Abstract
When a neutral mutation arises in an invading population, it quickly either dies out or ‘surfs’, i.e. it comes to occupy almost all the habitat available at its time of origin. Beneficial mutations can also surf, as can deleterious mutations over finite time spans. We develop descriptive statistical models that quantify the relationship between the probability that a mutation will surf and demographic parameters for a cellular automaton model of surfing. We also provide a simple analytic model that performs well at predicting the probability of surfing for neutral and beneficial mutations in one dimension. The results suggest that factors – possibly including even abiotic factors – that promote invasion success may also increase the probability of surfing and associated adaptive genetic change, conditioned on such success.
Keywords: ecological genetics, invasions, mutation surfing
Introduction
The genetic composition of a natural population is always changing because mutations inevitably arise and rise or fall in frequency due to selection or drift (i.e. stochasticity in finite populations). Genetic changes continue during invasions, when a population is entering and becoming established in new territory. Ecologists recognize that selection can play a crucial role in determining the success, speed and ultimate extent of invasions (Hoffmann and Sgrò 1995; Sakai et al. 2001; Holt et al. 2005). Even neutral genetic change during invasions is of high interest to researchers who use neutral markers to infer the migration history of populations (Semino et al. 2000; Sunnucks 2000; Schaal and Leverich 2001; Wang and Whitlock 2003; Kawiecki and Ebert 2004). This is because to test any hypothesis concerning the past spatial distribution of a population, it is necessary to specify a null model that incorporates explicit assumptions about demography and dispersal patterns. It is further necessary to understand the distribution of neutral allele frequencies under the specified model, in order to compare observed marker data with the distribution expected under the null hypothesis. Yet, despite the recognized importance of genetic changes during invasions, theoreticians have paid little attention to this topic until recently (work on the evolution of dispersal constitutes an important exception; see, for example, Holt 2003; Simmons and Thomas 2004).
Recently, several authors have used stochastic simulations to study the fate of a single mutant that arises during an invasion (Edmonds et al. 2004; Klopfstein et al. 2005; Travis et al. 2007; Hallatschek and Nelson 2008). These studies have focused on ‘mutation surfing’, in which mutants arise on or advance to the front of a traveling wave of population density and essentially shut out wild types (i.e. nonmutants) from a certain point in the invasion onwards (Edmonds et al. 2004). Early studies of this topic offered a number of descriptive statements about the fate of mutations in invasions, including a ‘midpoint rule’ for the location of the center of population density of a successful neutral mutation (Edmonds et al. 2004) and a proposal that a certain ‘lumped’ model parameter has a strong linear correlation with the ultimate fraction of the invading population that carries such a mutation (Klopfstein et al. 2005). More recent studies have treated surfing with selection, using mathematical models such as annihilating random walks, generalized diffusion equations and stochastic partial differential equations to explain the phenomena observed in surfing simulations and in vitro experiments (Hallatschek et al. 2007; Hallatschek and Nelson 2009) (Korolev et al., 2010). Recently, it has been argued that some genetic differences between human populations that had previously been attributed to selection in fact resulted from surfing by neutral alleles (Hofer et al. 2009).
One purpose of the present work is to provide a careful qualitative and quantitative description of neutral mutation surfing as seen in a stochastic model like those studied in Edmonds et al. (2004), Klopfstein et al. (2005), Travis et al. (2007) and Hallatschek and Nelson (2008). We focus on a model of a one-dimensional habitat but include some results for two-dimensional habitats. It should be noted that a one-dimensional habitat is a realistic model for certain types of invasions, such as invasion along a coastline or river (Lubina and Levin 1988; Speirs and Gurney 2001; Pachepsky et al. 2005). As a consequence, study of such models and comparisons between them and two-dimensional models can be practically as well as theoretically meaningful.
There are two main reasons to study the neutral case. First, neutrality is simpler than selection, and with so little existing theory, it is reasonable to study the simpler case first. Second, neutral genetic markers are of interest because they can provide information about the history of an invasion. Indeed, much of the original interest in mutation surfing was among researchers whose primary goal is to reconstruct range expansion (e.g. the spread of humans into Europe) with such markers (Edmonds et al. 2004).
On the other hand, adaptive change during invasions may be the genetic phenomenon of most practical interest to conservation biologists. Accordingly, we have begun to extend our models to the cases of beneficial and deleterious mutations, and we present some results here. Our main goals are to describe how the probability of surfing depends on model parameters (with or without selection), to explain heuristically the nature of this dependence, and to offer a simple model of the surfing process as a contribution to the development of analytic models that yield quantitative predictions about genetic change during invasions.
Our work is based on data obtained from a series of simulations of cellular automata. In what follows, we state the specifications of the simulations, use statistical methods (in particular, logistic regression) to describe the probability of surfing and to assess our analytic model, and offer likely explanations for the quantitative results we obtain.
Model and simulation specifications
The model we studied, following Edmonds et al. (2004), Klopfstein et al. (2005) and Travis et al. (2007), is a type of individual-based model known to mathematicians as a contact process (Liggett 1999). We simulated a contact process in which wild-type (i.e. nonmutant) and mutant individuals reproduce asexually and move between adjacent cells in a rectangular grid. For the neutral case, grid lengths l used were 100, 200 and 400 cells; grid widths w used were 1, 3, 7, 13 and 25 cells. For the case of selection, only 1×400 grids were used. We note that previous studies used only 25 × 100 grids (Edmonds et al. 2004; Klopfstein et al. 2005; Travis et al. 2007). We varied grid width in order to study the effect of dimensionality on the probability of surfing. We varied grid length in order to make sure that numerically ascertained probabilities of surfing on a finite grid came close to asymptotes, which we expect to correspond to probabilities of surfing on an infinitely long grid. Accordingly, all results below pertain to grids of length 400 unless otherwise specified.
As in Klopfstein et al. (2005), each simulation run began with a single wild-type individual placed at the center of the leftmost column of the grid. (This is not the only possible choice. For example, the founder could be placed along a side or in the middle of the grid to model colonization beginning other that at the mouth of a river. We have not yet extended our simulations to such cases.) Generations were discrete and comprised three steps. First, each individual was replaced in the same cell by a number of offspring chosen from a Poisson distribution with mean λ. For the neutral case, we used λ − 1 = 0.05, 0.1, 0.2, 0.4, 0.8, 7, 15 and 31; in each simulation run, λ was the same for mutants and wild types. For the case with selection, we used λwt − 1 = 0.1 and 0.4 for wild types and λmut − 1 = α(λwt − 1) for mutants, with α = 0.5, 0.95, 1.05 and 1.5. Next, each cell whose population N (of mutant and wild types combined) was greater than a carrying capacity K underwent culling, in which N−K randomly chosen individuals were discarded. All individuals within a cell, whether mutant or wild type, had equal probability of being culled. For the neutral case with λ < 2, we used carrying capacities K = 10, 50, 100 and 500; we used K = (λ − 1)/2 for all λ > 2. For the case with selection, we used K = 10 and 100.
The final step in each generation was migration. Each individual (whether mutant or wild type) had a probability m of migrating in each generation. For the neutral case with λ < 2, we used mean migration rates m = 0.05, 0.1, 0.2 and 0.4; with λ > 2, we used m = 0.1 and 0.2. For the case with selection, we used m = 0.05 and 0.2. Each individual chosen to migrate then moved to a cell chosen with equal probability from all those adjacent to its original cell. (Cells in the interior of the grid had four neighbors, those on the boundary had three and those in the corners had two; diagonal moves were not permitted).
Table 1.
Predictors, estimated coefficients with standard errors and 95% confidence interval lower and upper bounds for odds ratios for one-dimensional habitat with K = 10
| Predictor | Coefficient | SE | Lower odds | Higher odds |
|---|---|---|---|---|
| Constant | −0.90 | 0.13 | NA | NA |
| ln m | −0.47 | 0.057 | 0.56 | 0.70 |
| ln (λ−1) | 1.1 | 0.095 | 2.57 | 3.73 |
When at least one member of the population reached the 10th column from the left, one (wild-type) individual chosen randomly with equal probability from all individuals in the 10th column was replaced by a mutant. No other mutation events were included in the model. Thus, each offspring shared the type of its parent, with the sole exception being the original mutant.
A run was stopped after the first generation in which either the entire population went extinct, mutants went extinct after being introduced or at least one individual reached the rightmost column of the grid. After a run was stopped, a code for the state of the mutant and wild-type populations was saved. Three states were distinguished for each type: extinction (no individuals of a given type persisted), persistence without advancing (at least one individual remained, but no individuals of the specified type were found in the rightmost column) and advancing (at least one individual of the specified type was found in the rightmost column). A run was considered ‘unsuccessful’ if both mutants and wild types went extinct and ‘successful’ otherwise. Experimentation showed that wild types virtually never went extinct after a mutation event had occurred; so, runs with mutation events were counted as successful, even though they were usually aborted if the mutants subsequently went extinct. (As the total population size was close to 10K times the habitat width when the mutant was introduced, it is not surprising that extinction almost never occurred after this time.)
Surfing was considered to occur in runs where a mutant advanced to the rightmost column. When this occurred, it was extremely rare for wild types to also be found in the rightmost column. In addition to outcome codes for each run, we saved the state of the 7h subgrid centered on the 10th column for the three generations immediately following introduction of the mutant. We also saved the complete final grids for each run.
For the neutral case with a one-dimensional (1 × 400) grid, each possible combination of the model parameters λ, K and m was used in 600 simulation runs. For the case of selection, each combination of the model parameters was used in 10 000 runs. For each two-dimensional (w × 400 with w = 3, 7, 13 and 25) grid, each parameter was used in 50 simulation runs. We also performed small numbers of runs with parameter combinations other than those listed here; specifications for these runs are given when they are discussed below.
Unlike in Klopfstein et al. (2005), sequences of runs were not stopped when a predetermined number of instances of surfing was reached. This avoided bias in estimating the probability of surfing, as otherwise a negative binomial distribution would have had to be used to model this probability (Hogg and Tanis 1997).
Dependence of the probability of surfing on model parameters
Visual inspection of runs in progress gave a strong impression of a dichotomy of outcomes: mutants either vanished or flourished (see Figs 1 and 2). In many runs, mutants went extinct. In most runs (and in nearly all runs on grids of length 400) where mutants persisted without advancing, only a few mutants remained on the grid at the end of the simulation, and these few were found close to their point of origin. Longer habitat lengths led to more mutant extinctions and fewer cases of mutant persistence without advancing. This was apparently because runs on longer grids simply took longer, allowing more time for nonsurfing mutant populations to dwindle and ultimately die out.
Figure 1.
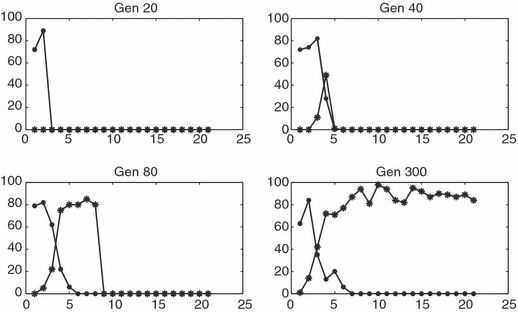
Wild-type (circle) and mutant (star) population densities as functions of longitude on a 1 × 400 grid after 20, 40, 80 and 300 generations in a run with mutant surfing. Parameters: m = 0.4, λ = 1.8, K = 100.
Figure 2.
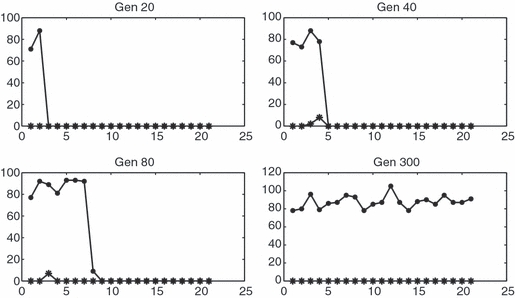
Wild-type (circle) and mutant (star) population densities as functions of longitude on a 1 × 400 grid after 20, 40, 80 and 300 generations in a run with mutant extinction. Parameters are as in Figure 2.
Visual inspection also showed that in virtually all runs where mutant surfing occurred, the mutants excluded wild types from the wavefront very soon after the initial mutation arose, and proceeded to nearly monopolize the habitat between the point of mutant introduction and the right-hand boundary. We thus viewed all successful runs as random trials with two possible outcomes, surfing or mutant extinction. For a given combination of parameters, we denote the probability of surfing (conditioned on nonextinction of the total population) by psurf.
We note that under neutrality, the left-hand boundary of the habitat occupied by surfing mutants remained close to the point at which the mutation first arose. By contrast, beneficial mutations ‘backfilled’ from the point of introduction to replace the wild types. Likewise, deleterious mutations that originally appeared to ‘surf’ subsequently lost habitat as wild types advanced to replace the less-fit types.
A major goal of work on mutation surfing is to relate the probability of surfing to model parameters such as cell carrying capacity K, migration rate m and growth rate λ (Edmonds et al. 2004; Klopfstein et al. 2005; Hallatschek and Nelson 2008). For the one-dimensional case, plots of mean (over all runs with given values of λ, m and K) psurf against model parameters showed an interaction between K and λ (Fig. 3). For each value of K, psurf increased with λ. However, the rate of increase was much greater for the smallest carrying capacity, K = 10, than for the higher values of K. This motivated dividing the runs into two groups, one with K = 10 and one with K = 50, 100, 500, for further analysis.
Figure 3.
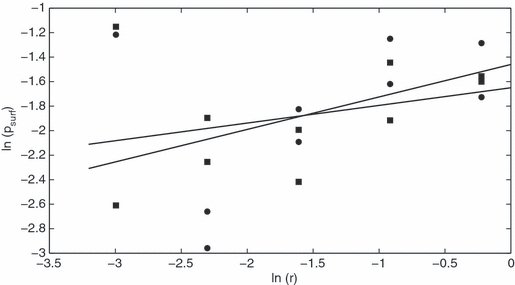
ln (psurf) versus ln (λ−1) for a one-dimensional habitat, with regression lines for K = 10 (circles) and 100 (squares).
To quantify the dependence of psurf on K, m and λ, we use logistic regression, a tool which is adapted to processes with binary outcomes for individual trials (Hogg and Tanis 1997). In our logistic regression, the response (or y) variable is the log-odds of surfing:
| (1) |
The predictor (or x) variables could in principle be any or all of the model or simulation parameters, or functions of those parameters.
In seeking an appropriate statistical model to describe the simulation output, we tried various functions of K, m and λ as predictor variables. We used three different goodness-of-fit tests (Pearson's chi-squared, deviance and Hosmer–Lemeshow) (Hosmer and Lemeshow 2000) to assess the suitability of each statistical model. We used three criteria to choose among the models that passed the goodness-of-fit tests at the 1% significance level. First, we sought a model in which every predictor was associated with an odds ratio that differed significantly from 1. Second, we sought a model with few or no interaction terms. Third, we sought a model with a low value of the Akaike information criterion (AIC) (Akaike 1974). We judged the most suitable model to be a function of ln m, ln (λ − 1) and ln K:
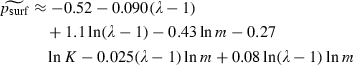 |
(2) |
(standard errors for the coefficients and confidence intervals for odds ratios are given in Table 2). We used McFadden's pseudo-R2 (ψR2) to assess the amount of variability explained by equation (2); we found ψR2 = 0.31, indicating reasonably strong explanatory power.
Table 2.
Predictors, estimated coefficients with standard errors and 95% confidence interval lower and upper bounds for odds ratios for one-dimensional habitat with K = 10, 100, 500
| Predictor | Coefficient | SE | Lower odds | Higher odds |
|---|---|---|---|---|
| Constant | −0.74 | 0.17 | NA | NA |
| ln m | −0.51 | 0.039 | 0.56 | 0.65 |
| ln (λ−1) | 0.82 | 0.053 | 2.04 | 2.51 |
| ln K | −0.28 | 0.030 | 0.72 | 0.81 |
For the same one-dimensional grid with K = 10, we did not find a satisfactory model. This may have been because values of psurf with K = 10 were typically very low, so that the relative sampling error in estimating psurf was high enough to obscure associations with the other model parameters; further simulations are planned to explore this possibility. It was possible to combine data for all values of K and find a model that satisfied our formal criteria, but this required including so many interaction terms that the model did not seem to be useful for gaining insights.
It is possible to explain heuristically the direction of the dependence of psurf on each predictor (λ, m, K) in equation (2). To do so, recall that just one mutant is initially present. It is well known for simpler demographic structures that the smaller the total population, the greater is the probability that a mutation initially present in a single copy will rise to high frequencies (Hartl and Clark 1997). Correspondingly, in our model the mutant should be more likely to surf if it initially appears in a cell with no other (wild-type) occupants. This is turn should be more likely if the cell carrying capacity K and the migration rate m are low because these conditions should lower the number of individuals at the wavefront, where the mutation occurs. Furthermore, cells at the wavefront should generally contain fewer individuals than cells further back in the wave; so, fewer offspring will be culled at the front, making the effective growth rate higher there. Again, as the mutant must arise at the front, this means that the probability of surfing should increase with λ (as for fixed m and K, a higher value of λ should allow more mutants to be born before wild types ‘catch up’ to the mutant population at the leading edge).
In equation (2), it is a straightforward calculus exercise to check that the derivatives 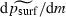 and
and 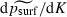 are both negative, while
are both negative, while 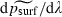 is positive, for the range of parameter values used. Thus, our statistical model predicts that the probability of surfing psurf will decline as m or K increases but rise as λ increases, which is consistent with our heuristic argument.
is positive, for the range of parameter values used. Thus, our statistical model predicts that the probability of surfing psurf will decline as m or K increases but rise as λ increases, which is consistent with our heuristic argument.
For the (neutral) two-dimensional case, we first considered only those runs with a 25 × 400 grid. Using the same criteria as for the one-dimensional case, we found a simple model to provide the best description of the relationship between psurf and the model parameters:
| (3) |
(standard errors for the three coefficients, in order, were 0.27, 0.063 and 0.13). We caution, however, that for this model ψR2 = 5%. As in the one-dimensional case, here 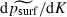 is negative and
is negative and 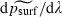 is positive. Unlike in the one-dimensional case, however, including functions of the migration rate m as a predictor did not improve the model in two dimensions. Equations (not shown) with coefficients close to those in equation (3) provided good fits to simulation data with grids of widths 13 or 7; the fit was less good for a 3 × 400 grid. However, all results for the two-dimensional case should be viewed as preliminary because of the small number of trials performed with each combination of parameters.
is positive. Unlike in the one-dimensional case, however, including functions of the migration rate m as a predictor did not improve the model in two dimensions. Equations (not shown) with coefficients close to those in equation (3) provided good fits to simulation data with grids of widths 13 or 7; the fit was less good for a 3 × 400 grid. However, all results for the two-dimensional case should be viewed as preliminary because of the small number of trials performed with each combination of parameters.
For the case with selection, surfing never occurred when λmut was less than 1; so, we considered only parameter combinations with λmut > 0 (these did include some deleterious mutations). We did not find a statistical model that simultaneously satisfied goodness-of-fit criteria and provided good explanatory power as measured by ψR2. The model
| (4) |
gave ψR2=0.23 but failed goodness-of fit tests (P < 0.001); standard errors for the coefficients, in order, were 0.061, 0.055, 0.030, 0.015 and .0093.
In equation (4), it is noteworthy that psurf depends on λmut much more than on λwt. This suggests that for the cases studied, absolute fitness was more important than relative fitness in determining surfing success (a similar effect was found in an island model analyzed by Holt and Gomulkiewicz (1997)). We also note that although the coefficients of ln m and ln K differ from those in the neutral case (2), their sign is the same. Thus, for the cases studied, the probability of mutant surfing varies inversely with m and K for both neutral and selected mutations.
Analytic models
Beyond merely describing (even quantitatively) the dependence of psurf on model parameters, it is clearly important to explain the form of this dependence in a way that sheds light on the process by which a mutation succeeds or fails. A partial explanation can be obtained from two observations; we consider the neutral case first. In this case, mutants and wild types in our model behave identically. They reproduce and migrate in identical ways, and ‘count’ identically toward the carrying capacity of a cell. The second observation, based on watching generation-by-generation plots of mutant and wild-type population density as a function of longitude, is that in virtually every case when surfing occurs, mutants take the lead and begin to fill the available unoccupied terrain within a very few generations after the original mutation occurs.
As mutant and wild-type individuals behave identically, we may draw a conclusion about the genealogies of individuals in the advancing wave of population density as follows. Imagine drawing a line at the longitude where the mutant first arises, or a short distance past this point. Then when a simulation run terminates, nearly all of the individuals to the right of this line will have descended from a single common ancestor in the generation when the mutant first appeared – regardless of whether mutation surfing occurred. For when surfing does occur, almost all of the relevant terrain will be occupied by mutants, which must have descended from the single original mutant. Therefore, as mutants and wild types are functionally identical, similar genealogies must arise even when surfing does not occur and the terrain is filled by wild types. We can thus recharacterize mutation surfing as the scenario in which the common ancestor of nearly all individuals in the population that ultimately invades happens to be the original mutant, rather than any of the wild types present at the time the mutation occurred. (This line of reasoning informs models of the coalescent with stepping-stone spatial structure; see, for example, Austerlitz et al. 1997; Wilkins and Wakeley 2002; Durrett and Restrepo 2008.)
Our second observation – that a mutation's fate appears to be sealed within a few generations of its appearance – suggests a simplified model of the process by which this common ancestor is ‘‘chosen’’. This model consists of an iteration of two steps. The first step occurs in a early generation (P, F1 or F2, where the generation including the original mutant is counted as P). In this step, one individual is chosen randomly from those in the rightmost cells as a potential common ancestor for the future population. Each individual in the rightmost cell, whether mutant or wild type, has the same probability of being chosen. In the second step, the chosen individual succeeds or fails to propagate with probabilities equal to the probabilities of survival or extinction at the very beginning of a run (when the population consists of exactly one individual, located at the left-hand boundary). Motivation for this step comes from the strong association between psurf and prun, the a priori probability that a run will not end in extinction, when λ is not very high (Fig. 4; R2 = 60.1% between ln psurf and ln prun). If the chosen individual fails to propagate (i.e. its progeny die out), the process begins again with a new choice of potential common ancestor from those which were present in the rightmost cells during the P generation (all of which are assumed to have survived).
Figure 4.
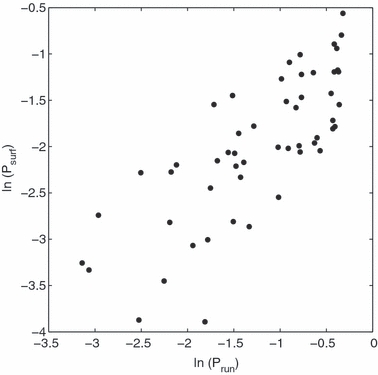
;ln (psurf) versus ln (prun) for a one-dimensional habitat and λ < 2.
This model leads to a testable quantitative prediction. To derive an estimate 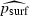 for psurf under this model, we let N denote the number of individuals present in the rightmost cell in generation P. Then
for psurf under this model, we let N denote the number of individuals present in the rightmost cell in generation P. Then
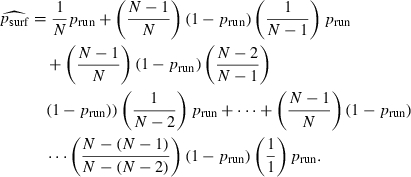 |
(5) |
The first term on the right-hand side of equation (5), (1/N)prun, is the probability that the mutant is the first candidate chosen and does succeed in propagating. The second term is the probability (N − 1)/N(1 − prun) that a wild-type individual is the first chosen but fails to propagate, times the probability 1/(N − 1)prun that the mutant is the second chosen and does propagate. The subsequent terms are derived similarly. Algebraic manipulation condenses equation (5) into the form
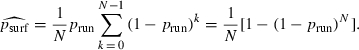 |
(6) |
To test whether equation (6) was a good fit to the one-dimensional simulation results, we first estimated prun and mean N separately for each combination of the parameters λ, m and K that yielded at least one run not ending in extinction (there were 70 such parameter combinations), then ran a linear regression of the observed values of ln (psurf) against the predicted values 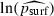 from equation (6). (Taking logarithms rendered the variance of observed psurf approximately equal for all values of
from equation (6). (Taking logarithms rendered the variance of observed psurf approximately equal for all values of 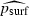 , a necessary condition for valid inference in linear regression.) We caution that in our simulations when λ was very high (i.e. λ≥8), prun was always very close to 1 and so a relationship between prun and psurf was no longer apparent; therefore, the following discussion considers only our neutral simulations with λ < 2.
, a necessary condition for valid inference in linear regression.) We caution that in our simulations when λ was very high (i.e. λ≥8), prun was always very close to 1 and so a relationship between prun and psurf was no longer apparent; therefore, the following discussion considers only our neutral simulations with λ < 2.
The regression equation was
| (7) |
(R2=56.1%), which is broadly consistent with prediction (6), except that the constant was lower than expected (standard errors for the constant and the coefficient of ln (prun) were 0.1766 and 0.1120 respectively). When the regression was run for each value of the migration rate m separately, the discrepancy between predicted and observed psurf was seen to occur only for very high migration rates. Indeed, the constant term and the coefficient of 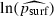 in the regression equation were not significantly different from the predicted values of 0 and 1, respectively, except for the highest value of m, namely 0.40. Agreement between the predicted and observed equations improved as m decreased (details are shown in Table 3).
in the regression equation were not significantly different from the predicted values of 0 and 1, respectively, except for the highest value of m, namely 0.40. Agreement between the predicted and observed equations improved as m decreased (details are shown in Table 3).
Table 3.
Coefficients for regression of observed on predicted ln (psurf)
| m | Constant | SE | Coefficient of ln (psurf) | SE |
|---|---|---|---|---|
| 0.40 | −1.5 | 0.35 | 0.43 | 0.24 |
| 0.20 | −0.31 | 0.38 | 1.5 | 0.28 |
| 0.10 | −0.24 | 0.31 | 1.2 | 0.20 |
| 0.05 | −0.22 | 0.19 | 0.99 | 0.11 |
We now briefly consider model (6) for large λ. When λ and K are both very large, prun approaches 1, and taking a limit of the right-hand side of equation (6) with K being a fixed multiple of λ − 1 (K = a(λ − 1)) and prun = 1 − e−λ suggests that psurf should approximately equal 1/λ. However, our simulations with large λ and K = (λ − 1)/2 show far higher values of psurf than this calculation would predict – for example, psurf ≍ 0.85 with λ = 32, K = 16 and m = 0.1. Furthermore, psurf did not appear to decrease as λ was increased. The explanation appears to be that even though most cells filled to capacity within one or two generations of colonization, the rightmost cell generally contained very few individuals – averaging fewer than two for all conditions simulated – and each of these individuals had a high probability of becoming the common ancestor of the future occupants of the grid from that point onwards. (It is possible that different behavior might be found if K were held constant as λ increased, however.)
Analogues of equation (6) proved to be almost completely unexplanatory in two-dimensional settings under neutrality, yielding R2 values of less than 5%. Indeed, there appeared to be no meaningful relationship between any powers of psurf and prun in two dimensions.
Under selection (i.e. with λmut ≠ λwt), the analogue of equation (6) is more complicated because the numbers of mutant and wild-type ‘candidates’ remaining after each unsuccessful attempt to propagate do not follow a simple linear pattern. Rather, if the rightmost cell in generation P is occupied by one mutant and N − 1 wild types, and if the number of candidates is still assumed to decrease by 1 after each failed attempt, then the expected numbers u(t) and w(t) of mutant and wild-type candidates t generations later (conditioned on mutant survival) are given by
| (8) |
| (9) |
When these expressions are used in an analogue of equation (6) that also includes different values of prun for mutants and wild types, the result is not readily simplified. However, the first two terms can be computed:
| (10) |
| (11) |
where 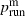 and
and 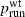 are the values of prun for mutants and wild types, respectively, and Ti is the probability of mutant surfing, conditioned on the event that the successful surfing attempt occurs i − 1 generations after generation P.
are the values of prun for mutants and wild types, respectively, and Ti is the probability of mutant surfing, conditioned on the event that the successful surfing attempt occurs i − 1 generations after generation P.
For the parameter combinations used in our simulations, mean T1 performed very well as a predictor of mean psurf for beneficial mutations (R2 = 96.8%), and mean T1 + T2 performed slightly better (R2 = 98.5%). Specifically, linear regression yielded the equations
| (12) |
with standard errors of 0.028 for the constant and 0.056 for the coefficient of 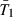 , and
, and
| (13) |
with standard errors of 0.019 for the constant and 0.037 for the coefficient of T1+T2. As equations (10) and (11) show that T1 and T2 are linear functions of 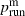 and
and 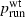 , this indicates that psurf itself is very close to a linear function of these two quantities. For deleterious mutations, however,
, this indicates that psurf itself is very close to a linear function of these two quantities. For deleterious mutations, however, 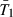 and T1+T2 drastically overestimated psurf, possibly because deleterious mutations that initially ‘surfed’ later lost ground to introgressing wild types.
and T1+T2 drastically overestimated psurf, possibly because deleterious mutations that initially ‘surfed’ later lost ground to introgressing wild types.
Discussion
We have developed descriptive statistical models that quantify the relationship between the probability psurf that a mutation will surf and the parameters λ, m and K of a frequently studied cellular automaton model (Edmonds et al. 2004; Klopfstein et al. 2005; Travis et al. 2007; Hallatschek and Nelson 2008). We have also proposed an analytic model that performs reasonably well at predicting psurf for the case of a neutral or beneficial mutation on a one-dimensional grid. Such simplified models are desirable because a completely rigorous explanation of the dependence of surfing probability on a full range of demographic and genetic parameters currently appears unobtainable. (This is not to say that the mathematical models that have been developed, such as those in Hallatschek and Nelson 2008, 2009, are uninformative, but simply that they do not yield self-contained formulas for psurf as a function of system parameters in all regimes.) Moreover, a typical rigorous result for an individual-based model of invasion dynamics may simply give conditions under which the probability of persistence is either positive or zero; see, for example, the recent result on surfing in E. Andjel, J. Miller and E. Pardoux, unpublished manuscript. In the absence of a rigorous theory, descriptive statistical models and simplified analytic models like those developed here can assess the strengths or weaknesses of heuristic explanations and may be of value in making predictions of invasiveness. Thus, such models remain crucial tools for checking and expanding upon nonquantitative verbal models of mutation dynamics during invasions.
For the neutral case, our statistical models differ in several respects from those proposed in earlier work (Klopfstein et al. 2005). One reason may simply be that previous authors used only grids of length 100 cells for their simulations (Edmonds et al. 2004; Klopfstein et al. 2005; Travis et al. 2007; Hallatschek and Nelson 2008). This may have inflated estimates of the probability of surfing, and especially of mutant survival without surfing (Travis et al. 2007 contains some discussion on this point). With the grids of length 400 used here, it is extremely rare for mutants to survive to the end of a simulation unless surfing has occurred. As a result, the bimodality of the final spatial distribution of mutants reported in Edmonds et al. (2004) essentially vanishes. Some experimentation (details not shown) suggested that extension to still longer grids would have little effect on observed probability of mutant survival and psurf.
We note that with the bimodality removed, it is easy to explain the ‘midpoint rule’ for locating the point of origin of a mutation proposed in Edmonds et al. (2004). Specifically, surviving mutants have almost invariably surfed. Furthermore, visual inspection of simulations in progress shows that surfing neutral mutants almost invariably occupy nearly all of the terrain from their point of origin to the rightmost boundary of the grid. It follows that the centroid of the final mutant population is halfway between the point of origin and the boundary, which is a rephrasing of the midpoint rule. In addition, as we have noted above, when beneficial mutants surf they also introgress backwards into territory previously colonized by wild types. The extent to which weak selective advantage or linkage to selected loci may bias midpoint-rule estimates therefore deserves study.
More substantively, it is important to address why equations (2) and (3), which we propose as statistical descriptions of the relationship between the log-odds of surfing 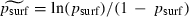 and the parameters λ, m and K, differ dramatically from the simple linear relationship between ‘mutant success’ (a proxy for psurf) and the ‘lumped’ parameter (λ − 1)/Km proposed in Klopfstein et al. (2005) (which, we note, is also consistent with the heuristics presented above). Indeed, equation (2) suggests that more appropriate lumped parameters for the one- and two-dimensional cases, respectively, might be (λ−1)/K1/4m1/2 and (λ − 1)1/2/K1/4. The answer appears to be that the analysis in Klopfstein et al. (2005) depends on a linear regression of psurf on (λ − 1)/Km. Linear regression is generally not appropriate for response variables, like psurf, that are constrained to lie between 0 and 1 (Hogg and Tanis 1997). Furthermore, from fig. 3 in Klopfstein et al. (2005) it appears that much of the apparently high R2 attributed to this regression is due to one or a very few influential data points. Thus, we believe that equations (2) and (3) are better justified as statistical descriptions of the dependence of
and the parameters λ, m and K, differ dramatically from the simple linear relationship between ‘mutant success’ (a proxy for psurf) and the ‘lumped’ parameter (λ − 1)/Km proposed in Klopfstein et al. (2005) (which, we note, is also consistent with the heuristics presented above). Indeed, equation (2) suggests that more appropriate lumped parameters for the one- and two-dimensional cases, respectively, might be (λ−1)/K1/4m1/2 and (λ − 1)1/2/K1/4. The answer appears to be that the analysis in Klopfstein et al. (2005) depends on a linear regression of psurf on (λ − 1)/Km. Linear regression is generally not appropriate for response variables, like psurf, that are constrained to lie between 0 and 1 (Hogg and Tanis 1997). Furthermore, from fig. 3 in Klopfstein et al. (2005) it appears that much of the apparently high R2 attributed to this regression is due to one or a very few influential data points. Thus, we believe that equations (2) and (3) are better justified as statistical descriptions of the dependence of 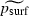 (and hence psurf) on λ, m and K.
(and hence psurf) on λ, m and K.
However, perhaps the most noteworthy difference between our statistical model (3) for the two-dimensional case and that proposed in Klopfstein et al. (2005) is that equation (3) does not include the migration rate m. In fact, a logistic regression of 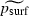 on ln (m) for a 25 × 100 grid did show a marginally statistically significant inverse dependence [
on ln (m) for a 25 × 100 grid did show a marginally statistically significant inverse dependence [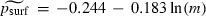 , with a standard error of 0.1077 for the coefficient of ln (m) (P = 0.090)]. This agrees with the inverse dependence noted in Klopfstein et al. (2005), which was not quantified. However, the important point is the weakness of the dependence. This is also a major difference between equation (3) and our proposed model (2) for the one-dimensional case. It is reasonable to ask what factors account for the decreased importance of m in the two-dimensional setting.
, with a standard error of 0.1077 for the coefficient of ln (m) (P = 0.090)]. This agrees with the inverse dependence noted in Klopfstein et al. (2005), which was not quantified. However, the important point is the weakness of the dependence. This is also a major difference between equation (3) and our proposed model (2) for the one-dimensional case. It is reasonable to ask what factors account for the decreased importance of m in the two-dimensional setting.
One such factor may be the irregular profile of the wavefront in two dimensions (see Fig. 5). This irregularity may dampen the effect of migration rate in two ways. First, individuals in cells surrounded by empty or nearly empty cells (i.e. where the wavefront has positive curvature) are expected to have more surviving offspring than individuals in cells surrounded by relatively full cells (i.e. where the wavefront has negative curvature). Thus, the probability of surfing will be greater for mutations that happen to arise in reasons of positive curvature. Second, the irregular profile will result in competition between outcroppings of the wavefront at different latitudes in the grid. Unlike in the one-dimensional case, an outcropping of mutants at one latitude may be rivalled by an outcropping of wild types at another latitude. The mutant and wild types will eventually meet, and only one type will then fill the remaining open terrain (this has been discussed in detail in Hallatschek et al. 2007). Thus, the irregular, two-dimensional wavefront introduces two additional random factors affecting the probability of surfing that do not have clear relationships to the migration rate m.
Figure 5.
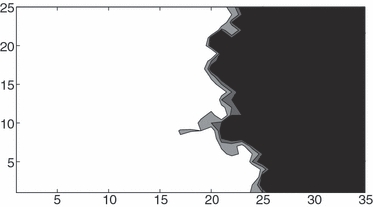
Contour plot of total population density in part of a 25 × 100 grid. Black region contains 0 individuals per cell, white region contains eight or more individuals per cell. Parameter values are λ = 1.8, m = 0.4, K = 10.
A point related to the relative unimportance of m in two dimensions concerns equation (6), which predicts the probability of surfing in the one-dimensional case. As noted in the Analytic models section, the model performed better at smaller values of the migration rate m. This may be because the model ignores all individuals present on the grid except for those in the leading cell. In reality, individuals behind the wavefront do have some chance of filling the remaining open terrain with their progeny. To do so, however, their progeny must advance to the wavefront, and the likelihood of this occurring should decrease with m. An analogy may be drawn to solutions of Fisher's equation,
| (14) |
a classical model of travelling waves of population density (Fisher 1937). The diffusion coefficient D in equation (14) is one-half the mean-squared distance traveled by an individual during one generation; so, for us D = m2/2. It is straightforward to check that near the leading edge, traveling wave solutions of equation (15) have shapes proportional to the exponential function 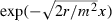 . Thus, the number of individuals originally present one cell behind the leading cell (say) whose progeny will travel forward a given distance in a given short span of time is proportional to
. Thus, the number of individuals originally present one cell behind the leading cell (say) whose progeny will travel forward a given distance in a given short span of time is proportional to 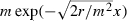 . This quantity increases with m. Therefore, referring back to our simplified model, when an individual originally in the leading cell fails to propagate, we expect fewer candidates from cells further back to be available at the wavefront as potential propagators. As a result, ignoring all cells other than the leading cell makes equation (6) a better approximation of psurf when m is small.
. This quantity increases with m. Therefore, referring back to our simplified model, when an individual originally in the leading cell fails to propagate, we expect fewer candidates from cells further back to be available at the wavefront as potential propagators. As a result, ignoring all cells other than the leading cell makes equation (6) a better approximation of psurf when m is small.
The simulations in the present work are necessarily limited in scope; even for the neutral case, they do not represent all possible scenarios of biological interest. In our simulations, density regulation precedes migration in each generation. In many natural populations this ordering is reversed. A change in the ordering of life cycle events may change the conclusions we draw from the simulations, and it will therefore be necessary to vary this ordering in future work. Likewise, our simulations assume nearest neighbor migration in a stepping-stone model with a selection regime that is spatially and temporally invariant. If such a scenario is used as a null model for hypothesis tests regarding a population's migration history, other spatial structures and migration patterns may need to be considered as alternatives.
The present work modeled only mutations arising at the leading edge of an invasion. Some authors have modeled mutations arising further back in an invasion as well (Klopfstein et al. 2005; Travis et al. 2007; Hallatschek and Nelson 2008). It was noted in both Klopfstein et al. (2005) and Travis et al. (2007) that the probability of surfing for the model considered here drops off sharply for neutral mutations arising even a few cells behind the front; in Burton and Travis (2008b), it was shown that this holds true for deleterious and, to a lesser extent, beneficial mutations as well (but different boundary conditions can alter this behavior). As mutants and wild types behave identically in our neutral model, this helps explain why our simplified analytic model achieves good predictive power, even though it assumes that the probability of propagation is zero for individuals behind the front.
In all the simulations discussed here, a single mutant was introduced at a fixed location once the wild-type invasion had advanced to the 10th grid column from the left. One could ask how varying mutation times and locations might affect the results. As extinction almost never occurred after the mutant was introduced, we suspect that increasing the longitude of the mutation would have little effect on psurf; however, this remains to be checked. On the other hand, changing the model to include multiple mutations, perhaps at random times, would shed light on a number of questions. Serial neutral mutations might lead to a ‘banding’ of the colonized territory by genotype (R. Holt, personal communication), allowing for more precise tracking of a population's migration history and possibly providing the basis for a null distribution in tests for selection at other loci. Simulations with serial selected mutations could be used to study adaptive changes in quantitative traits during invasion. Among other questions, they could be used to study the distribution of effects of quantitative trait loci (QTL) in a trait subject to selection during an invasion, extending existing theory which does not take spatial population dynamics into account (Mackay 2001; Orr 2005).
As distance from the front strongly influences the probability of surfing for neutral mutations, one might ask why we have chosen to study only mutations arising at the front. The explanation is that we have done so precisely in order to focus on what other factors may affect the probability of surfing. Analogously, one might study a group known to be at high risk of contracting a certain disease, in order to learn what additional factors might predispose an individual to contract the disease or to stay healthy. On the other hand, Hallatschek and Nelson have shown that as population density is (almost by definition) low at the wavefront, if mutation rates are constant over space then the modal point of origin of surfing mutations will be slightly behind the front (Hallatschek and Nelson 2008). Thus, studies of adaptive genetic change during invasions via the accumulation of weakly beneficial mutations need to vary their points of origin; a two-locus model which includes this variation has already been studied in Burton and Travis (2008a,b).
As have previous studies of mutation surfing (Edmonds et al. 2004; Klopfstein et al. 2005; Travis et al. 2007; Hallatschek and Nelson 2008), the present work considers only nearest-neighbor migration, in which an individual can move at most one cell length per generation. This results in diffusive movement, which can be modeled deterministically by a reaction–diffusion equation; such equations are the basis for the analysis of surfing in Vlad et al. (2002) (cited in Edmonds et al. 2004) and Hallatschek and Nelson (2008). However, it is well established that models assuming diffusive movement can yield inaccurate predictions when applied to many real invasions. This may be especially true when the movement rate m is relatively high for a given cell carrying capacity K. Not every combination of m and K may be well described by a limiting process on a continuous spatial domain (see, for example, Durrett and Neuhauser 1999). A future task will be to assess, in the first instance through simulation, how patterns that have been observed for discrete mutation surfing models converge or fail to converge to limiting relationships as the scaled island model approaches a continuum.
For accurate modeling, ‘heavy-tailed’ dispersal kernels that model a greater frequency of long-distance dispersal events are often necessary (see, for example, Kot et al. 1996 and references therein). An important area for future work will be to extend studies of mutation surfing to invasion models that incorporate heavy-tailed kernels (e.g. Laplace, or reflected exponential, kernels) and rare long-range dispersal events (this was noted in Burton and Travis 2008b as well). Yet, as stochastic effects inevitably will be even more important in such models, it may be extremely difficult to develop a rigorous theory that will yield useful quantitative predictions of psurf under these conditions. Simplified models that yield good agreement with simulation and, ultimately, experimental data will be especially necessary in these settings.
The bulk of the present work concerns neutral mutations, but we have obtained some results for beneficial and deleterious mutations (in one-dimensional habitats) as well. Equation (4) describes the dependence of psurf on m, K and nonidentical reproduction rates λmut and λwt. As noted above, the direction of the dependence on m and K was the same under neutrality and selection. In addition, the psurf was much more sensitive to the value of λmut than to λwt, so that absolute fitness was a better predictor of surfing success than relative fitness. Consistent with this finding, equation (13), which includes 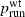 as a predictor of psurf, performs only slightly better than equation (12), which does not.
as a predictor of psurf, performs only slightly better than equation (12), which does not.
As it is difficult to identify consistent predictors of establishment success from empirical studies (Kolar and Lodge 2001; Hayes and Barry 2008), one might ask why analytic models that include prun (whether for mutants or wild types) as a predictor of psurf are informative. We offer two answers. First, our results indicate that finding the determinants of surfing and of invasiveness may not be two separate hard problems but only one. Second, they lead to a novel and (in principle) empirically testable prediction about adaptive change during invasions, as follows.
Our simulations indicate a strong positive association between the probability of establishment (which in this model leads to invasion) 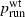 and the probability psurf of surfing, conditioned on establishment, for neutral and beneficial mutations in one dimension. (However, this association was not found for parameter regimes in which
and the probability psurf of surfing, conditioned on establishment, for neutral and beneficial mutations in one dimension. (However, this association was not found for parameter regimes in which 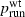 was very close to 1.) In these simulations, the only difference between mutants and wild types was fecundity, as measured by the growth rate λ. For most of the parameter combinations studied here, when λm and λwt are held fixed and cell carrying capacity K and migration rate m are varied, higher
was very close to 1.) In these simulations, the only difference between mutants and wild types was fecundity, as measured by the growth rate λ. For most of the parameter combinations studied here, when λm and λwt are held fixed and cell carrying capacity K and migration rate m are varied, higher 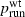 is associated with higher psurf. This suggests that in the scenarios modeled here, conditions that favor the establishment of a population in a novel habitat might in and of themselves also favor the fixation of beneficial mutations once establishment has occurred and an invasion has begun. It has been noted previously that high intrinsic growth rates and small deme sizes at the front may promote surfing, and that ‘genetic revolutions’ during invasions can occur because of surfing due to increased drift at the sparsely populated front (Klopfstein et al. 2005; Travis et al. 2007; Excoffier and Ray 2008; Hallatschek and Nelson 2008, 2009), but the finding that factors other than λ and population density can promote surfing in some invasions is novel, to our knowledge.
is associated with higher psurf. This suggests that in the scenarios modeled here, conditions that favor the establishment of a population in a novel habitat might in and of themselves also favor the fixation of beneficial mutations once establishment has occurred and an invasion has begun. It has been noted previously that high intrinsic growth rates and small deme sizes at the front may promote surfing, and that ‘genetic revolutions’ during invasions can occur because of surfing due to increased drift at the sparsely populated front (Klopfstein et al. 2005; Travis et al. 2007; Excoffier and Ray 2008; Hallatschek and Nelson 2008, 2009), but the finding that factors other than λ and population density can promote surfing in some invasions is novel, to our knowledge.
As K and m do not depend solely on organismal traits (whether genetic or nongenetic) but also on features of a given habitat, our results suggest that even abiotic factors that promote establishment might also promote the survival and spread of beneficial mutations (as well as neutral ones), conditioned on establishment. This finding is complementary to studies that have identified propagule size and number of introductions as the factors with the strongest currently known association with establishment success (Kolar and Lodge 2001; Hayes and Barry 2008). Further simulation and empirical studies seem warranted to assess the generality of this effect and its importance in explaining adaptive genetic change during invasions.
Acknowledgments
This research was partially supported by the National Science Foundation grant DMS-0818727. J.M. thanks two anonymous referees for comments and Bob Holt for editorial comments that strengthened the paper.
Literature cited
- Akaike H. A new look at the statistical model identification. IEEE Transactions on Automatic Control. 1974;19:716–723. [Google Scholar]
- Austerlitz F, Jung-Muller B, Godelle B, Gouyon P-H. Evolution of coalescence times, genetic diversity and structure during colonization. Theoretical Population Biology. 1997;51:148–164. [Google Scholar]
- Burton OJ, Travis JMJ. The frequency of fitness peak shifts is increased at expanding range margins due to mutation surfing. Genetics. 2008a;174:941–950. doi: 10.1534/genetics.108.087890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton OJ, Travis JMJ. Landscape structure and boundary effects determine the fate of mutations occurring during range expansions. Heredity. 2008b;101:329–340. doi: 10.1038/hdy.2008.56. [DOI] [PubMed] [Google Scholar]
- Durrett R, Neuhauser C. Particle systems and reaction–diffusion equations. Annals of Probability. 1999;22:289–333. [Google Scholar]
- Durrett R, Restrepo M. One-dimensional stepping stone models, sardine genetics and Brownian local time. Annals of Applied Probability. 2008;18:334–358. [Google Scholar]
- Edmonds CA, Lillie AS, Cavalli-Sforza LL. Mutations arising in the wave front of an expanding population. Proceedings of the National Academy of Sciences. 2004;101:975–979. doi: 10.1073/pnas.0308064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Ray N. Surfing during population expansions promotes genetic revolutions and structuration. Trends in Ecology and Evolution. 2008;23:347–351. doi: 10.1016/j.tree.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The wave of advance of advantageous genes. Annals of Eugenics. 1937;7:355–369. [Google Scholar]
- Hallatschek O, et al. Genetic drift at expanding frontiers promotes gene segregation. Proceedings of the National Academy of Sciences. 2007;104:19926–19930. doi: 10.1073/pnas.0710150104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallatschek O, Nelson DR. Gene surfing in expanding populations. Theoretical Population Biology. 2008;73:158–170. doi: 10.1016/j.tpb.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Hallatschek O, Nelson D. Life at the front of an expanding population. Evolution. 2009;64:193–206. doi: 10.1111/j.1558-5646.2009.00809.x. [DOI] [PubMed] [Google Scholar]
- Hartl DL, Clark AG. Principles of population genetics. 3rd edn. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- Hayes KR, Barry SC. Are there any consistent predictors of invasion success? Biological Invasions. 2008;10:483–506. [Google Scholar]
- Hofer T, Ray N, Wegmann D, Excoffier L. Large allele frequency differences between human continental groups are more likely to have occurred by drift during range expansions than by selection. Annals of Human Genetics. 2009;73:95–108. doi: 10.1111/j.1469-1809.2008.00489.x. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Sgrò CM. Ecological population genetics: the interface between genes and the environment. Annual Review of Genetics. 1995;29:349–370. doi: 10.1146/annurev.ge.29.120195.002025. [DOI] [PubMed] [Google Scholar]
- Hogg RV, Tanis EA. Probability and statistical inference. Upper Saddle River, NJ: Prentice-Hall; 1997. [Google Scholar]
- Holt RD. On the evolutionary ecology of species’ ranges. Evolutionary Ecology Research. 2003;5:159–178. [Google Scholar]
- Holt RD, Gomulkiewicz R. How does immigration influence local adaptation? A reexamination of a familiar paradigm. American Naturalist. 1997;149:563–572. [Google Scholar]
- Holt RD, Barfield M, Gomulkiewicz R. Theories of niche conservatism and evolution: could exotic species be potential tests? In: Sax DF, Stachowicz JJ, Gaines SD, editors. Species invasions: insights into ecology, evolution and biogeography. Sunderland, MA: Sinauer Associates; 2005. pp. 259–290. [Google Scholar]
- Hosmer DW, Lemeshow S. Applied logistic regression. New York: Wiley; 2000. [Google Scholar]
- Kawiecki TJ, Ebert D. Conceptual issues in local adaptation. Ecology Letters. 2004;7:1225–1241. [Google Scholar]
- Klopfstein S, Currat M, Excoffier L. The fate of mutations surfing on the wave of a range expansion. Molecular Biology and Evolution. 2005;23:482–490. doi: 10.1093/molbev/msj057. [DOI] [PubMed] [Google Scholar]
- Kolar CS, Lodge DM. Progress in invasion biology: predicting invaders. Trends in Ecology & Evolution. 2001;16:199–204. doi: 10.1016/s0169-5347(01)02101-2. [DOI] [PubMed] [Google Scholar]
- Korolev KS, Avlund M, Hallatschek O, Nelson DR. Genetic demixing and evolution in linear stepping stone models. Reviews of Modern Physics. 2010 doi: 10.1103/RevModPhys.82.1691. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kot M, Lewis MA, Van Den Driessche P. Dispersal data and the spread of invading organisms. Ecology. 1996;77:2027–2042. [Google Scholar]
- Liggett TM. Grundlehren der Mathematischen Wissenschaften [Fundamental Principles of Mathematical Sciences] Berlin: Springer-Verlag; 1999. Stochastic interacting systems: contact, voter and exclusion processes; p. 324. [Google Scholar]
- Lubina JA, Levin SA. The spread of a reinvading species: range expansion in the California Sea otter. American Naturalist. 1988;131:526–543. [Google Scholar]
- Mackay TFC. The genetic architecture of quantitative traits. Annual Review of Genetics. 2001;35:303–339. doi: 10.1146/annurev.genet.35.102401.090633. [DOI] [PubMed] [Google Scholar]
- Orr HA. The genetic theory of adaptation: a brief history. Nature Reviews Genetics. 2005;6:119–127. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- Pachepsky E, Lutscher F, Nisbet R, Lewis MA. Persistence, spread and the drift paradox. Theoretical Population Biology. 2005;67:61–73. doi: 10.1016/j.tpb.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Sakai AK, et al. The population biology of invasive species. Annual Review of Ecology and Systematics. 2001;32:305–332. [Google Scholar]
- Schaal BA, Leverich WJ. Plant population biology and systematics. Taxon. 2001;50:679–695. [Google Scholar]
- Semino O, et al. The genetic legacy of paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science. 2000;290:1155–1159. doi: 10.1126/science.290.5494.1155. [DOI] [PubMed] [Google Scholar]
- Simmons AD, Thomas CD. Changes in dispersal during species’ range expansions. American Naturalist. 2004;164:378–395. doi: 10.1086/423430. [DOI] [PubMed] [Google Scholar]
- Speirs DC, Gurney WSC. Population persistence in rivers and estuaries. Ecology. 2001;82:1219–1237. [Google Scholar]
- Sunnucks P. Efficient genetic markers for population biology. Trends in Ecology and Evolution. 2000;15:199–203. doi: 10.1016/s0169-5347(00)01825-5. [DOI] [PubMed] [Google Scholar]
- Travis JMJ, et al. Deleterious mutations can surf to high densities on the wave front of an expanding population. Molecular Biology and Evolution. 2007;24:2334–2343. doi: 10.1093/molbev/msm167. [DOI] [PubMed] [Google Scholar]
- Vlad MO, et al. Neutrality condition and response law for nonlinear reaction–diffusion equations, with application to population genetics. Physical Review E. 2002;65:061110. doi: 10.1103/PhysRevE.65.061110. [DOI] [PubMed] [Google Scholar]
- Wang JL, Whitlock MC. Estimating effective population size and migration rates from genetic samples over space and time. Genetics. 2003;163:429–446. doi: 10.1093/genetics/163.1.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JF, Wakeley J. The coalescent in a continuous, finite, linear population. Genetics. 2002;161:873–888. doi: 10.1093/genetics/161.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]


