Abstract
Reproductive traits are key parameters for the evolution of invasiveness in weedy crop–wild hybrids. In Beta vulgaris, cultivated beets hybridize with their wild relatives in the seed production areas, giving rise to crop–wild hybrid weed beets. We investigated the genetic structure, the variation in first-year flowering and the variation in mating system among weed beet populations occurring within sugar beet production fields. No spatial genetic structure was found for first-year populations composed of F1 crop–wild hybrid beets. In contrast, populations composed of backcrossed weed beets emerging from the seed bank showed a strong isolation-by-distance pattern. Whereas gametophytic self-incompatibility prevents selfing in wild beet populations, all studied weed beet populations had a mixed-mating system, plausibly because of the introgression of the crop-derived Sf gene that disrupts self-incompatibility. No significant relationship between outcrossing rate and local weed beet density was found, suggesting no trends for a shift in the mating system because of environmental effects. We further reveal that increased invasiveness of weed beets may stem from positive selection on first-year flowering induction depending on the B gene inherited from the wild. Finally, we discuss the practical and applied consequences of our findings for crop-weed management.
Keywords: Beta vulgaris, genetic structure, invasive species, outcrossing rate, timing of flowering, weediness
Introduction
Recent biological invasions provide insights into the roles that the fundamental evolutionary forces of selection, gene flow and genetic drift play in processes such as local adaptation and successful spread of detrimental species in highly disturbed, human-altered environments (Barrett et al. 2008; Lee and Gelembiuk 2008; Wilson et al. 2009). In plants, reproductive traits and the mating system are key parameters for establishment in anthropogenic habitats because they drive many processes, such as dispersal through seed and pollen, the timing of flowering and the genetic structure within and among populations (Hamrick and Godt 1996; Kalisz et al. 2004; Cheptou and Schoen 2007; Michalski and Durka 2007; Barrett et al. 2008). In this respect, invasive crop weeds, which evolve rapidly in response to crop cultivation and the associated selection pressures, have recently attracted the attention of evolutionary ecologists in applied crop-weed management (Neve et al. 2009; Ridley and Ellstrand 2010).
In domesticated plants, crop selection often leads to significant changes in life history compared with wild relatives, especially for reproductive traits (e.g. Guillemin et al. 2008). Successful crop-wild hybridization and subsequent multiple introductions in a new habitat associate large amounts of genetic diversity with novel genetic combinations (Dlugosch and Parker 2008; Wilson et al. 2009). This provides opportunities to increase the adaptability of populations to a new niche on contemporary timescales. Likewise, the evolution of invasive weeds from crop–wild hybrids is a recurring pattern of adaptive evolution that entails numerous changes in life history, morphology and ecology (Baker 1974; Ellstrand and Schierenbeck 2000; Campbell et al. 2009; Ridley and Ellstrand 2010). Among these changes, knowledge of the impact of reproductive traits on the successful establishment and subsequent spread of weedy lineages in agricultural landscapes is of crucial importance for understanding micro-evolutionary changes.
Weed beets are a model of choice for investigating adaptive evolution on contemporary time scales. They are the result of accidental cross-fertilization that takes place between crop lineages (Beta vulgaris ssp. vulgaris) used as seed parents and ruderal wild relatives (Beta vulgaris ssp. maritima) found in close proximity to seed production fields, as in southwestern France (Van Dijk and Desplanque 1999). During the past few decades, widespread occurrence of weed beets within sugar beet fields throughout Europe suggests that hybridization between cultivated beets and wild relatives leads to increased weediness, i.e. the ability of a plant to colonize a disturbed habitat and compete with cultivated species (Ellstrand and Schierenbeck 2000). Indeed, crop–wild hybrid seeds are indistinguishable from beet cultivar seeds and are inadvertently sown in sugar beet fields. They result in individuals that bolt, called weed beets, and occur at very low densities at this first stage of infestation. While sugar beet, harvested for sucrose before flowering, is a biennial crop, weed beets can bolt and flower as of the first year. If not removed in due time, weed beets can set large amounts of seed. Therefore, under lax weeding practices, F1 crop–wild hybrids give rise to a long-lived seed bank and weed beets recur in subsequent years (Desplanque et al. 2002). Once a seed bank has been established, weed beets germinate spontaneously between the sowing lines, with a strong potential increase in population size and density over the years, depending on agricultural practices (Van Dijk and Desplanque 1999; Sester et al. 2006). Owing to the resurgence of dormant seed banks established from previous sugar beet crops, changes in genetic diversity and spatial genetic structure may be expected among weed beet populations (Viard et al. 2002).
The key process implicated in the occurrence of weed beets is the introgression of a wild genetic background into the crop gene pool. Among the numerous weedy traits, first-year flowering induction is inherited from a Mendelian gene whose dominant B allele cancels any vernalization requirement, i.e. the process by which a period of cold temperatures is necessary for the plant to switch from the vegetative to the reproductive stage (Boudry et al. 1994; Van Dijk 2009). The B allele is introgressed from wild beets to cultivar seed parents when accidental hybridizations occur during the cultivar seed production process.
Another key reproductive parameter selected for in crops may account for the successful establishment of weed beets in sugar beet fields: the Sf gene, a Mendelian self-fertility factor widely used by breeders to produce inbred lines, found in cultivated beet germplasm (Owen 1942). This factor behaves as a dominant gene, and allows the gametophytic self-incompatibility system to be circumvented (Maletski and Weisman 1978). If inherited during crop–wild hybridization, self-fertilization is thus likely to confer a selective advantage for weed beets, at least in the first stage of field infestation when the density of bolting F1 crop–wild hybrids is low. If mate availability is limited because of low plant density and pollen limitation, reproductive assurance through selfing is often advocated as a factor driving mating system evolution, especially in weed species that colonize human-disturbed environments (Baker 1974; Kalisz et al. 2004; Elam et al. 2007; Dornier et al. 2008).
This study aimed at investigating the evolution of weed beet populations during the course of an invasion. We surveyed four different sugar beet fields subjected to identical agricultural practices and suitable for weed establishment, but with contrasting weed beet density because of seed bank resurgence, and asked the following questions:
Are there differences in spatial genetic structure among weed beet populations? F1 crop–wild hybrids combine genetically differentiated gene pools. We thus expect to find high genetic diversity and no significant spatial genetic structure within a first-year contaminated field where few F1 crop-wild weed beets occur. In contrast, in late-stage contaminated fields, we expect stronger genetic structure owing to the kin-structured seed bank that accumulates over growing seasons.
How does first-year flowering vary among weed beet populations? Van Dijk (2009) recently showed a strong potential for natural selection on flowering time at metapopulation or regional scales in wild beets. Therefore, it is expected that strong selective pressures promote first-year flowering in weed beets, allowing them to reproduce before being eliminated during the harvest in autumn.
Are there significant genetic signatures of selfing events within weed beet populations and is there variation in the outcrossing rate among populations? The expectation would be to find selfed progenies in first-year contaminated fields, suggesting the escape of the crop-derived gene Sf in weedy lineages. Because B. vulgaris is an obligatory outcrossing self-incompatible and wind-pollinated species in the wild, we also investigated whether inbreeding depression (the decline in fitness of inbred versus outbred individuals, Charlesworth and Charlesworth 1987) could occur in inbred populations.
Materials and methods
The species
Cultivated beets, wild beets (mostly found along the coastline) and weed beets (defined as flowering individuals found within sugar beet fields) all belong to the same species, Beta vulgaris L. In the wild, B. vulgaris is strictly self-incompatible, with up to four gametophytic S loci, and has a purely outcrossing mating system that depends on wind pollination (Owen 1942; Maletski and Weisman 1978; De Cauwer et al. 2010). However, the Sf gene introduced by breeders to maintain selfed, near-isogenic lines (type O) and produce male-sterile seed bearers can override self-incompatibility. Variable frequencies of the Sf gene can thus be found in the male-sterile seed bearers used to produce certified seeds in seed production fields (Darmency et al. 2009).
There is no vegetative reproduction, and thus dispersal can only occur through seeds and/or pollen movement. Seeds are aggregated in a seedball that contains 1–8 seeds. This seedball has no particular dispersal mechanism and is primarily dispersed by gravity or by water movements during high tide in wild populations (Fievet et al. 2007). Therefore, except for rare, human-mediated, long-distance dispersal events in disturbed environments (see Arnaud et al. 2003), seed movements usually show a short-range pattern of dispersal, in contrast to pollen which is the most efficient source of gene flow through wind dispersal (Fénart et al. 2007; De Cauwer et al. 2010).
While cultivated beets are biannual and harvested for their roots before flowering, wild and weed beets can bolt, flower and reproduce in a single crop season provided they carry the B allele that cancels the vernalization requirement. As a result, weed beets efficiently compete with cultivated beets for resources and have been identified as a serious agronomic problem because they cause yield losses, decreased quality and mechanical problems during harvest (Sester et al. 2006). As weed beets belong to the same species as sugar beets, the weed beets cannot be eliminated by herbicides in sugar beet crops and the only efficient way to manage their spread is by mechanical or traditional hand-weeding. If weedy F1 crop–wild hybrids are left in sugar beet crops at the initial stage of infestation, a large amount of long-lived dormant seeds are then buried in the soil. This can give rise to a very large weed beet population in subsequent sugar beet crops that are cultivated 2–4 years later depending on crop rotation (Van Dijk 2004; Sester et al. 2006). Following Viard et al. (2002), two classes of flowering weed beets can be defined: (1) the first includes seeds sown and emerging within the sowing line; these ‘in-row’ weeds correspond mostly to F1 crop–wild hybrids sired by nearby populations of wild beets during the seed production process (Boudry et al. 1993; Arnaud et al. 2009) and (2) the second class of flowering weed beets grows outside the sowing lines and originates from the seed bank; these ‘out-row’ weed beets are progeny from crosses between flowering individuals in previous sugar beet fields.
Sampling
The study area is located in sugar beet production fields in Northern France (Fig. S1). Throughout this study, it is important to keep in mind that gene flow only occurs among weed beets, which are the only flowering individuals within a sugar beet field. Crop–wild hybridization can only take place in the seed production area located in southwestern France while the studied sugar beet fields are located in Northern France, far (150 km) from the shoreline where wild beets occur. Weed beet individuals were collected in summer 2003 from four populations named A, B, C and D, located in four sugar beet fields in which pollen dispersal distribution has been studied (Fénart et al. 2007). These four fields were located in a restricted geographical area, from 500 m to 8 km apart (Fig. S1). Based on field observations and personal communication from farmers, these sugar beet fields did not differ either in their suitability for weed beets or in their agricultural features (no detectable herbivore damage, identical fertilizers). Weed beet populations were characterized by contrasting occurrence of weedy individuals. Populations A and B were composed of spatially dispersed weed beet individuals found within the sowing lines in two neighbouring fields (400 m apart) that had very low levels of infestation (50 weed beet individuals; 500 ind. ha−1 for both fields). In both populations, most weed beet individuals were growing within the row of cultivation: 87.5% and 90% of weed beets were classified as ‘in-row’ bolters, in populations A and B respectively (see Table 1). Population C exhibited an intermediate level of individual density of 2000 ind. ha−1 across the field, of a total of 150 ind. with 35% of ‘in-row’ bolters. Population D showed the highest density of weed beets (3000 ind. ha−1, for a population size of 330 ind.) and featured spatially delimited clusters of weed beets completely localized outside sowing lines, only one out 75 weed beets was localized within the row of cultivation (Table 1).
Table 1.
Summary of genetic diversity estimated from the four populations of weed beets at 10 nuclear microsatellite loci and over all loci
| Population A (n = 40) | Population B (n = 40) | Population C (n = 40) | Population D (n = 75) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Ar | He | FIS | P-value | Ar | He | FIS | P-value | Ar | He | FIS | P-value | Ar | He | FIS | P-value |
| Bvm3 | 11.610 | 0.799 | 0.165 | 0.001 | 9.923 | 0.787 | 0.023 | 0.052 | 8.882 | 0.789 | 0.050 | 0.279 | 8.982 | 0.694 | 0.154 | 0.002 |
| FDSB1027 | 9.538 | 0.707 | −0.051 | 0.666 | 7.764 | 0.746 | 0.003 | 0.471 | 8.790 | 0.798 | 0.154 | 0.001 | 8.663 | 0.742 | 0.119 | 0.104 |
| Caa1 | 14.360 | 0.675 | −0.025 | 0.566 | 9.615 | 0.671 | 0.083 | 0.096 | 9.764 | 0.694 | 0.243 | <10−3 | 4.462 | 0.601 | −0.220 | 0.795 |
| Gcc1 | 2.923 | 0.514 | −0.198 | 0.927 | 3.923 | 0.567 | 0.045 | 0.359 | 4.799 | 0.564 | −0.152 | 0.038 | 2.964 | 0.519 | 0.050 | 0.315 |
| Gtt1 | 3.923 | 0.580 | −0.106 | 0.664 | 3.923 | 0.420 | −0.161 | 0.963 | 3.991 | 0.572 | −0.135 | 0.780 | 4.000 | 0.534 | −0.024 | 0.558 |
| Gaa1 | 2.923 | 0.145 | −0.058 | 1.000 | 2.923 | 0.100 | −0.031 | 1.000 | 3.973 | 0.145 | 0.310 | 0.002 | 2.462 | 0.090 | 0.261 | 0.098 |
| SB04 | 6.000 | 0.746 | −0.117 | 0.940 | 8.923 | 0.758 | −0.183 | 0.949 | 7.946 | 0.792 | 0.010 | 0.668 | 7.180 | 0.681 | 0.021 | 0.493 |
| SB06 | 5.918 | 0.703 | 0.015 | 0.054 | 4.923 | 0.688 | 0.031 | 0.558 | 4.947 | 0.662 | 0.285 | <10−3 | 5.324 | 0.592 | 0.055 | 0.014 |
| SB07 | 10.610 | 0.727 | 0.083 | 0.085 | 6.913 | 0.655 | 0.099 | 0.271 | 9.391 | 0.670 | 0.142 | 0.026 | 4.990 | 0.665 | 0.077 | 0.488 |
| SB15 | 7.947 | 0.786 | −0.004 | 0.397 | 6.918 | 0.794 | −0.034 | 0.765 | 9.838 | 0.823 | 0.040 | 0.336 | 6.890 | 0.787 | 0.001 | 0.436 |
| All loci | 7.575 | 0.638 | −0.018 | 0.051 | 6.5748 | 0.619 | −0.008 | 0.561 | 7.2321 | 0.651 | 0.083 | <10−4 | 5.5917 | 0.591 | 0.033 | <10−4 |
| Geographical coordinates | Latitude N | Longitude E | Latitude N | Longitude E | Latitude N | Longitude E | Latitude N | Longitude E | ||||||||
| 50.5458 | 2.8232 | 50.5421 | 2.8303 | 50.5836 | 2.9067 | 50.5801 | 2.8697 | |||||||||
| Spatial position of weed beets | In-row | Out-row | In-row | Out-row | In-row | Out-row | In-row | Out-row | ||||||||
| No. of individuals | 35 | 5 | 36 | 4 | 14 | 26 | 1 | 74 | ||||||||
Geographical coordinates of sampled populations and the number of weed beets located within (in-row bolter) or outside (out-row bolter) the sowing line are also indicated for each population. Significant heterozygote deficiencies are presented in the P-value column (score test).
n, number of genotyped individuals; Ar, allelic richness; He, expected heterozygosity (gene diversity); FIS, Intra-Population Fixation Index, a measurement of departure from panmixia.
In populations A, B and C, 40 weed beet individuals per population were sampled for molecular analyses (Table 1). For each of these populations, half of the sampled adults were used as maternal plants for mating system analysis with 24 seeds per individual randomly chosen for progeny analyses (Table 2). To ensure representative sampling of the most highly infested sugar beet field, a total of 75 weed beets were sampled and all used as maternal plants for progeny analyses (24 seeds per adult) for population D. Seeds used for progeny analyses were germinated in a greenhouse and grown until seedlings had several leaves to get enough leaf tissue for total DNA extraction.
Table 2.
Estimates of mating system parameters for weed beet progenies from four weed populations sampled within sugar beet fields
| Population | A | B | C | D |
|---|---|---|---|---|
| N | 20 | 20 | 20 | 75 |
| n | 480 | 480 | 480 | 1800 |
| Ft − 1 | 0.068 (0.003) | –0.041 (0.011) | 0.036 (0.006) | 0.042 (0.001) |
| Ft | 0.150 (0.006) | 0.095 (0.003) | 0.076 (0.003) | 0.097 (0.004) |
| δ | 0.503 (0.035) | 0.581 (0.024) | 0.562 (0.030) | 0.031 (0.061) |
| tm | 0.560 (0.011) | 0.629 (0.009) | 0.718 (0.010) | 0.809 (0.006) |
| ts | 0.537 (0.014) | 0.615 (0.012) | 0.697 (0.014) | 0.695 (0.007) |
| tm− ts | 0.023 (0.011) | 0.014 (0.010) | 0.021 (0.011) | 0.114 (0.008) |
| rp | 0.179 (0.022) | 0.087 (0.016) | 0.293 (0.024) | 0.262 (0.015) |
| rs | 0.813 (0.021) | 0.855 (0.029) | 0.787 (0.027) | 0.777 (0.020) |
| 1/rp | 5.6 | 11.5 | 3.4 | 3.8 |
Standard errors, estimated using 1000 bootstraps, are indicated in parentheses.
N, number of adult plants for which progeny arrays were analysed; n, number of progeny; Ft − 1, mean inbreeding coefficient of maternal parents; Ft, mean inbreeding coefficient of progenies; δ, indirect estimates of inbreeding depression following Ritland (1990); tm, mean multilocus population outcrossing rate; ts, mean single-locus population outcrossing rate; (tm − ts), estimation of biparental inbreeding; rp, multilocus correlated paternity within maternal sibships; rs, correlation of selfing among families; 1/rp, approximation of number of males contributing to the paternal mating pool.
Bolting ability
First-year flowering, i.e. bolting ability of seedlings from sampled mother plants, was tested under controlled nonvernalizing conditions for each population. Three seeds per mother plant from populations A, B and C and two seeds per mother plant from population D were sown in a greenhouse. The 330 plants obtained were grown for 28 weeks under nonvernalizing conditions at temperatures ranging from 18.5 to 30.5°C and a 16 h/8 h day/night photoperiod in a greenhouse. Negative controls of bolting were obtained by sowing 10 seeds from each of five sugar beet varieties under the same conditions. Because each tested seedling was genetically related to the mother plant, the statistical analysis of significant differences in bolting rates among populations was carried out, using r version 2.7.1, by means of generalized estimating equations (GEE) to determine parameter estimations for correlated data (Liang and Zeger 1986). As the dependent variable of this analysis is binomial (i.e. a seedling bolts or not), this statistical modelling assumed binomial error and a logit link function.
Genetic data collection
Extraction and purification of total DNA from leaf tissues of the 195 sampled individuals together with their 3240 offspring was performed using a DNeasy 96 Plant Kit (Qiagen Inc., Hilden, Germany) as described in Fénart et al. (2007). All individuals were examined for nuclear genetic variation using 10 microsatellite loci named GAA1, GTT1, GCC1, BVM3, CAA1 (Mörchen et al. 1996; Viard et al. 2002), SB04, SB06, SB07, SB15 (Richards et al. 2004) and FDSB1027 (McGrath et al. 2007). Polymerase chain reaction conditions and allele sizing procedures can be found in Fénart et al. (2007, 2008).
Population genetic structure and mating system analysis
For each population, nuclear genetic polymorphism was described using measurements of allelic richness (Ar), genetic diversity (He) and the unbiased intra-population fixation index [FIS, measuring departures from Hardy–Weinberg (HW) equilibrium within populations] for each locus and over all loci using fstat version 2.9.3.2. (Goudet 1995). Then, deviations from HW equilibrium within each population were tested using the score test implemented in genepop version 3.3 (Raymond and Rousset 1995). Genetic differentiation between populations was quantified using pairwise FST values between populations and their significance was tested by randomly permuting multilocus genotypes among samples (10 000 permutations) using the G log-likelihood statistic (Goudet et al. 1996).
Spatial genetic structure within each population was assessed using the multilocus estimate of the kinship coefficient (Fij) between individuals as described in Loiselle et al. (1995). The average Fij value was computed for 10 distance classes. Confidence intervals of the average Fij values under the null hypothesis of no spatial genetic structure was assessed by permutation tests, in which spatial distances were permuted randomly among pairs of individuals (10 000 permutations). To quantify and compare the strength of spatial genetic structure among populations without arbitrarily setting distance intervals (which can introduce bias), we used the statistic Sp proposed by Vekemans and Hardy (2004). This statistic is independent of the sampling scheme and is defined as the ratio −bF/(1 − F(N)), where bF is the regression slope of Fij against spatial distance between individuals and F(N) is the mean kinship coefficient between neighbouring individuals for the first distance class. All computations were performed using the software SPAGeDi v1.2 (Hardy and Vekemans 2002).
Mating system parameters were estimated at the population level using a maximum-likelihood approach under a mixed-mating system model (Ritland 2002). Average single-locus outcrossing rates (ts), multilocus outcrossing rates (tm), correlation of outcrossed paternity (rp) and correlation of selfing (rs) among families were computed following the numeric Newton-Raphson algorithm and population gene frequencies, using the mltr version 3.2 software package (Ritland 2002). rp Gives an estimation of the proportion of full sibs within an outcrossed progeny array and rs is an estimate of normalized variance in selfing rate among families within a given population, i.e. whether some maternal plants are more prone to increased selfing. The difference between multilocus and single outcrossing rates (tm − ts) provides an estimation of the fraction of apparent selfing because of biparental inbreeding, and the number of effective pollen donors (Nep) siring with success a mother plant can be approximated by 1/rp (e.g. Mimura and Aitken 2007; Sampson and Byrne 2008). Standard errors of estimates were calculated with 1000 bootstraps using progeny arrays as resampling units within families and pairwise population differences were considered significantly different when their confidence intervals of bootstrap distributions did not overlap. Multilocus individual-level estimates of outcrossing rate were also calculated at the family level using a method-of-moment estimator described in Ritland (2002), by using mltr version 3.2. A Mann–Whitney U-test was carried out to test for differences in individual outcrossing rates between ‘in-row’ and ‘out-row’ bolters. The relationship between individual outcrossing rates and local population density was further examined using logistic regressions (r version 2.7.1). The local population density was measured as the number of individuals within a radius of 20 m around each focal maternal plant used for progeny analyses.
As B. vulgaris is a strictly outcrossing species in the wild, selfed progenies of weed beets may be subject to large amounts of inbreeding depression. Direct experimental measurement of inbreeding depression (δ) is challenging and is often underestimated under greenhouse conditions compared with field situations (Husband and Schemske 1996; Cheptou and Schoen 2007). An alternative approach is to estimate δ from the observed selfing rate and the change in inbreeding coefficient between two generations (Goodwillie et al. 2005). An indirect method-of-moments estimator of the relative inbreeding depression acting on selfed progenies was then calculated as δ = 1 − 2tmF′t/[s(1 + Ft − 1 − 2F′t)] where Ft − 1 and F′t denote the inbreeding coefficient of parents and their offspring respectively, and s (= 1 − tm) the selfing rate (Ritland 1990). This estimation of the relative inbreeding depression is based on the change in F between two generations where some selfing occurs, i.e. this method cannot be used to estimate inbreeding depression in a completely outcrossing population. This indirect approach does not assume inbreeding equilibrium and allows the magnitude of inbreeding depression to be conveniently compared among populations in evolutionary and ecological studies (Ritland 1990; Michalski and Durka 2007; Tamaki et al. 2009).
Results
Within and among population genetic structure
Levels of genetic diversity as measured by allelic richness (Ar) and expected heterozygosity (He) were similar to what is usually observed in wild beet populations (see Fievet et al. 2007; Fénart et al. 2008) and can be found in Table 1. Across loci and populations Ar and He ranged from 2.46 to 14.36 and from 0.09 to 0.82 respectively. No significant differences were found among populations in terms of mean levels of genetic diversity (two-tailed t-test, all at P > 0.05). Nonetheless, contrasting results were found for genotypic structure under HW equilibrium: populations A and B showed no significant heterozygote deficiencies (mean FIS = −0.018 and −0.008 respectively), whereas both populations C and D significantly departed from HW expectations with mean FIS values of 0.083 and 0.033 respectively, both at P < 10−4 (Table 1). With respect to genetic differentiation occurring among the four populations, all pairwise comparisons showed highly significant differentiation, except between populations A and B (FST = 0.002; P = 0.064).
Spatial genetic structure differed greatly among populations. No clear relationships were found between pairwise genetic relatedness and geographical location of individuals within populations A and B (Fig. 1). In contrast, populations C and D were genetically structured with stronger spatial genetic structure (Sp = 0.041 and 0.042 respectively, P < 10−3). As a result, we found a significant decrease in genetic similarity with geographical distance between individuals in populations C and D, as expected under isolation by distance when genetic diversity reflects genetic drift and short-range gene flow (Fig. 1). For population D, the correlogram mirrored high genetic distinctiveness between geographically differentiated clusters of weed beets, involving family structure coming from different seed bank sources (Bayesian clustering of individuals, data not shown).
Figure 1.
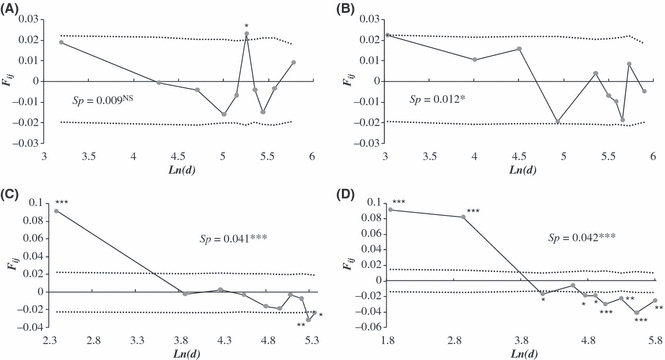
Variation in the average kinship coefficient (Fij) between pairs of weed beet individuals according to (log-transformed) geographical distance for populations A, B, C and D. Ten distance classes were defined in such a way that the number of pairwise comparisons within each distance interval was constant. Dashed lines depict the 95% (two-tailed) confidence interval for the null hypothesis of complete spatial randomness of genotypes. Sp statistics and their significance are also indicated for each population. NS, nonsignificant; *P < 0.05; **P < 0.01; ***P < 0.001.
Variation in first-year flowering
Seeds from cultivated sugar beets (N = 50) used as negative control to test for first-year flowering did not bolt during the 28 weeks of the experiment, ensuring that nonvernalizing conditions were successfully applied in the greenhouse. Thus, the induction of flowering the first year after germination may be as a result of the presence of the dominant B allele in studied weed beet populations. Variations in first-year flowering (bolting rates) in each population are depicted in Fig. 2A. The GEE model showed that there were significant differences among populations for bolting rate (χ23 = 20.38; P < 10−3). While populations A, B and C were not significantly different from each other and displayed the lowest bolting rates, population D was characterized by the highest bolting rate and was significantly different from all the other populations (β = 1.53, SE = 0.39, P < 10−4; Fig. 2A). Given the genetic similarity between populations A and B, these two datasets were pooled. We still found a significant effect of the population on bolting rates (χ22 = 19.72; P < 10−4). Significantly higher bolting rates were found for progenies belonging to population D (β = 1.40, SE = 0.32, P < 10−5) compared with population C and the pooled data set of populations A and B, but population C and the grouping of populations A and B were not significantly different (β = 0.69, SE = 0.38, P = 0.06). With regard to the spatial location of each weed beet individual within the fields, the bolting rate was found to be significantly lower for ‘in-row’ weed beets sampled within the sowing line compared with ‘out-row’ weed beets sampled outside the sowing line (χ21 = 5.10; P < 10−3; Fig. 2B).
Figure 2.
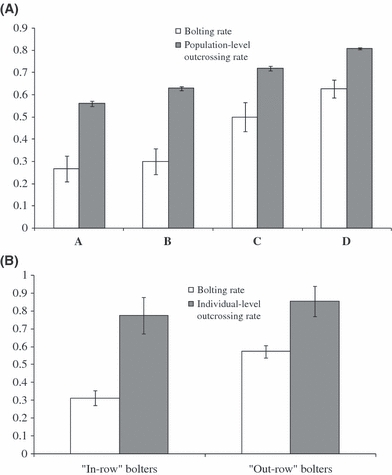
(A) Variation in first-year flowering (bolting rate) and population-level outcrossing rate (tm) in weed beets for populations A, B, C and D. Bolting rates were estimated in the greenhouse after 28 weeks in noninductive conditions (temperature ranging from 18.5 to 30.5°C; 16 h/8 h day/night period). The vertical error bars refer to the standard errors for both rates. (B) Variation in bolting rate and individual-level outcrossing rate (tm) according to weed beet classification: ‘in-row bolters’ found within the row of cultivation and ‘out-row bolters’ found outside the row of cultivation. The vertical error bars refer to the standard errors for both rates.
Mating system analyses
Results from the progeny array analyses are reported in Table 2 and Fig. 2. Large family samples provided estimates with very low standard errors. All populations showed significant departures from complete outcrossing with multilocus estimates of selfing rate s (= 1 − tm) varying from 0.19 (population D) to 0.44 (population A) (Fig. 2A and Table 2). Each population differed significantly from other populations in its propensity for selfing as assessed by nonoverlapping bootstrap estimates, with population D exhibiting the highest outcrossing rate (tm). Selfing rates varied greatly among families, as shown by high levels of correlation of selfing (rs) for all studied populations (Table 2).
We then further performed analyses at the level of individuals: individual family estimates ranged from pure selfing (tm = 0) to pure outcrossing (tm = 1) with 25% of individual tm values ranging from 0 to 0.64 (mean = 0.78; SE = 0.09) and from 0 to 0.51 (mean = 0.76; SE = 0.11) for population A and B respectively. In contrast, population C and D were preferentially composed of purely outcrossed families, with more than 75% of families with individual tm values higher than 0.90; mean (SE) of individual tm being of 0.81 (0.11) and 0.86 (0.07) for populations C and D respectively. No significant relationships between individual tm values and local density (measured as the number of individuals within a radius of 20 m) were observed, as shown in Fig. 3. Likewise, no significant trends were obtained using a radius of 10 m and/or by removing the individuals assumed to be unlikely to carry the Sf gene, i.e. the individuals characterized by a purely outcrossing mating (data not shown). Using the spatial location of each weed beet, individual tm were, however, slightly lower for ‘in-row’ weed beets sampled within the sowing line compared with ‘out-row’ weeds sampled outside the sowing line (Mann–Whitney U-test, P = 0.042; see Fig. 2B). By further examining separately the individual tm between ‘in-row’ and ‘out-row’ weed beets within population C and within populations A and B (pooled together to increase sample size), no significant differences were found in both cases (P = 0.46 and 0.40 respectively).
Figure 3.
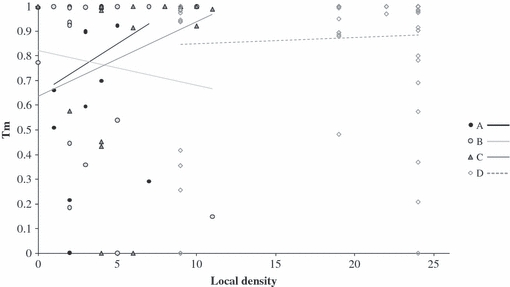
Relationship between individual-level outcrossing rates (tm) and the local density measured as the number of flowering plants within a radius of 20 m around each individual within populations A, B, C and D. Correlation coefficients associated with regression lines are equal to 0.201, −0.132, 0.268 and 0.062 for population A, B, C and D respectively. Logistic regressions suggested no effect of local density on individual outcrossing rates (all at P > 0.05 with χ21 = 0.51, 0.23, 0.82 and 0.18 for population A, B, C and D respectively).
The rate of consanguineous mating because of biparental inbreeding was significantly different from zero for population D only (tm − ts = 0.11). Nonetheless, when biparental inbreeding was investigated by accounting for the occurrence of fine-scaled clusters of genetically distinct weed beets, all tm − ts values dropped, giving estimates not significantly different from 0 (data not shown). The level of correlated paternity rp, the probability that a mother plant draws two male gametes from the same pollen donor, varied from 0.087 to 0.293 (Table 2). This gave rise to a number of sires ranging from 3.4 (population C) to 11.5 (population B). Indirect two-generation estimates of inbreeding depression δ, inferred from inbreeding coefficient of parents and progeny and observed selfing rates, suggested that mother plants were generally much less inbred than expected in the absence of inbreeding depression. As a result, δ estimates were high for all populations (>0.5) with the exception of population D for which δ was close to 0 (Table 2).
Discussion
Keeping in mind that our results must be interpreted with caution because of the limited number of populations studied, patterns of variation in population genetic structure and mating system in relation to weed population characteristics can be drawn from this study. Weed beet populations usually suffer from rapid turnover owing to farming practices, namely crop rotation and weed management (Desplanque et al. 2002; Arnaud et al. 2003). Once a long-lived seed bank has been established, weed beet populations can reappear despite stochastic disturbance. These weed beets can be kept at very low population sizes through efficient and careful hand-weeding (Desplanque et al. 2002). Populations A and B (i) were characterized by low densities of weed beets, most of them being localized within the sowing line and thus originating from the sown beet crop seeds, (ii) were not genetically differentiated from each other, (iii) showed no or very moderate spatial structure of nuclear genetic diversity and (iv) displayed a very large heterozygosity level often found in F1 crop–wild hybrids (Prentis et al. 2008). As a result of admixture among previously isolated lineages, F1 crop–wild hybrids are expected to exhibit high levels of genotypic diversity and recent weed populations with a common history of hybridization are not likely to be strongly genetically differentiated from each other (Viard et al. 2002). Therefore, both the location of individuals and the within-population genetic structure support the hypothesis that weed populations A and B may be in their first year of establishment and are mainly composed of F1 crop–wild hybrids coming from the sown crop seed. The lack of genetic differentiation also suggests that they share a common history of invasion, arising from similar sugar beet seedlots containing F1 crop–wild hybrid seeds that had been accidentally sired by wild pollen donors in the seed production area. However, the occurrence of both a geographical and a genetic proximity for populations A and B do not mean that the genetic similarity among weed beet populations is dependent on the geographical distance separating them. Indeed, weed populations generally arise independently and from different seedlots. Furthermore, no isolation-by-distance genetic structure has been found to date among populations of weed beets (Arnaud et al. 2003; Viard et al. 2004; Fénart et al. 2008).
Our findings provide further evidence that admixture between individuals from disparate sources can result in high levels of genetic diversity similar to what can be found in wild beet populations (Viard et al. 2004; Arnaud et al. 2009). This departs from classical models of natural dispersal where genetic bottlenecks arise because of founder events during the colonization process (Born et al. 2008), and supports the fact that, contrary to conventional expectations, significantly reduced genetic diversity is relatively unusual in plant invasions (reviewed in Dlugosch and Parker 2008; Wilson et al. 2009). Because of the domestication process and the ensuing breeding selection, sugar beet varieties generally show a strongly reduced nuclear and cytoplasmic genetic variation compared with wild populations (Fénart et al. 2008). In the particular case of crop–wild hybrid weed beets, most of the observed nuclear polymorphism is thus likely to come from the wild ruderal beet populations.
In contrast, strong spatial genetic structure associated with significant departures from HW equilibrium was detected in populations C and D, both of which were characterized by a high proportion of weed beet individuals found outside the row of cultivation. These ‘out-row’ weed beets originate from the seed bank and are likely to be backcrossed crop–wild hybrids from crosses between flowering weed beets in previous sugar beet crops, cultivated at least 2–4 years ago depending on the crop rotation management. A long-lived seed bank is a key factor for successful establishment and subsequent re-emergence of weed beet populations in later years (Sester et al. 2006). In Beta vulgaris, the unit of dispersal is a multi-seeded fruit dispersed by gravity which provides opportunities for a strong kin-genetic structuring involving sib groups (Fievet et al. 2007). Both stronger within-population spatial genetic structure and significant among-population differentiation are then expected in weed beet populations emerging from a seed bank (Viard et al. 2002). We suggest that this hypothesis may be relevant for populations C and D for which significant within and among spatial genetic structure occurred, and for which most weed beets were found to bolt and flower outside the sowing line. The increase in isolation by distance, as portrayed by correlograms, actually reflects the result of independent resurgence events that involve genetically related individuals dispersed in family groups. Accordingly, the significant FIS values found within populations C and D may stem from a Wahlund effect involving the resurgence of genetically differentiated seed sources rather than as a consequence of the mating system, which was found to be highly outcrossing compared with populations A and B (see below). However, we did not detect significant differences in genetic diversity among the studied populations. This implies that once established weed beet populations can sustain a high level of genetic variation.
Variation of first-year flowering
Among the mechanisms that can be involved in rapid evolutionary change, a ruderal trait such as first-year flowering is a good candidate as an adaptive response to a novel and changing environment (Campbell et al. 2009). Moreover, characteristics of particular source populations within initial native ranges are likely to shape the evolution of invasive populations (Lee and Gelembiuk 2008). This is particularly true in our case study as inland wild beets that sired the cultivated seed bearers in seed production fields are already adapted to the margins of arable fields and other anthropogenic habitats through short life-span and first-year flowering (Van Dijk and Desplanque 1999; Arnaud et al. 2009). In Beta vulgaris the induction of first-year flowering mainly depends on the Mendelian bolting gene B (Van Dijk 2009). F1 crop–wild hybrids are likely to show the genotype Bb as a result of the hybridization between cultivated lines (bb) selected for strong vernalization requirement and wild beets that express the dominant B allele in high frequencies in the seed production area in southwestern France (Van Dijk 2004). Weed populations characterized by a high occurrence of F1 crop–wild hybrids showed the lowest bolting rates, whereas offspring from population D, resulting from the seed bank resurgence, exhibited a significantly higher bolting rate. Accordingly, a higher mean level of bolting was found in ‘out-row’ weed beets, compared with ‘in-row’ weed beets. Thus, first-year flowering appeared to be positively selected for in later stages of weed beet populations. This finding was expected given the current agricultural practices: weed beets that start flowering and reproducing 1 year earlier are selected for in response to selective pressures driving rapid and massive seed production before harvest. Through outcrossing recombination events, the mating system is also likely to accelerate the loss of crop traits towards increased weediness inherited from the wild (Van Dijk and Desplanque 1999). However, despite intensive artificial selection to avoid flowering, a low quantitative vernalization requirement can allow the flowering of cultivar bb genotypes (Van Dijk 2004). This is likely to slow down the selection of the B gene. It may also explain the large difference in density of ‘in-row’ bolters for population D compared with populations A, B and C. This difference could be related to (i) cultivars coming from different seedlots showing differential vernalization requirements and/or (ii) to differences in sowing dates, with sugar beets sown too early in spring being more likely to be subject to cold conditions. Alternatively, differences among studied populations in frequencies of ‘in-row’ weed beets may reflect, over space and time, fluctuating contamination rates of seedlots by pollen from wild ruderal populations in the vicinity of seed production areas (Viard et al. 2002; Arnaud et al. 2009).
Evolutionary outcomes of mating system variation
We found a mixed-mating system and a significant departure from complete outcrossing within all studied weed beet populations. The progeny array analysis indicated that biparental inbreeding was marginal and provided evidence that the estimated selfing rates were not biased by mating among relatives. Therefore, this genetic signature of selfing events indirectly suggests the introgression of the Sf gene into a mixed crop-wild genetic background in weed beet lineages. Nonetheless, the mating system varied significantly among populations, with predominantly outcrossed progenies occurring in large populations.
When considering ecological and demographical factors, the mating system can be influenced by effective neighbourhood size, density and isolation (Goodwillie et al. 2005; Cheptou and Schoen 2007; Michalski and Durka 2007; Mimura and Aitken 2007). Indeed, while the mating system in animal-pollinated species can be the result of complex interactions between plants and their associated pollinators, in wind-pollinated species the outcrossing rate is often positively correlated with population size and density because of spatially restricted pollen dispersal (Robledo-Arnuncio et al. 2004; Friedman and Barrett 2008; Zhao et al. 2009). Populations A and B were not genetically differentiated from each other at neutral markers but differed significantly in their selfing rates. While this difference may arise from environmental effects, we did not depict any significant relationship between the individual outcrossing rate and the local density for either population. One equally likely alternative explanation implicates sampling effects associated with a variable frequency of the Sf gene in male-sterile seed bearers selected in breeding programs and used in the seed production fields.
The joint occurrence of the Sf gene and low population density should limit successful outcrossing events during the first stages of weed colonization. It may also explain why populations A and B composed of F1 crop–wild hybrids exhibited the highest selfing rates, whereas higher individual tm values were found for ‘out-row’ weed beets resurging from the seed bank. Although the reproductive assurance hypothesis enjoys little empirical support as a successful selective determinant for the breakdown of self-incompatibility (e.g. Elam et al. 2007; Lafuma and Maurice 2007), it may be relevant for newly established weed beets because selfing confers an advantage by ensuring offspring production when outcrossed pollen is limiting (Baker 1974; Kalisz et al. 2004; Dornier et al. 2008). However, we did not find any significant relationship between individual outcrossing rates and local density for any of the studied populations. Beyond limited statistical power because of the small number of studied populations, this may be as a result of the production of large amounts of pollen able to disperse over long distances in agricultural landscapes (Fénart et al. 2007; Darmency et al. 2009). Accordingly, the higher number of effective pollen donors found for low-density weed beet populations A and B could thus be ascribed to long-distance pollination events. A long-lived seed bank could also theoretically offset the advantage of reproductive assurance by increasing the effective population size, thereby providing more opportunities for outcrossing events (Pannell and Barrett 1998).
Gametophytic self-incompatibility prevents selfing and reproduction with genetically related individuals in wild beet populations (Maletski and Weisman 1978; De Cauwer et al. 2010). In this study, the two-generation δ estimates suggested that inbreeding depression was >0.5 in weed populations that exhibited the highest selfing rates. Assuming that inbreeding depression is negatively correlated with historical levels of selfing in a population (Charlesworth and Charlesworth 1987; Ritland 1990; Goodwillie et al. 2005), higher inbreeding depression is expected in less inbred populations. Crop–wild hybridization could explain this discrepancy: populations composed of F1 crop–wild hybrid beets are de facto less inbred (because of crossing between genetically differentiated lineages). Some of these hybrids are likely to carry the crop-like Sf gene that offsets self-incompatibility, but they may suffer from inbreeding depression in their selfed progeny because of a genetic load coming from the wild gene pool. This situation is far from classical equilibrium state and should be validated with additional population sampling and greenhouse experiments. Moreover, these indirect measurements of δ, estimated from molecular progeny analyses have some limits because the strong inbreeding depression acting on the earliest stages of development and preventing seed emergence cannot be accounted for (Goodwillie et al. 2005). This could explain why the δ estimate was close to zero for population D, while a moderate, but significant, proportion of selfing events (s = 0.19) still occurred. Therefore, any inbreeding depression that occurs before assaying seedlings for selfing can only be estimated by measuring the decrease in seed set in artificial self-pollination experiments.
Conclusion
We found positive selection for first-year flowering, high genetic diversity and a mixed-mating system in weed beet populations. However, there was no clear evidence for a mating system shift towards increased selfing as would be expected under a reproductive assurance hypothesis. From an applied and practical point of view, our results highlight the need to carefully survey and manage both the sugar beet fields and the seed production area. In sugar beet fields, low-density weed beet populations may be difficult to detect and considered harmless by farmers. Nonetheless, they should be eradicated in due time before the onset of flowering season in mid-July to avoid giving rise to dormant seed banks in which first-year flowering ability has been positively selected (Van Dijk and Desplanque 1999). Moreover, farmers should avoid sowing sugar beet seeds too early in spring to limit the exposure to cold weather which is likely to favour an additional flowering of cultivars with low vernalization requirement. Likewise, in the light of the development of genetically modified (GM) crops, GM herbicide-tolerant beet lines could be potentially put on the market. Careful destruction of all bolting beets is thus of particular concern and should be a compulsory measure to avoid the occurrence of transgenic herbicide-tolerant weed beets and to prevent any gene flow between conventional and GM sugar beet fields (Ellstrand 2003; Darmency et al. 2009). Similarly, the seed production area is a theatre of recurrent crop–wild hybridization giving rise to weed beets. Although breeders take extreme precautions to eradicate all wild or weedy beets to avoid any contamination of seedlots by crop–wild hybrids, more efficient management of wild beet populations occurring in the vicinity of seed production fields is still needed (Desplanque et al. 2002; Arnaud et al. 2009).
From an evolutionary point of view, agricultural weeds evolve in response to selective pressures associated with crop cultivation. The central importance of evolutionary ecology for understanding weed invasion, persistence and management has recently been emphasized by Neve et al. (2009). In this regard, an efficient weeding could also promote pollen limitation of seed production, which would indirectly generate significant levels of selfing and low reproductive success of weed beets through inbreeding depression. However, this hypothesis would require further validation because high selfing rates, in the long term, could also lead to a purging of genetic load which could favour crop–wild hybrids. In this respect, additional weed populations with known population history and further comparative experiments to measure progeny fitness along with assessments of seed set are needed to gain further insights into the effects of inbreeding depression and pollen limitation on the invasive ability of weedy beets.
Acknowledgments
We would like to thank V. Castric, M. Dufay, C.R. Engel, I. Olivieri, E. Petit, P. Touzet, H. Van Dijk and two anonymous referees for insightful comments on the manuscript. We also wish to express our gratitude to S. Billiard, I. De Cauwer and Y. Outreman for helpful advices in statistical analyses and to R. Dron and E. Schmitt for technical assistance in the greenhouse. This work was funded by the ‘Contrat de Plan État/Région Nord-Pas-de-Calais’. S. Fénart was supported by an INRA/Région Nord-Pas-de-Calais fellowship. J.-F. Arnaud is grateful to the CNRS for supporting him as a full-time research scientist during the 2008–2009 academic year.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Graphical representation of the geographical location of the sugar beet seed production area, the sugar beet production area, and the geographical distribution where inland wild beet and wild sea beet occur in France.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Literature cited
- Arnaud J-F, Viard F, Delescluse M, Cuguen J. Evidence for gene flow via seed dispersal from crop to wild relatives in Beta vulgaris (Chenopodiaceae): consequences for the release of genetically modified crop species with weedy lineages. Proceedings of the Royal Society of London B. 2003;270:1565–1571. doi: 10.1098/rspb.2003.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud J-F, Fénart S, Godé C, Deledicque S, Touzet P, Cuguen J. Fine-scale geographical structure of genetic diversity in inland wild beet populations. Molecular Ecology. 2009;18:3201–3215. doi: 10.1111/j.1365-294X.2009.04279.x. [DOI] [PubMed] [Google Scholar]
- Baker HG. The evolution of weeds. Annual Review of Ecology & Systematics. 1974;5:1–24. [Google Scholar]
- Barrett SCH, Colautti RI, Eckert CG. Plant reproductive systems and evolution during biological invasion. Molecular Ecology. 2008;17:373–383. doi: 10.1111/j.1365-294X.2007.03503.x. [DOI] [PubMed] [Google Scholar]
- Born C, Kjellberg F, Chevallier M-H, Vignes H, Dikangadissi J-T, Sanguié J, Wickings EJ. Colonization processes and the maintenance of genetic diversity: insights from a pioneer rainforest tree, Aucoumea klaineana. Proceedings of the Royal Society of London B. 2008;275:2171–2179. doi: 10.1098/rspb.2008.0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudry P, Mörchen M, Saumitou-Laprade P, Vernet P, Van Dijk H. The origin and evolution of weed beets: consequences for the breeding and release of herbicide resistant transgenic sugar beets. Theoretical and Applied Genetics. 1993;87:471–478. doi: 10.1007/BF00215093. [DOI] [PubMed] [Google Scholar]
- Boudry P, Wieber R, Saumitou-Laprade P, Pillen K, Van Dijk H, Jung C. Identification of RFLP markers closely linked to the bolting gene B and their significance for the study of the annual habit in beets (Beta vulgaris L.) Theoretical and Applied Genetics. 1994;88:852–858. doi: 10.1007/BF01253996. [DOI] [PubMed] [Google Scholar]
- Campbell LG, Snow AA, Sweeney PM, Ketner JM. Rapid evolution in crop-weed hybrids under artificial selection for divergent life histories. Evolutionary Applications. 2009;2:172–186. doi: 10.1111/j.1752-4571.2008.00051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. Inbreeding depression and its evolutionary consequences. Annual Review of Ecology & Systematics. 1987;18:237–268. [Google Scholar]
- Cheptou P-O, Schoen DJ. Combining population genetics and demographical approaches in evolutionary studies of plant mating systems. Oikos. 2007;116:271–279. [Google Scholar]
- Darmency H, Klein EK, Gestat De Garambé T, Gouyon P-H, Richard-Molard M, Muchembled C. Pollen dispersal in sugar beet production fields. Theoretical and Applied Genetics. 2009;118:1083–1092. doi: 10.1007/s00122-009-0964-y. [DOI] [PubMed] [Google Scholar]
- De Cauwer I, Dufay M, Cuguen J, Arnaud J-F. Effects of fine-scale genetic structure on male mating success in gynodioecious Beta vulgaris ssp. maritima. Molecular Ecology. 2010;19 doi: 10.1111/j.1365-294X.2010.04586.x. (in press) [DOI] [PubMed] [Google Scholar]
- Desplanque B, Hautekèete N-C, Van Dijk H. Transgenic weed beets: possible, probable, avoidable? Journal of Applied Ecology. 2002;39:561–571. [Google Scholar]
- Dlugosch KM, Parker IM. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Molecular Ecology. 2008;17:431–449. doi: 10.1111/j.1365-294X.2007.03538.x. [DOI] [PubMed] [Google Scholar]
- Dornier A, Munoz F, Cheptou P-O. Allee effect and self-fertilization in hermaphrodites: reproductive assurance in a structured metapopulation. Evolution. 2008;62:2558–2569. doi: 10.1111/j.1558-5646.2008.00464.x. [DOI] [PubMed] [Google Scholar]
- Elam DR, Ridley CE, Goodell K, Ellstrand NC. Population size and relatedness affect fitness of a self-incompatible invasive plant. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:549–552. doi: 10.1073/pnas.0607306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellstrand NC. Current knowledge of gene flow in plants: implications for transgene flow. Philosophical Transactions of the Royal Society of London B. 2003;358:1163–1170. doi: 10.1098/rstb.2003.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellstrand NC, Schierenbeck KA. Hybridization as a stimulus for the evolution of invasiveness in plants? Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7043–7050. doi: 10.1073/pnas.97.13.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fénart S, Austerlitz F, Cuguen J, Arnaud J-F. Long distance pollen-mediated gene flow at a landscape level: the weed beet as a case study. Molecular Ecology. 2007;16:3801–3813. doi: 10.1111/j.1365-294X.2007.03448.x. [DOI] [PubMed] [Google Scholar]
- Fénart S, Arnaud J-F, De Cauwer I, Cuguen J. Nuclear and cytoplasmic genetic diversity in weed beet and sugar beet accessions compared to wild relatives: new insights into the genetic relationships within the Beta vulgaris complex species. Theoretical and Applied Genetics. 2008;116:1063–1077. doi: 10.1007/s00122-008-0735-1. [DOI] [PubMed] [Google Scholar]
- Fievet V, Touzet P, Arnaud J-F, Cuguen J. Spatial analysis of nuclear and cytoplasmic DNA diversity in wild sea beet (Beta vulgaris ssp. maritima) populations: do marine currents shape the genetic structure? Molecular Ecology. 2007;16:1847–1864. doi: 10.1111/j.1365-294X.2006.03208.x. [DOI] [PubMed] [Google Scholar]
- Friedman J, Barrett SCH. High outcrossing in the annual colonizing species Ambrosia artemisiifolia (Asteraceae) Annals of Botany (London) 2008;101:1303–1309. doi: 10.1093/aob/mcn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwillie C, Kalisz S, Eckert CG. The evolutionary enigma of mixed mating systems in plants: occurrence, theoretical explanations, and empirical evidence. Annual Review of Ecology, Evolution & Systematics. 2005;36:47–79. [Google Scholar]
- Goudet J. FSTAT (Version 1.2). A computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Goudet J, Raymond M, De Meeüs T, Rousset F. Testing differentiation in diploid populations. Genetics. 1996;144:1933–1940. doi: 10.1093/genetics/144.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemin M-L, Faugeron S, Destombe C, Viard F, Correa JA, Valero M. Genetic variation in wild and cultivated populations of the haploid-diploid red alga Gracilaria chilensis: how farming practices favor asexual reproduction and heterozygosity. Evolution. 2008;62:1500–1519. doi: 10.1111/j.1558-5646.2008.00373.x. [DOI] [PubMed] [Google Scholar]
- Hamrick JL, Godt MJW. Effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society of London B. 1996;351:1291–1298. [Google Scholar]
- Hardy OJ, Vekemans X. SPAGEDI: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes. 2002;2:618–620. [Google Scholar]
- Husband BC, Schemske DW. Evolution of the magnitude and timing of inbreeding depression in plants. Evolution. 1996;50:54–70. doi: 10.1111/j.1558-5646.1996.tb04472.x. [DOI] [PubMed] [Google Scholar]
- Kalisz S, Vogler DW, Hanley KM. Context-dependent autonomous self-fertilization yields reproductive assurance and mixed mating. Nature. 2004;430:884–887. doi: 10.1038/nature02776. [DOI] [PubMed] [Google Scholar]
- Lafuma L, Maurice S. Increase in mate availability without loss of self-incompatibility in the invasive species Senecio inaequidens (Asteraceae) Oikos. 2007;116:201–208. [Google Scholar]
- Lee CE, Gelembiuk GW. Evolutionary origins of invasive populations. Evolutionary Applications. 2008;1:427–448. doi: 10.1111/j.1752-4571.2008.00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- Loiselle BA, Sork VL, Nason J, Graham C. Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae) American Journal of Botany. 1995;82:1420–1425. [Google Scholar]
- Maletski SI, Weisman NJ. A population genetic analysis of self- and cross-incompatibility in sugar beet (Beta Vulgaris L.) Theoretical and Applied Genetics. 1978;52:21–28. doi: 10.1007/BF00273762. [DOI] [PubMed] [Google Scholar]
- McGrath JM, Trebbi D, Fenwick A, Panella L, Schulz B, Laurent V, Barnes SR, et al. An open-source first-generation molecular genetic map from a sugarbeet × table beet cross and its extension to physical mapping. Crop Science. 2007;47(S1):27–44. [Google Scholar]
- Michalski SG, Durka W. High selfing and high inbreeding depression in peripheral populations of Juncus atratus. Molecular Ecology. 2007;16:4715–4727. doi: 10.1111/j.1365-294X.2007.03547.x. [DOI] [PubMed] [Google Scholar]
- Mimura M, Aitken SN. Increased selfing and decreased effective pollen donor number in peripheral relative to central populations in Picea sitchensis (Pinaceae) American Journal of Botany. 2007;94:991–998. doi: 10.3732/ajb.94.6.991. [DOI] [PubMed] [Google Scholar]
- Mörchen M, Cuguen J, Michaelis G, Hanni C, Saumitou-Laprade P. Abundance and lenght polymorphism of microsatellite repeats in Beta vulgaris L. Theoretical and Applied Genetics. 1996;92:326–333. doi: 10.1007/BF00223675. [DOI] [PubMed] [Google Scholar]
- Neve P, Vila-Aiub M, Roux F. Evolutionary-thinking in agricultural weed management. New Phytologist. 2009;184:783–793. doi: 10.1111/j.1469-8137.2009.03034.x. [DOI] [PubMed] [Google Scholar]
- Owen FV. Inheritance of cross- and self-sterility and self-fertility in Beta vulgaris. Journal of Agricultural Research. 1942;64:679–698. [Google Scholar]
- Pannell JR, Barrett SCH. Baker's law revisited: reproductive assurance in a metapopulation. Evolution. 1998;52:657–668. doi: 10.1111/j.1558-5646.1998.tb03691.x. [DOI] [PubMed] [Google Scholar]
- Prentis PJ, Wilson JRU, Dormontt EE, Richardson DM, Lowe AJ. Adaptive evolution in invasive species. Trends in Plant Science. 2008;13:288–294. doi: 10.1016/j.tplants.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (Version 1.2): a population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Richards CM, Brownson M, Mitchell SE, Kresovich S, Panella L. Polymorphic microsatellite markers for inferring diversity in wild and domesticated sugar beet (Beta vulgaris. Molecular Ecology Notes. 2004;4:243–245. [Google Scholar]
- Ridley CE, Ellstrand NC. Rapid evolution of morphology and adaptive life history in the invasive California wild radish (Raphanus sativus) and the implications for management. Evolutionary Applications. 2010;3:64–76. doi: 10.1111/j.1752-4571.2009.00099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritland K. Inferences about inbreeding depression based on changes of the inbreeding coefficient. Evolution. 1990;44:1230–1241. doi: 10.1111/j.1558-5646.1990.tb05227.x. [DOI] [PubMed] [Google Scholar]
- Ritland K. Extensions of models for the estimation of mating systems using n independent loci. Heredity. 2002;88:221–228. doi: 10.1038/sj.hdy.6800029. [DOI] [PubMed] [Google Scholar]
- Robledo-Arnuncio JJ, Alia R, Gil L. Increased selfing and correlated paternity in a small population of a predominantly outcrossing conifer, Pinus sylvestris. Molecular Ecology. 2004;13:2567–2577. doi: 10.1111/j.1365-294X.2004.02251.x. [DOI] [PubMed] [Google Scholar]
- Sampson JF, Byrne M. Outcrossing between an agroforestry plantation and remnant native populations of Eucalyptus loxophleba. Molecular Ecology. 2008;17:2769–2781. doi: 10.1111/j.1365-294X.2008.03779.x. [DOI] [PubMed] [Google Scholar]
- Sester M, Dürr C, Darmency H, Colbach N. Evolution of weed beet (Beta vulgaris L.) seed bank: quantification of seed survival, dormancy, germination and pre-emergence growth. European Journal of Agronomy. 2006;24:19–25. [Google Scholar]
- Tamaki I, Ishida K, Setsuko S, Tomaru N. Interpopulation variation in mating system and late-stage inbreeding depression in Magnolia stellata. Molecular Ecology. 2009;18:2365–2374. doi: 10.1111/j.1365-294X.2009.04195.x. [DOI] [PubMed] [Google Scholar]
- Van Dijk H. Gene exchange between wild and crop in Beta vulgaris: how easy is hybridization and what will happen in later generations? In: Den Nijs HCM, Bartsch D, Sweet J, editors. Introgression from Genetically Modified Plants into Wild Relatives and its Consequences. Oxfordshire: CABI publishers, Inc; 2004. pp. 53–69. [Google Scholar]
- Van Dijk H. Evolutionary change in flowering phenology in the iteroparous herb Beta vulgaris ssp. maritima: a search for the underlying mechanisms. Journal of Experimental Botany. 2009;60:3143–3155. doi: 10.1093/jxb/erp142. [DOI] [PubMed] [Google Scholar]
- Van Dijk H, Desplanque B. European Beta: crops and their wild and weedy relatives. In: Van Raamsdonk LWD, Den Nijs JCM, editors. Plant Evolution in Man-Made Habitats. Amsterdam: Hugo de Vries Laboratory; 1999. pp. 257–270. [Google Scholar]
- Vekemans X, Hardy OJ. New insights from fine-scale spatial genetic structure analyses in plant populations. Molecular Ecology. 2004;13:921–935. doi: 10.1046/j.1365-294x.2004.02076.x. [DOI] [PubMed] [Google Scholar]
- Viard F, Bernard J, Desplanque B. Crop-weed interactions in the Beta vulgaris complex at a local scale: allelic diversity and gene flow within sugar beet fields. Theoretical and Applied Genetics. 2002;104:688–697. doi: 10.1007/s001220100737. [DOI] [PubMed] [Google Scholar]
- Viard F, Arnaud J-F, Delescluse M, Cuguen J. Tracing back seed and pollen flow within the crop-wild Beta vulgaris complex: genetic distinctiveness versus hot spots of hybridization over a regional scale. Molecular Ecology. 2004;13:1357–1364. doi: 10.1111/j.1365-294X.2004.02150.x. [DOI] [PubMed] [Google Scholar]
- Wilson JRU, Dormontt EE, Prentis PJ, Lowe AJ, Richardson DM. Something in the way you move: dispersal pathways affect invasion success. Trends in Ecology & Evolution. 2009;24:136–144. doi: 10.1016/j.tree.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Zhao R, Xia H, Lu BR. Fine-scale genetic structure enhances biparental inbreeding by promoting mating events between more related individuals in wild soybean (Glycine soja; Fabaceae) populations. American Journal of Botany. 2009;96:1138–1147. doi: 10.3732/ajb.0800173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


