Abstract
Biological invasions are generally thought to occur after human aided migration to a new range. However, human activities prior to migration may also play a role. We studied here the evolutionary genetics of introduced populations of the invasive ant Wasmannia auropunctata at a worldwide scale. Using microsatellite markers, we reconstructed the main routes of introduction of the species. We found three main routes of introduction, each of them strongly associated to human history and trading routes. We also demonstrate the overwhelming occurrence of male and female clonality in introduced populations of W. auropunctata, and suggest that this particular reproduction system is under selection in human-modified habitats. Together with previous researches focused on native populations, our results suggest that invasive clonal populations may have evolved within human modified habitats in the native range, and spread further from there. The evolutionarily most parsimonious scenario for the emergence of invasive populations of the little fire ant might thus be a two-step process. The W. auropunctata case illustrates the central role of humans in biological change, not only due to changes in migration patterns, but also in selective pressures over species.
Keywords: biological invasion, introduction routes, parthenogenesis, reproduction system, Wasmannia auropunctata
Introduction
Biological invasions are recognized as a major component of global change (Vitousek et al. 1997). The successful establishment and spread of species across previously unoccupied habitats has been shown to cause biodiversity declines (e.g. Clavero and Garcia-Berthou 2005), to disrupt ecosystems functions (e.g. Crooks 2002) and to incur severe socio-economic losses around the World (e.g. Shogren and Tschirhart 2005). The role of human activity has become a primary driver in the current displacement of species at the global scale (McKinney and Lockwood 1999; Foley et al. 2005). When displaced, species are introduced in new environments where they probably lack specific adaptations, and may also undergo bottleneck events (Sakai et al. 2001). Invasive species, which thrive in their introduced environment, thus overcome this hypothetical lack of adaptation, despite a probable low size and genetic variability of the introduced propagule. Some authors therefore argue that biological invasions are paradoxical events (Sax and Brown 2000; Frankham 2005a). However, recent studies refute this ‘paradoxical’ vision of bioinvasions, mostly through the demonstration of high propagule pressure and maintained, or even increased, genetic variability in the introduced populations (Kolbe et al. 2004; Lavergne and Molofsky 2007; Roman and Darling 2007; Dlugosch and Hays 2008; Facon et al. 2008).
Five ant species appear in the list of the 100 world's worst invasive organisms (Lowe et al. 2000), and these ant species have long been recognized to pose important threats to biodiversity and human activities (Holway et al. 2002). Our study focuses on one of the least studied of these particularly harmful invasive species, the little fire ant, Wasmannia auropunctata (Roger, 1863) (Formicidae: Myrmicinae). This species originates from Central and South America and is successfully spreading over the World tropics since the beginning of the last century. Its introduced range now encompasses many Caribbean islands, Florida, several West-African countries, and a large number of Pacific islands (Wetterer and Porter 2003). It also recently established populations in the Mediterranean zone, in Israel, which is raising concerns about its potential distribution range outside the tropics (Vonshak et al. 2009b).
Previous studies demonstrated that two types of populations coexist within the native range of W. auropunctata (Foucaud et al. 2009b; Orivel et al. 2009). In French Guiana, natural forest habitats, and especially floodplains along creeks, are occupied by low density, mostly sexually reproducing populations. On the contrary, human-disturbed habitats of the native range are generally occupied by high density, dominant, populations (see Orivel et al. 2009; Foucaud et al. 2009b for details). These latter populations generally display an extraordinary ‘clonal’ reproduction system, where males are produced clonally, female queens are parthenogens and workers are produced sexually (see Fig. S1; Fournier et al. 2005a). These clonal populations also display a specific mating pattern, where the mated male and female tend to harbor more divergent genotypes than in sexually reproducing populations (Foucaud et al. 2009b). The sexually produced worker offspring of these clonal couples is therefore highly heterozygous. This particular genetic architecture may be selected for in native human-disturbed habitats, and may provide the opportunity to invade other human-disturbed areas (Foucaud et al. 2009b).
In agreement with this, Foucaud et al. (2006) hypothesized that the introduced populations of W. auropunctata originated from the dominant, clonally reproducing populations of the native range. This hypothesis is, however, based on the study of a single introduction area, New Caledonia (Foucaud et al. 2006). A first attempt of biogeographical study of native and introduced populations of W. auropunctata invasion has recently been completed (Mikheyev and Mueller 2007). Using a single mitochondrial region (between cytochrome oxidase subunits I and II), Mikheyev & Mueller provided useful insights into the species biogeography, but the low level of variation and the mode of inheritance of the markers used did not make it possible to address the evolutionary processes at play during the invasion.
The present study, which is based on an extensive worldwide set of molecular data obtained at 12 microsatellite loci, aimed at answering two main questions. First, what are the main routes of introduction of W. auropunctata around the globe? Second, what evolutionary processes could have enabled some W. auropunctata populations to invade remote areas? To address both questions, we deciphered the reproduction system and genotypic patterns of introduced populations, and compared these data with data gathered from previous studies focusing on the native range of the species (Foucaud et al. 2009b). This, in turn, enabled us to construct a parsimonious scenario for the worldwide invasion of W. auropunctata.
Methods
Field collection
Field work was conducted in 16 countries belonging to the introduced range of W. auropunctata (Fig. 1; Table 1). A total of 251 nests (i.e. an aggregation of workers, brood and/or queens within a woodstick, under stones or between dead leaves) belonging to 60 sites of the introduced range of W. auropunctata were collected from 1997 to 2007. The number of collected nests per site varied from one to 25 nests (Mean ± SD: 5 ± 4 nests). For most of the nests (except Dominican, Cuban, Galapagos and Cocos Island nests), a large number of workers and most if not all of the reproductives were collected. The distance between sampled nests was always larger than two meters, to avoid sampling neighboring nests that most probably exchange workers and hence underestimating the genetic diversity within the sampled sites. Most of the samples (140 nests from 15 countries) were specifically collected for the present study. The Gabonese, Hawaiian and Floridian samples are different from the samples analyzed in Mikheyev et al. (2009). The New Caledonian and Israeli samples (82 and 29 nests, respectively) were used in previous genetic studies (Foucaud et al. 2006 and Vonshak et al. 2009a, respectively). For addressing some questions, we also used data from previously published population samples collected within the native range of W. auropunctata (Foucaud et al. 2007, 2009b).
Figure 1.
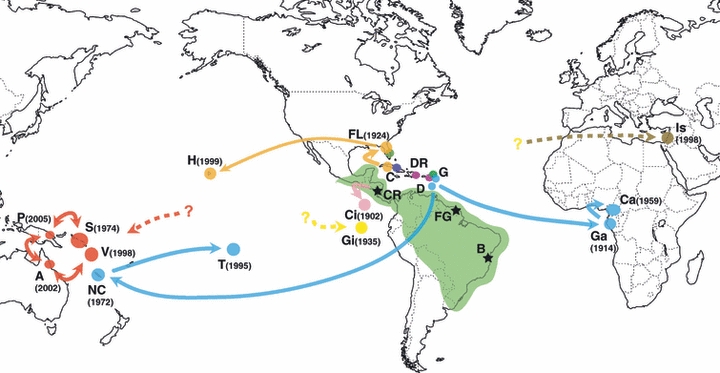
Routes of introduction of Wasmannia auropunctata. Note: Colored dots represent genetically distinct introduced clonal populations. The dark green area represents the native range of W. auropunctata (Wetterer and Porter 2003), and black stars represent areas where populations from the native range have been sampled. The code letter for each introduced country is given in Table 1. Estimated dates of introduction are indicated between brackets when available (see Table S1 for detailed references).
Table 1.
Number of sampled nests and genotyped queens, males and workers, for each surveyed country
| Number of genotyped individuals | |||||||
|---|---|---|---|---|---|---|---|
| Range | Country | Code letter | Sampled nests | Queens | Males | Workers | References |
| Native | Brazil | B | 66 | 250 | 135 | 527 | Foucaud et al. 2009b |
| Native | French Guiana | FG | 103 | 255 | 210 | 1107 | Foucaud et al. 2009b |
| Native | Costa Rica | CR | 8 | 25 | 6 | 63 | This study |
| Total | 177 | 530 | 351 | 1697 | |||
| Introduced | Cameroon | Ca | 55 | 196 | 79 | 250 | This study |
| Introduced | Gabon | Ga | 19 | 59 | 45 | 150 | This study |
| Introduced | Israel | Is | 29 | 56 | 44 | 229 | Vonshak et al. 2009a |
| Introduced | Florida | FL | 11 | 26 | 8 | 88 | This study |
| Introduced | Cuba | Cu | 2 | 5 | 0 | 23 | This study |
| Introduced | Guadeloupe | G | 10 | 2 | 0 | 75 | This study |
| Introduced | Dominican Republic | DR | 1 | 0 | 0 | 8 | This study |
| Introduced | Dominica | DR | 1 | 0 | 0 | 8 | This study |
| Introduced | Cocos Island | Ci | 1 | 2 | 0 | 7 | This study |
| Introduced | Galapagos Islands | Gi | 1 | 0 | 0 | 8 | This study |
| Introduced | New Caledonia | NC | 82 | 580 | 208 | 702 | Foucaud et al. 2006 |
| Introduced | Tahiti | T | 9 | 69 | 45 | 71 | This study |
| Introduced | Hawaii | H | 9 | 16 | 0 | 71 | This study |
| Introduced | Vanuatu | V | 10 | 18 | 2 | 71 | This study |
| Introduced | Australia | A | 7 | 14 | 10 | 54 | This study |
| Introduced | Papua New Guinea | P | 3 | 4 | 0 | 23 | This study |
| Introduced | Solomons | S | 1 | 13 | 0 | 32 | This study |
| Total | 251 | 1060 | 441 | 1870 | |||
Microsatellite genotyping
The microsatellite genotyping was carried out as described in Foucaud et al. (2006). Briefly, for each sampled nest, DNA was extracted from at least seven workers and most if not all collected reproductives. Our microsatellite genotyping data set includes 3371 individuals collected in the introduced range of W. auropunctata, genotyped at 12 microsatellite loci (Fournier et al. 2005b). For certain comparisons, we additionally used another microsatellite genotyping data set including 2578 individuals from the native range of W. auropunctata, genotyped at 12 microsatellite loci (Table 1). This latter microsatellite data set corresponds to that published in recent studies focusing on native populations of W. auropunctata (Foucaud et al. 2009b), except for eight additional nests collected in Costa Rica in 1997.
Reproduction system and relationships between genotypes
We characterized the reproductive systems and the relationships between genotypes by investigating individual microsatellite genotypes visually and using two programs we developed in the Pascal object programming language (inquiries about details of the programs should be sent to the corresponding author). The first program was used to identify clones (i.e. identical multilocus genotypes) in a given sample of genotypes. The second program was used to construct dendrograms from individual genotypes (queens, males or workers) using the Neighbor-Joining algorithm (Saitou and Nei 1987). The genetic distance used to construct the dendrograms was a variant of Chakraborty and Jin's allele-shared distance (Chakraborty and Jin 1993), as defined in Fournier et al. (2005a).
The identification of the main routes of introduction turned out to be relatively simple due to the introduction of almost entirely clonal queen and male genotypes in each invaded area (see Results section). In particular, we could directly assess the relationships between two areas when they shared a common queen and/or male clonal genotype. We considered two genotypes to be clonal when they shared identical multilocus genotypes at 12 microsatellite loci, or when they differed either (i) by only one dinucleotide repeat at one of the 12 genotyped loci (as this pattern is likely to correspond to one mutational event at a microsatellite locus) or (ii) by homozygosity for one allele at a single heterozygous locus of the clonal queen genotype (as this pattern probably corresponds to a recombination or gene conversion event during thelytoky; Foucaud et al. 2006). The genotypes used to infer introduction routes were either the genotypes of the males and queens collected in the field, or the genotypes of queens and males inferred from the genotypes of collected workers (cf. workers are sexually produced; Fournier et al. 2005a; Foucaud et al. 2006, 2007, 2009a). The ‘direction’ of the identified routes of introduction was assessed by the estimated dates of introduction in the given countries (i.e. from the oldest to the most recently invaded country). Historical information regarding dates of introduction of W. auropunctata invasion was gathered from Wetterer and Porter (2003) and from various experts and local people (see Fig. 1 and Table S1 for details).
Genotypic patterns
Previous studies that focused on the native populations of W. auropunctata found that clonal couples (i.e. male/queen mating pairs) differed significantly from sexual couples regarding their heterozygosity and difference in microsatellite allelic size (see below for definitions; Foucaud et al. 2009b). Both statistics were significantly higher in clonal couples, indicating a trend for outbreeding in these native populations (i.e. mating with genetically distant individuals). These clonal mating pairs result in significantly more heterozygous workers in clonal populations than sexual populations in the native range of W. auropunctata. We here investigated the same statistics in the introduced populations of the species. The differences between the queen and male genotypes of a given couple were assessed using a personal program that computes basic population genetic statistics (i.e. observed heterozygosity and mean difference in allele size within and between multilocus genotypes). Within-individual heterozygosity, How, was computed as the number of loci of an individual genotype showing different alleles. Heterozygosity of a queen-male couple, Hob, was computed as the mean number of times the male allele was different from each queen allele at a given locus. Within-individual difference in allelic size, DSw, was computed as the difference in base pairs between the two alleles at a given locus of a single individual genotype. Difference in allelic size of a queen-male couple, DSb, was computed as the mean difference between the male allele and the two queen alleles at a given locus. Because microsatellite sequences mutate under a stepwise model (Estoup et al. 2002), the differences in allele size between two microsatellite DNA copies measured either within or between individuals is related to the coalescence time and hence the level of divergence between the two compared genomes. Ho and DS statistics were computed for every locus, and we calculated their means for every population, nested in a single ‘type’ of population (three types: Native Sexual, Native Clonal, Introduced Clonal; see Results section). We then tested for statistical differences in mean Ho and DS values for couples or workers: (i) between all three types of populations using a non-parametrical Friedman test, and (ii) between each pair of populations’ type using non-parametrical Wilcoxon sign rank tests. We used loci as statistical units for all statistical tests.
Results
Reproduction system of introduced populations
The first result of our study is that most if not all introduced populations of W. auropunctata are reproducing clonally. All the 110 nests where both queens and males were collected showed direct proofs of clonal reproduction by both queens and males (i.e. groups of identical queen genotypes and groups of identical male genotypes). One hundred and two of the 103 nests where only queens were collected provided direct proof of clonal reproduction by queens (i.e. groups of identical queen genotypes). In all those nests, males were directly or indirectly shown to reproduce clonally (i.e. male genotypes inferred from queens’ spermathecal contents or from workers were identical to known clonal male genotypes, respectively). Thirty-four of the 38 nests that lacked reproductives at the time of collection provided indirect proof of clonal reproduction by both queens and males (i.e. inferred queen genotypes identical to known clonal queen genotypes and inferred male genotypes identical to known clonal male genotypes). It was not possible to determine the type of reproduction system for only five of the 251 nests collected in the introduced range of W. auropunctata (three in Gabon, one in Guadeloupe and one in Florida). In those nests, the sampled or inferred genotypes of reproductives were neither identical nor close to known clonal genotypes. Furthermore, those nests were genetically monogynous and monoandrous. Hence, the male and queen of these nests could have reproduced sexually or clonally. Altogether, the proportion of nests sampled in the introduced range of W. auropunctata where both male and queens reproduce clonally (i.e. clonal nests) was over 98%. We did not find any introduced nest reproducing uniquely via sexuality, as was found in the native range of the species, where entire populations (composed of many nests) are either sexual or clonal (Foucaud et al. 2009b). In this previous study of the native range, we found around one-third of sampled nests to be exclusively sexual, which contrasts with the figure of 98% obtained here in the introduced range.
The overwhelming preponderance of the clonal reproduction in the introduced range of W. auropunctata does not necessarily imply that clonal reproductives never reproduce sexually. Some rare sexual reproduction events, already observed in New Caledonia (Foucaud et al. 2006) and Gabon (Mikheyev et al. 2009), were also apparent here in introduced populations of Tahiti, Cameroon and other Gabonese populations. In these countries, some new clonal queen lineages were indeed derived from sexual recombination by local clonal queen and male lineages (see Fig. S2 and Foucaud et al. 2006 for a detailed description of derived lineages). However we did not detect any new clonal male lineage arising in those countries. We could therefore distinguish between two types of clonal couples within the introduced range of W. auropunctata. First, the original couples of clones are likely composed of the originally introduced male and queen genotypes (showing distinct genotypes without recombination events). Second, the derived couples of clones are composed of the original male genotype and a queen genotype deriving from a sexual reproduction event between the original male and queen genotypes.
Routes of introduction
Since virtually all introduced populations were clonal, we directly assessed the relationships among the queen and male genotypes from the introduced and the native range of W. auropunctata and hence infer on introduction routes. Our results show two major types of introduction pattern.
First, the Caribbean zone has been invaded by multiple couples of clonal queens and males (Figs 1 and 2). Several Caribbean countries share clonal queen genotypes, including Guadeloupe and Dominica, Guadeloupe and Dominican Republic, and Cuba and Florida (i.e. on Fig. 2A: Guadeloupe 1 – Dominica 2; Guadeloupe 2 – Dominican Republic; Cuba 2 – Florida 4). The slight differences between some of these genotypes are likely due to single mutational or recombination events during thelytoky. Interestingly enough, we observed a greater diversity of male genotypes in the Caribbean zone, as only Cuba and Florida share a clonal male genotype (i.e. on Fig. 2B: Cuba 2 – Florida 3). All other introductions were founded by single couples of clonal queen and male genotypes.
Figure 2.
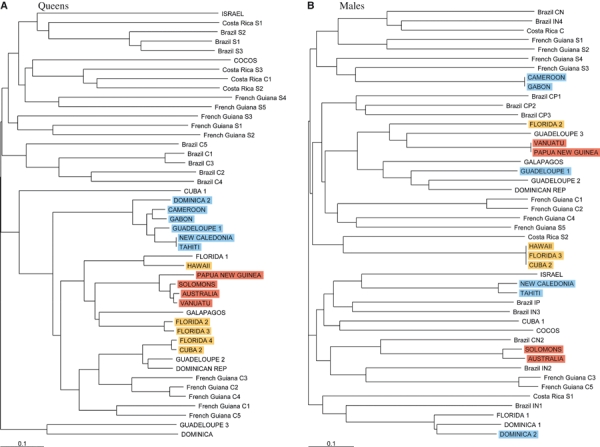
Neighbor-Joining dendrograms of the microsatellite (allele-shared) distances between individual queen (A) and male (B) genotypes. Note: Groups of introduced queens and males present in more than one country that share clonal genotypes for queens, males or both, were highlighted with colors similar to Fig. 1. Individual genotypes from the introduced and native ranges of Wasmannia auropunctata are written in upper case and lower case letters, respectively. All introduced and, due to space limitation, a randomly chosen subset of native genotypes were included for both sexes. Similar results were obtained when using all individual genotypes (not shown).
We could directly retrace the introduction histories for three groups of invaded countries. First, the clonal queen genotype shared by Guadeloupe and Dominica is also shared with Gabon, Cameroon, New Caledonia and Tahiti (Fig. 2A). This clonal queen genotype is mated to four distinct male genotypes: one in Guadeloupe, one in Dominica, one shared between Gabon and Cameroon, and one shared between New Caledonia and Tahiti (Fig. 2B). Second, the clonal male shared by Cuba and Florida is also shared with Hawaii (Fig. 2B). It is also worth pointing that the clonal Hawaiian queen genotype mated to this clonal male is also related to a Floridian queen genotype (Fig. 2A). Finally, one clonal queen genotype is shared between the Melanesian populations from the Solomon Islands, Vanuatu, Papua New Guinea and Australia (Fig. 2A). This clonal queen genotype is mated to two distinct male genotypes: one shared between Vanuatu and Papua New Guinea, and one shared between the Solomon Islands and Australia (Fig. 2B).
Treating together our microsatellite data set from the introduced range and our previous data set from the native range revealed two additional clusters of related genotypes. While other genetic distances led to very similar topologies, these two clusters must however be analyzed cautiously because of the low bootstrap values of some of the nodes of the queen tree (a feature expected when bootstrapping individual tree over loci). The first cluster included the clonal queen genotypes found in the Caribbean zone and some of the clonal queen genotypes found in French Guiana (i.e. genotypes French Guiana C1 to C5; Fig. 2A). Two alternative hypotheses may explain this cluster: (i) the clonal genotypes introduced in the Caribbean Islands originate from the northern part of South America, and (ii) the clonal populations of French Guiana are a re-introduction of W. auropunctata from the Caribbean zone. While the definitive data to distinguish these two hypotheses are lacking, the first hypothesis seems more probable because Guianese clonal queen genotypes share a large proportion of their alleles at each locus with the neighboring sexual populations and hence likely originated from these sexual populations (Foucaud et al. 2007). A larger sample from the northern coast of the native range is needed to further disentangle these two hypotheses. The second cluster included the queen genotypes from the native populations of Costa Rica and the introduced populations of Cocos Island (belonging to the Costa Rican state). It is interesting to note that this relationship in our microsatellite data set parallels previous findings based on mitochondrial DNA (Mikheyev and Mueller 2007).
We could not infer any routes of introduction for the clonal populations of Galapagos Islands and Israel, as well as any origin for the Melanesian clonal populations. These couples of clonal queen and male genotypes did not match or showed tight relationships with any other genotype from the native range of W. auropunctata. These introductions probably correspond to three independent routes that may originate from the native area or the Caribbean area (e.g. the hypothesized Brazilian origin of the Israeli population; Vonshak et al. 2009b).
Genotypic patterns of introduced populations
The characterization of the genotypic patterns of native populations provides some useful insights into the native origin of introduced populations of W. auropunctata. As a matter of fact, heterozygosity and difference in allele size differ markedly between sexual and clonal populations of the native range (see Methods section; Foucaud et al. 2009a,b). We thus compared the heterozygosity and difference in allele size between the male and queen genotypes of three distinct types of couples: sexual couples of the native range, clonal couples of the native range and clonal couples of the introduced range. When treating altogether all three types of couples, we found significant differences for both statistics (Friedman tests: Hob: F2 = 18; P < 10−3; DSb: F = 7.16; P = 0.027; Table 2). When considering the types of couples by pair, the males and females genotypes from clonal couples from both the introduced and native areas had significantly higher level of between-individuals heterozygosity and difference in allele size than those from native sexual couples (Wilcoxon sign rank tests: all P-values <0.05; Table 2). On the other hand, there was no significant difference between introduced and native clonal couples for both statistics (Wilcoxon sign rank tests: all P > 0.23).
Table 2.
Heterozygosities (Ho) and differences in allele size (DS) in couples and workers from native sexual, native clonal and introduced clonal populations
| Native sexual | Native clonal | Introduced clonal | Wilcoxon tests | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SE | Mean | SE | Mean | SE | Friedman test | NaS-NaC | NaS-lntC | NaC-lntC | |
| Couples | ||||||||||
| Ho | 0.668 | 0.093 | 0.897 | 0.021 | 0.890 | 0.019 | *** | ** | ** | NS |
| DS | 7.502 | 1.604 | 11.992 | 0.852 | 9.768 | 0.512 | * | ** | * | NS |
| Workers | ||||||||||
| Ho | 0.710 | 0.032 | 0.852 | 0.026 | 0.808 | 0.018 | * | ** | 0.08 | NS |
| DS | 7.859 | 0.761 | 11.214 | 1.043 | 8.485 | 0.400 | NS | * | NS | NS |
Note: Levels of significativity of Friedman and Wilcoxon sign rank tests have been included, where NS: P > 0.10, ***P < 10−3, **P < 10−2 and *P < 0.05. Native sexual, native clonal and introduced clonal populations are designated by NaS, NaC and IntC respectively.
When considering the same statistics measured within individual worker genotypes, we found significant differences for heterozygosity, but not for difference in allele size (Friedman tests: How: F2 = 7.17; P = 0.028; DSw: F = 3.50; P = 0.17; Table 2). Pairwise analyses revealed that workers from clonal populations from both the introduced and native areas had significantly (or nearly for introduced populations) higher level of heterozygosity than those from native sexual populations (Table 2). There was no significant difference between workers of introduced and native clonal populations for both statistics (Wilcoxon sign rank tests: all P > 0.11).
Examining our data in detail, we found that the non-significant differences between workers of the introduced and native sexual populations were at least partly due to the presence of sexually derived female clones in some of the introduced areas (in New Caledonia, Tahiti and Western Africa). It is worth stressing that the genotypes of these derived female clones display some alleles of the genotype of their male mate, resulting in couples (and hence workers) with lower heterozygosity and difference in allele size (see Foucaud et al. 2006 for details). When removing the derived clonal genotypes from our data set, heterozygosity, but not difference in allele size, was significantly higher in the worker offspring of the originally introduced clonal couples than in the worker offspring of the native sexual populations (Wilcoxon sign rank test: How: Z = 2.43; P = 0.015). The worker offspring of the originally introduced clonal couples remained similar to workers from native clonal populations for both statistics (all P-values >0.39).
Altogether, we found that introduced clonal populations were similar to native clonal populations and strongly dissimilar to native sexual populations with regards to heterozygosity and difference in allele size of couples and workers.
Discussion
The present worldwide study enables us to reconstruct some of the main routes of introduction of W. auropunctata and provides some insights into the ecological and evolutionary factors that may have favored the current expansion of its distribution range around the globe. This in turn suggests some general mechanisms that could be involved in other, potentially numerous, cases of biological invasions.
Routes of introduction
Our study highlights the fact that the key factor explaining the current distribution of W. auropunctata over the world is trade and historical shipping routes. While this idea has been proposed long ago (Passera 1994; Wetterer and Porter 2003), our study illustrates it in a striking way (Fig. 1). Overall, we can distinguish several ‘human cultural routes’ of introduction of W. auropunctata. First, our study pinpoints the Caribbean zone as being an introduced area of primary importance. To date, Caribbean populations were classified among native populations of W. auropunctata (Mikheyev and Mueller 2007), with, however, substantial doubts (Wetterer and Porter 2003). Our study indicates that Caribbean populations are most likely introduced and not native. All Caribbean populations are indeed clonal, similarly to other introduced populations, whereas native populations are either clonal or sexual. Moreover, male and female clonal genotypes were highly dispersed throughout the Caribbean area, a situation that is parsimoniously explainable by human-mediated dispersal. Altogether, our microsatellite data set clearly shows that the Caribbean zone has undergone multiple introductions of W. auropunctata, maybe from the northern coast of South America, represented here by our Guianese samples. The diversity of male and female clones that encompasses all the Caribbean area indicates that introductions are most probably ancient and/or frequent events. This situation probably arose through the extensive human exchange between South and Central America and the Caribbean islands that followed the European colonization in the XVIth century. This latter result parallels those of a previous study based on a mitochondrial DNA marker (Mikheyev and Mueller 2007).
Our study also shows that the Caribbean zone has been an important platform for secondary long-distance introductions all over the World. We found two main routes of introduction connected to the Caribbean zone (Fig. 1). First, a ‘French’ introduction route connects the French Caribbean island Guadeloupe to former (Gabon, Cameroon) and present French overseas territories (New Caledonia, Tahiti). Second, we also demonstrate the existence of an ‘Hispano-American’ introduction route linking sequentially Cuba to Florida, and Florida to Hawaii. The latter link between Floridian and Hawaiian introduced population, recently proposed by Mikheyev et al. (2009), is here clearly evidenced. The geographical and socio-economical proximity of the Caribbean archipelago to the tropical American mainland (i.e. the native area of W. auropunctata), together with its ongoing history of strong connection with other tropical areas worldwide through a variety of ‘cultural’ networks, are the probable causes of the intermediate position of the Caribbean populations in the worldwide invasion of W. auropunctata.
Two other introduction routes illustrate the ‘cultural’ component of the invasion of W. auropunctata. First, we show that Australia, Vanuatu, Papua New Guinea and the Solomon Islands have been invaded by only one clonal queen and two clonal male genotypes. The most probable explanation for this strong link between these populations is the introduction of W. auropunctata through the traditional exchange of plants and goods between Melanesian peoples. We could not further trace back the origin of these introduced populations, but mtDNA data indicate that they probably originate from the Caribbean area (Mikheyev and Mueller 2007). Finally, our microsatellite data suggest, without strong statistical support, an introduction of the Cocos Island population from the Costa Rican mainland, a scenario already suspected in previous studies (Solomon and Mikheyev 2005; Mikheyev and Mueller 2007).
That cultural and commercial networks represent key factors in the current distribution and origins of introduced populations has already been shown in others invasive species, including other invasive ants. Trade explains much of the current distribution of Solenopsis invicta (Tschinkel 2006; Caldera et al. 2008; Zhang et al. 2009), Linepithema humile (Suarez et al. 2001; Corin et al. 2007; Sunamura et al. 2009a) and other invasive ant species (McGlynn 1999). Cultural and economical hubs, such as the Caribbean area, are also ‘invasive species hubs’, as recently illustrated by the presence of three distinct supercolonies of L. humile in the port of Kobe, Japan (Sunamura et al. 2009b).
Eco-evolutionary pathways to invasion
The present study does not strictly link any known introduced population to a known native population. This result was somewhat expected given the high genetic diversity and strong structure of native W. auropunctata populations (Foucaud et al. 2009b). Recent studies pointed that high-density, dominant populations were present in the native geographical range of W. auropunctata (Orivel et al. 2009). The large majority of these native dominant populations were headed by clonal queens and males displaying a specific genetic pattern of outbreeding (i.e. clonal male and queen from a given couple tend to possess divergent genotypes; Foucaud et al. 2009b). We found here that virtually all introduced populations were also clonal, and that these introduced populations stemmed from clonal couples associating male and queen genotypes that also produced particularly heterozygous workers. Dominant populations from the native range and invasive population from the introduced range are therefore similar in term of reproduction system and specific mating patterns, and distinct from the native sexual populations. This work hence provides indirect evidences that the introduced populations of W. auropunctata originate from the native dominant populations.
It is worth pointing that all native dominant populations are located in human-disturbed habitats, in sharp contrast to native non-dominant populations, located in natural habitats such as primary forests. It has therefore been hypothesized that the high heterozygosity of workers from native clonal populations might help them to cope with the particular biotic and abiotic conditions of human-disturbed habitats (Foucaud et al. 2009b). Several studies have suggested that the maintenance of highly heterozygous combinations of genes might be advantageous to maintain viable populations in habitats where abiotic conditions are extreme or changing (Kearney and Shine 2004; Frankham 2005b; Ferreira and Amos 2006), as found for temperature and humidity in human-disturbed habitats of the native range of W. auropunctata (Orivel et al. 2009). Alternatively, highly heterozygous combinations of genes may enable individuals to better exploit their environment, particularly when resources are abundant (Reznick et al. 2000; Vorburger 2005), which is probably the case in human-disturbed habitats such as plantations (Delabie et al. 1994). If the maintenance of specific gene combinations is required to maintain viable populations in human-disturbed habitats, then the male and female clonal reproduction system of W. auropunctata is expected to be advantageous in these habitats, because it lacks recombination, contrary to a sexual reproduction system. The results we obtained here on introduced populations, which were almost only sampled in invaded human-disturbed habitats, are consistent with this hypothesis. Laboratory experiments are needed to test for fitness differences between sexual and clonal populations from both native and introduced ranges using abiotic conditions specific to human-disturbed habitats.
An additional advantage of the male and female clonal reproduction system of W. auropunctata is expected during remote introduction events. Even if the number of initial founders is small, this system indeed prevents the rapid erosion of the introduced population genetic diversity through drift, thus limiting genetical side effects of bottlenecks (i.e. inbreeding depression for sexually reproducing populations, Keller et al. 1994; Haag et al. 2002).
Scenario of the little fire ant worldwide invasion
A traditional vision of biological invasions, illustrated in Fig. 3A, assumes that it is a single step process, where an ‘exotic’ species from a ‘distant’ native area establishes and spreads into an introduced area (Richardson et al. 2000; Sakai et al. 2001; Colautti and MacIsaac 2004). In agreement with this, most current definitions of bioinvasions explicitly insist on the occurrence of a long-distance transport between the native and introduced ranges (Colautti and MacIsaac 2004; Vermeij 2005; Falk-Petersen et al. 2006; but see Valéry et al. 2009). In the case of W. auropunctata, our data, together with previous studies (Foucaud et al. 2009b), suggests that a two-step process as illustrated in Fig. 3B is actually more parsimonious.
Figure 3.
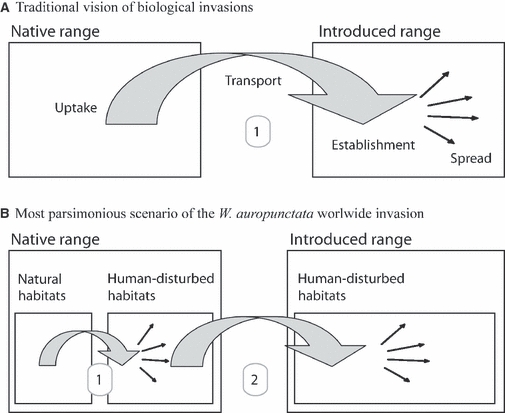
Schematic representation of the traditional vision of bioinvasions (A) and of the most parsimonious scenario of the worldwide invasion of Wasmannia auropunctata (B).
The first step occurs within the native range of the species, where mostly clonal populations, likely originating from natural habitats, spread to and dominate some human-disturbed habitats (Foucaud et al. 2009b). Because natural and human-disturbed habitats are often spatially adjacent within the native range of the species, it is likely that the propagule pressure exerted from the natural on the human-disturbed habitats is significantly higher than traditionally supposed between ‘distant’ native and introduced ranges. This is expected to favor the emergence of dominant populations adapted to human-disturbed habitats.
The second step is the transfer, establishment and local spread of populations from the native to the remote introduced areas of the species. The most parsimonious hypothesis is that these introduced invasive populations stem from the native clonal populations of the human-disturbed habitats, for at least three reasons. First, our study shows that the main vector of W. auropunctata long-distance dispersal is trade, in accordance with previous results on this species (Mikheyev and Mueller 2007) as well as many other pests (McGlynn 1999; Mack et al. 2000; Sunamura et al. 2009b). Because human-modified areas are nowadays extensively connected on a global scale (Rahel 2007; Tatem and Hay 2007), it is therefore highly probable that the propagule uptake from the native range of W. auropunctata is several orders higher from the high-density populations typical of human-disturbed habitats than from low-density populations of natural habitats. Second, it is now widely recognized that one of the major impacts of human activities on Earth's ecosystems is biotic and abiotic homogenization (McKinney and Lockwood 1999; Tilman et al. 2001; Olden et al. 2004; Foley et al. 2005; Ewers et al. 2009). It may therefore not be necessary, or at least less difficult, for introduced populations to adapt to new environmental conditions (i.e. human-disturbed habitats of the introduced range), as long as they come from human-disturbed habitats of the native range. Finally, putative genetic costs of introductions of small size propagules (e.g. inbreeding depression) can be avoided by clonal populations but not by sexual populations of the native range of W. auropunctata (Sakai et al. 2001).
The two-step scenario illustrated in Fig. 3 might apply to a substantial proportion of invasive species, including ants. In S. invicta for instance, populations introduced in the USA originate from native areas that are disturbed naturally or by human activities and where the species is ecologically dominant (Calcaterra et al. 2008). Second, S. invicta was most probably dispersed from ports from the Buenos Aires region into Port Mobile, Alabama (Lofgren 1986). Finally, it has been shown that S. invicta is subsequently favored by human-induced ecological change in its introduced range (King and Tschinkel 2008). Other invasive species that are found in close contact with humans within their native range could also comply with our two-step scenario as was noticed in previous studies (e.g. Sakai et al. 2001).
Conclusions
Our study of the worldwide invasion by W. auropunctata illustrates the central role of human-induced biological change. This human-induced change does not seem to only modify species migration patterns, but also selective pressures over species in both their native and introduced ranges. In the case of W. auropunctata, it is likely that the dramatic biological shifts that putatively occurred during the transition from natural to human-disturbed habitats within its native range is the basis of the worldwide invasion success of the species.
The W. auropunctata case also illustrates the arbitrary aspect of the use of geographical factors (i.e. native/introduced ranges) as a conceptual basis in the study of biological invasions (see Valéry et al. 2009). We argue that invasion biologists should rather use objective ecological factors (i.e. habitats and niches) as a basis to decipher the evolution of invasiveness in wild populations. The need to root invasion biology deeper into ecology and evolution has already been underlined in several seminal publications (Heger and Trepl 2003; Facon et al. 2006; Lee and Gelembiuk 2008).
Acknowledgments
We would like to thank Barbara Gerber for her great help in the lab, Tommy Thompson for sampling the Hawai'ian Wasmannia population, and Benoît Facon and Ruth Hufbauer for helping us improve earlier versions of this manuscript. This work was supported by a grant from the French Ministère de l'Ecologie et du Développement Durable – appel d'offre ECOTROP to AE and JO and by a grant CORUS no 02 412 062 du Ministère des Affaires Etrangères to MT. JHCD acknowledges his research grant by CNPq. Data used in this work were (partly) produced through molecular genetic analysis technical facilities of the IFR119 Montpellier Environnement Biodiversité.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Schematic representation of the gene transmission between two consecutive generations in the two types of reproduction system found in Wasmannia auropunctata.
Figure S2. Schema (A) and example drawn from the Cameroon sample (B) of the diversification of clonal queen genotypes in several populations of the introduced range.
Table S1. Estimated dates of introduction of Wasmannia auropunctata in the sampled introduced range.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Literature cited
- Calcaterra L, Livore J, Delgado A, Briano J. Ecological dominance of the red imported fire ant, Solenopsis invicta, in its native range. Oecologia. 2008;156:411–421. doi: 10.1007/s00442-008-0997-y. [DOI] [PubMed] [Google Scholar]
- Caldera E, Ross K, DeHeer C, Shoemaker D. Putative native source of the invasive fire ant Solenopsis invicta in the USA. Biological Invasions. 2008;10:1457–1479. [Google Scholar]
- Chakraborty R, Jin L. Determination of relatedness between individuals using DNA fingerprinting. Human Biology. 1993;65:875–895. [PubMed] [Google Scholar]
- Clavero M, Garcia-Berthou E. Invasive species are a leading cause of animal extinctions. Trends in Ecology & Evolution. 2005;20:110. doi: 10.1016/j.tree.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Colautti RI, MacIsaac HJ. A neutral terminology to define ‘invasive’ species. Diversity and Distributions. 2004;10:135–141. [Google Scholar]
- Corin SE, Lester PJ, Abbott KL, Ritchie PA. Inferring historical introduction pathways with mitochondrial DNA: the case of introduced Argentine ants (Linepithema humile) into New Zealand. Diversity and Distributions. 2007;13:510–518. [Google Scholar]
- Crooks JA. Characterizing ecosystem-level consequences of biological invasions: the role of ecosystem engineers. Oikos. 2002;97:153–166. [Google Scholar]
- Delabie JHC, Encarnação AMV, Carzola IM. Relations between the little fire ant, Wasmannia auropunctata, and its associated mealybug Planococcus citri in Brazilian coca farms. In: Williams DF, editor. Exotic Ants: Biology, Impact, and Control of Introduced Species. Boulder, CO: Westview Press; 1994. pp. 91–103. [Google Scholar]
- Dlugosch KM, Hays CG. Genotypes on the move: some things old and some things new shape the genetics of colonization during species invasions. Molecular Ecology. 2008;17:4583–4585. doi: 10.1111/j.1365-294X.2008.03932.x. [DOI] [PubMed] [Google Scholar]
- Estoup A, Jarne P, Cornuet JM. Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Molecular Ecology. 2002;11:1591–1604. doi: 10.1046/j.1365-294x.2002.01576.x. [DOI] [PubMed] [Google Scholar]
- Ewers RM, Scharlemann JPW, Balmford A, Green RE. Do increases in agricultural yield spare land for nature? Global Change Biology. 2009;15:1716–1726. [Google Scholar]
- Facon B, Genton BJ, Shykoff J, Jarne P, Estoup A, David P. A general eco-evolutionary framework for understanding bioinvasions. Trends in Ecology & Evolution. 2006;21:130–135. doi: 10.1016/j.tree.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Facon B, Pointier J-P, Jarne P, Sarda V, David P. High genetic variance in life-history strategies within invasive populations by way of multiple introductions. Current Biology. 2008;18:363–367. doi: 10.1016/j.cub.2008.01.063. [DOI] [PubMed] [Google Scholar]
- Falk-Petersen J, Bøhn T, Sandlund O. On the numerous concepts in invasion biology. Biological Invasions. 2006;8:1409–1424. [Google Scholar]
- Ferreira AGA, Amos W. Inbreeding depression and multiple regions showing heterozygote advantage in Drosophila melanogaster exposed to stress. Molecular Ecology. 2006;15:3885–3893. doi: 10.1111/j.1365-294X.2006.03093.x. [DOI] [PubMed] [Google Scholar]
- Foley JA, DeFries R, Asner GP, Barford C, Bonan G, Carpenter SR, Chapin FS, et al. Global consequences of land use. Science. 2005;309:570–574. doi: 10.1126/science.1111772. [DOI] [PubMed] [Google Scholar]
- Foucaud J, Jourdan H, Le Breton J, Loiseau A, Konghouleux D, Estoup A. Rare sexual reproduction events in the clonal reproduction system of introduced populations of the little fire ant. Evolution. 2006;60:1646–1657. [PubMed] [Google Scholar]
- Foucaud J, Fournier D, Orivel J, Delabie JHC, Loiseau A, Le Breton J, Kergoat GJ, et al. Sex and clonality in the Little Fire Ant. Molecular Biology and Evolution. 2007;24:2465–2473. doi: 10.1093/molbev/msm180. [DOI] [PubMed] [Google Scholar]
- Foucaud J, Estoup A, Loiseau A, Rey O, Orivel J. Thelytokous parthenogenesis, male clonality and genetic caste determination in the little fire ant: new evidence and insights from the lab. Heredity. 2009a doi: 10.1038/hdy.2009.169. doi: 10.1038/hdy.2009.169. [DOI] [PubMed] [Google Scholar]
- Foucaud J, Orivel J, Fournier D, Delabie JHC, Loiseau A, Le Breton J, Cerdan P, et al. Reproductive system, social organization, human disturbance and ecological dominance in native populations of the little fire ant, Wasmannia auropunctata. Molecular Ecology. 2009b;18:5059–5073. doi: 10.1111/j.1365-294X.2009.04440.x. [DOI] [PubMed] [Google Scholar]
- Fournier D, Estoup A, Orivel J, Foucaud J, Jourdan H, Le Breton J, Keller L. Clonal reproduction by males and females in the little fire ant. Nature. 2005a;435:1230–1235. doi: 10.1038/nature03705. [DOI] [PubMed] [Google Scholar]
- Fournier D, Foucaud J, Loiseau A, Cros-Arteil S, Jourdan H, Orivel J, Le. Breton J, et al. Characterization and PCR multiplexing of polymorphic microsatellite loci for the invasive ant Wasmannia auropunctata. Molecular Ecology Notes. 2005b;5:239–242. [Google Scholar]
- Frankham R. Resolving the genetic paradox in invasive species. Heredity. 2005a;94:385–385. doi: 10.1038/sj.hdy.6800634. [DOI] [PubMed] [Google Scholar]
- Frankham R. Stress and adaptation in conservation genetics. Journal of Evolutionary Biology. 2005b;18:750–755. doi: 10.1111/j.1420-9101.2005.00885.x. [DOI] [PubMed] [Google Scholar]
- Haag CR, Hottinger JW, Riek M, Ebert D. Strong inbreeding depression in a Daphnia metapopulation. Evolution. 2002;56:518–526. [PubMed] [Google Scholar]
- Heger T, Trepl L. Predicting biological invasions. Biological Invasions. 2003;5:313–321. [Google Scholar]
- Holway DA, Lach L, Suarez AV, Tsutsui ND, Case TJ. The causes and consequences of ant invasions. Annual Review of Ecology and Systematics. 2002;33:181–233. [Google Scholar]
- Kearney M, Shine R. Developmental success, stability, and plasticity in closely related parthenogenetic and sexual lizards (Heteronotia, Gekkonidae) Evolution. 2004;58:1560–1572. doi: 10.1111/j.0014-3820.2004.tb01736.x. [DOI] [PubMed] [Google Scholar]
- Keller LF, Arcese P, Smith JNM, Hochachka WM, Stearns SC. Selection against inbred song sparrows during a natural population bottleneck. Nature. 1994;372:356–357. doi: 10.1038/372356a0. [DOI] [PubMed] [Google Scholar]
- King JR, Tschinkel WR. Experimental evidence that human impacts drive fire ant invasions and ecological change. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20339–20343. doi: 10.1073/pnas.0809423105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbe JJ, Glor RE, Rodriguez Schettino L, Lara AC, Larson A, Losos JB. Genetic variation increases during biological invasion by a Cuban lizard. Nature. 2004;431:177–181. doi: 10.1038/nature02807. [DOI] [PubMed] [Google Scholar]
- Lavergne S, Molofsky J. Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3883–3888. doi: 10.1073/pnas.0607324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CE, Gelembiuk GW. Evolutionary origins of invasive populations. Evolutionary Applications. 2008;1:427–448. doi: 10.1111/j.1752-4571.2008.00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofgren CS. History of imported fire ants in the United States. In: Lofgren CS, Vander Meer RK, editors. Fire Ants and Leaf Cutting Ants: Biology and Management. Boulder, CO: Westview Press; 1986. pp. 36–49. [Google Scholar]
- Lowe S, Browne M, Boudjelas S. 100 of the world's worst invasive alien species. Aliens. 2000;12S:1–12. [Google Scholar]
- Mack RN, Simberloff D, Lonsdale WM, Evans H, Clout M, Bazzaz FA. Biotic invasions: causes, epidemiology, global consequences, and control. Ecological Applications. 2000;10:689–710. [Google Scholar]
- McGlynn TP. The worldwide transfer of ants: geographical distribution and ecological invasions. Journal of Biogeography. 1999;26:535–548. [Google Scholar]
- McKinney ML, Lockwood JL. Biotic homogenization: a few winners replacing many losers in the next mass extinction. Trends in Ecology & Evolution. 1999;14:450–453. doi: 10.1016/s0169-5347(99)01679-1. [DOI] [PubMed] [Google Scholar]
- Mikheyev AS, Mueller UG. Genetic relationships between native and introduced populations of the little fire ant Wasmannia auropunctata. Diversity and Distributions. 2007;13:573–579. [Google Scholar]
- Mikheyev AS, Bresson S, Conant P. Single-queen introductions characterize regional and local invasions by the facultatively clonal little fire ant Wasmannia auropunctata. Molecular Ecology. 2009;18:2937–2944. doi: 10.1111/j.1365-294X.2009.04213.x. [DOI] [PubMed] [Google Scholar]
- Olden JD, LeRoy Poff N, Douglas MR, Douglas ME, Fausch KD. Ecological and evolutionary consequences of biotic homogenization. Trends in Ecology & Evolution. 2004;19:18–24. doi: 10.1016/j.tree.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Orivel J, Grangier J, Foucaud J, Le Breton J, Andrès F-X, Jourdan H, Delabie JHC, et al. Ecologically heterogeneous populations of the invasive ant Wasmannia auropunctata within its native and introduced ranges. Ecological Entomology. 2009;34:504–512. [Google Scholar]
- Passera L. Characteristics of tramp species. In: Williams DF, editor. Exotic Ants: Biology, Impact, and Control of Introduced Species. Boulder, CO: Westview Press; 1994. pp. 23–43. [Google Scholar]
- Rahel FJ. Biogeographic barriers, connectivity and homogenization of freshwater faunas: it's a small world after all. Freshwater Biology. 2007;52:696–710. [Google Scholar]
- Reznick D, Nunney L, Tessier A. Big houses, big cars, superfleas and the costs of reproduction. Trends in Ecology & Evolution. 2000;15:421–425. doi: 10.1016/s0169-5347(00)01941-8. [DOI] [PubMed] [Google Scholar]
- Richardson DM, Pysek P, Rejmanek M, Barbour MG, Panetta FD, West CJ. Naturalization and invasion of alien plants: concepts and definitions. Diversity and Distributions. 2000;6:93–107. [Google Scholar]
- Roman J, Darling JA. Paradox lost: genetic diversity and the success of aquatic invasions. Trends in Ecology & Evolution. 2007;22:454–464. doi: 10.1016/j.tree.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, Baughman S, et al. The population biology of invasive species. Annual Review of Ecology and Systematics. 2001;32:305–332. [Google Scholar]
- Sax DF, Brown JH. The paradox of invasion. Global Ecology and Biogeography. 2000;9:363–371. [Google Scholar]
- Shogren JF, Tschirhart J. Integrating ecology and economics to address bioinvasions. Ecological Economics. 2005;52:267–271. [Google Scholar]
- Solomon SE, Mikheyev AS. The ant (Hymenoptera: Formicidae) fauna of Cocos island, Costa Rica. Florida Entomologist. 2005;88:415–423. [Google Scholar]
- Suarez AV, Holway DA, Case TJ. Patterns of spread in biological invasions dominated by long-distance jump dispersal: insights from Argentine ants. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:1095–1100. doi: 10.1073/pnas.98.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunamura E, Espadaler X, Sakamoto H, Suzuki S, Terayama M, Tatsuki S. Intercontinental union of Argentine ants: behavioral relationships among introduced populations in Europe, North America, and Asia. Insectes Sociaux. 2009a;56:143–147. [Google Scholar]
- Sunamura E, Hatsumi S, Karino S, Nishisue K, Terayama M, Kitade O, Tatsuki S. Four mutually incompatible Argentine ant supercolonies in Japan: inferring invasion history of introduced Argentine ants from their social structure. Biological Invasions. 2009b;11:2329–2339. [Google Scholar]
- Tatem AJ, Hay SI. Climatic similarity and biological exchange in the worldwide airline transportation network. Proceedings of the Royal Society B: Biological Sciences. 2007;274:1489–1496. doi: 10.1098/rspb.2007.0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D, Fargione J, Wolff B, D'Antonio C, Dobson A, Howarth R, Schindler D, et al. Forecasting agriculturally driven global environmental change. Science. 2001;292:281–284. doi: 10.1126/science.1057544. [DOI] [PubMed] [Google Scholar]
- Tschinkel WR. The Fire Ants. Cambridge, MA: Harvard University Press; 2006. [Google Scholar]
- Valéry L, Fritz H, Lefeuvre J-C, Simberloff D. Invasive species can also be native. Trends in Ecology & Evolution. 2009;24:585. doi: 10.1016/j.tree.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Vermeij GJ. Invasion as expectation: a historical fact of life. In: Sax DF, Stachowicz JJ, Gaines SD, editors. Species Invasions: Insights into Ecology, Evolution, and Biogeography. Sunderland: Sinauer Associates, Inc; 2005. pp. 315–339. [Google Scholar]
- Vitousek PM, D'Antonio CM, Loope LL, Rejmanek M, Westbrooks R. Introduced species: a significant component of human-caused global change. New Zealand Journal of Ecology. 1997;21:1–16. [Google Scholar]
- Vonshak M, Dayan T, Foucaud J, Estoup A, Hefetz A. The interplay between genetic and environmental effects on colony insularity in the clonal invasive little fire ant Wasmannia auropunctata. Behavioral Ecology and Sociobiology. 2009a;63:1667–1677. [Google Scholar]
- Vonshak M, Dayan T, Ionescu-Hirsh A, Freidberg A, Hefetz A. The little fire ant Wasmannia auropunctata: a new invasive species in the Middle East and its impact on the local arthropod fauna. Biological Invasions. 2009b doi: 10.1007/s10530-009-9593-2. [Google Scholar]
- Vorburger C. Positive genetic correlations among major life-history traits related to ecological success in the aphid Myzus persicae. Evolution. 2005;59:1006–1015. [PubMed] [Google Scholar]
- Wetterer JK, Porter SD. The little fire ant, Wasmannia auropunctata: distribution, impact, and control. Sociobiology. 2003;42:1–41. [Google Scholar]
- Zhang R, Li Y, Liu N, Porter SD. An overview of the red imported fire ant (Hymenoptera: Formicidae) in mainland China. Florida Entomologist. 2009;90:723–731. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


