Abstract
Habitat fragmentation affects the integrity of many species, but little is known about species-specific sensitivity to fragmentation. Here, we compared the genetic structure of four freshwater fish species differing in their body size (Leuciscus cephalus; Leuciscus leuciscus; Gobio gobio and Phoxinus phoxinus) between a fragmented and a continuous landscape. We tested if, overall, fragmentation affected the genetic structure of these fish species, and if these species differed in their sensitivity to fragmentation. Fragmentation negatively affected the genetic structure of these species. Indeed, irrespective of the species identity, allelic richness and heterozygosity were lower, and population divergence was higher in the fragmented than in the continuous landscape. This response to fragmentation was highly species-specific, with the smallest fish species (P. phoxinus) being slightly affected by fragmentation. On the contrary, fish species of intermediate body size (L. leuciscus and G. gobio) were highly affected, whereas the largest fish species (L. cephalus) was intermediately affected by fragmentation. We discuss the relative role of dispersal ability and effective population size on the responses to fragmentation we report here. The weirs studied here are of considerable historical importance. We therefore conclude that restoration programmes will need to consider both this societal context and the biological characteristics of the species sharing this ecosystem.
Keywords: cultural context, cyprinids, dams, genetic structure, historical monument, meta-analysis, river restoration, umbrella species, weirs
Introduction
Habitat fragmentation is probably the most pervasive effect humans impose on wild species (Vitousek et al. 1997; Lawler et al. 2006). It is also among the most highly studied phenomenon of the process of global change, and many studies have improved our knowledge on the ecological and evolutionary outcomes of fragmented populations (Fahrig 2003; Ewers and Didham 2006; Lawler et al. 2006). Typically, at the meta-population scale, habitat fragmentation causes habitat patches to be reduced in size and to be isolated from one another, hence decreasing gene flow between patches (Fahrig 2003). For most species, this spatial rearrangement also decreases the effective population size as well as genetic diversity at the patch scale, through the processes of genetic drift and inbreeding (Frankham 1998; Couvet 2002; DiBattista 2008). Ultimately, in the better cases, changes in the genetic structure of connected populations modify their evolutionary trajectory (by changes in life-history characteristics for instance Johansson et al. 2007; Waples et al. 2008), or in the worst cases, lead to local extinction (Spielman et al. 2004).
An important question that needs to be resolved for predicting the ecological and evolutionary damages caused by habitat fragmentation is as follows: is the response to habitat fragmentation species-specific? Answering this question is essential for ecological managers as it allows the identification of ‘umbrella species’ (i.e. species whose environmental requirements encapsulate the needs of most other species) that are used to define the appropriate ways landscapes must be managed for preserving diversity (Lambeck 1997). Indeed, several phenotypic traits can make a species more or less susceptible to habitat fragmentation. Body size is one such trait because it correlates with several characteristics of individuals or populations (e.g. dispersal, effective population size or trophic status) that are known to affect species sensibility to fragmentation (Davies et al. 2000; Henle et al. 2004; Ewers and Didham 2006). For instance, all things being equal, theory predicts that large-bodied species should have a higher dispersal capability enabling them to rescue or re-colonize distant patches and hence to support patch isolation, even in a highly fragmented habitat (reviewed in Ewers and Didham 2006; Henle et al. 2004). Alternatively, large-bodied species often have a small population size (Cotgreave 1993; Blackburn and Gaston 1997), and rare species are expected to be more sensitive to fragmentation, probably because of their low effective population size (Ne) (Davies et al. 2000). However, empirical studies testing the hypothesis that body size underlies species-specific responses to fragmentation remain scant because most studies target single species (but see Davies et al. 2000).
Fragmentation caused by humans is particularly prevalent in freshwater ecosystems (Nilsson et al. 2005). Indeed, for a very long time, humans have built dams for managing irrigation, producing energy or for recreational purposes. Dams are so widespread that the water flow of over half the large river systems in the world is affected by them (Nilsson et al. 2005). The ecological and evolutionary impacts of dams are obvious and range from the loss of biodiversity to the malfunctioning of the whole ecosystem (Loot et al. 2007; Poulet 2007; Grenouillet et al. 2008; Maloney et al. 2008). At the genetic level, the negative effects of dams have been unambiguously demonstrated for many fish species (Alo and Turner 2005; Wofford et al. 2005; Raeymaekers et al. 2008). Based on these results, a straightforward restoration tool consists in ‘un-damming’ rivers (Bednarek 2001; Palmer et al. 2005, 2008). Although environmentally beneficial, this solution can however prove undesirable when dams are also part of the historical and cultural heritage of a region or when they provide benefits for society. For example, many small to medium European rivers are scattered with water mills associated to small weirs (2–3 m high) that, at least partially, impede the movement of fish (Ovidio and Philippart 2002; Raeymaekers et al. 2008). Many of these man-made structures date back to the 15th century, and are thus part of the cultural heritage of local populations (see Fig. 1 and Raeymaekers et al. 2009). In such cases, adopting management practices requires consideration of both the ecological and social dimensions of these structures.
Figure 1.

Traditional mill weir of the river Viaur (the fragmented landscape). They are made of rocks and most date back to the Middle Ages (12th–15th centuries). They are hence part of the local and cultural heritage, but they also impede the dispersal of several fish species. More than 50 such weirs are scattered along the river Viaur. Copyright: Loïc Tudesque.
In this study, we present a comparative study aimed at evaluating the effects of anthropogenic river fragmentation on the genetic integrity of four widespread fish species (the European chub, Leuciscus cephalus; the rostrum dace, Leuciscus leuciscus; the gudgeon, Gobio gobio and the European minnow, Phoxinus phoxinus). These fish species differ mostly with respect to their maximal body size and we hypothesized that they may display species-specific responses to river fragmentation (Davies et al. 2000). To test this hypothesis, we sampled and genotyped (using microsatellite loci) these species along two nearby river basins (i.e. landscapes); one being highly fragmented (the river Viaur) and the other being a continuous landscape (the river Célé). We first compared the spatial patterns of genetic diversity between the landscapes and among the fish species using allelic richness (AR) and heterozygosity as two measures of genetic diversity. Irrespective of the species, we expected to find a lower genetic diversity in the fragmented landscape because of the stronger influence of patch isolation on genetic drift and inbreeding (Frankham 1998; Couvet 2002; Keller and Waller 2002). We also expected the smallest fish species to exhibit the greatest response to fragmentation if dispersal ability is the most important characteristic explaining species sensitivity to fragmentation (Ewers and Didham 2006). The reverse is expected (i.e. the largest species are more sensitive) if, alternatively, effective population size is the best explanation for sensitivity to fragmentation (Davies et al. 2000). By combining the use of F-statistics (Wright 1951) and the isolation-by-distance (IBD) framework (hereafter IBD Hutchison and Templeton 1999; Slatkin 1993), we then compared the population genetic structure of each species between the two landscapes. We expect a higher level of genetic structure (i.e. higher Fst values) for all species in the fragmented landscape because weirs might partially impede dispersal between patches (Raeymaekers et al. 2008). Particularly, the smallest or the largest species should be more strongly affected if dispersal ability or effective population size respectively explain sensitivity to fragmentation. We also expect species-specific responses on IBD patterns. Specific responses would depend upon the initial equilibrium between genetic drift and gene flow of each species in the absence of fragmentation (Hutchison and Templeton 1999; Templeton et al. 2001). For all species, IBD in the fragmented river basin should tend towards a pattern whereby genetic drift is higher and gene flow is lower. Finally, we discuss our results in the light of the cultural and historical status of the particular human fragmentation studied here (Fig. 1).
Material and methods
Study area
The area studied is located in Southwestern France and encompasses two river basins: the River Célé and the River Viaur (Fig. 2). We focused on these two river basins because (i) they are geographically very close to one another (their sources are roughly 50 km apart) and (ii) they both belong to the Adour-Garonne river drainage, which suggest a shared geological, colonization and evolutionary histories (Costedoat et al. 2006). These two rivers are very similar in terms of hydrological and chemical characteristics (G. Loot and S. Blanchet, unpublished data). Respectively, the River Célé and the River Viaur are 136 and 168 km long, their drainage areas cover 1350 and 1530 km² and their annual mean flow ranges between 7–30 m³/s and 8–25 m³/s.
Figure 2.
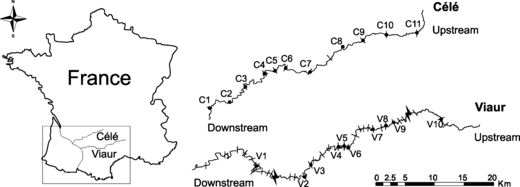
Map showing the geographical position of the river Célé (the continuous landscape) and the river Viaur (the fragmented landscape). Both rivers belong to the Adour-Garonne basin drainage. This map also shows the sampling sites (black dots) within each river. Weirs (small lines) and hydro-electric dams (large lines in the river Viaur) are also shown. However, note that not all weirs (over 50) are represented on the river Viaur.
However, these two rivers differ with regards to the level of anthropogenic fragmentation. The river Viaur is highly fragmented and will be hereafter referred to as the ‘fragmented landscape’. More than 50 small mill weirs (2–3 m high; see Fig. 1) are scattered along its main channel (approximately one weir every 2–3 km; Grenouillet et al. 2008; Poulet 2007). Most of these structures were built during the 15th century (it seems that weir building even began during the 12th century, see also Raeymaekers et al. 2009) and very few of them have been equipped with fish ladders. Based on non-standardized interviews, these weirs (even if often non-functional) are often viewed as parts of the local culture and heritage (G. Loot and S. Blanchet, unpublished data). In the first half of the 20th century, two hydroelectric dams (30 m high) were built; one located 30 km from the source and the other located 80 km from the source of the river (Fig. 2). The River Célé is weakly fragmented and will be hereafter referred to as the ‘continuous landscape’. Ten small weirs (2–3 m high) were constructed on the River Célé during the 20th century but most were equipped with fish ladders, enabling nearly all of the fish species to disperse between river segments (Fig. 2).
Sampling design
We sampled 11 sites on the river Célé (from C1 to C11; Fig. 2) and 10 sites on the river Viaur (from V1 to V10; Fig. 2). Because the genetic structure of freshwater fish is often spatially structured along the upstream–downstream gradient (Hanfling and Weetman 2006; Raeymaekers et al. 2008), the sites were directly on the main channel of each river and were chosen at regular distances along the gradient to cover this whole upstream–downstream gradient (Fig. 2). The riparian distance between each sampling site and the river source was calculated using a geo-referenced map (ArcView v.9©, ESRI, Redlands, CA, USA). A maximum of 24 adults per species and per site were caught by electrofishing (Table 1). For each individual, a small piece of pelvic fins was collected and preserved in 70% ethanol. Two fish species (L. leuciscus and L. cephalus) were absent from some sampling sites (i.e. the upper sampling sites, Table 1) while the other two species (G. gobio and P. phoxinus) were present all along the sampled gradient. At each site we also estimated the density of each species following a standardized two-pass electrofishing method (Bohlin et al. 1989).
Table 1.
Sampling characteristics of the study
| Proportion of occupied sites (mean sampling size ± 95% CI) | Mean density (ind/100m²) (mean ± 95% CI) | Mean effective population size (mean ± 95% CI) | Mean body length (mm) (mean ± 95% CI) | |||||
|---|---|---|---|---|---|---|---|---|
| Species | Fragmented | Continuous | Fragmented | Continuous | Fragmented | Continuous | Fragmented | Continuous |
| Leuciscus cephalus | 8/10 (17.5 ± 0.96) | 9/11 (18.44 ± 2.79) | 1.43 ± 1.08 | 0.94 ± 0.49 | 33.31 ± 31.97 | 77.05 ± 55.43 | 212.30 ± 25.93 | 182.63 ± 25.27 |
| Leuciscus leuciscus | 8/10 (18.37 ± 1.97) | 10/11 (15.12 ± 3.69) | 0.94 ± 1.17 | 2.64 ± 2.99 | 65.38 ± 21.54 | 75.87 ± 46.90 | 185.68 ± 10.55 | 165.72 ± 38.82 |
| Gobio gobio | 10/10 (20.00 ± 0.00) | 11/11 (19.72 ± 0.72) | 20.12 ± 8.55 | 18.61 ± 9.70 | 123.60 ± 62.99 | 113.27 ± 74.40 | 83.11 ± 5.81 | 83.19 ± 7.11 |
| Phoxinus phoxinus | 10/10 (20.44 ± 0.92) | 11/11 (19.81 ± 0.69) | 26.29 ± 19.12 | 11.44 ± 5.85 | 126.30 ± 67.80 | 115.00 ± 81.92 | 59.54 ± 2.89 | 57.59 ± 4.01 |
This table shows, for each landscape (fragmented or continuous), the proportion of sampling sites that was occupied, the mean sample size, the mean population density, the mean effective population size, and the mean body length (total body length in mm) for each of the four sampled species (95% CI, 95% confidence interval).
Biological models
All four fish species are widely distributed in Europe and belong to the Cyprinids family, which is the most diverse family in Europe (Reyjol et al. 2007). These species varied strikingly according to their maximal body length (Keith and Allardi 2001). The largest is L. cephalus which can reach a maximum body length of 600 mm, followed by L. leuciscus (400 mm), G. gobio (200 mm) and P. phoxinus (140 mm) (Keith and Allardi 2001). The average body lengths of the sampled fish were highly correlated with the maximal body lengths reported in the literature (r2 = 0.96, n = 4; see also Table 1). All these species are mainly insectivorous, with some feeding in the water column (L. cephalus and P. phoxinus) and the other preferentially feeding on the bottom (L. leuciscus and G. gobio) (Keith and Allardi 2001). By focusing on species belonging to the same trophic level and the same family, we should limit potential comparative biases inherent to phylogenetic constraints and to phenotypic attributes other than fish body size (Henle et al. 2004). Finally, by focusing on fish species that are not targeted by anglers, we limit the possibility of stocking effects and uninformed translocation between river drainages.
In fish, as in most animals, body length is positively correlated to dispersal ability at the interspecific level, and body size can hence be viewed as a good proxy of dispersal capability (Hugueny 1989; Jenkins et al. 2007). Although we lack of precise data on the dispersal ability of each of these four species, there are evidences that L. cephalus and L. leuciscus are good dispersers as they are both able to perform long-distance upstream movements (up to 10 km) and to migrate into section upstream weirs (Clough and Beaumont 1998; Lucas 2000; Bolland et al. 2008; De Leeuw and Winter 2008). On the contrary, the two other species (G. gobio and P. phoxinus) seems to have lower dispersal ability (Holthe et al. 2005; Knaepkens et al. 2007), which is not surprising given their relatively small body sizes. Moreover, a negative relationship between species body size and population density is expected at the interspecific level (Cotgreave 1993; Blackburn and Gaston 1997). Accordingly, the two largest species (L. cephalus and L. leuciscus) had lower densities than the two smallest species (G. gobio and P. Phoxinus) (see details in Table 1), which suggested a lower effective population size for these two largest species. Effective population sizes were estimated using microsatellites data (see below the section Descriptive genetic analyses). Both dispersal ability and effective population size are considered as important predictors of vulnerability to fragmentation; the relative role of both characteristics will be considered further in the discussion.
Genetic analyses
Genomic DNA was extracted from the pelvic fins using a salt-extraction protocol (Aljanabi and Martinez 1997). We employed a cross-amplification approach for selecting the loci for each species. Using an initial set of 40 markers developed for target and sister species (see Table S1), we cross-amplified the 40 markers for each of the four species and we conserved only the markers that displayed highly readable and repeatable profiles. These cross-amplified markers were then tested for the presence of null-alleles using the software micro-checker 2.3 (Van Oosterhout et al. 2004). The loci that showed evidence of the presence of null-alleles were dropped out from our final choice. This approach resulted in the selection of a set of 8–15 loci according to the species [L. cephalus (n = 10); L. leuciscus (n = 15); G. gobio (n = 8), P. phoxinus (n = 8); Table S1]. The selected loci were co-amplified using the QIAGEN® Multiplex PCR Kit (Qiagen, Valencia, CA, USA). Polymerase chain reaction (PCR) reactions were carried out in a 10 μL final volume containing 5–20 ng of genomic DNA, 5 μL of 2× QIAGEN Multiplex PCR Master Mix, and locus-specific optimized combination of primers (detailed recipes are available upon request). PCR amplifications were performed in a Mastercycler PCR apparatus (Eppendorf®, Hauppauge, NY, USA) under the following conditions: 15 min at 95°C followed by 30 cycles of 1 min at 94°C, 1 min at 60°C and 1 min at 72°C and finally followed by a 60 min final elongation step at 72°C. Amplified fragments were then separated on an ABI PRISM™ 3730 automated capillary sequencer (Applied Biosystems). Allelic sizes were then scored using GENEMAPPER™ v.4.0 (Applied Biosystems, Foster City, CA, USA).
Statistical analyses
Descriptive analyses
For each species, locus-by-locus heterozygosity (observed and expected) as well as Fis estimates were calculated using GENETIX version 4.05.2 (Belkhir et al. 2002). Departure from the Hardy–Weinberg equilibrium was tested using GENEPOP version 3.4 (Raymond and Rousset 1995). Linkage disequilibrium between all pairs of loci was tested in the program Fstat version 2.9.3.2 (Goudet 1995). We used the software NeEstimator 1.3 (Queensland Government, Brisbane, Australia) to estimate the effective population size for each species. We used a point estimation method based on linkage/gametic disequilibrium (Hill 1981). This method was preferred over the ‘heterozygote excess method’ (Pudovkin et al. 1996) because the later was less effective for converging. It was applied on each sampling site independently and we calculated a mean effective population size over all sampling sites. Point estimation methods must be interpreted cautiously and more sophisticated methods involving computationally intensive algorithms and/or multiple cohorts are often preferred to estimate properly effective population size (Waples 1989). The results presented here will therefore be used only for inter-species comparison but not for inter-landscapes comparison.
In addition, we used unpaired t-test (two-tails) to test if, within-species, density, effective population size and mean body size differed between the two landscapes.
Effect of fragmentation on genetic diversity
Two measures of genetic diversity were computed for each sampling site and for each species. AR corrected for the minimum sampling size was computed using Fstat version 2.9.3.2 (Goudet 1995), and observed heterozygosity (Hobs) was computed using GENETIX version 4.05.2 (Belkhir et al. 2002). For each species we computed generalized linear models (GLM) with either AR or Hobs as the dependent variables, landscape status (fragmented or continuous landscapes) as a categorical predictor and the position of each sampling site from the river source as a continuous predictor. As observed elsewhere (e.g. Raeymaekers et al. 2008) we expected both AR and Hobs to be positively correlated to the distance from the river's source. We also fitted the two-way interaction to test for a significant effect of the landscape status on the slope of the relationship between either AR or Hobs and the distance from the source. For all models we assumed a Gaussian error-term distribution and the sampling site was the replicate unit.
In addition to testing for a significant effect, we also used a meta-analytic framework to investigate how the magnitude of the effect of fragmentation (i.e. the effect size; Nakagawa and Cuthill 2007) differed between species. From mean and standard deviation data, we calculated the Hedges’d-value for each species and each measure of genetic diversity (see Rosenberg et al. 2000 for formulas). The Hedges’d-value is a standardized effect size that is comprised between −∞ and +∞, with a negative value representing effects where the control group (here the continuous landscape) attains a greater value than the experimental group (here the fragmented landscape) (Rosenberg et al. 2000). We then calculated the cumulative effect size (Ē) and the 95% confidence interval for all pooled species. This procedure allows the calculation of a mean effect size weighted for differential sampling sizes between species (Rosenberg et al. 2000). We then verified whether the effect size calculated for each species fell within the 95% confidence interval calculated for all species (i.e. did an individual effect significantly differ from the mean effect?). Finally, we evaluated if the set of effect sizes calculated for each species was homogeneous or not. The total heterogeneity of a sample (Qt) was calculated as described in Rosenberg et al. (2000) and its significance was tested using chi-square statistics.
Effect of fragmentation on genetic structure
We first investigated the population genetic structure by estimating the Fst (Weir and Cockerham 1984). We calculated global Fst values for each locus and each species using the program Fstat version 2.9.3.2 (Goudet 1995). For each species, we computed a GLM with Fst as the dependent variable and landscape status (fragmented or continuous landscapes) as a categorical predictor. For all models. we assumed a Gaussian error-term distribution and the locus was the replicate unit. The meta-analytic framework we described above was used to compare the magnitude of the effect of fragmentation on the Fst values for each species.
We then investigated the spatial genetic structure using the IBD framework (Hutchison and Templeton 1999; Templeton et al. 2001). Pairwise Fst between all pairs of populations were computed for each landscape and each species separately using GENETIX version 4.05.2 (Belkhir et al. 2002). Pairwise riparian distances between sites were computed using the Geographical Information System. We first graphically inspected the linear relationship between Fst/(1 − Fst) and the distance between pairs of sampling sites as proposed by Hutchison and Templeton (1999) to infer the migration drift equilibrium for each species in each landscape. Because of the non-independence of the replicate unit (i.e. a pair of sampling sites), we assessed the strength (as the Pearson coefficient of correlation) and the significance of each linear relationship using a re-sampling procedure (i.e. procedure analogous to the Mantel test; Manly 1997). We then investigated the effect of fragmentation on the slope of the linear relationship between Fst/(1 − Fst) and the distance between a pair of sampling sites using another re-sampling procedure in which the slope of the fragmented landscape for a given species was compared with the slope of the continuous landscape for this species (Manly 1997, #2831; see also Epps et al. 2005). If gene flow is reduced between sub-populations, we would expect a significantly higher slope in the fragmented than in the continuous landscape.
Results
Descriptive analyses
We did not find evidence for any significant heterozygous deficit (after Bonferroni correction) for any of the loci that were considered (Table S1). No pair of loci displayed significant linkage disequilibrium (results not shown). As expected, population density correlates quite strongly to effective population size (r2 = 0.766, n = 8, P < 0.005; see Table 1). Roughly, the two largest bodied-species (L. cephalus and L. leuciscus) tended to have lower effective population size than the two smallest-bodied species (G. gobio and P. phoxinus) (Table 1). However, within-species, we detected no significant differences between landscapes in term of mean body length, population density and effective population size (t-test, all P > 0.05, see Table 1).
Effect of fragmentation on genetic diversity
In case of the AR, we found significant differences between the fragmented versus the continuous landscape for all the species except P. phoxinus (Table 2; Fig. 3A). In all significant cases, the AR was lower in the fragmented landscape (Fig. 3A). It is noteworthy that the AR (irrespective of the landscape) increases as the maximum body size of the considered species decreases (Fig. 3A, see also Frankham 1996). Over all species, the magnitude of the effect imposed by fragmentation was significantly different from 0 (see the 95% confidence interval in Fig. 4A), but was significantly heterogeneous among species (Qt = 24.44, d.f. = 3, P < 0.001). This means that not all species respond equally to river fragmentation. Specifically, the effect sizes for L. leuciscus and G. gobio were greater than the mean whereas the effect size was lower than the mean for P. phoxinus (Fig. 4A). For one species (G. gobio), the AR was not affected by the distance of the sampling site from the river's source (Table 2D). For L. leuciscus and P. phoxinus, there was a significant positive correlation between AR and distance from the river's source (Table 2D; see also Fig. S1C,G). We also found a significant interaction between ‘fragmented status’ and ‘distance from the river's source’ for L. cephalus (Table 2A). This indicated that the relationship between AR and distance from the source was positive and significant for the continuous landscape (r = 0.75, n = 8, P = 0.033) but not for the fragmented landscape (r = 0.03, n = 8, P = 0.936; see Fig. S1A).
Table 2.
Results of generalized linear models aimed at testing the effects of fragmentation (fragmented of continuous) and of the distance of each site from the source on allelic richness and observed heterozygosity
| Allelic richness | Observed heterozygosity | ||||||
|---|---|---|---|---|---|---|---|
| Fish species | d.f. | Deviance | F-value | P-value | Deviance | F-value | P-value |
| (A) Leuciscus cephalus | |||||||
| Null model | 4.764 | 0.054 | |||||
| Fragmented status | 1,14 | 1.907 | 32.086 | <0.001 | 0.031 | 12.637 | 0.004 |
| Distance to the source | 1,13 | 1.645 | 2.944 | 0.111 | 0.029 | 0.391 | 0.543 |
| Distance × status | 1,12 | 1.068 | 6.489 | 0.026 | 0.022 | 3.928 | 0.070 |
| (B) Leuciscus leuciscus | |||||||
| Null model | 9.168 | 0.011 | |||||
| Fragmented status | 1,14 | 1.244 | 207.306 | <0.001 | 0.007 | 10.546 | 0.006 |
| Distance to the source | 1,13 | 0.483 | 19.894 | <0.001 | 0.006 | 1.806 | 0.203 |
| Distance × status | 1,12 | 0.458 | 0.649 | 0.435 | 0.005 | 0.094 | 0.763 |
| (C) Gobio gobio | |||||||
| Null model | 28.820 | 0.032 | |||||
| Fragmented status | 1,19 | 5.073 | 99.614 | <0.001 | 0.024 | 7.019 | 0.017 |
| Distance to the source | 1,18 | 4.256 | 3.423 | 0.081 | 0.023 | 0.003 | 0.958 |
| Distance × status | 1,17 | 4.052 | 0.85 | 0.367 | 0.022 | 0.762 | 0.394 |
| (D) Phoxinus phoxinus | |||||||
| Null model | 13.190 | 0.053 | |||||
| Fragmented status | 1,19 | 12.779 | 1.075 | 0.314 | 0.047 | 5.232 | 0.035 |
| Distance to the source | 1,18 | 7.681 | 13.332 | 0.002 | 0.031 | 14.170 | 0.001 |
| Distance × status | 1,17 | 6.500 | 3.087 | 0.096 | 0.019 | 10.997 | 0.004 |
Results are shown for each of the four species independently [from (A) to (D), in decreasing order of body length, see Table 1)]. Bold P-values are significant (P < 0.05).
Figure 3.
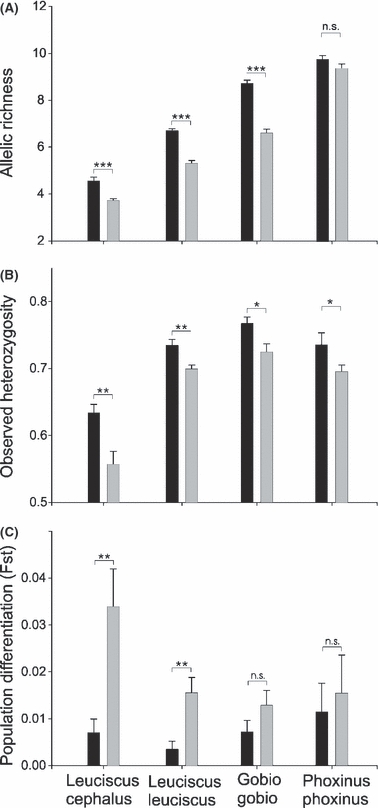
Bar plots showing the mean allelic richness (A), the mean observed heterozygosity (B) and the mean Fst (C) for each of the four species studied independently and each landscape (continuous landscape, black bars; fragmented landscape, grey bars). The four species are ranked from the largest to the smallest from left to right (see also Table 1). The stars (*P < 0.05; **P < 0.01; ***P < 0.001) indicated significant differences (generalized linear models; see Table 2) between the two landscapes. n.s., non-significant differences. Error bars are standard errors.
Figure 4.
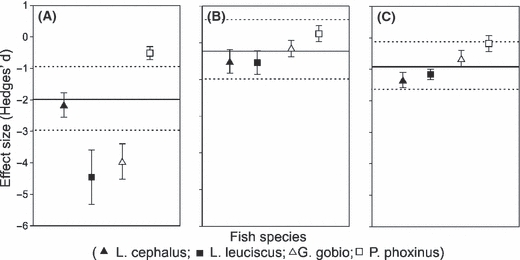
Effect sizes (measured as the Hedges’d ratio) measured for each genetic indices [(A) allelic richness; (B) observed heterozygosity; (C) Fst] and for each of the four species studied independently (Leuciscus cephalus, Leuciscus leuciscus, Gobio gobio and Phoxinus phoxinus). Error bars are standard deviations. For each genetic index, the mean effect size has been summarized across species using the cumulative effect size (Ē, full horizontal lines). The 95% confidence interval around the cumulative effect size has been added (dotted horizontal lines). Low values of effect size indicate strong effects of fragmentation.
Concerning observed heterozygosity, we found significant differences between the fragmented versus the continuous landscape for all species (Table 2; Fig. 3B). In all cases, the observed heterozygosity was lower in the fragmented landscape (Fig. 3B). Over all species, the magnitude of the effect imposed by fragmentation was significantly different from 0 (Fig. 4B), and did not differ significantly among species (Qt = 1.58, d.f. = 3, P = 0.661). The effect sizes for P. phoxinus tended to be slightly lower than for the other species (Fig. 4B). The effect of the distance of the sampling site from the river's source on Hobs was significant only for P. phoxinus (Table 2D). For this species, there was also a significant interaction between ‘fragmented status’ and ‘distance from the river's source’ (Table 2D). This showed that the relationship between AR and distance from the source was positive for both the continuous and the fragmented landscape, but the slope of the relationship was steeper in the continuous landscape (Fig. S1H).
Effect of fragmentation on genetic structure
Concerning Fst, we found significant differences between the continuous and fragmented landscape for the two largest species [L. cephalus (F1,18 = 9.828, P = 0.006) and L. leuciscus (F1,28 = 10.844, P = 0.003)] but not for the two smallest species [G. gobio (F1,14 = 1.964, P = 0.182) and P. phoxinus (F1,14 = 0.159, P = 0.696)]. In both significant cases, the level of population differentiation (i.e. Fst) was higher in the fragmented landscape (Fig. 3C). Over all species, the magnitude of the effect was significantly different from zero (Fig. 4C), and did not differ significantly between species (Qt = 3.518, d.f. = 3, P = 0.320). There was a positive tendency between the maximum body size of each species and the magnitude of the effect (Fig. 4C).
Concerning IBD, there were two species (L. cephalus and P. phoxinus) for which there were significant relationships between genetic differentiation and geographical distance for both the continuous and the fragmented landscapes (Table 3; Fig. 5A,D). In both cases, we detected no significant differences between the slope of the continuous landscape and the slope of the fragmented landscape (re-sampling tests, L. cephalus, P = 0.711; P. phoxinus, P = 0.118; Fig. 5A,D). For the other two species (L. leuciscus and G. gobio), we found a significant relationship between genetic differentiation and geographical distance only for the fragmented landscape (Table 3; Fig. 5B,C). The pattern was not significant for the continuous landscape (Table 3; Fig. 5B,C) indicating that distance between sampling sites had no influence on the genetic structure of these species in the continuous landscape. In both cases, there was a significant difference between the slope of the continuous landscape and the slope of the fragmented landscape (re-sampling tests, L. leuciscus, P = 0.014; G. gobio, P = 0.002; Fig. 5B,C).
Table 3.
Results of bootstrap-based test aimed at assessing the relationships between a measure of genetic dissimilarity [Fst/(1 − Fst)] and the riparian distance between pairs of sites
| Fragmentation status | ||
|---|---|---|
| Fish species | Continuous | Fragmented |
| (A) Leuciscus cephalus | 0.571 (0.071) 0.002 | 0.586 (0.074) 0.001 |
| (B) Leuciscus leuciscus | 0.033 (0.092) 0.565 | 0.648 (0.094) < 0.001 |
| (C) Gobio gobio | 0.106 (0.094) 0.182 | 0.637 (0.039) < 0.001 |
| (D) Phoxinus phoxinus | 0.587 (0.057) < 0.001 | 0.680 (0.035) < 0.001 |
The first line of each row reports the coefficient of correlation (Pearson r, 1000 iterations) of the relationships and of its standard deviation (in brackets). The second line of each row is the P-value (italic) associated to such coefficients. Bold P-values are significant (P < 0.05). Results are shown for each landscape (fragmented or continuous) and each of the four species [from (A) to (D), in decreasing order of body length; see Table 1] separately.
Figure 5.
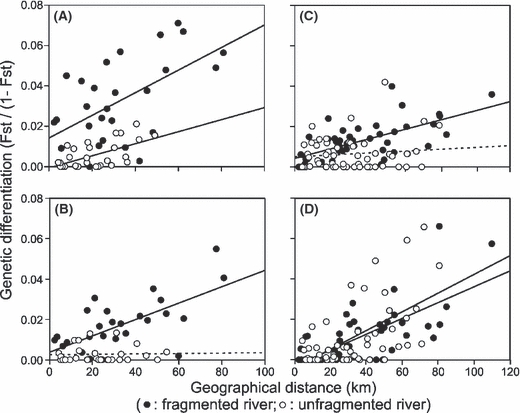
Univariate plots showing patterns of isolation by distance, i.e. relationships between the geographical distance between pairs of sampling sites and a measure of genetic dissimilarity [Fst/(1 − Fst)]. Relationships are shown for each species [(A) Leuciscus cephalus; (B) Leuciscus leuciscus; (C) Gobio gobio; (D) Phoxinus phoxinus] and each landscape (continuous landscape, white dots; fragmented landscape, black dots) separately. Continuous lines indicate significant univariate relationships (P < 0.05; see Table 3), while dotted lines are not significant.
Discussion
Using a meta-analytic framework, we highlight significant differences between fragmented and continuous landscapes, both for genetic diversity and the genetic structure of the set of species studied. In streams and rivers, gene flow is thought to be quasi-unidirectional as it is mainly driven by the direction of the water flow (Fraser et al. 2004; Hanfling and Weetman 2006, Crispo et al. 2006). This downstream-biased gene flow can impede our ability to disentangle the effects of historical colonization, man-made migration barriers and downstream-biased gene flow (Raeymaekers et al. 2008). We ought to bypass this difficulty by using a synchronic comparison, i.e. a comparison between a fragmented and a continuous landscape that share similar evolutionary and geological histories (Hoehn et al. 2007; Hendry et al. 2008). Although we can not totally discard that the pattern observed here are a hangover from the past or downstream-biased gene flow (Hanfling and Weetman 2006), such a comparison makes it likely that the between-landscape differences reported here are (at least in part) the result of anthropogenic fragmentation. Thus, our results support numerous studies on many animal species showing that human fragmentation modifies the genetic integrity of natural populations, notably in decreasing gene flow among populations and genetic diversity within the remnant population patch (e.g. Van den Bussche et al. 2003; Alo and Turner 2005; Trizio et al. 2005; Hoehn et al. 2007; Schiffer et al. 2007). It is worth noting that our preliminary results on the ecological characteristics of these fish populations (see Table 1) indicate that, from an ecological point of view, these populations have not suffered intensively from the changes in genetic diversity and structure we report here. Therefore, it is possible that these species have undergone evolutionary changes that allow them to cope with fragmentation (and the associated genetic changes), and/or that genetic consequences reported here are not strong enough to influence significantly the biological fitness of populations.
More interestingly, we detected obvious species-specific responses to fragmentation. This important result is consistent with previous papers showing that not all species are equally affected by fragmentation (Davies et al. 2000; Henle et al. 2004; Ewers and Didham 2006; Hoehn et al. 2007), and hence suggest that the effect of fragmentation in streams and rivers can not be generalized from one fish species to another. Contrary to certain expectations (Ewers and Didham 2006), we failed to verify the hypothesis that large-bodied species are less sensitive to habitat fragmentation. On the contrary, all genetic indices clearly indicated that the smallest-bodied species (P. phoxinus; Table 1) was the least affected by fragmentation. We further provide evidence that species with intermediate body size might in fact be the most affected by fragmentation, at least at the genetic level. Species differences in the responses to fragmentation were obvious and significant for two discriminant genetic indices; the AR and IBD patterns. For these two genetic indices, L. leuciscus and G. gobio (which are the two intermediate-bodied species; Table 1) were most affected by the presence of weirs. Indeed, in the case of AR, these two species had an effect size that was significantly lower than the mean (Fig. 4A). With regard to IBD, these two species showed patterns that significantly differed between landscapes: a pattern evocative of high gene flow and low genetic drift in the continuous landscape, and a pattern evocative of a regional equilibrium between gene flow and drift in the fragmented landscape (Hutchison and Templeton 1999). As expected, this latter result suggests that barriers limit gene flow between populations and favour genetic drift within them. Very interestingly, this demonstrates that human fragmentation has disturbed the initial balance between genetic drift and gene flow, and has constrained all these fish species in a similar population structure whereby an equilibrium between genetic drift and gene flow is attained (Raeymaekers et al. 2008). We can hence conclude that (i) not all species react similarly to fragmentation, (ii) the smallest sized species (P. phoxinus) is the least affected by fragmentation and (iii) there is a tendency for the intermediate-bodied species (L. leuciscus and G. gobio) to be the more affected.
Many traits have been proposed as potential predictors of species sensitivity to fragmentation (reviewed in Ewers and Didham 2006; Henle et al. 2004). Predictions regarding body size depend upon the level of organization being considered. For instance, it is often claimed that large-bodied species are at greater risk of extinction than small-bodied species because large-bodied species are often at higher trophic levels and hence, have lower population size and more unstable population dynamics (Gaston and Blackburn 1996; Henle et al. 2004). However, we control for this possibility by selecting species of the same trophic level, which orientates our prediction towards the hypothesis that large-bodied species are better dispersers and are therefore less sensitive to fragmentation (see Fig. 2Q,R in Ewers and Didham 2006). As stated above, this hypothesis was not supported, as the smallest-bodied species was the least sensitive to fragmentation. In fish, small-bodied species generally have a narrower habitat range than large-bodied species (Pyron 1999; Rosenfield 2002). Therefore, we can speculate that small-bodied species are not affected by fragmentation because their home range is smaller than the distance separating two barriers. A non-mutually exclusive speculation relates to the possibility that small-bodied species also have large effective population size (which is verified in our dataset; Table 1), which might blur the effect of genetic drift within remnant population patches (Lawton et al. 1994; Davies et al. 2000). Because of the strong covariance between body size and several other traits such as home range, population size and dispersal ability, it is currently difficult to ascertain which mechanisms underlie the observed pattern for the smallest-bodied species of our dataset (Henle et al. 2004; Theodorou et al. 2009).
Nevertheless, we claim that effective population size cannot explain why the two intermediate-bodied species are the most impacted by human fragmentation. Indeed, because these two species varied greatly in their respective abundance and effective population size (Table 1) while being equally affected by fragmentation, it is more likely that this common pattern arises from other traits such as dispersal ability or behaviour. The idea that species with intermediate dispersal ability might be more affected by fragmentation than species with either high or low dispersal has already been predicted theoretically (Fahrig 1998). Empirically, this has been demonstrated in a butterfly community in which species with intermediate mobility were more likely to decline in abundance following habitat fragmentation than butterflies with either low or high mobility (Thomas 2000). Although this remains to be tested, empirical data, however, support the idea that L. leuciscus and G. gobio display different dispersal ability (see the Biological models section; Knaepkens et al. 2007; Lucas 2000). It is therefore highly probable that movement behaviour and life-history strategies linked to feeding habits for instance (L. leuciscus and G. gobio feed on the bottom while the other two species feed on the water column) interact with dispersal ability to explain species sensitivity to fragmentation. For instance, L. cephalus and L. leuciscus are known to have similar dispersal ability (Lucas 2000; De Leeuw and Winter 2008; Ovidio et al. 2008), but are however differentially affected by weirs (our study). We have shown that in absence of weirs, L. cephalus population display a significant IBD pattern in the unfragmented landscape, which might indicate that they are more sedentary than L. leuciscus. This means that, in that case, dispersal behaviour rather than dispersal ability per se could explain species discrepancy in sensitivity to weirs. Given the descriptive nature of our study, further experimental work is needed to sort out the relative role of dispersal ability, dispersal behaviour and effective population size as drivers of species sensitivity to fragmentation. However, this means that certain phenotypic traits make species more or less sensitive to fragmentation, and that evolutionary processes (e.g. local adaptation by natural selection) can themselves differentially affect species.
Implications for management
That fragmentation differentially affects fish species means that not all populations are concerned by fragmentation, and hence that programmes of restoration can be prioritized. For instance, programmes aimed at avoiding the genetic erosion of species (e.g. hatchery and stocking programmes) do not need to be undertaken for all species at the same time, and it is here clear that the first measures that are needed must concern L. leuciscus and G. gobio. Being able to prioritize conservation efforts is very important, notably when funding is limited.
In case of river fragmentation by dams or weirs, four practical management solutions can be undertaken to restore the genetic integrity of populations: (i) the stocking of fish produced in a hatchery, (ii) the translocation of individuals from site to site, (iii) the construction of fish passages and (iv) the un-damming of rivers (Bednarek 2001; Palmer et al. 2005, 2008; Raeymaekers et al. 2009). Stocking is often associated with other evolutionary problems that make this solution undesirable (Blanchet et al. 2008; Frankham 2008). Translocation is too time-consuming to be efficient. For weirs such as those in the river Viaur (the fragmented landscape), un-damming is not a solution because these weirs date back to the Middle Ages (see also Raeymaekers et al. 2009) and are thus part of the local heritage and culture. However, there is some evidence that building fish passages can be an efficient restoration tool to preserve both the genetic integrity of fish species and the authenticity of the weirs (Raeymaekers et al. 2009). In the river Viaur, there are more than 70 weirs that have been identified. Because of the high financial cost of constructing fish passages on all the weirs, a priority must be given to weirs that have a high impact on the fish genetic structure, and the passages must be efficient (i.e. all fish can benefit from the passage). Prioritization in weirs restoration can be evaluated qualitatively and quantitatively by using the same procedure as the one used in Raeymaekers et al. (2009). According to the ‘umbrella species’ principle (Lambeck 1997), L. leuciscus and G. gobio appear as good candidates species for investigating prioritization in weirs restoration in the near future. The construction of fish passages should, however, follow the biological requirements of most species inhabiting such river systems. Following the recommendations made by Raeymaekers et al. (2009) in a similar system, we can expect that priority will be given to the highest weirs. However, the construction of fish passages should follow the biological requirements of most species inhabiting such river systems. Following the recommendations made by Raeymaekers et al. (2009) in a similar system, we propose that priority will be given to the highest weirs.
It is worth remembering that the river Viaur also contains two recently built hydro-electrics dams that are more than 30-m high. The effect of these dams (relative to the effect of weirs) on the evolution and ecology of fish populations is currently not known, and it should be the focus of our future studies. However, the flow rate of the river Viaur has been considerably altered by these two dams, being severely decreased downstream of the dams (i.e. a major part of the flow is diverted for agricultural purposes), which has increased sedimentation above small weirs (S. Blanchet and G. Loot personal observation). If fish ladders are installed in the river Viaur, they will need to be built according to the flow rate imposed by dams to ensure that these ladders function well (Larinier et al. 2002).
The approach used here for evaluating the effect of fragmentation on fish populations differs from previous studies in that (i) it focuses on four fish species and (ii) it uses a synchronic sampling design (Hendry et al. 2008). These characteristics make this study highly valuable from a conservation perspective, as it allows inferring the effect of fragmentation at a ‘community-wide’ level. Furthermore, it allows detecting subtle effects that can be blurred when more traditional approaches are used (i.e. when only a single-fragmented landscape is considered). However, this approach implicitly assumes that differences observed between landscapes are mainly the result of human fragmentation. This assumption is hard to ascertain and as a consequence, our study should be viewed as a basis for future studies considering simultaneously the effects of historical and contemporary histories on the genetic structure of wild fish populations. The use of a larger spatial scale sampling design and/or different genetic markers could be use for reaching this goal. This improvement should get a more complete picture of the relative role of humans in shaping the nowadays genetic structure of wild fish populations, and hence should improve management policies.
Conclusion
To conclude, our study demonstrates the importance of considering several species when investigating the effect of fragmentation mediated by humans. We have shown that a biological trait such as body size is important to understand the influence of fragmentation on the genetic integrity of fish populations. This means that the evolutionary outcomes of fragmentation will depend upon the traits that characterize species. Moreover, this implies that restoration programmes will need to be prioritized and targeted according to the specificity of each species. Future experimental studies are needed to ascertain the traits that affect the genetic sensitivity to fragmentation, and to test restoration programmes that can benefit a spectrum of species that are functionally dissimilar.
Acknowledgments
The authors thank Julie Turgeon for comments on an early draft. Gaël Grenouillet, Loïc Tudesque, Muriel Gevrey, Laetitia Buisson, Sébastien Brosse, Leslie Faggiano and Fabien Leprieur are thanking for their help on the field. The authors also thank the Agence de l'Eau Adour-Garonne for financial support and the Genopole Toulouse for their help with genotyping. Joost Raymaekers as well as two anonymous referees are thanked for their constructive comments.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Relationships between distance from the source and genetic diversity (AR and Obs H) for the four species studied here.
Table S1. Description of the microsatellites loci used in this study.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Literature cited
- Aljanabi SM, Martinez I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Research. 1997;25:4692–4693. doi: 10.1093/nar/25.22.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alo D, Turner TF. Effects of habitat fragmentation on effective population size in the endangered Rio Grande silvery minnow. Conservation Biology. 2005;19:1138–1148. [Google Scholar]
- Bednarek AT. Undamming rivers: a review of the ecological impacts of dam removal. Environmental Management. 2001;27:803–814. doi: 10.1007/s002670010189. [DOI] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. genetix 4.05.2, Logiciel sous Windows TM pour la génétique des populations. France: University of Montpellier; 2002. Laboratoire Genome Population, Interactions: CNRS UMR 5000. [Google Scholar]
- Blackburn TM, Gaston KJ. A critical assessment of the form of the interspecific relationship between abundance and body size in animals. Journal of Animal Ecology. 1997;66:233–249. [Google Scholar]
- Blanchet S, Paez DJ, Bernatchez L, Dodson JJ. An integrated comparison of captive-bred and wild Atlantic salmon (Salmo salar): implications for supportive breeding programs. Biological Conservation. 2008;141:1989–1999. [Google Scholar]
- Bohlin T, Hamrin S, Heggberget TG, Rasmussen G, Saltveit SJ. Electrofishing – theory and practice with special emphasis on salmonids. Hydrobiologia. 1989;173:9–43. [Google Scholar]
- Bolland JD, Cowx IG, Lucas MC. Movements and habitat use of wild and stocked juvenile chub, Leuciscus cephalus (L.), in a small lowland river. Fisheries Management and Ecology. 2008;15:401–407. [Google Scholar]
- Clough S, Beaumont WRC. Use of miniature radio-transmitters to track the movements of dace, Leuciscus leuciscus (L.) in the River Frome, Dorset. Hydrobiologia. 1998;372:89–97. [Google Scholar]
- Costedoat C, Chappaz R, Barascud B, Guillard O, Gilles A. Heterogeneous colonization pattern of European Cyprinids, as highlighted by the dace complex (Teleostei: Cyprinidae) Molecular Phylogenetics and Evolution. 2006;41:127–148. doi: 10.1016/j.ympev.2006.04.022. [DOI] [PubMed] [Google Scholar]
- Cotgreave P. The relationship between body-size and population abundance in animals. Trends in Ecology & Evolution. 1993;8:244–248. doi: 10.1016/0169-5347(93)90199-Y. [DOI] [PubMed] [Google Scholar]
- Couvet D. Deleterious effects of restricted gene flow in fragmented populations. Conservation Biology. 2002;16:369–376. [Google Scholar]
- Crispo E, Bentzen P, Reznick DN, Kinnison MT, Hendry AP. The relative influence of natural selection and geography on gene flow in guppies. Molecular Ecology. 2006;15:49–62. doi: 10.1111/j.1365-294X.2005.02764.x. [DOI] [PubMed] [Google Scholar]
- Davies KF, Margules CR, Lawrence KF. Which traits of species predict population declines in experimental forest fragments? Ecology. 2000;81:1450–1461. [Google Scholar]
- De Leeuw JJ, Winter HV. Migration of rheophilic fish in the large lowland rivers Meuse and Rhine, the Netherlands. Fisheries Management and Ecology. 2008;15:409–415. [Google Scholar]
- DiBattista JD. Patterns of genetic variation in anthropogenically impacted populations. Conservation Genetics. 2008;9:141–156. [Google Scholar]
- Epps CW, Palsboll PJ, Wehausen JD, Roderick GK, Ramey RR, McCullough DR. Highways block gene flow and cause a rapid decline in genetic diversity of desert bighorn sheep. Ecology Letters. 2005;8:1029–1038. [Google Scholar]
- Ewers RM, Didham RK. Confounding factors in the detection of species responses to habitat fragmentation. Biological Reviews. 2006;81:117–142. doi: 10.1017/S1464793105006949. [DOI] [PubMed] [Google Scholar]
- Fahrig L. When does fragmentation of breeding habitat affect population survival? Ecological Modelling. 1998;105:273–292. [Google Scholar]
- Fahrig L. Effects of habitat fragmentation on biodiversity. Annual Review of Ecology Evolution and Systematics. 2003;34:487–515. [Google Scholar]
- Frankham R. Relationship of genetic variation to population size in wildlife. Conservation Biology. 1996;10:1500–1508. [Google Scholar]
- Frankham R. Inbreeding and extinction: island populations. Conservation Biology. 1998;12:665–675. [Google Scholar]
- Frankham R. Genetic adaptation to captivity in species conservation programs. Molecular Ecology. 2008;17:325–333. doi: 10.1111/j.1365-294X.2007.03399.x. [DOI] [PubMed] [Google Scholar]
- Fraser DJ, Lippe C, Bernatchez L. Consequences of unequal population size, asymmetric gene flow and sex-biased dispersal on population structure in brook charr (Salvelinus fontinalis) Molecular Ecology. 2004;13:67–80. doi: 10.1046/j.1365-294x.2003.02038.x. [DOI] [PubMed] [Google Scholar]
- Gaston KJ, Blackburn TM. Global scale macroecology: interactions between population size, geographic range size and body size in the Anseriformes. Journal of Animal Ecology. 1996;65:701–714. [Google Scholar]
- Goudet J. fstat (version 1.2): a computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Grenouillet G, Brosse S, Tudesque L, Lek S, Baraille Y, Loot G. Concordance among stream assemblages and spatial autocorrelation along a fragmented gradient. Diversity and Distributions. 2008;14:592–603. [Google Scholar]
- Hanfling B, Weetman D. Concordant genetic estimators of migration reveal anthropogenically enhanced source-sink population structure in the River Sculpin, Cottus gobio. Genetics. 2006;173:1487–1501. doi: 10.1534/genetics.105.054296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry AP, Farrugia TJ, Kinnison MT. Human influences on rates of phenotypic change in wild animal populations. Molecular Ecology. 2008;17:20–29. doi: 10.1111/j.1365-294X.2007.03428.x. doi: 10.1111/j.1365-1294X.2007.03428.x. [DOI] [PubMed] [Google Scholar]
- Henle K, Davies KF, Kleyer M, Margules C, Settele J. Predictors of species sensitivity to fragmentation. Biodiversity and Conservation. 2004;13:207–251. [Google Scholar]
- Hill WG. Estimation of effective population size from data on linkage disequilibrium. Genetical Research. 1981;38:209–216. [Google Scholar]
- Hoehn M, Sarre SD, Henle K. The tales of two geckos: does dispersal prevent extinction in recently fragmented populations? Molecular Ecology. 2007;16:3299–3312. doi: 10.1111/j.1365-294X.2007.03352.x. [DOI] [PubMed] [Google Scholar]
- Holthe E, Lund E, Finstad B, Thorstad EB, McKinley RS. A fish selective obstacle to prevent dispersion of an unwanted fish species, based on leaping capabilities. Fisheries Management and Ecology. 2005;12:143–147. [Google Scholar]
- Hugueny B. Biogéographie et structure des peuplements de poissons d'eau douce d'Afrique de l'Ouest : approches quantitatives. Paris: Université Paris-VII; 1989. PhD Thesis. [Google Scholar]
- Hutchison DW, Templeton AR. Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution. 1999;53:1898–1914. doi: 10.1111/j.1558-5646.1999.tb04571.x. [DOI] [PubMed] [Google Scholar]
- Jenkins DG, Brescacin CR, Duxbury CV, Elliott JA, Evans JA, Grablow KR, Hillegass M, et al. Does size matter for dispersal distance? Global Ecology and Biogeography. 2007;16:415–425. [Google Scholar]
- Johansson M, Primmer CR, Merila J. Does habitat fragmentation reduce fitness and adaptability? A case study of the common frog (Rana temporaria. Molecular Ecology. 2007;16:2693–2700. doi: 10.1111/j.1365-294X.2007.03357.x. [DOI] [PubMed] [Google Scholar]
- Keith P, Allardi J. Atlas des poissons d'eau douce de France. Patrimoines Naturels. 2001;47:387. [Google Scholar]
- Keller LF, Waller DM. Inbreeding effects in wild populations. Trends in Ecology and Evolution. 2002;117:230–241. [Google Scholar]
- Knaepkens G, Maerten E, Eens M. Performance of a pool-and-weir fish pass for small bottom-dwelling freshwater fish species in a regulated lowland river. Animal Biology. 2007;57:423–432. [Google Scholar]
- Lambeck RJ. Focal species: a multi-species umbrella for nature conservation. Conservation Biology. 1997;11:849–856. [Google Scholar]
- Larinier M, Travade F, Porcher JP. 2002. Fishways: Biological Basis, Design Criteria and Monitoring. Bulletin Francais de la Peche et de la Pisciculture 364.
- Lawler JJ, Aukema JE, Grant JB, Halpern BS, Kareiva P, Nelson CR, Ohleth K, et al. Conservation science: a 20-year report card. Frontiers in Ecology and the Environment. 2006;4:473–480. [Google Scholar]
- Lawton JH, Daily G, Newton I. Population–dynamic principles. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences. 1994;344:61–68. [Google Scholar]
- Loot G, Reyjol Y, Poulet N, Simkova A, Blanchet S, Lek S. Effects of small weirs on fish parasite communities. Parasitology Research. 2007;101:1265–1276. doi: 10.1007/s00436-007-0632-6. [DOI] [PubMed] [Google Scholar]
- Lucas MC. The influence of environmental factors on movements of lowland-river fish in the Yorkshire Ouse system. Science of the Total Environment. 2000;251:223–232. doi: 10.1016/s0048-9697(00)00385-5. [DOI] [PubMed] [Google Scholar]
- Maloney KO, Dodd HR, Butler SE, Wahl DH. Changes in macroinvertebrate and fish assemblages in a medium-sized river following a breach of a low-head dam. Freshwater Biology. 2008;53:1055–1068. [Google Scholar]
- Manly BFJ. Randomization, Bootstrap and Monte Carlo Methods in Biology. London: Chapman et Hall; 1997. [Google Scholar]
- Nakagawa S, Cuthill IC. Effect size, confidence interval and statistical significance: a practical guide for biologists. Biological Reviews. 2007;82:591–605. doi: 10.1111/j.1469-185X.2007.00027.x. doi: 10.1111/j.1469-185X.2007.00027.x. [DOI] [PubMed] [Google Scholar]
- Nilsson C, Reidy CA, Dynesius M, Revenga C. Fragmentation and flow regulation of the world's large river systems. Science. 2005;308:405–408. doi: 10.1126/science.1107887. [DOI] [PubMed] [Google Scholar]
- Ovidio M, Philippart JC. The impact of small physical obstacles on upstream movements of six species of fish – synthesis of a 5-year telemetry study in the River Meuse basin. Hydrobiologia. 2002;483:55–69. [Google Scholar]
- Ovidio M, Capra H, Philippart JC. Regulated discharge produces substantial demographic changes on four typical fish species of a small salmonid stream. Hydrobiologia. 2008;609:59–70. [Google Scholar]
- Palmer MA, Bernhardt ES, Allan JD, Lake PS, Alexander G, Brooks S, Clayton S, et al. Standards for ecologically successful river restoration. Journal of Applied Ecology. 2005;42:208–217. [Google Scholar]
- Palmer MA, Liermann CAR, Nilsson C, Florke M, Alcamo J, Lake PS, Bond N. Climate change and the world's river basins: anticipating management options. Frontiers in Ecology and the Environment. 2008;6:81–89. [Google Scholar]
- Poulet N. Impact of weirs on fish communities in a piedmont stream. River Research and Applications. 2007;23:1038–1047. [Google Scholar]
- Pudovkin AI, Zaykin DV, Hedgecock D. On the potential for estimating the effective number of breeders from heterozygote excess in progeny. Genetics. 1996;144:383–387. doi: 10.1093/genetics/144.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyron M. Relationships between geographical range size, body size, local abundance, and habitat breadth in North American suckers and sunfishes. Journal of Biogeography. 1999;26:549. [Google Scholar]
- Raeymaekers JAM, Maes GE, Geldof S, Hontis I, Nackaerts K, Volckaert FAM. Modeling genetic connectivity in sticklebacks as a guideline for river restoration. Evolutionary Applications. 2008;1:475–488. doi: 10.1111/j.1752-4571.2008.00019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeymaekers JAM, Raeymaekers D, Koizumi I, Geldof S, Volckaert FAM. Guidelines for restoring connectivity around water mills: a population genetic approach to the management of riverine fish. Journal of Applied Ecology. 2009;46:562–571. [Google Scholar]
- Raymond M, Rousset F. genepop (version-1.2) – population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Reyjol Y, Hugueny B, Pont D, Bianco PG, Beier U, Caiola N, Casals F, et al. Patterns in species richness and endemism of European freshwater fish. Global Ecology and Biogeography. 2007;16:65–75. [Google Scholar]
- Rosenberg MS, Adams DC, Gurevitch J. MetaWin: Statistical Software for Meta-Analysis, Version 2.0. Sunderland: Sinauer Associates, Inc; 2000. [Google Scholar]
- Rosenfield JA. Pattern and process in the geographical ranges of freshwater fishes. Global Ecology and Biogeography. 2002;11:323–332. [Google Scholar]
- Schiffer M, Kennington WJ, Hoffmann AA, Blacket MJ. Lack of genetic structure among ecologically adapted populations of an Australian rainforest Drosophila species as indicated by microsatellite markers and mitochondrial DNA sequences. Molecular Ecology. 2007;16:1687–1700. doi: 10.1111/j.1365-294X.2006.03200.x. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution. 1993;39:53–65. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- Spielman D, Brook BW, Frankham R. Most species are not driven to extinction before genetic factors impact them. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15261–15264. doi: 10.1073/pnas.0403809101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Robertson RJ, Brisson J, Strasburg J. Disrupting evolutionary processes: the effect of habitat fragmentation on collared lizards in the Missouri Ozarks. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5426–5432. doi: 10.1073/pnas.091093098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorou K, Souan H, Couvet D. Metapopulation persistence in fragmented landscapes: significant interactions between genetic and demographic processes. Journal of Evolutionary Biology. 2009;22:152–162. doi: 10.1111/j.1420-9101.2008.01634.x. [DOI] [PubMed] [Google Scholar]
- Thomas CD. Dispersal and extinction in fragmented landscapes. Proceedings of the Royal Society of London Series B-Biological Sciences. 2000;267:139–145. doi: 10.1098/rspb.2000.0978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trizio I, Crestanello B, Galbusera P, Wauters LA, Tosi G, Matthysen E, Hauffe HC. Geographical distance and physical barriers shape the genetic structure of Eurasian red squirrels (Sciurus vulgaris) in the Italian Alps. Molecular Ecology. 2005;14:469–481. doi: 10.1111/j.1365-294X.2005.02428.x. [DOI] [PubMed] [Google Scholar]
- Van den Bussche RA, Hoofer SR, Wiedenfeld DA, Wolfe DH, Sherrod SK. Genetic variation within and among fragmented populations of lesser prairie-chickens (Tympanuchus pallidicinctus. Molecular Ecology. 2003;12:675–683. doi: 10.1046/j.1365-294x.2003.01755.x. [DOI] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- Vitousek PM, Mooney HA, Lubchenco J, Melillo JM. Human domination of earth's ecosystems. Science. 1997;277:494–499. [Google Scholar]
- Waples RS. A generalized approach for estimating effective population size from temporal changes in allele frequency. Genetics. 1989;121:379–391. doi: 10.1093/genetics/121.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples RS, Zabel RW, Scheuerell MD, Sanderson BL. Evolutionary responses by native species to major anthropogenic changes to their ecosystems: Pacific salmon in the Columbia River hydropower system. Molecular Ecology. 2008;17:84–96. doi: 10.1111/j.1365-294x.2007.03510.x. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wofford JEB, Gresswell RE, Banks MA. Influence of barriers to movement on within-watershed genetic variation of coastal cutthroat trout. Ecological Applications. 2005;15:628–637. [Google Scholar]
- Wright S. The gnetical structure of populations. Annals of Eugenics. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


