Abstract
The understanding of migration patterns can significantly contribute to conservation and management. The spawning migrations of Atlantic salmon (Salmo salar) cover thousands of kilometers from the feeding areas at sea to their natal rivers to reproduce. Migrating salmon are exposed to intensive harvest, but little is known of the population-specific differences in migration behavior. In this study, timing of return migration was investigated among one-sea-winter Atlantic salmon within a river system. By utilizing knowledge of the genetic population structure, population of origin was reliably identified for c. 1500 fish caught in mixed stock fisheries after adopting an approach to minimize the complications arising from potential nonsampled populations. Results demonstrated significant and temporally stable differences among populations as well as between sexes. Generally, female salmon from tributary populations entered fresh water first. Run timing was not however related to in-river migration distance. Rather, one-sea-winter salmon from larger populations and with a higher proportion of multi-sea-winter females arrived later in the season. These findings are a significant step toward a more thorough understanding of the salmon migration behavior and behavioral ecology, providing concrete tools for the management and conservation of the remaining indigenous Atlantic salmon stocks.
Keywords: behavior, exploitation, fisheries management, genetic stock identification, life history, Salmo salar, spawning migration
Introduction
Migration is a common behavioral, ecological, and evolutionary phenomenon taking many forms in animal kingdom (Dingle and Drake 2007). Annual migrations, where individuals make long-distance migrations from breeding grounds to more favorable environments in pursuit of better food resources and growth possibilities or to avoid unfavorable environmental conditions or predation are well known (Northcote 1978; Dingle 1996). Return migration to natal site may in turn be connected to certain life stages, often to ensure successful reproduction. It has been recognized that variability in this migration behavior has a crucial role in ecology and evolution of populations as physical condition or date of arrival can explain important variation in reproductive success and annual survival of individuals (Webster et al. 2002). However, migratory phase of a life-cycle may also expose animals to diverse risks, including intensive human exploitation and migratory species may therefore be more vulnerable to extinction threats than resident species (Pimm et al. 1998). This is widely recognized and a number of behavioral biologists have acknowledged that insights to both individual and population level variability in behavior can significantly contribute to conservation biology and management (Nordqvist 1924; Dingle et al. 1997; Shumway 1999; Caro 2007; Angeloni et al. 2008).
The spawning migrations covering thousands of kilometers from feeding areas at sea to the natal river to reproduce is a key element in the life history of many Atlantic and Pacific salmon populations (Dittman and Quinn 1996). Because of this accurate natal homing, salmon inhabiting different rivers are largely reproductively isolated from each other and therefore have accumulated significant inter-population genetic variation. High levels of differentiation have been shown even at a sub-basin level between tributary populations of Atlantic salmon (Vähä et al. 2007). Divergent selection on heritable variation of traits enhancing survival and reproductive success of individuals under particular physical and biotic determinants has led also to significant variation in many morphological and life-history traits as well as in behavioral characteristics both within and among populations (reviewed in Garcia de Leaniz et al. 2007). For example, variation in age at smolting (Englund et al. 1999) and in sea-age at maturity (Niemelä 2004) among fish from neighboring tributaries within a river system has been documented. Such traits have a significant impact on the reproductive success of individuals as mature salmon may vary 30-fold in size (weight) depending on the duration of sea migration (1–5 years). Although phenotypic plasticity from environmental variation probably plays a significant role in shaping populations, there is much evidence also for the heritable, additive component in trait variation (reviewed in Garcia de Leaniz et al. 2007).
Large multi-sea-winter (MSW salmon), having spent 2–5 years in the sea, often arrive to fresh water earlier in the season than smaller one-sea-winter (1SW, ‘grilse’) salmon (Shearer 1990; Niemelä et al. 2006). Despite the essential role of spawning migration in the life-cycle of Atlantic salmon, very few studies have investigated population-specific differences in run timing apart from distinguishing variation between sea-age groups. An exceptional study by Stewart et al. (2002) addressed the within-season variation in run timing of salmon originating from two tributaries of River Tay in Scotland. Utilizing a mark–recapture and a transplantation approach, they found a significant and persistent difference between the two populations in timing of arrival to coastal waters. Findings of Stewart et al. (2002) with results of other similar investigations (Jonsson et al. 1990; Hansen and Jonsson 1991) suggest that run timing, a behavioral trait, may have a considerable genetic component. There is currently no satisfactory explanation for the variation in timing of arrival to breeding grounds in Atlantic salmon, but it has been proposed that early run timing is likely to carry a high cost in terms of lost feeding opportunities, reduced growth and ultimately reduced reproductive success (Fleming 1996). On the other hand, earlier arrival on the breeding grounds, through processes such as optimal territory acquisition, could increase subsequent breeding success (Currie et al. 2000; Gill et al. 2001).
It is widely acknowledged that to optimize the yield of fisheries in sustainable manner, each contributing population should be managed separately (Begg et al. 1999). In this respect, the migratory phase of a life cycle imposes a great challenge as fisheries harvesting returning salmon are generally exploiting mixed stock fisheries. From the conservation and management perspective, knowledge of the behavioral characteristics of populations is important as the duration of sea migration and seasonal structuring of run timing may have significant effects on the prosperity and evolutionary trajectory of the population depending on timing of human exploitation patterns (Thorley and Youngson 2007; Hard et al. 2008). This manifests especially within large river systems where different fisheries operating in the estuary and in the lower main stem may strongly exploit salmon originating from several genetically distinct populations. Thus, potential population and/or sex-specific variation in timing of fresh water entry together with potential size differences of migrating individuals might lead to differential exploitation rates among populations and introduce directional selection pressure on life-history traits.
The river Teno (Tana in Norwegian) in northernmost Europe is one of the few remaining large river systems that still support abundant Atlantic salmon stocks. As a large part of the salmon fishery in the river is mixed stock fisheries, insights to variability in migration behavior can significantly contribute to conservation and management of Teno salmon. In our previous studies we have found that Atlantic salmon within the Teno River is structured into a number of demographically independent, genetically distinct and stable population segments (Vähä et al. 2007, 2008). To investigate the potential population and sex-specific differences in migration behavior of 1SW Atlantic salmon homing to a large river system, we utilized genetic stock identification (GSI) of systematically collected, documented and archived scale samples from catches of net fisheries in the lowermost part of the main stem of the Teno river system. The specific aims of the present study were to address the following questions: (i) Do 1SW salmon originating from different parts of the large river system show temporal variation in timing of river entry? (ii) Is the pattern similar across years with varying river flows and fish abundance? (iii) Is the date of arrival related to the in-river migration distance?
Materials and methods
Study area
The mainstem of large subarctic River Teno (70°N 28°E, catchment area 16 386 km2) runs between northern Norway and Finland and drains into the Barents Sea (Fig. 1). The total length of the river Teno is 351 km, and with tributaries included there is more than 1200 km of waterway passable by adult salmon. Salmon production in the river system is entirely dependent on natural reproduction; release of reared fish and eggs is forbidden. There are 20–30 tributaries in the river system, where salmon reproduce, but the sizes of the tributaries and spawning populations vary considerably, from hundreds to several thousand spawners. Among males, 1SW salmon generally outnumber other sea-age groups (9:1), and also female spawners in many tributaries mainly consist of 1SW salmon (Niemelä 2004). However, MSW salmon form an important component of the female spawning stock in some tributaries as well as in the mainstem and all three large headwater rivers, Kárášjohka, Iešjohka and Inarijoki (Niemelä 2004). The average long-term proportion of repeat spawners among the spawning stocks has been 5%, but has increased in the last decade (Niemelä et al. 2006). Spawning takes place from September to early October probably depending on the local water temperature. Ice typically covers the river system from November until mid-May.
Figure 1.
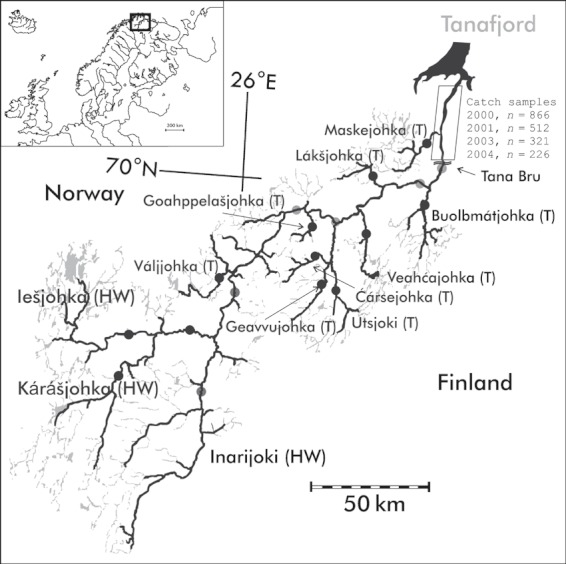
Map of the River Teno in northernmost Europe. Distribution of adult Atlantic salmon within the river system is shown with thick line. Locations of the sampled baseline populations are indicated with circles (dark gray – Teno ms upper, light gray – Teno ms lower). The first riffle area (Tana Bru) and the study site with numbers of fisheries catch samples for each year are also shown.
Teno salmon fisheries
The mean salmon catch in the River Teno is 133 t (range 70–250 t in 1972–2008), accounting for up to 20% of the annual freshwater salmon harvest in Europe (ICES 2002). The salmon stocks are exploited by both local fishermen and by c. 10 000 tourist anglers visiting the River Teno annually. In addition to recreational rod and line, some local people practise commercial and subsistence fishery with drift nets, weirs and gill nets in the river. The most intensive area of harvesting is the lowermost 100-km section of the main stem, where the mixed stock fishery probably includes salmon originating from the main stem as well as from the numerous tributaries.
The fishing season commences on May 20 and ceases August 20 for tourists, and August 31 for local people. Drift net fishing is limited to the period of May 20 to June 15, but the use of gill nets and weirs is allowed until end of August. Throughout the season, all fishing is closed from Sunday to Monday and fishing with nets is permitted only from Monday to Thursday.
Catch samples
For this study, we selected archived scale samples of 1SW salmon collected by local fishermen from drift net, weir and gill net fisheries at the lowermost slow flowing 37-km section of the river that the salmon pass without stopping within a mean of 26 h (13–70 h) (Økland et al. 2001; Karppinen et al. 2004). Therefore, we treated the time of capture as a reasonable approximation for actual time of fresh water entry. Fishermen had recorded the catch date, place and gear as well as length (cm) and sex when storing the scale samples in paper envelopes. Scale reading, detecting growth pattern differences as described by Fiske et al. (2005), was then performed to identify the sea-age class as well as age at smoltification (juvenile leaving the fresh water). For GSI, we selected 1925 salmon from the fishing seasons 2000 (n = 866), 2001 (n = 512), 2003 (n = 321) and 2004 (n = 226). These years represent seasons of high and low salmon abundance (estimated 1SW salmon catches: 2000: 40 000; 2001: 20 000; 2003: 13 000; 2004: 4000; Niemelä et al. 2006).
DNA extraction and PCR-amplification
DNA was extracted from specimen by digesting two scales with proteinase K, followed by purification of nucleic acids on silica fines in 96-well filter plates (0.45-μm GHP; Pall Life Sciences) using a protocol slightly modified from Elphinstone et al. (2003). In this study, genotypes of 32 microsatellite markers were utilized of which 30 were the same as described in Vähä et al. (2007), where the detailed information and their original references are available. In addition, we utilized the data from two additional loci to maximize the power to assign individuals to populations: an MHC I-linked microsatellite locus (Grimholt et al. 2002) and an MHC II-linked minisatellite locus (Stet et al. 2002).
All microsatellite loci were amplified by multiplex PCR. Simultaneous amplification of up to nine loci were carried out in 7.5 μL reaction volume, which consisted of 1 μL of extracted DNA elute, 0.9x QIAGEN Multiplex PCR Master Mix solution (QIAGEN) and varying concentrations (0.03–0.35 μm) of primers (details available at http://users.utu.fi/jpvaha). Thermal cycling profiles for the multiplex protocols were as follows: 15 min at 95°C, followed by 36 cycles of 30 s at 94°C, 90 s at 58°C and 60 s at 72°C, followed by final extension step of 10 min at 72°C. Even volumes of the PCR amplified products were pooled and 0.09 μL of GS600LIZ size standard (Applied Biosystems) was added as an internal size standard to each sample. Electrophoresis was then performed on ABI 3130xl (Applied Biosystems). Example electropherograms of all loci are available from http://users.utu.fi/jpvaha. Allele scoring was performed with genemapper V3.7 (Applied Biosystems) followed by manual corrections. The amplification procedure was only slightly modified from Vähä et al. (2008), where the genotyping error rate for the original procedure was estimated to be low (<0.4%).
Baseline material
Our previous studies exploring population structure of Teno salmon with genetic clustering methods indicated that each tributary (T) sample constitutes a separate genetic cluster, while a collection of samples from the mainstem and headwater rivers (MS/HW) constituted four genetic clusters: Teno main stem lower (TmsL), Teno main stem Upper (TmsU), Iešjohka and Kárášjohka (Vähä et al. 2007). For baseline data, we selected individuals assigned to these clusters/populations (Table 1 in Vähä et al. 2008) with the following modifications. We excluded the upper Utsjoki 1982–1984 (n = 75), River Maskejohka 1985 (n = 29) and 1SW salmon collected from Tana Bru in 2003 (n = 15, treated here as mixed fishery samples). In turn, the baseline data were supplemented with adult salmon samples from rivers Iešjohka (n = 12), Lákšjohka (n = 19) and Maskejohka (n = 12) collected in 1998. In total, the baseline sample consisted of 1780 salmon from 10 tributary populations, two mainstem (TmsL and TmsU) and two headwater populations (Iešjohka and Kárášjohka) (Fig. 1). The two Lake Buolbmátjohka populations were treated in the GSI results as a single reporting unit.
Table 1.
Proportions (%) of salmon assigned to the 14 baseline populations (single reporting unit for two Lake Buolbmátjohka populations) and four ‘dummy clusters’ representing potential nonsampled populations (see Materials and methods for details). Genetic stock identification solutions after filtering individuals originating from populations not included in the baseline data are presented in the lower part of the table (solution used in subsequent analyses is shown in bold)
| Mainstem and headwaters | Tributaries | Dummy clusters | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K | Lower | Upper | Iešjohka | Kárášjohka | Maskej. | Buolbmátj. | Lákšj. | Veahća | Ćársej. | Geavvuj. | Utsjoki | Goahppelašj. | Váljj. | 1 | 2 | 3 | 4 | Total assigned | n assigned | |
| oncor | 14 | 21.3 | 22.4 | 13.6 | 12.2 | 8.9 | 2.0 | 2.7 | 5.8 | 2.6 | 2.7 | 0.9 | 1.6 | 2.3 | – | – | – | – | 98.9 | 1900 |
| Structure | 14 | 16.2 | 16.9 | 10.8 | 11.8 | 9.9 | 2.0 | 2.8 | 6.0 | 2.7 | 2.4 | 0.9 | 2.3 | 3.0 | – | – | – | – | 87.7 | 1685 |
| Structure | 15 | 15.4 | 16.7 | 10.5 | 11.7 | 8.1 | 2.0 | 2.8 | 5.9 | 2.7 | 2.4 | 0.9 | 2.2 | 2.9 | 3.1 | – | – | – | 87.2 | 1676 |
| Structure | 16 | 10.6 | 16.0 | 10.4 | 11.2 | 8.1 | 2.0 | 2.8 | 5.9 | 2.7 | 2.4 | 0.9 | 2.0 | 2.9 | 2.8 | 4.1 | – | – | 84.7 | 1628 |
| Structure | 17 | 13.1 | 9.4 | 9.9 | 9.8 | 8.1 | 2.0 | 2.8 | 5.6 | 2.7 | 2.4 | 0.8 | 2.0 | 2.9 | 2.2 | – | 2.8 | 3.6 | 80.2 | 1541 |
| Structure | 18 | 8.5 | 10.0 | 9.8 | 10.2 | 7.9 | 2.0 | 2.8 | 5.6 | 2.8 | 2.4 | 0.8 | 2.1 | 2.8 | 2.3 | 3.6 | 3.0 | 2.7 | 79.4 | 1525 |
| oncor | 14 | 16.2 | 18.4 | 13.0 | 11.6 | 8.1 | 2.0 | 2.7 | 5.7 | 2.6 | 2.6 | 0.9 | 1.6 | 2.3 | Filtered | 87.6 | 1683 | |||
| Structure | 14 | 12.2 | 14.2 | 10.7 | 11.6 | 8.6 | 2.0 | 2.8 | 5.9 | 2.7 | 2.4 | 0.9 | 2.3 | 3.0 | Filtered | 79.2 | 1521 | |||
A total of 474 alleles were identified in 32 microsatellite loci with number of alleles per locus varying from 4 to 32. The overall level of genetic differentiation was significantly higher (P = 0.002) among tributary populations (FST = 0.11) than among MS/HW populations (FST = 0.02). Allelic richness (12 genes) in turn was significantly higher (P = 0.003) among MS/HW populations (AR = 4.9) than among tributary populations (AR = 4.1). Genetic characteristics were calculated with the program Fstat 2.9.3. (Goudet 2001).
Data analyses
Individual assignment methods
Generally, mixed stock analysis–methods such as implemented in the programs oncor (S. Kalinowski, available at: http://www.montana.edu/kalinowski) and CBayes (Neaves et al. 2005) have been applied to mixed stock fisheries samples for both assessing the stock composition of the sample and assigning individuals to the population of origin (e.g. Beacham et al. 2005).
oncor implements the method of Rannala and Mountain (1997) to estimate baseline allele frequency distributions. The method is tailored to assign individuals in a mixture sample to the baseline population having the highest probability of producing the given genotype in the mixture. Individual assignment of unknown individual is then performed by estimating the probability of each baseline population producing the given genotype taking in to account the estimated stock composition of the mixture sample.
The program CBayes uses Bayesian mixture model of Pella and Masuda (2001) for calculating the stock proportions starting with an uninformative uniform prior using baseline and mixture sample information to update allele frequency distributions of the baseline stocks. Each mixture individual is then assigned to the source stock whose average posterior source probability is a maximum. This method has been shown to outperform classical individual assignments such for both estimating stock composition and identifying individuals’ sources (Koljonen et al. 2005).
However, neither of the two above methods is capable of appropriately dealing with incomplete baseline sample. As we could not exclude the possibility of our fishery samples including individuals from nonsampled populations, we performed analyses also with the program Structure (Pritchard et al. 2000). Population information was used for the baseline data and ancestries of unknown fishery individuals were updated according to the admixture model with correlated allele frequencies (Falush et al. 2003).
Assessing the performance of the methods
As our inferences from the data are built upon the foundation set by the accomplishment in assigning individuals to their populations of origin, we performed, through simulations, an assessment of general accuracy that could potentially be achieved with the current set of markers and baseline. All three assignment methods provide a posterior probability for an individual originating from each of the baseline populations. For comparison of the methods, we defined ‘efficiency’ as the proportion of assigned fish of the total number of samples and ‘accuracy’ as the proportion of correctly assigned fish out of all assigned fish.
In the first round of power analysis, we compared the methods oncor and CBayes to assess their relative performance and to determine the method of choice for subsequent analyses. For this, 10 mixture files containing 150 individuals from each baseline population were simulated using the simulation methods implemented in the program oncor. For comparison, we also performed estimations using the leave-one-out cross-validation method developed by Anderson et al. (2008), which has been showed to yield essentially unbiased estimates of GSI accuracy. Simulations were performed with gsi_sim software (written by Eric Anderson) to assess the level of accuracy assigning 1500 individuals from each population.
In the second round of power analysis, the effect of a missing baseline population on the assignments was explored by removing each tributary population in turn from the baseline data and scoring the assignments for the above-simulated samples. This was performed only with the program oncor, as it performed slightly better than CBayes (Fig. S1).
Third round of power analysis was performed only using the program Structure. In this assessment, each tributary population baseline sample was in turn treated as of unknown origin, i.e. as fishery sample, and analyzed collectively with our actual fishery catch samples. In addition to monitoring where tributary individuals were most likely assigned, this approach also allowed us to assess if Structure was able to find a clustering solution corresponding to the missing population. The individual assignments of fishery samples were explored with different clustering solutions: setting the K equal to number of baseline populations up to having four ‘dummy’ populations (K + 4).
Assessment of the simulated mixtures and the efficiency of methods
Assessment of the general assignment success that could be achieved with the methods oncor and CBayes in a full baseline data scenario indicated very high levels of efficiency and accuracy (Fig. S1). Assignment success appeared particularly high for the tributary populations, which all were assigned with more than 98% efficiency and accuracy with oncor independent of the used probability cut-off value. Although the assignment success for the MS/HW samples were generally lower, more than 97% of the MS/HW samples were assigned with nearly 98% accuracy using oncor with probability cut-off value 0.7. Accuracy estimates obtained using the leave-one-out cross-validation method of Anderson et al. (2008) were not markedly lower adding credibility to the obtained estimates for the general level of accuracy. However, analyses performed with the incomplete baseline data, where each tributary population was in turn removed from the baseline data revealed a potential source of error.
As expected, a population missing from the baseline data introduced a significant bias to the results: 81% of the samples originating from a population not included in the baseline were incorrectly assigned to a population in the baseline data. Of the false assignments 76% were assigned to the four large mainstem/headwater (MS/HW) populations, while samples of only one tributary population (Ćársejohka) were systematically assigned to another tributary (Geavvujohka). Apart from Ćársejohka samples, only 5% of the tributary fish without a baseline data were assigned to another tributary of which Veahćajohka and Maskejohka appeared to absorb the most: 2% and 1.5% of all false assignments, respectively. These results thus implied that at least the proportions of MS/HW populations may be greatly inflated in mixed stock analyses if fish from unknown populations were present in the fisheries samples.
As we expected not to have a complete baseline population data from the system, we tested if Structure could be used to overcome the problem. Exercises with Structure program indicated that the proportion of false assignments due to missing baseline data could be minimized by allowing a small number of ‘dummy clusters’ in the analysis. Structure converged finding a cluster corresponding to a missing baseline population with runs K + 1 in all but one case: when Ćársejohka samples were treated as unknown fishery samples they were assigned to a neighboring Geavvujohka population. However, a partitioning solution with K + 3 assigned nearly all (99%) Ćársejohka samples in to a separate, correct cluster (cut-off value 0.5). As these exercises indicated that using ‘dummy’ clusters (e.g. K + 1, K + 2…) may lower the number of false assignments in a missing baseline data scenario, we adopted this approach in our GSI of mixed stock fishery sample from Teno River.
Statistical methods
Catch accumulation for each population was used as an approximation of the timing of the spawning run. For comparing run timing among groups we used Wilcoxon rank scores [i.e. Mann–Whitney–Wilcoxon test for two-sample data and Kruskal–Wallis (K–W) test in one-way ANOVA statistic]. Analyses were performed using the npar1way procedure of sas statistical package (9.2) with Enterprise Guide 4.1 (SAS Institute Inc).
As we had data on individual salmon with sex, date of arrival and population of origin, we also applied linear models to test statistically and explore the behavioral differences among groups of salmon. For ease of interpreting and understanding the model and to increase the number of observations in each category, samples were pooled by the river type in two groups (MS/HW and tributary), excluding the individuals assigned to the lower most tributary, Maskejohka. As traditional linear models are inapplicable when the response variable is discrete or has a skewed distribution, we applied generalized linear models, which have been designed to cover other distributions besides the normal distribution (McCullagh and Nelder 1989). Through generalization the relevant distribution for the data is identified together with a ‘link function’ connecting the observations to the values predicted by the linear model. Analyses were performed using the genmod procedure of sas statistical package (9.2 with Enterprise Guide 4.1; SAS Institute Inc.) with a logit link function and a binomial error term. The value predicted by our model is the log of odds (=log [π/(1-π)]), where π is the binomial probability value.
General linear modeling was also used to assess if median date of arrival of a population was related to in-river migration distance, to the population size or sea-age class structure of the spawning population. In-river migration distances were measured as a waterway distance (km) from the sea to the tributary outlet or the estimated central rearing area in main stem stretches. Surrogate estimates of population sizes were obtained from the population proportions (male data only) in the final GSI solution. The variation in sea-age at maturity at each rearing site was measured as the proportion of MSW (≥2 sea years, including repeat spawners) female salmon in the spawning population. Records for sea-age at maturity in various parts of the River Teno were collected as part of the long-term monitoring project of Teno fisheries (Niemelä 2004, updated 2008). As the estimate for date of arrival of Utsjoki population was based only on 17 individuals, related analyses were also performed without this population. Analyses were performed using the glm procedure in sas statistical package (9.2) with Enterprise Guide 4.1 (SAS Institute Inc).
Results
Assignment of mixed stock fishery samples
Genetic stock identification of 1SW salmon (n = 1925) caught in the net fishery at the lower most stretch of the Teno River was performed with baseline population data available from 14 populations (K = 14). oncor and Structure provided very similar results in assigning individuals to tributary populations, but the number of individuals assigned to the MS/HW populations was substantially higher with oncor (Table 1). This difference was reflected also in the overall assignment efficiency: oncor assigned 13% more individuals to populations than Structure.
Structure analysis with 0–4 dummy clusters (K, K + 1…K + 4, see Materials and methods) revealed that the assignments to all but one tributary population remained practically unaffected despite increasing the number of K (Table 1). While the assignments to headwater populations (Iešjohka and Kárášjohka) were relatively invariable, the assignments to the two mainstem populations were more sensitive to the number of K in the analysis. As the number of K was set to include four dummy clusters, the proportion of assigned salmon to Teno mainstem lower and upper populations decreased by 48% and 40%, respectively. This indicated that, as expected, individuals from nonsampled populations were probably present in our fishery samples and if not accounted for the proportions of the two mainstem populations in the fishery samples might be significantly over-estimated.
To minimize the effect of individuals from nonsampled populations in the final GSI, individuals assigned to the four dummy clusters (n = 224) were filtered out, after which individual assignments were defined from K = 14 partitioning solution. This approach provided an overall individual assignment efficiency of 79% with Structure and 87.6% with oncor. For subsequent analyses of run timing, the GSI solution of Structure was chosen, in which population of origin was defined successfully for 1521 individuals with (Table 1).
Variation in run timing
Variation in run timing among 1SW salmon was first evaluated with samples pooled across the years for each population. Highly significant variation in spawning migration behavior was observed among populations (K–W χ212 = 392, P < 0.0001; Fig. 2). Generally, 1SW salmon originating from the tributaries arrived significantly earlier than those originating from the MS/HW (K–W χ21 = 108.1, P < 0.0001). A closer inspection of the median dates of arrival across years indicated that salmon of the tributaries Lake Buolbmátjohka, Goahppelašjohka, Lákšjohka, Váljjohka and Ćársejohka arrived earliest (June 20–22), followed by Geavvujohka and Veahćajohka fish (June 26). There were no significant differences in run timing of 1SW salmon from Teno mainstem upper and headwater populations (Iešjohka and Kárášjohka), which appeared to enter fresh water 2 weeks later (July 3) than those from the tributaries. 1SW salmon from the two populations closest to the estuary (tributary Maskejohka and Teno mainstem lower) appeared to arrive latest in early July (July 5 and 8, respectively).
Figure 2.
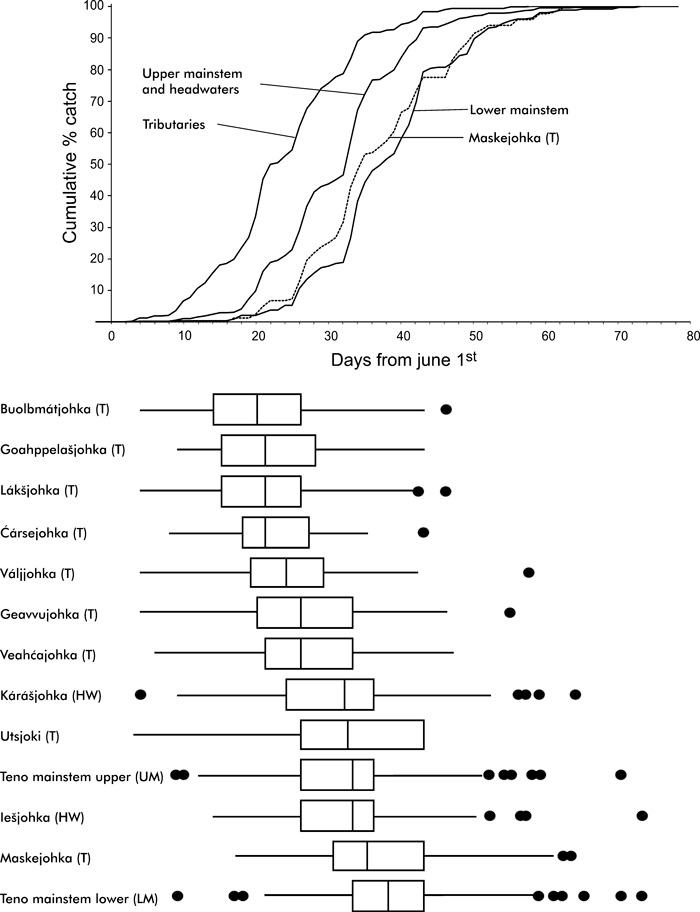
Variation in run timing of one-sea-winter salmon returning to the Teno River. (A) Accumulated catch percentage of tributary, upper mainstem and headwaters, the lowermost tributary Maskejohka and the lower mainstem salmon. (B) Variation in run timing of each population with a box plot (median and 25th and 75th percentiles) with whiskers to the most extreme point within 1.5 inter-quartile ranges and extreme values.
Although there was some variation across years in the sequential order of populations entering fresh water (populations ordered by the median date of arrival of 1SW salmon, data not shown), the above pattern was tenable. Significant annual variation in timing of river entry was observed only in Teno mainstem lower (K–W χ23 = 18.85, P = 0.0003), Maskejohka (K–W χ23 = 28.84, P < 0.0001), Veahćajohka (K–W χ2 = 19.65, d.f. = 3, P = 0.0002) and Goahppelašjohka (K–W χ23 = 9.51, P = 0.0232) when tested separately for each population.
There were also significant differences between sexes in arrival dates of 1SW salmon females originating from tributary populations ascended fresh water significantly earlier than their male counterparts (females = 21, males = 28 days after June 1; Z = −6.06, P < 0.0001). However, there were no significant differences between females and males among 1SW salmon from the MS/HW populations (females = 33, males = 34 days after June 1; Z = −0.1203, P = 0.90).
In the generalized linear approach, we continued the analysis of run timing by simultaneously taking into account date of arrival, year and sex of the fish in a model predicting the origin (Tributary or MS/HW) of 1SW salmon. All the variables (‘date of arrival’, ‘year’ and ‘sex’) and a ‘date of arrival’–‘year’ interaction were significant in the best model (Table 2). The model indicated that until June 20, a 1SW salmon entering Teno is more likely to originate from a tributary than from mainstem/headwater population and the probability for females was always higher than for males (Fig. 3). Extrapolating to the opening of the fishing season (May 20) revealed that during the first 2 weeks of the season a 1SW salmon entering Teno has more than 90% probability to originate from a tributary, although the interaction term in the model indicated that the pattern might not be as extreme each year (year 2003 in Fig. 3).
Table 2.
Generalized linear models for predicting the origin of one-sea-winter salmon: tributaries (excluding Maskejohka) or from the mainstem and headwaters. Chi-squared value with the respective significance level (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001) is given for each explanatory variable: date of arrival, sex, year and their interactions
| d.f. | Model 1 | Model 2 | |
|---|---|---|---|
| Date | 1 | 108.34*** | 126.90*** |
| Sex | 1 | 1.95 | 18.10*** |
| Year | 3 | 9.09* | 7.25 |
| Date × Year | 3 | 10.53* | 9.29* |
| Date × Sex | 1 | 0.16 | |
| Sex × Year | 3 | 7.47 | |
| Date × Sex × Year | 3 | 4.94 | |
| Log likelihood | −687.9097 | −692.8824 |
Figure 3.
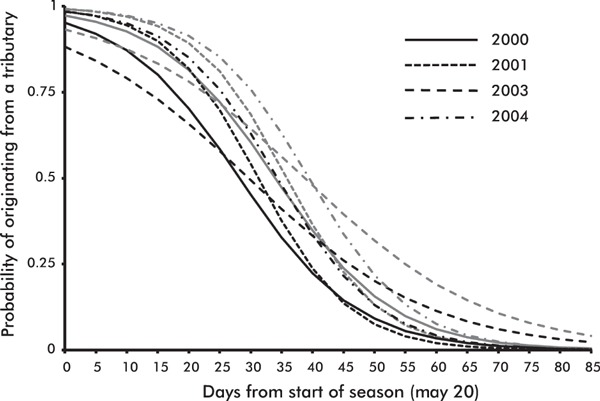
Probability of an individual originating from a tributary as a function of time (days from start of season May 20) according to the model 2 presented in Table 2. Predicted probability curves are shown for females (gray) and males (black) each year.
Finally, the date of arrival of 1SW salmon was not related to the distance of the natal rearing habitat from the sea (R2 = 0.02, F1,11 = 0.2, P = 0.64). Instead, a significant proportion of variance in date of arrival was explained by the population size (R2 = 0.61, F1,11 = 17.6, P = 0.0017; w/o Utsjoki: R2 = 0.86, F1,10 = 63.2, P < 0.0001) as well as by the proportion of MSW females in the spawning stock of the natal population (R2 = 0.64, F1,11 = 19.1, P = 0.0011; w/o Utsjoki: R2 = 0.61, F1,10 = 15.9, P = 0.0026) (Fig. 4). Although population size explained a significant proportion of the variation of MSW females in a population (R2 = 0.39, F1,11 = 7.0, P = 0.022; w/o Utsjoki: R2 = 0.68, F1,10 = 21.4, P = 0.001), both variables were significant (population size, F2,10 = 5.60, P = 0.040; proportion of MSW females F2,10 = 6.76, P = 0.027: interaction F3,9 = 2.6, P = 0.15) when concurrently included in the model explaining variation in date of arrival of 1SW salmon (R2 = 0.77, F2,10 = 16.4, P = 0.0007).
Figure 4.
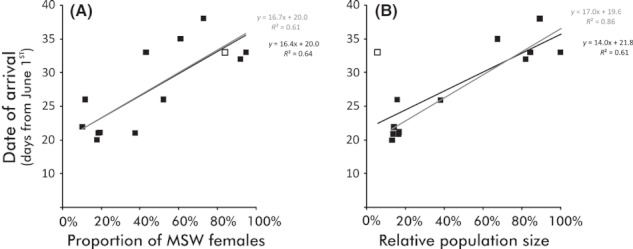
Relationships between the date of arrival and (A) the proportion of multi-sea-winter females in a population and (B) the relative population size (scaled to the largest). Open squares denote estimates for the Utsjoki population with the corresponding regression lines and equations in gray.
Discussion
We examined run timing of 1SW Atlantic salmon originating from multiple, genetically distinct populations within a single large river system. To the best of our knowledge, this study represents the first detailed study of return migration behavior of Atlantic salmon applying GSI approach as a ‘tagging’ method. The results demonstrated significant and temporally stable differences in timing of arrival with 1SW salmon originating from tributary populations entering fresh water earlier than their counterparts from headwater and mainstem populations. There were also differences between sexes whereby females arrived earlier than males within tributary populations, whereas no such differences were found for 1SW salmon originating from headwater and mainstem populations. The observed differences in run timing among populations were related to population size and sea-age group structure of the spawning stock, but not to in-river migration distances.
In addition to the usability of archived tissue samples, a major advantage of the GSI approach is that large sample sizes can easily be obtained as unlike conventional tagging studies it is not dependent on the recapture rate of marked individuals. However, the GSI approach is not without impediments. While the genetic structure of the system and the used methodology are important in determining the individual identification success, an important, but difficult to assess and therefore often neglected potential source of error in mixed stock assignment studies is the adequacy of the baseline data. In the following, we will first discuss the issues related to genetic tagging method and the accuracy of GSI of Teno salmon. Thereafter, we proceed to discuss our findings in more detail and provide implications for management.
Methodological aspects: genetic stock identification
Power analyses of expected accuracy of GSI employing 32 microsatellite loci (mean 14.8 alleles per locus) within the Teno population complex (global FST = 0.07) indicated very high individual assignment success: near 100% for tributary salmon and over 90% success for the MS/HW salmon. The higher success in assigning tributary fish was expected as they are genetically more diverged (FST = 0.1) and less diverse (GD = 60–80%) compared to MS/HW fish (FST = 0.02). In general, the levels of assignment success were in concordance with expectations from a number of simulation studies indicating that a near 100% assignment success can be achieved by using ≥10 moderately variable microsatellite loci in a set of 10 populations with FST ≥ 0.1 (Cornuet et al. 1999; Manel et al. 2002) and in some cases very high assignment success (∼90%) can be achieved even with lower level of differentiation FST ≥ 0.02 and fewer loci (Hauser et al. 2006). However, the high level of GSI accuracy with e.g. oncor and CBayes methods is expected only in situations with the baseline data available from each potential source population (Anderson et al. 2008).
Indeed, our simple power analyses with a population sample missing from the baseline data suggested that performing GSI using e.g. oncor, nearly all individuals originating from populations not included in the baseline were incorrectly assigned to one of the four most diverse and less diverged MS/HW populations. This indicated that the relative contribution of these populations to the fisheries catch sample would be inflated by the relative contribution of populations not in the baseline. In our study, this was a conceivable source of error if not considered as there are at least three tributaries draining to the mainstem of Teno with considerable 1SW salmon stock that were not sampled for the baseline.
Although, the magnitude of error due to having individuals from nonsampled populations in the mixture sample cannot be systematically accounted for in any simulations of accuracy (Anderson et al. 2008), an approach where Structure analysis was performed using additional ‘dummy’ clusters, i.e. K + 4 clusters [where K was the number of populations (14) in the baseline data], was found a potential strategy to minimize the error resulting from incomplete baseline data. This was demonstrated in our test runs with Structure, where individuals of each tributary population were in turn treated as of unknown origin, and analysis performed with up to three additional clusters (K + 3): nearly all individuals from the ‘unknown’ populations were correctly identified and assigned to these dummy clusters indicating that individuals of unknown origin could potentially be filtered out by using ‘dummy’ clusters.
Performing Structure analyses with four ‘dummy’ clusters (K = 18) on our mixed stock fisheries catch samples, 224 individuals were excluded from the data as originating from nonsampled populations. The majority of these individuals were assigned to the MS/HW populations with oncor (92%) and with Structure (84%; K = 14) according to expectations from the simulations. In our final GSI solution, a further 176 individuals had ambiguous genotypes and were not assigned to any of the clusters. These individuals may have been inter-population hybrids, inbred or from other nonreferenced populations (Baudouin et al. 2004). At this point, we do not have the means to assess the true accuracy of the individual assignments, but based on our simulations and test runs we expect the deduced GSI solution to be appropriate. A more comprehensive baseline data from the more remote areas in the headwaters should enable us to assess the accuracy of our GSI solution and the ‘dummy’ cluster approach, in addition to performing a more exhaustive genetic structure assessment of these areas.
Nevertheless, the adopted ‘dummy’ cluster approach represents a potential alternative to exclusion methods, where the genotype of each individual is compared to a set of simulated individuals and excluded if its probability of appearing falls below a set threshold, e.g. as implemented in GeneClass (Cornuet et al. 1999). The adopted approach, however, is likely more efficient than the exclusion method, as CBayes and Structure have been shown to outperform GeneClass in assigning individuals to populations (Manel et al. 2002; Koljonen et al. 2005; but see Hauser et al. 2006). However, the performance of Structure and GeneClass in excluding individuals from populations of unknown origin should be thoroughly tested with a view to identifying also inter-population hybrids and inbred individuals (Baudouin et al. 2004).
One of the advantages of the GSI approach is that large sample sizes can be obtained with relatively little effort compared to conventional tagging studies. For example, during six consecutive years, Stewart et al. (2002) tagged more than 30 000 salmon of which they obtained <700 returns, yielding a ∼2% recapture rate. In this study within the Teno river system, the ‘recapture rate’ was ∼79%, nearly 40 times of what would have been expected from the conventional tagging studies. It must be noted, however, that in mark–recapture studies accuracy of individual assignment is without error assuming that individual was marked at natal site. An alternative tagging method, marking individuals on their way to natal sites using radio-tags, has been successfully applied on Pacific salmon (e.g. Keefer et al. 2004). In this approach, population of origin is identified by last observation or the actual breeding event of an individual, and the accuracy is related to the population straying rate. In the case of genetic tagging of Teno salmon, expanding the baseline data to cover all populations contributing to the mixed stock fishery are expected to increase both the accuracy and the ‘recapture rate’ of GSI analyses with relatively small genotyping effort.
Variation in run timing
This study demonstrated significant differences in run timing among 1SW Atlantic salmon originating from different populations within the Teno river system. The results also confirmed a remarkable level of temporal stability in migration pattern despite varying water flow regimes and fish abundance. Generally, the 1SW salmon from tributaries entered freshwater first, on average nearly 2 weeks earlier than their counterparts from headwater and upper mainstem populations. Interestingly, the 1SW salmon from the two populations closest to estuary, Teno mainstem lower and Maskejohka (T) arrived latest, but in general, timing of freshwater entry was not related to in-river migration distance. Instead, it was significantly and positively related to population size; 1SW salmon originating from small populations arrive first, and to sea-age structure of the spawning stock; the higher the proportion of larger MSW females in the population, the later the 1SW salmon arrive. These insights to the migration behavior of Atlantic salmon are interesting from the biological as well as from the management perspective.
Although the time gap between runs of tributary 1SW salmon and mainstem/headwater 1SW salmon appears small (2–3 weeks) compared to e.g. ‘fall’ and ‘spring’ runs in Pacific salmon (Keefer et al. 2004; Anderson and Beer 2009) or that reported by Stewart et al. (2002) for the Scottish Atlantic salmon, the observed difference may nonetheless play a significant role in optimal timing of arrival to a sub-arctic river (70°N). The functional argument for the adaptive significance of the trait is that salmon would not waste resources in feeding areas to change its behavior unless it was adaptive. As earlier studies have demonstrated the genetic basis of the timing of salmon arrival (Hansen and Jonsson 1991; Stewart et al. 2002), the first condition of Taylor's (1991) three prerequisites for demonstrating local adaptation is fulfilled. But what is the mechanism of selection responsible for the maintenance of the trait and is differential expression of the trait associated with differential reproductive capability?
One potential mechanism of selection for the earlier run timing of tributary 1SW salmon compared to the MS/HW is the water level and the accessibility of the spawning site. Characteristic to tributaries, apart from Utsjoki, is the lack of lakes. Consequently, the breakage of ice in mid/late May is followed by a very short spring flood period after which the water discharge and level rapidly decreases. As a result, tributaries become shallow compared to the MS/HW and in some years the tributaries may even occasionally be inaccessible to salmon later in the season. Thus, in tributaries late arrival may strongly be selected against, while in the MS/HW run timing is not constrained by water level.
Contrary to the tributaries, postponing arrival may be a better strategy for 1SW salmon breeding in the MS/HW. Our result showing that the proportion of MSW females in the spawning stock was significantly and positively related to run timing of 1SW salmon (Fig. 4) suggests that sea-age structure of the spawning population may have played a significant role in shaping the migration behavior. In spawning areas with a low proportion of larger MSW females, i.e. majority of the tributaries, 1SW salmon can compete and gain control over the best spawning sites, making early arrival beneficial in terms of breeding success, while at sites with a higher proportion of MSW salmon, postponing arrival and remaining at the feeding areas to increase in size at maturity may be a better strategy for 1SW salmon.
Among tributary 1SW salmon, sex had a significant effect on the timing of river entry: females arrived significantly earlier than their male counterparts. Among MS/HW 1SW salmon, there was no significant difference in the timing of arrival between sexes although in Kárášjohka (HW) females appeared to arrive earlier than males. These results are in accordance with earlier studies reporting earlier accumulation of females in the total catch of 1SW salmon (Niemelä et al. 2006). However, our results highlight the importance of appropriate management units, since general trends, as e.g. 1SW females arriving earlier than males, may be driven by population rather than sex-specific differences in run timing. At least in the Teno, the earlier ascendance of tributary populations may drive the earlier accumulation of females in the total catch of the mainstem mixed stock fishery, as 1SW females are common in tributaries, but not in the MS/HW. But why do females leave feeding areas at sea earlier than males?
According to theory, the intensity of sexual selection drives the arrival timing of individuals on breeding grounds and e.g. in migratory birds, both male-biased adult sex ratios and high levels of sperm competition both produce protandry, i.e. males arriving first (Kokko et al. 2006). Similarly, the importance of territorial control may cause females to precede males in arriving to breeding grounds. In the Teno, tributaries are generally dominated by 1SW salmon, although 2SW females are not uncommon, while in the MS/HW there are up to seven and eight different sea-age groups (including previous spawners) for females and males, respectively (Niemelä et al. 2006). As females are known to choose the breeding location where they dig redds to lay eggs in, with males competing for access to females (Fleming 1996), territorial control is characteristic behavior of Atlantic salmon. Interestingly, the aggressive behavior of females for a territory does not begin until close to the start of breeding season, late September. This is supported by snorkeling observations and telemetry studies in tributaries of Teno suggesting that females and males rest, often side-by-side, in deeper pools of the tributaries (Orell and Erkinaro 2007; P. Orell, unpublished data). The distance from the resting or holding site may often be hundreds of meters from the actual spawning site. Why then, is there a difference in the run timing between sexes, when there is more than 2 months before the start of the spawning season? Telemetry studies have indicated that immediately following the ascendance to natal river, salmon show searching behavior by migrating up and down the river for several kilometers before coming to a holding phase (Økland et al. 2001). Females finding and evaluating possible spawning sites before the low water level and rise of temperature, which both limit swimming performance and influence the physiological response to exhaustive exercise (reviewed by Kieffer 2000), is a potential hypothesis to be tested for explaining this behavior.
An alternative explanation to variation in timing of return migration is that arrival to fresh water is simply associated with the use of different feeding grounds in the open sea. The use of different oceanic regions has been previously related to variation in run timing among sea-age classes (Shearer 1992), but Jutila et al. (2003) further showed that salmon of same sea-age, but with different backgrounds (wild versus hatchery) also utilized different areas in the Baltic Sea basin. They proposed that differences in the sea distribution of postsmolts may result from the availability of suitable prey in relation to the size of a smolt.
Finally, as the samples used in this study were collected by local fishermen from drift net, weir and gill net fisheries, and net fisheries in general are known to be size selective (Reddin 1986), it is important to consider how well such a sampling procedure represents the natural 1SW salmon run of Teno River. As the smallest net size allowed in the mainstem of Teno is 58 mm (knot to knot), it is possible that the smallest 1SW salmon may pass through a net without being caught. Given that tributary 1SW salmon are generally 20–25% smaller in size (mean weight 1.2 kg) than their mainstem counterparts (Länsman and Niemelä 2010), it is possible that their contribution to the total 1SW stock may be underestimated in our study. However, a simple size selective fishery could not generate the observed pattern of run-timing behavior. While we acknowledge the potential for nonrandom sampling, this is unlikely to change the conclusions of the study regarding run-timing differences and its relationship to the sea-age structure of the spawning populations. On the contrary, if a fraction of the smaller tributary fish escape the fishery early in the season, our main result, that the proportion of tributary fish is higher early in the season, would actually be conservative.
Implications for management
The discovery of clear differences in run timing of 1SW salmon originating from different populations within a river system has considerable implications for the estuary and in-river management of salmon. Operating net fisheries are often seen as size selective and their potential to introduce directional selection pressure on size at maturity or growth rate is attested (Conover and Munch 2002; Ernande et al. 2004; Swain et al. 2007) and now widely accepted. Much less appreciated and only very recently realized is the potentiality for fisheries induced directional selection and/or differential exploitation rates among populations owing to variation in migration behavior (Quinn et al. 2007; Hard et al. 2008; Anderson and Beer 2009). Salmon fisheries are often temporally regulated with high fishing effort at use during a short fishing season (Crozier et al. 2004; Siira et al. 2009), but in the light of this study, such a strategy is likely to result in heavily biased harvest of 1SW fish of particular populations and/or sex. For example, allowing the mixed stock fisheries in Teno to operate intensively the first month of the fishing season would result in a relative overexploitation of tributary salmon. From this perspective, harvest pressure should be distributed evenly across the whole migration window of 1SW salmon by controlling effort. This should minimize the potential evolutionary response (Quinn et al. 2007; Hutchings and Fraser 2008).
From the perspective of total effective size of the meta-population, considerable gain can be made through a harvest strategy adjusted prioritizing between different subpopulations varying in size and degree of isolation (Tufto and Hindar 2003). To increase the effective population size (Ne), the optimal harvesting strategy in terms of maximizing the total yield, is to harvest relatively less isolated and/or smallest populations because these contribute more to the total effective size with a relatively small reduction in the total yield (Tufto and Hindar 2003). In the light of this theory, allowing 1SW targeted fisheries in the mainstem to operate only from late June allowing salmon from tributary populations to escape fisheries would lead to an increase in the total Ne, i.e. conserving genetic variation, while minimizing the reduction in the harvesting yield.
Management of salmon fisheries on both sides of the Atlantic is in general, through procedures recommended by the North Atlantic Salmon Conservation Organization (NASCO; http://www.nasco.int), moving toward the use of management targets as a tool for evaluating stock status. First-generation spawning targets have recently been developed also for some parts of the Teno river system (Hindar et al. 2007). Although the present study provides important information on the return migration behavior of salmon from different populations, which in itself is useful to the managers, several factors need to be considered and investigated in order to reach specific spawning targets in the Teno. This study investigated only 1SW salmon in the Teno River, which comprise about 60% of the total number, but only 20% of the total weight of salmon catches. The MSW component of the salmon stock is thus more important in terms of catch yield. Differences in run timing of MSW salmon originating from different parts of Teno may also exist and unraveling the contribution of each population to MSW fisheries during the fishing season should be investigated. Furthermore, although radio-telemetry studies have suggested high exploitation rates for the Teno salmon fisheries in general (up to 60–70%; Erkinaro et al. 1999; Karppinen et al. 2004), practically nothing is known about the relative population-specific harvest rates. As the in-river salmon fisheries in Teno include commercial, presumably size selective methods such as weir (19% of total catch), gill net (10%) and drift net (12%), in addition to the recreational rod and line (58%) the selectivity of different fisheries for different populations should be also investigated in order to build a comprehensive and adaptive management strategy of Teno salmon. Similarly, there is an obvious need to study the exploitation of various returning populations of the Teno system in the coastal mixed stock salmon fishery in Norway.
Acknowledgments
We thank Anti Vasemägi and two anonymous referees for valuable comments on the manuscript. We thank Jari Haantie for scale archive mining and scale ring pattern analyses and Suvi Pakkala for laboratory assistance. The project was funded by the Finnish Ministry of Agriculture and Forestry (MAKERA trust), Turku University Foundation and the Academy of Finland.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Results of the first round of power analysis assessing the general assignment success expected with the current genetic marker system and the level of genetic differentiation (overall for tributaries and mainstem/headwater populations).
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Literature cited
- Anderson J, Beer WN. Oceanic, riverine, and genetic influences on spring Chinook salmon migration timing. Ecological Applications. 2009;19:1989–2003. doi: 10.1890/08-0477.1. [DOI] [PubMed] [Google Scholar]
- Anderson E, Waples R, Kalinowski S. An improved method for predicting the accuracy of genetic stock identification. Canadian Journal of Fisheries and Aquatic Sciences. 2008;65:1475–1486. [Google Scholar]
- Angeloni L, Schlaepfer M, Lawler J, Crooks K. A reassessment of the interface between conservation and behavior. Animal Behaviour. 2008;75:731–737. [Google Scholar]
- Baudouin L, Piry S, Cornuet JM. Analytical Bayesian approach for assigning individuals to populations. Journal of Heredity. 2004;95:217–224. doi: 10.1093/jhered/esh035. [DOI] [PubMed] [Google Scholar]
- Beacham TD, Candy JR, McIntosh B. Estimation of stock composition and individual identification of sockeye salmon on a Pacific Rim basis using microsatellite and major histocompatibility complex variation. Transactions of the North American Fisheries Society. 2005;134:1124–1146. [Google Scholar]
- Begg GA, Friedland KD, Pearce JB. Stock identification and its role in stock assessment and fisheries management: an overview. Fisheries Research. 1999;43:1–8. [Google Scholar]
- Caro T. Behavior and conservation: a bridge too far? Trends in Ecology and Evolution. 2007;22:394–400. doi: 10.1016/j.tree.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Conover DO, Munch SB. Sustaining fisheries yields over evolutionary time scales. Science. 2002;297:94–96. doi: 10.1126/science.1074085. [DOI] [PubMed] [Google Scholar]
- Cornuet JM, Piry S, Luikart G, Estoup A, Solignac M. New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics. 1999;153:1989–2000. doi: 10.1093/genetics/153.4.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozier WW, Schon P, Chaput G. Managing Atlantic salmon (Salmo salar L.) in the mixed stock environment: challenges and considerations. ICES Journal of Marine Science. 2004;61:1344–1358. [Google Scholar]
- Currie D, Thompson D, Burke T. Patterns of territory settlement and consequences for breeding success in the Northern Wheatear (Oenanthe oenanthe. Ibis. 2000;142:389–398. [Google Scholar]
- Dingle H. Migration: The Biology of Life on the Move. New York: Oxford University Press; 1996. [Google Scholar]
- Dingle H, Drake VA. What is migration? BioScience. 2007;57:113–121. [Google Scholar]
- Dingle H, Carroll SP, Loye JE, By JR. Conservation, behavior, and 99% of the world's biodiversity: is our ignorance really bliss? In: Clemmons JR, Buchholz R, editors. Behavioral Approaches to Conservation in the Wild. Cambridge: Cambridge University Press; 1997. pp. 72–92. [Google Scholar]
- Dittman A, Quinn T. Homing in Pacific salmon: mechanisms and ecological basis. Journal of Experimental Biology. 1996;199:83–91. doi: 10.1242/jeb.199.1.83. [DOI] [PubMed] [Google Scholar]
- Elphinstone M, Hinten G, Anderson M, Nock C. An inexpensive and high-throughput procedure to extract and purify total genomic DNA for population studies. Molecular Ecology Notes. 2003;3:317–320. [Google Scholar]
- Englund V, Niemela E, Lansman M, Heino M. Variations in Atlantic salmon (Salmo salar L.) smolt age in tributaries of the River Teno, Finland. Fisheries Management and Ecology. 1999;6:83–86. [Google Scholar]
- Erkinaro J, Økland F, Moen K, Niemelä E. Return migration of Atlantic salmon in the River Tana - distribution and exploitation of radio-tagged multi-sea-winter salmon. Boreal Environment Research. 1999;4:115–124. [Google Scholar]
- Ernande B, Dieckmann U, Heino M. Adaptive changes in harvested populations: plasticity and evolution of age and size at maturation. Proceedings of the Royal Society of London B: Biological Sciences. 2004;271:415–423. doi: 10.1098/rspb.2003.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiske P, Lund RA, Hansen LP. Identifying fish farm escapees. In: Cadrin SX, Friedland KD, Waldman JR, editors. Stock Identification Methods; Applications in Fishery Science. Amsterdam: Elsevier Academic Press; 2005. pp. 659–680. [Google Scholar]
- Fleming IA. Reproductive strategies of Atlantic salmon: ecology and evolution. Reviews in Fish Biology and Fisheries. 1996;6:379–416. [Google Scholar]
- Garcia de Leaniz C, Fleming IA, Einum S, Verspoor E, Jordan WC, Consuegra S, Aubin-Horth N, et al. A critical review of adaptive genetic variation in Atlantic salmon: implications for conservation. Biological Reviews of the Cambridge Philosophical Society. 2007;82:173–211. doi: 10.1111/j.1469-185X.2006.00004.x. [DOI] [PubMed] [Google Scholar]
- Gill JA, Norris K, Potts PM, Gunnarsson TG, Atkinson PW, Sutherland WJ. The buffer effect and large-scale population regulation in migratory birds. Nature. 2001;412:436–438. doi: 10.1038/35086568. [DOI] [PubMed] [Google Scholar]
- Goudet J. 2001. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Updated from Goudet (1995). http://www2.unil.ch/popgen/softwares/fstat.htm (accessed April 2010)
- Grimholt U, Drabløs F, Jørgensen SM, Høyheim B, Stet RJM. The major histocompatibility class I locus in Atlantic salmon (Salmo salar L.): polymorphism, linkage analysis and protein modelling. Immunogenetics. 2002;54:570–581. doi: 10.1007/s00251-002-0499-8. [DOI] [PubMed] [Google Scholar]
- Hansen LP, Jonsson B. Evidence of a genetic component in the seasonal return pattern of Atlantic salmon (Salmo salar L.) Journal of Fish Biology. 1991;38:251–258. [Google Scholar]
- Hard JJ, Gross MR, Heino M. Evolutionary consequences of fishing and their implications for salmon. Evolutionary Applications. 2008;1:388–408. doi: 10.1111/j.1752-4571.2008.00020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser L, Seamons TR, Dauer M, Naish KA, Quinn TP. An empirical verification of population assignment methods by marking and parentage data: hatchery and wild steelhead (Oncorhynchus mykiss) in Forks Creek, Washington, USA. Molecular Ecology. 2006;15:3157–3173. doi: 10.1111/j.1365-294X.2006.03017.x. [DOI] [PubMed] [Google Scholar]
- Hindar K, Diserud O, Fiske P, Forseth T, Jensen AJ, Ugedal O, Jonsson N, et al. 2007. Gytebestandsmål for laksebestander i Norge (In Norwegian with an English abstract). NINA Rapport 226, 78 pp.
- Hutchings JA, Fraser DJ. The nature of fisheries- and farming induced evolution. Molecular Ecology. 2008;17:294–313. doi: 10.1111/j.1365-294X.2007.03485.x. [DOI] [PubMed] [Google Scholar]
- ICES. 2002. Report of the Working Group on North Atlantic Salmon. ICES CM 2002/ACFM: 14. http://www.ices.dk/CMdocs/2002/ACFM/Acfm1402.pdf.
- Jonsson N, Jonsson B, Hansen LP. Partial segregation in the timing of migration of Atlantic salmon of different ages. Animal Behavior. 1990;40:313–321. [Google Scholar]
- Jutila E, Jokikokko E, Kallio-Nyberg I, Saloniemi I, Pasanen P. Differences in sea migration between wild and reared Atlantic salmon (Salmo salar L.) in the Baltic Sea. Fisheries Research. 2003;60:333–343. [Google Scholar]
- Karppinen P, Erkinaro J, Moen K, Niemelä E, Økland F. Return migration of Atlantic salmon in the River Tana: distribution, exploitation and migration pattern of radio-tagged 1SW salmon. Journal of Fish Biology. 2004;64:1179–1192. [Google Scholar]
- Keefer ML, Peery CA, Jepson MA, Tolotti K, Bjornn TC. Stock-specific migration timing of adult spring-summer chinook salmon in the Columbia River basin. North American Journal of Fisheries Management. 2004;24:1145–1162. [Google Scholar]
- Kieffer JD. Limits to exhaustive exercise in fish. Comparative Biochemistry and Physiology Part A, Molecular and Integrative Physiology. 2000;126:161–179. doi: 10.1016/s1095-6433(00)00202-6. [DOI] [PubMed] [Google Scholar]
- Kokko H, Gunnarsson TG, Morrell LJ, Gill JA. Why do female migratory birds arrive later than males? Journal of Animal Ecology. 2006;75:1293–1303. doi: 10.1111/j.1365-2656.2006.01151.x. [DOI] [PubMed] [Google Scholar]
- Koljonen M, Pella J, Masuda M. Classical individual assignments versus mixture modeling to estimate stock proportions in Atlantic salmon (Salmo salar) catches from DNA microsatellite data. Canadian Journal of Fisheries and Aquatic Sciences. 2005;62:2143–2158. [Google Scholar]
- Länsman M, Niemelä E. 2010. Lohenkalastus Tenojoen sivuvesissä (In Finnish with an English abstract). Finnish Game and Fisheries Research Institute Report 2, 26 pp. http://www.rktl.fi/www/uploads/pdf/uudet%20julkaisut/selvityksia_2_2010.pdf.
- Manel S, Berthier P, Luikart G. Detecting wildlife poaching: identifying the origin of individuals with Bayesian assignment tests and multilocus genotypes. Conservation Biology. 2002;16:650–659. [Google Scholar]
- McCullagh P, Nelder JA. Generalized Linear Models. London: Chapman & Hall; 1989. [Google Scholar]
- Neaves PI, Wallace CG, Candy JR, Beacham TD. 2005. CBayes: computer program for mixed stock analysis of allelic data, version 3.0 http://www.pac.dfo-mpo.gc.ca/science/facilities-installations/pbs-sbp/mgl-lgm/index-eng.htm (accessed April 2010)
- Niemelä E. 2004. Variation in the yearly and seasonal abundance of juvenile Atlantic salmon in a long-term monitoring programme: methodology, status of stocks and reference pointsPhD thesis, Acta Universitatis Ouluensis A415.
- Niemelä E, Orell P, Erkinaro J. Previously spawned Atlantic salmon ascend a large subarctic river earlier than their maiden counterparts. Journal of Fish Biology. 2006;69:1151–1163. [Google Scholar]
- Nordqvist O. Times of entering of the Atlantic salmon (Salmo salar L.) in the rivers. Rapports et Proce’s-Verbaux. Conseil Permanent International pour L'Exploration de la Me’r. 1924;33:5–58. [Google Scholar]
- Northcote TG. Migratory strategies and production in freshwater fishes. In: Gerking SD, editor. Ecology of Freshwater Fish Production. Oxford: Blackwell Scientific Publications; 1978. pp. 326–359. [Google Scholar]
- Økland F, Erkinaro J, Moen K, Niemelä E, Fiske P, McKinley R, Thostad E. Return migration of Atlantic salmon in the River Tana: phases of migratory behaviour. Journal of Fish Biology. 2001;59:862–874. [Google Scholar]
- Orell P, Erkinaro J. Snorkeling as a method for assessing spawning stock of Atlantic salmon, Salmo salar. Fisheries Management and Ecology. 2007;14:199–208. [Google Scholar]
- Pella J, Masuda M. Bayesian methods for analysis of stock mixtures from genetic characters. Fishery Bulletin. 2001;99:151–167. [Google Scholar]
- Pimm S, Jones H, Diamond J. On the risk of extinction. American Naturalist. 1998;132:757–785. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn TP, Hodgson S, Flynn L, Hilborn R, Rogers DE. Directional selection by fisheries and the timing of sockeye salmon (Oncorhynchus nerka) migrations. Ecological Applications. 2007;17:731–739. doi: 10.1890/06-0771. [DOI] [PubMed] [Google Scholar]
- Rannala B, Mountain JL. Detecting immigration by using multilocus genotypes. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:9197–9201. doi: 10.1073/pnas.94.17.9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddin DG. Effects of different mesh sizes on gill-net catches of Atlantic salmon in Newfoundland. North American Journal of Fisheries Management. 1986;6:209–215. [Google Scholar]
- Shearer WM. The Atlantic salmon (Salmo salar L.) of the North Esk with particular reference to the relationship between river and sea age and time of return to home waters. Fisheries Research. 1990;10:93–123. [Google Scholar]
- Shearer WM. The Atlantic Salmon: Natural History, Exploitation and Future Management. New York: Halsted Press; 1992. [Google Scholar]
- Shumway CA. A neglected science: applying behavior to aquatic conservation. Environmental Biology of Fishes. 1999;55:183–201. [Google Scholar]
- Siira A, Erkinaro J, Suuronen P, Jounela P. Run timing and migration routes of returning Atlantic salmon in the Northern Baltic Sea; implications for fisheries management. Fisheries Management and Ecology. 2009;16:177–190. [Google Scholar]
- Stet RJM, De Vries B, Mudde K, Hermsen T, Van Heerwaarden J, Shum BP, Grimholt U. Unique haplotypes of co-segregating major histocompatibility class II A and class II B alleles in Atlantic salmon (Salmo salar) give rise to diverse class II genotypes. Immunogenetics. 2002;54:320–331. doi: 10.1007/s00251-002-0477-1. [DOI] [PubMed] [Google Scholar]
- Stewart DC, Smith GW, Youngson AF. Tributary-specific variation in timing of return of adult Atlantic salmon (Salmo salar) to fresh water has a genetic component. Canadian Journal of Fisheries and Aquatic Sciences. 2002;59:276–281. [Google Scholar]
- Swain DP, Sinclair AF, Mark Hanson J. Evolutionary response to size-selective mortality in an exploited fish population. Proceedings of the Royal Society of London B: Biological Sciences. 2007;274:1015–1022. doi: 10.1098/rspb.2006.0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor EB. A review of local adaptation in salmonidae, with particular reference to Pacific and Atlantic salmon. Aquaculture. 1991;98:185–207. [Google Scholar]
- Thorley JL, Youngson AF, Laughton R. Seasonal variation in rod recapture rates indicates differential exploitation of Atlantic salmon, Salmo salar, stock components. Fisheries Management and Ecology. 2007;14:191–198. [Google Scholar]
- Tufto J, Hindar K. Effective size in management and conservation of subdivided populations. Journal of Theoretical Biology. 2003;222:273–281. doi: 10.1016/s0022-5193(03)00018-3. [DOI] [PubMed] [Google Scholar]
- Vähä JP, Erkinaro J, Niemela E, Primmer CR. Life-history and habitat features influence the within-river genetic structure of Atlantic salmon. Molecular Ecology. 2007;16:2638–2654. doi: 10.1111/j.1365-294X.2007.03329.x. [DOI] [PubMed] [Google Scholar]
- Vähä JP, Erkinaro J, Niemela E, Primmer CR. Temporally stable genetic structure and low migration in an Atlantic salmon population complex: implications for conservation and management. Evolutionary Applications. 2008;1:137–154. doi: 10.1111/j.1752-4571.2007.00007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MS, Marra PP, Haig S, Bensch S, Holmes RT. Links between worlds: unraveling migratory connectivity. Trends in Ecology and Evolution. 2002;17:76–83. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


