Over the past 10 years, defects in mitochondrial oxidative phosphorylation (OXPHOS) have been implicated in a wide variety of neurodegenerative and neuromuscular diseases. Mutations in the mitochondrial tRNA and mRNA genes have been associated with various forms of epilepsy, spasticity (1, 2), stroke-like episodes (3, 4), and sensory neural deafness (5, 6), to mention a few. One neurological symptom that has definitely been associated with OXPHOS is the movement disorder dystonia. A specific missense mutation in the mtDNA complex I (NADH dehydrogenase) gene, MTND6, has been linked to maternally inherited dystonia along with the companion phenotype, Leber’s hereditary optic neuropathy (LHON) (7). In the current issue of these Proceedings, the association between the mitochondria and dystonia is extended by demonstrating that a mutation in a nuclear DNA (nDNA) gene encoding a protein involved in the import of mitochondrial inner membrane carrier proteins can also cause dystonia (8). These observations raise the intriguing possibility that, like dystonia, other movement disorders might have a mitochondrial etiology.
The mitochondrial OXPHOS components are assembled from genes distributed between the mtDNA and the nDNA. These genes include those encoding the structural proteins that form the mitochondrial inner membrane OXPHOS enzyme complexes (I, II, III, IV, and V), and the associated substrate and product carriers such as the adenine nucleotide translocator (ANT), as well as the proteins necessary for mitochondrial biogenesis, the apparatus to import cytoplasmically synthesized mitochondrial proteins, and the proteins necessary for mitochondrial assembly and turnover (9).
LHON was the first neurodegenerative disease to be associated with a mtDNA missense mutation, an A-to-G transition at nucleotide position (np) 11778 in the MTND4 gene of complex I. LHON is characterized by midlife, sudden-onset blindness due to atrophy of the optic nerve. The 11778 mutation converts the highly conserved arginine at codon 340 to a histidine (1). Subsequently, the LHON phenotype was observed to segregate in a large maternal pedigree along with early-onset generalized dystonia. The dystonic patients exhibited rigidity, ataxia, dysarthria, short stature, reduced intelligence, and a neuropathology characterized by bilateral basal ganglia degeneration, originally designated bilateral striatal necrosis (10). Molecular analysis of this pedigree revealed that both the LHON and the dystonia were the result of a mutation in the mtDNA complex I gene MTND6, resulting in the conversion of the highly conserved alanine codon at position 72 to a valine (7). This same mutation in different families can be associated with either LHON or dystonia or both (11). Cells containing this mutation have a 55% reduction in respiratory complex I activity, which can be transferred from one cell to another along with the mtDNA via the cytoplasmic (cybrid) transfer technique (12). Defects in complex I have also been associated with other cases of dystonia. A survey of the mitochondrial OXPHOS complexes in the blood platelet mitochondria revealed that patients with generalized or segmental dystonia had an average of a 62% reduction in complex I activity, whereas patients with focal dystonia had an average of a 37% reduction in complex I activity (13).
The current Proceedings paper (8) reports the function of the gene responsible for the chromosome X-linked Mohr–Tranebjaerg syndrome, which presents in early childhood with sensory neural hearing loss that can progress to dystonia, spasticity, mental deterioration, paranoia, and cortical blindness. Those patients who develop the movement disorder characteristically exhibit progressive degeneration of the basal ganglia, corticospinal tract, and brain stem. The gene responsible for Mohr–Tranebjaerg syndrome was identified through a patient with a deletion of the locus. The identity of the gene was confirmed in two additional families, one harboring a 10-bp deletion in exon 2 and the second resulting from a 1-bp deletion in exon 1. This gene, designated DFN-1, generates a 1,167-bp cDNA encoding a 97-amino acid, 11-kDa polypeptide, designated DDP1. The predicted polypeptide has a high similarity to a Schizosaccharomyces pombe gene of unknown function (14).
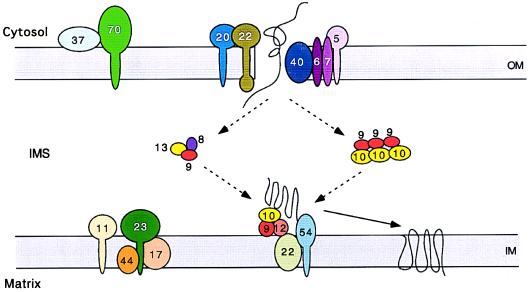
The function of the Mohr–Tranebjaerg syndrome DFN-1 gene was discovered through a totally different line of investigation. While studying the mitochondrial protein import pathway for multispanning carrier proteins such as ANT (AAC in yeast) into the mitochondrial inner membrane of Saccharomyces cerevisiae, Koehler, Schatz, and co-workers discovered two essential proteins of the mitochondrial intermembrane space, Tim10p and Tim12p. Tim10p is required for transport of carrier proteins through the mitochondrial outer membrane transport complex (TOM) and into the intermembrane space. Tim12p is involved in inserting the carrier proteins into the inner membrane (15). Tim10p is associated with another polypeptide, Tim9p, in a 300-kDa complex bound to the outer face of the inner membrane. This complex also includes Tim54p, Tim22p, and Tim12p. Tim9p and Tim10p are also associated in a soluble 70-kDa complex in the intermembrane space that can be cross-linked to the partially translocated carrier proteins (16). This 70-kDa complex is proposed to transfer the carrier from the outer membrane to the inner membrane. Tim9p appears to be associated with a second soluble intermembrane 70-kDa complex that assists in carrier protein manipulation. This complex includes two additional polypeptides, Tim8p and Tim13p (Fig. 1) (8). Tim8p, Tim9p, Tim10p, Tim12p, and Tim13p are all similar to each other, and they contain a distinctive duplicated C(N)3C motif.
Figure 1.
Model of the mitochondrial import apparatus, showing the proposed function for the Tim8p family of proteins: Tim8p, Tim9p, Tim10p, Tim12p, and Tim13p. OM, outer membrane; IMS, intermembrane space; IM, inner membrane. Adapted from Koehler et al., refs. 15 and 16 and personal communication.
Surprisingly, the DDP1 protein of the Mohr–Tranebjaerg syndrome was found to be similar to Tim8p, Tim9p, Tim10p, and Tim12p, with the highest similarity to Tim8p. Moreover, synthetic DDP1 is incorporated into the mitochondrial intermembrane space in yeast, and hapten-tagged DDP1 is specifically localized to yeast and mammalian mitochondria. Hence, it appears that the deafness and dystonia associated with Mohr–Tranebjaerg syndrome is due to a defect in the import of carrier proteins into the mitochondria and insertion into the mitochondrial inner membrane (8).
The demonstration that the DDP1 protein is probably involved in the import of the mitochondrial proteins implies that the underlying defect of the Mohr–Tranebjaerg syndrome is a defect in mitochondrial OXPHOS, specifically due to deficiencies in carrier proteins like AAC/ANT. This hypothesis that Mohr-Tranebjaerg syndrome is caused by an OXPHOS defect is particularly satisfying, since the phenotypes associated with the systemic OXPHOS defects resulting from mutations in the mtDNA give an array of clinical symptoms that nicely overlap with those of the Mohr–Tranebjaerg syndrome. Hence, deafness and dystonia associated with basal ganglia degeneration can now be linked to mitochondrial defects resulting from nDNA as well as mtDNA mutations.
Nuclear OXPHOS gene mutations have recently been identified in a variety of other neurodegenerative diseases. Leigh’s syndrome is a frequently lethal childhood disease associated with optic atrophy, ophthalmoplegia, nystagmus, ataxia, hypotonia, spasticity, and developmental delay or regression. Leigh’s patients frequently show characteristic bilateral basal ganglia degeneration with capillary proliferation. Leigh’s disease has been linked to mutations in the E1α subunit gene of the pyruvate dehydrogenase complex (17), the flavoprotein of respiratory complex II (18), and the NDUFS8 subunit of respiratory complex I (19). Leigh’s syndrome associated with respiratory complex IV (cytochrome c oxidase or COX) deficiency has recently been linked to mutations in the SURF1 gene on chromosome 9. By analogy with yeast, SURF1 may play a role in the assembly of complex IV (20, 21). Hereditary spastic paraplegia, which is generally associated with progressive weakness and spasticity, but can also encompass mental retardation, peripheral neuropathy, amyotrophy, ataxia, retinitis pigmentosa, optic atrophy, deafness, and ichthyosis, has been linked to a defect in a mitochondrial ATPase protease encoded by a gene located on chromosome 16q. This protein might be involved in the turnover of mitochondrial proteins (22). Hence, neurodegenerative diseases associated with mitochondrial dysfunction have now been identified in mtDNA-encoded polypeptide, tRNA, and rRNA genes, and in the nDNA-encoded mitochondrial OXPHOS polypeptide genes, respiratory complex assembly genes, mitochondrial protein turnover genes, and mitochondrial import apparatus genes.
The extensive array of neurological and neuromuscular symptoms associated with the mitochondrial encephalomyopathies, resulting from both nDNA and mtDNA mutations, raises the possibility that mitochondrial defects may account for other enigmatic degenerative diseases showing Mendelian inheritance. If this is the case, then all of the nDNA genes involved in mitochondrial bioenergetics and biogenesis become candidate genes for neurodegenerative diseases. One method to identify these nDNA mitochondrial genes is by homology with comparable yeast genes, as demonstrated by the current Proceedings article (8). Unfortunately, many genes involved in mitochondrial biology have diverged sufficiently between yeast and humans that the true mammalian homologues are difficult if not impossible to identify. However, an alternative method for identifying the nDNA genes involved in mitochondrial biogenesis has recently become available.
Patients with severe mitochondrial encephalomyopathy show a characteristic muscle pathology involving the degeneration of the muscle fibers and the massive proliferation of abnormal mitochondria. This mitochondrial myopathy is routinely detected by staining frozen muscle sections with Gomori modified trichrome. The weakened muscle fibers tear on sectioning, giving them a ragged appearance, and the aggregates of abnormal mitochondria stain red, giving the characteristic ragged red fibers (RRFs). Analysis of OXPHOS gene expression in muscle with RRFs has revealed that both mtDNA and nDNA transcripts are up-regulated, presumably in an effort to produce more mitochondria to compensate for the bioenergetic defect caused by the mtDNA mutation (23, 24). This same phenomenon has been recapitulated in a mouse model of mitochondrial disease, created by the genetic inactivation of the heart-muscle isoform of ANT (ANT1). The ANT carrier proteins exchange the mitochondrially synthesized ATP for the spent cytosolic ADP across the mitochondrial inner membrane, thus providing the door by which mitochondrial ATP is made available to the cell. Mice have two ANT isoforms, ANT1, which is expressed primarily in the heart and skeletal muscle, and ANT2, which is expressed in all tissues except the skeletal muscle. Hence, in the ANT1-deficient mouse the skeletal muscle is blocked from access to the mitochondrial ATP. Like humans with mitochondrial disease, the ANT1-deficient mice develop mitochondrial myopathy associated with RRFs and the massive proliferation of giant mitochondria. They also develop a hypertrophic cardiomyopathy and exhibit severe fatigability and exercise intolerance (25).
Because ANT1-deficient mice show massive proliferation of skeletal muscle mitochondria, it follows that genes necessary for mitochondrial biogenesis must be up-regulated. Consequently, a careful determination of all of the genes for which the mRNA levels are increased in the skeletal muscle of ANT1-deficient mice should provide a good sampling of the different kinds of genes that are required to assemble a functional mitochondrion. This expectation has been confirmed by isolating the cDNAs of mRNAs that are up-regulated in ANT1-deficient mice by using differential display (26). The first set of differential display experiments revealed 38 products whose mRNA levels appeared to be up-regulated in the mutant muscle. Seventeen of these mRNA/cDNAs were sufficiently abundant that their transcriptional induction could be confirmed by filter hybridization. Of these 17, 7 were mtDNA transcripts, including 4 of the mtDNA genes for complex I, 1 of the subunits of complex IV, the large rRNA, and a cluster of tRNAs. The remaining 9 up-regulated transcripts included a variety of known nDNA energy metabolism genes as well as several novel transcripts. The known energy metabolism proteins included mitochondrial malate dehydrogenase, glycogen phosphorylase, two subunits of complex I (subunits 18 kDa and B8), and two subunits of complex IV (COXVa and COXVb). In addition, three novel genes were also found to be up-regulated: Mcl-1, the muscle homologue of Bcl-2; WS-3; and SKD3 (D. G. Murdock, B. E. Boone, K. E. Esposito, and D.C.W., unpublished results).
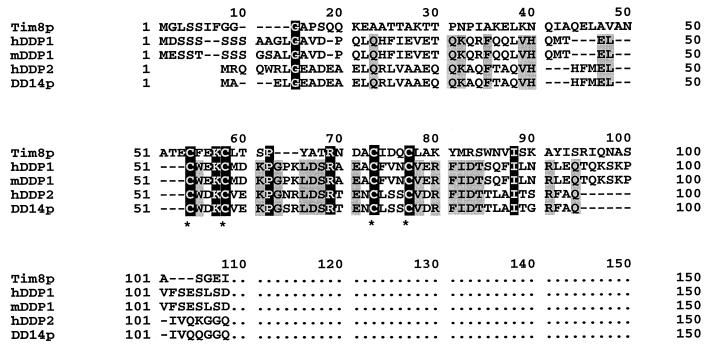
The SKD3 protein is of particular interest in the context of dystonia. This protein had been previously identified on the basis of its ability to suppress a yeast K+ transporter mutant, which could also be complemented by two other genes, both encoding members of the NSF family of proteins known to be involved in membrane fusion and organelle biogenesis (26). The SKD3 protein is a 76-kDa protein with the C-terminal domain having homology to the Clp/HSP104 family of proteins. This family includes Hsp78 of S. cerevisiae, which appears to cooperate with the mitochondrial matrix protein Hsp70 in mitochondrial maintenance. SKD3 also contains four ankyrin-like domains and a putative amino-terminal mitochondrial targeting domain (D. G. Murdock, B. E. Boone, L. E. Esposito, and D.C.W., unpublished results). Surprisingly, SKD3 shows significant similarity to the early-onset torsion dystonia gene (DYT1) product, torsin A. This is the most severe and common form of hereditary dystonia and is inherited in an autosomal dominant fashion with almost all cases resulting from a 3-bp deletion (GAG) that removes one glutamate codon (27). This association between dystonia and an nDNA-encoded mitochondrial gene up-regulated in ANT1-deficient mice was extended by comparison of the lower-abundance products identified by differential display with the Tim8p and DDP1 proteins of the current dystonia paper. The predicted protein product of one mRNA identified by differential display, DD14, showed high similarity to a human paralogue of DDP1, DDP2 (Fig. 2). Thus DD14 represents the mouse homologue of the gene encoding DDP2, a paralogous gene to Tim8p and the other members of its gene family: Tim9p, Tim10p, Tim12p, and Tim13p.
Figure 2.
Alignment of sequences related to the human DDP1 (hDDP1) protein responsible for the Mohr–Tranebjaerg syndrome. The hDDP2 protein is encoded by a closely linked homologous human gene (14), Tim8p is the yeast homologue reported in this Proceedings (8), mDDP2p was deduced from sequence identified in the mouse expressed sequence tag (EST) database, and mDD14 is the mouse paralogue isolated because it is up-regulated in the skeletal muscle of mice with mitochondrial myopathy, where the mitochondria proliferate to compensate for muscle mitochondrial energy deficiency (26). Amino acids conserved in all five proteins are in black boxes, and amino acids conserved in the four mammalian proteins are in gray boxes. Cysteines belonging to the twin C(N)3C motifs are marked with asterisks.
These observations raise an exciting possibility. Perhaps the nDNA-encoded mitochondrial genes will open a new window into the pathophysiological and molecular basis of neurodegenerative diseases, such as the movement disorders. If so, the further isolation and characterization of the nDNA genes involved in mitochondrial biogenesis promises to pay large dividends in unraveling some of the most perplexing problems in human genetics.
Footnotes
The companion to this Commentary begins on page 2141.
References
- 1.Wallace D C, Singh G, Lott M T, Hodge J A, Schurr T G, Lezza A M, Elsas L J, Nikoskelainen E K. Science. 1988;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 2.Shoffner J M, Lott M T, Lezza A M, Seibel P, Ballinger S W, Wallace D C. Cell. 1990;61:931–937. doi: 10.1016/0092-8674(90)90059-n. [DOI] [PubMed] [Google Scholar]
- 3.Goto Y, Nonaka I, Horai S. Nature (London) 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi Y, Momoi M Y, Tominaga K, Momoi T, Nihei K, Yanagisawa M, Kagawa Y, Ohta S. Biochem Biophys Res Commun. 1990;173:816–822. doi: 10.1016/s0006-291x(05)80860-5. [DOI] [PubMed] [Google Scholar]
- 5.Fischel-Ghodsian N, Prezant T R, Bu X, Oztas S. Am J Otolaryngol. 1993;14:399–403. doi: 10.1016/0196-0709(93)90113-l. [DOI] [PubMed] [Google Scholar]
- 6.Hutchin T, Haworth I, Higashi K, Fischel-Ghodsian N, Stoneking M, Saha N, Arnos C, Cortopassi G. Nucleic Acids Res. 1993;21:4174–4179. doi: 10.1093/nar/21.18.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jun A S, Brown M D, Wallace D C. Proc Natl Acad Sci USA. 1994;91:6206–6210. doi: 10.1073/pnas.91.13.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koehler C M, Leuenberger D, Merchant S, Renold A, Junne T, Schatz G. Proc Natl Acad Sci USA. 1999;96:2141–2146. doi: 10.1073/pnas.96.5.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace D C, Lott M T, Brown M D. In: Human Gene Mapping 1995, a Compendium. Cuticchia A J, Chipperfield M A, Foster P A, editors. Baltimore: Johns Hopkins Univ. Press; 1996. pp. 1280–1331. [Google Scholar]
- 10.Novotny E J, Singh G, Wallace D C, Dorfman L J, Louis A, Sogg R L, Steinman L. Neurology. 1986;36:1053–1060. doi: 10.1212/wnl.36.8.1053. [DOI] [PubMed] [Google Scholar]
- 11.Shoffner J M, Brown M D, Stugard C, Jun A S, Pollok S, Haas R H, Kaufman A, Koontz D, Kim Y, Graham J, et al. Ann Neurol. 1995;38:163–169. doi: 10.1002/ana.410380207. [DOI] [PubMed] [Google Scholar]
- 12.Jun A S, Trounce I A, Brown M D, Shoffner J M, Wallace D C. Mol Cell Biol. 1996;16:771–777. doi: 10.1128/mcb.16.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benecke R, Strumper P, Weiss H. Ann Neurol. 1992;32:683–686. doi: 10.1002/ana.410320512. [DOI] [PubMed] [Google Scholar]
- 14.Jin H, May M, Tranebjaerg L, Kendall E, Fontan G, Jackson J, Subramony S H, Arena F, Lubs H, Smith S, et al. Nat Genet. 1996;14:177–180. doi: 10.1038/ng1096-177. [DOI] [PubMed] [Google Scholar]
- 15.Koehler C M, Jarosch E, Tokatlidis K, Schmid K, Schweyen R J, Schatz G. Science. 1998;279:369–373. doi: 10.1126/science.279.5349.369. [DOI] [PubMed] [Google Scholar]
- 16.Koehler C M, Merchant S, Oppliger W, Schmid K, Jarosch E, Dolfini L, Junne T, Schatz G, Tokatlidis K. EMBO J. 1998;17:6477–6486. doi: 10.1093/emboj/17.22.6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthews P M, Marchington D R, Squier M, Land J, Brown R M, Brown G K. Ann Neurol. 1993;33:652–655. doi: 10.1002/ana.410330616. [DOI] [PubMed] [Google Scholar]
- 18.Bourgeron T, Rustin P, Chretien D, Birch-Machin M, Bourgeois M, Viegas-Pequignot E, Munnich A, Rotig A. Nat Genet. 1995;11:144–149. doi: 10.1038/ng1095-144. [DOI] [PubMed] [Google Scholar]
- 19.Loeffen J, Smeitink J, Triepels R, Smeets R, Schuelke M, Sengers R, Trijbels F, Hamel B, Mullaart R, van den Heuvel L. Am J Hum Genet. 1998;63:1598–1608. doi: 10.1086/302154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Z, Yao J, Johns T, Fu K, De Bie I, Macmillan C, Cuthbert A P, Newbold R F, Wang J, Chevrette M, et al. Nat Genet. 1998;20:337–343. doi: 10.1038/3804. [DOI] [PubMed] [Google Scholar]
- 21.Tiranti V, Hoertnagel K, Carrozzo R, Galimberti C, Munaro M, Granatiero M, Zelante L, Gasparini P, Marzella R, Rocchi M, et al. Am J Hum Genet. 1998;63:1609–1621. doi: 10.1086/302150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casari G, De Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, De Michele G, Filla A, Cocozza S, Marconi R, et al. Cell. 1998;93:973–983. doi: 10.1016/s0092-8674(00)81203-9. [DOI] [PubMed] [Google Scholar]
- 23.Heddi A, Lestienne P, Wallace D C, Stepien G. J Biol Chem. 1993;268:12156–12163. [PubMed] [Google Scholar]
- 24.Heddi A, Lestienne P, Wallace D C, Stepien G. Biochim Biophys Acta. 1994;1226:206–212. doi: 10.1016/0925-4439(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 25.Graham B, Waymire K, Cottrell B, Trounce I A, MacGregor G R, Wallace D C. Nat Genet. 1997;16:226–234. doi: 10.1038/ng0797-226. [DOI] [PubMed] [Google Scholar]
- 26.Perier F, Radeke C M, Raab-Graham K F, Vandenberg C A. Gene. 1995;152:157–163. doi: 10.1016/0378-1119(94)00697-q. [DOI] [PubMed] [Google Scholar]
- 27.Ozelius L J, Hewett J W, Page C E, Bressman S B, Kramer P L, Shalish C, de Leon D, Brin M F, Raymond D, Corey D P, et al. Nat Genet. 1997;17:40–48. doi: 10.1038/ng0997-40. [DOI] [PubMed] [Google Scholar]