Miller JR, Di Fonzo CD. Corn phenology influences Diabrotica virgifera virgifera emigration and visitation to soybean in laboratory assays. Environmental Entomology. 2004;33:35–44. [Google Scholar]
- Onstad DW, Joselyn MG, Isard SA, Levine E, Spencer JL, Bledsoe LW, Edwards CR, et al. Modeling the spread of western corn rootworm (Coleoptera: Chrysomelidae) populations adapting to soybean-corn rotation. Environmental Entomology. 1999;28:188–194. [Google Scholar]
- Onstad DW, Spencer JL, Guse CA, Levine E, Isard SA. Modeling evolution of behavioral resistance by an insect to crop rotation. Entomologia Experimentalis et Applicata. 2001;100:195–201. [Google Scholar]
- Onstad DW, Crowder DW, Isard SA, Levine E, Spencer JL, O'Neal ME, Ratcliffe ST, et al. Does landscape diversity slow the spread of rotation-resistant western corn rootworm (Coleoptera: Chrysomelidae)? Environmental Entomology. 2003;32:992–1001. [Google Scholar]
- Pauchet Y, Freitak D, Heidel-Fischer HM, Heckel DG, Vogel H. Glucanase activity in a glucan-binding protein family from Lepidoptera. Journal of Biological Chemistry. 2009;284:2214–2224. doi: 10.1074/jbc.M806204200. [DOI] [PubMed] [Google Scholar]
- Pereira HS, Sokolowski MB. Mutations in the larval foraging gene affect adult locomotory behavior after feeding in Drosophila melanogaster. Proceedings of the National Academy of Sciences United States of America. 1993;90:5044–5046. doi: 10.1073/pnas.90.11.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce CMF, Gray ME. Seasonal oviposition of a western corn rootworm, Diabrotica virgifera virgifera LeConte (Coleoptera: Chrysomelidae), variant in east central Illinois commercial maize and soybean fields. Environmental Entomology. 2006;35:676–683. [Google Scholar]
- Pierce CMF, Gray ME. Population dynamics of a western corn rootworm (Coleoptera: Chrysomelidae) variant in east central Illinois commercial maize and soybean fields. Journal of Economic Entomology. 2007;100:1104–1115. doi: 10.1603/0022-0493(2007)100[1104:pdoawc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Roeder T. Tyramine and octopamine: ruling behavior and metabolism. Annual Review of Entomology. 2005;50:447–477. doi: 10.1146/annurev.ento.50.071803.130404. [DOI] [PubMed] [Google Scholar]
- Rondon SI, Gray ME. Captures of western corn rootworm (Coleoptera: Chrysomelidae) adults with Pherocon AM and vial traps in four crops in east central Illinois. Journal of Economic Entomology. 2003;96:737–747. doi: 10.1093/jee/96.3.737. [DOI] [PubMed] [Google Scholar]
- Rondon SI, Gray ME. Ovarian development and ovipositional preference of the western corn rootworm (Coleoptera: Chrysomelidae) variant in east central Illinois. Journal of Economic Entomology. 2004;97:390–396. doi: 10.1093/jee/97.2.390. [DOI] [PubMed] [Google Scholar]
- Sappington TW, Siegfried BD, Guillemaud T. Coordinated Diabrotica genetics research: accelerating progress on an urgent insect pest problem. American Entomologist. 2006;52:90–97. [Google Scholar]
- SAS Institute. User's Manual, Version 9.1. Cary, NC: SAS Institute; 2004. [Google Scholar]
- Schroeder JB, Ratcliffe ST, Gray ME. Effect of four cropping systems on variant western corn rootworm (Coleoptera: Chrysomelidae) adult and egg densities and subsequent larval injury in rotated maize. Journal of Economic Entomology. 2005;98:1587–1593. doi: 10.1093/jee/98.5.1587. [DOI] [PubMed] [Google Scholar]
- Scott JG, Liu N, Wen Z. Insect cytochromes P450: diversity, insecticide resistance, and tolerance to plant toxins. Comparative Biochemistry and Physiology C. 1998;121:147–155. doi: 10.1016/s0742-8413(98)10035-x. [DOI] [PubMed] [Google Scholar]
- Siegfried BD, Waterfield N, Ffrench-Constant RH. Expressed sequence tags from Diabrotica virgifera virgifera midgut identify a coleopteran cadherin and a diversity of cathepsins. Insect Molecular Biology. 2005;14:137–143. doi: 10.1111/j.1365-2583.2005.00538.x. [DOI] [PubMed] [Google Scholar]
- Spencer JL, Levine E. Resistance to crop rotation. In: Onstad DW, editor. Insect Resistance Management: Biology, Economics and Prediction. San Diego, CA: Academic Press; 2008. pp. 153–183. [Google Scholar]
- Spencer JL, Isard SA, Levine E. Free flight of western corn rootworm (Coleoptera: Chrysomelidae) to corn and soybean plants in a walk-in wind tunnel. Journal of Economic Entomology. 1999;92:146–155. [Google Scholar]
- Tribolium Genome Sequencing Consortium. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Whitfield CW, Band MR, Bonaldo MF, Kumar CG, Liu L, Pardinas JR, Robertson HM, et al. Annotated expressed sequence tags and cDNA microarrays for studies of brain and behavior in the honey bee. Genome Research. 2002;12:555–566. doi: 10.1101/gr.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield CW, Cziko AM, Robinson GE. Gene expression profiles in the brain predict behavior in individual honey bees. Science. 2003;302:296–299. doi: 10.1126/science.1086807. [DOI] [PubMed] [Google Scholar]
- Zou Z, Evans JD, Lu ZQ, Zhao PC, Williams M, Sumathipala N, Hetru C, et al. Comparative genomic analysis of the Tribolium immune system. Genome Biology. 2007;8:R177. doi: 10.1186/gb-2007-8-8-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]


