Abstract
The level of genetic variation in natural populations influences evolutionary potential, and may therefore influence responses to selection in the face of future environmental changes. By combining long-term monitoring of marked individuals with genetic pedigree reconstruction, we assessed whether habitat loss influenced genetic variation in a lemon shark (Negaprion brevirostris) population at an isolated nursery lagoon (Bimini, Bahamas). We also tracked changes in the strength and direction of natural selection. Contrary to initial expectations, we found that after the habitat loss neutral genetic variation increased, as did additive genetic variance for juvenile morphological traits (body length and mass). We hypothesize that these effects might result from philopatric behavior in females coupled with a possible influx of male genotypes from other nursery sites. We also found changes in the strength of selection on morphological traits, which weakened considerably after the disturbance; habitat loss therefore changed the phenotypes favored by natural selection. Because such human-induced shifts in the adaptive landscape may be common, we suggest that conservation biologists should not simply focus on neutral genetic variation per se, but also on assessing and preserving evolutionary parameters, such as additive genetic variation and selection.
Keywords: additive genetic variance, evolutionary potential, heritability, heterozygosity, human disturbance, selection
Introduction
Humans are increasingly modifying the environment experienced by natural populations. These modifications likely reduce the degree to which populations are adapted to their local conditions, thereby decreasing mean fitness and possibly compromising population productivity or persistence (Bürger and Lynch 1995; Gomulkiewicz and Holt 1995; Stockwell et al. 2003; Frankham 2005; Both et al. 2006; Kinnison and Hairston 2007). Ongoing adaptation can potentially save these otherwise maladapted populations from extinction, but this potential depends on a host of factors related to population connectivity, initial population size, mortality rates, adaptive plasticity (or maternal effects), genetic variation, and the strength and form of selection (Bürger and Lynch 1995; Gomulkiewicz and Holt 1995; Boulding and Hay 2001; Price et al. 2003; Räsänen and Kruuk 2007; Orr and Unckless 2008; Bell and Gonzalez 2009). Adding to this complexity, many of these factors will interact and feedback on each other. As just one example, reduced population sizes can increase the expression of recessive deleterious mutations (Lynch et al. 1995; Crnokrak and Roff 1999) and reduce overall genetic variation (England et al. 2003; Leimu et al. 2006). Through these effects, environmental change that causes maladaptation can impede the ability of populations to evolve adaptively in response to that same (or future) environmental change (Stockwell et al. 2003; Kinnison and Hairston 2007).
Some of the above effects have been investigated in natural populations facing environmental change. First, anthropogenic disturbances have been shown to alter genetic variation in natural populations (see Caizergues et al. 2003; DiBattista 2008). Second, environmental changes, particularly those caused by humans, have been shown to cause the apparently adaptive evolution of phenotypic traits (reviews: Hendry and Kinnison 1999; Reznick and Ghalambor 2001; Stockwell et al. 2003; Hendry et al. 2008; Darimont et al. 2009). Third, these adaptive trait changes appear to improve individual fitness (Kinnison et al. 2007; Gordon et al. 2009) and also influence population dynamics (Hanski and Saccheri 2006; Pelletier et al. 2007). One limitation of this existing work is that it does not involve the simultaneous consideration of multiple factors that likely influence adaptive responses to environmental change (as described above). We attempt to advance this goal by measuring the number of breeding adults, mortality rates, maternal effects, genetic variation, and natural selection in a population facing dramatic environmental change. This analysis is made possible by a long-term data set that happens to be bisected by a major anthropogenic disturbance.
Anthropogenic disturbances can take many forms (e.g., hunting and harvesting, habitat fragmentation or loss, pollution, invasive species, and climate change), some of which clearly influence selection, adaptation, and genetic variation (Caizergues et al. 2003; Stockwell et al. 2003; Carroll 2008; DiBattista 2008; Hendry et al. 2008; McClure et al. 2008; Darimont et al. 2009). We here focus on habitat loss, where the immediate consequences of environmental change might include increasing isolation, reduced carrying capacity, increased environmental variation, and shifts in local trait optima (Carvajal-Rodríguez et al. 2005; McClure et al. 2008). Any of these changes might decrease population size and alter selection, which might then cause maladaptation and reduced genetic variation (Johansson et al. 2007; McClure et al. 2008). We test for these and other effects by comparing demographic and evolutionary parameters in the same natural population from before to after habitat loss.
We track several key variables. The first is the number of adults successfully producing offspring at a nursery site. Changes in this parameter might indicate whether adults are avoiding the site, or at least failing to reproduce successfully. Such changes might suggest the possibility of reductions in genetic variation. The second variable is the mortality rate of juveniles, which might indicate whether habitat loss is having a demographic cost through reduced offspring fitness. The third variable is neutral genetic variation, which might reflect a combination of the above effects that reduce census and effective population size (Frankham 1996; Godt et al. 1996; Leimu et al. 2006). The fourth variable involves several quantitative genetic parameters, which here include additive genetic variation, trait heritability, and maternal effects (which also include genetic dominance effects). These variables in particular should indicate whether or not evolutionary potential has been altered by habitat loss. The fifth variable is the strength and form of natural selection, which should indicate whether the need for adaptive change is likely to be an important part of the population's immediate future. Interestingly, no studies appear to have explicitly compared selection from before to after habitat loss, although such information seems critical to interpreting potential adaptive responses in the affected population (see Stockwell et al. 2003).
Lemon sharks and a mega-resort
Our study focuses on the lemon shark (Negaprion brevirostris), a large and placentally-viviparous coastal species found throughout the tropical western Atlantic, on the west coast of Africa, and in the Pacific from Mexico to Colombia (Compagno 1984; Schultz et al. 2008). Adult females of this species use shallow lagoons for both mating and parturition (Feldheim et al. 2002). These nursery areas are then critical for the survival of juveniles because they provide protection from predators, as well as appropriate foraging opportunities (Branstetter 1990; Rountree and Able 1996; Franks 2007). Both of these properties are important because juvenile sharks must fend for themselves immediately after birth (i.e., no parental care, Pratt and Casey 1990), and they tend to remain highly attached to their nursery (i.e., site fidelity, Morrissey and Gruber 1993; DiBattista et al. 2007; Franks 2007).
Our study site is Bimini, Bahamas (25°44′N, 79°16′W), a mangrove-fringed chain of islands located on the north-western edge of the Great Bahama Bank, 85 km east of Miami, Florida. The North Sound at Bimini (see Figs 1 and 2) is the only well characterized lemon shark nursery site in the Bahamas, and one of few studied throughout the Atlantic. Lemon sharks are born into this nursery from April to May of each year and remain there for at least 3 years (<90 cm total length; Morrissey and Gruber 1993; DiBattista et al. 2007; Franks 2007), during which time juveniles have daily home ranges of no more than a few hundred square meters (Morrissey and Gruber 1993). After leaving the nursery habitat, juvenile sharks then expand their home range and disperse into a wider variety of deeper habitats around the islands as they become less vulnerable to predators and seek out larger prey (Morrissey and Gruber 1993; Franks 2007). Although a few smaller, ‘satellite’ nursery areas around the Bimini islands do exist (i.e., South Bimini nursery), there is no effective exchange of juvenile individuals among them, and adult females are philopatric to specific sites around Bimini (J. D. DiBattista unpublished data).
Figure 1.
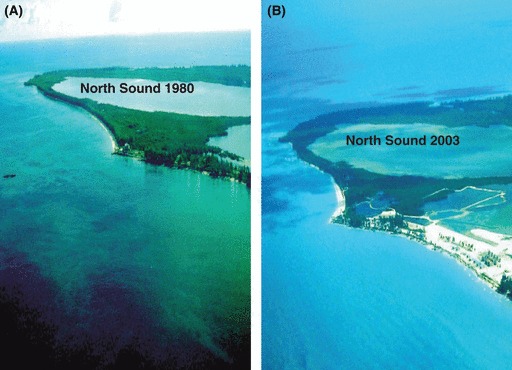
Aerial photograph of the North Sound at Bimini, Bahamas, in 1980 (A), and again after resort development in 2003 (B; Photo credits: S. Kessel).
Figure 2.

Aerial photograph of the North Sound at Bimini, Bahamas in 2003 (A), and this same area after further mangrove removal in 2005 (B; Photo credits: S. Kessel).
The North Sound nursery has recently been subject to large-scale mangrove removal on its western shores as part of a development project (Figs 1 and 2; Gruber and Parks 2002). As of August 2007, approximately 30% of the mangrove habitat (or 120 ha) surrounding the nursery had been removed, in addition to 750 000 m3 of sandfill and 18% of the seagrass (Thalassia testudinum; Jennings et al. 2008). Additional habitat loss has since occurred and is ongoing (S. Gruber personal communication). Although excavation was initiated in 1999, the most intensive dredging took place in March 2001 (Gruber and Parks 2002). In the first few years after this development began at Bimini, we saw a modest rise in the mortality rate of juvenile lemon sharks (Jennings et al. 2008; also see Fig. 3), although these effects have since levelled off. It should be noted that changes in mortality rates were not, however, observed over the same time period in an adjacent nursery area (i.e.,‘control’ site located 6 km away along the exposed, mangrove-lined coast of South Bimini; Jennings et al. 2008).
Figure 3.
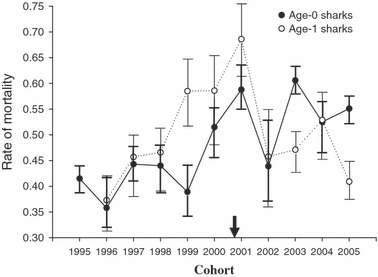
The proportion of age-0 lemon sharks not surviving their first year (black circles), or age-1 lemon sharks not surviving to age-2 (open circles), from 1995 to 2005. The black arrow indicates the approximate onset of disturbance on the x-axis. Values are means ± 1 SEM.
The present analysis was based on long-term, intensive sampling of the juvenile lemon shark population at Bimini. By reconstructing pedigrees at this site, these data allow the accurate estimation of the number of breeding adults (Feldheim et al. 2004; DiBattista et al. 2009), the mortality rate of juveniles (DiBattista et al. 2007), neutral genetic variation (DiBattista et al. 2008a), quantitative genetic parameters (DiBattista et al. 2009), and the strength and form of natural selection (DiBattista et al. 2007). Because these data come from a long-term data set, we can here examine whether demographic and evolutionary parameters change from before (1995–2000) to after (2001–2007) the start of habitat loss. Although there are a number of published genetic studies from this system, most deal with data collected prior to the disturbance (i.e.,Feldheim et al. 2001, 2002, 2004; DiBattista et al. 2007, 2008b). Moreover, studies that include data collected after the disturbance were either focused on mating patterns at other populations (DiBattista et al. 2008a), long-term estimates of quantitative genetic parameters and their statistical robustness (DiBattista et al. 2009), or sub-adult sharks that no longer show site fidelity to this nursery (Chapman et al. 2009). The present analysis therefore represents the first test of the effects of disturbance on genetic variation and selection in this system.
Materials and methods
Study site and sample collection
The Bimini Islands enclose a 21 km2 lagoon (0–120 cm deep at low tide) that serves as a nursery area for approximately 250 juvenile and sub-adult lemon sharks (DiBattista et al. 2009). Each year since 1995, we have captured almost 99% of the juveniles in the North Sound (Gruber et al. 2001; DiBattista et al. 2009), with a high proportion of these individuals recaptured in subsequent years (i.e., recapture probabilities ranged from 0.67 to 0.85 through age-3; for more details see DiBattista et al. 2007). Our sampling always took place between May 21st and June 25th, which is just after parturition by females. Newborn and juvenile sharks were captured in 180-m long by 2-m deep gill nets (Manire and Gruber 1991), and net locations were standardized across years within the lagoon. In addition to standardized netting, some sub-adult (males and females: 90–230 cm) and adult (mature > 230 cm, Brown and Gruber 1988) lemon sharks were captured opportunistically over the course of the study by rod and reel or long-line fishing gear.
The first time each shark was captured, it was measured for precaudal length (PCL, tip of snout to precaudal pit in mm; Compagno 1984), weighed (kg, when feasible), and tagged intramuscularly with an individually-coded passive integrated transponder tag (PIT). Each time a tagged shark was subsequently recaptured, its tag number, PCL, and mass (when feasible) were recorded. The growth rate of individuals was calculated as the change in body length between recapture events, expressed as cm per year. Ages were assigned to most individuals using methods described in our earlier work (Feldheim et al. 2004; DiBattista et al. 2008a,b).
Pedigree reconstruction
A small piece of fin tissue (2 mm2) was clipped from every captured shark, and genomic DNA was extracted with a salting-out protocol (Sunnucks and Hales 1996). Samples were genotyped with 11 species-specific microsatellite primer pairs (for methodological details see Feldheim et al. 2002, 2004; DiBattista et al. 2008a,b). Multilocus genotypes were obtained for a minimum of nine loci for all individuals, and genotyping error rates were deemed very low in this system based on a subset of samples that were re-run (DiBattista et al. 2008a). Each loci considered here also conformed to Hardy-Weinberg equilibrium, with no evidence for linkage disequilibrium between loci (for more details see DiBattista et al. 2008a,b).
Pedigree reconstruction was based on microsatellite data analyzed in the maximum-likelihood program CERVUS v. 3.0 (Marshall et al. 1998; also see Kalinowski et al. 2007). With this program, we assigned individual offspring to: (i) the few candidate parents (19 adult females and 11 adult males) that we were able to catch, (ii) sub-adult sharks (n = 175) that might have produced offspring in subsequent years, and (iii) parents (n = 40 females and 81 males) genetically inferred from offspring, but never physically sampled (see Feldheim et al. 2004; DiBattista et al. 2009). Assignment to these potential parents was done under a strict confidence level of 95% (for more details see Feldheim et al. 2004; DiBattista et al. 2008a,b, 2009). For the offspring not assigned with the above procedures (out of a total of 1501 offspring, 394 lacked assigned mothers and 830 lacked assigned fathers), we inferred sibling groups based on maximum-likelihood with COLONY v. 1.2 (Wang 2004; also see DiBattista et al. 2008a), thereby reconstructing genotypes for most of the remaining parents. In brief, we ran groups of age-0 sharks in COLONY, separated by year of birth (i.e., cohorts), to identify possible within-year sibling groups. Age-0 sharks from each cohort were also run separately with cohorts from every other year to identify potential between-year sibling groups. From these data, parental genotypes of unsampled parents were genetically reconstructed at individual loci with 95% confidence. Based on all of the above procedures, our final dataset included 1304 offspring assigned to fathers and 1351 offspring assigned to mothers. This reconstructed pedigree, along with the measured phenotype of each juvenile shark at first capture, formed the basis of all downstream analyses (n = 1351 distinct individuals).
It should be noted that our samples did not meet the assumptions and sample sizes necessary for calculating the ‘effective population size’, Ne (for review see Waples 2005; Fraser et al. 2007), but our reconstructed pedigree did allow us to identify the number of breeding adults at Bimini as a potential contributor to changes in genetic variance. We therefore estimated the number of parents successfully producing offspring annually. Juvenile mortality, on the other hand, was estimated as the proportion of age-0 (age-1) sharks tagged (caught) in each year that was never recaptured (see DiBattista et al. 2007); parameter uncertainty (i.e., standard error) was assessed using the capture-recapture program MARK (White and Burnham 1999). See DiBattista et al. (2007) for more details on the testing of discrete models and model averaging in MARK.
Neutral genetic variation assessment
The present analysis focuses on all newborn sharks sampled at Bimini between 1995 and 2007 (n = 1131). We specifically compare samples from before (1995–2000, n = 449) to after (2001–2007, n = 682) the disturbance with respect to several metrics of variation: (i) observed heterozygosity (Ho), (ii) expected heterozygosity (He), and (iii) the mean number of alleles per locus (A). For the last of these, a rarefaction procedure was used to correct for uneven sample sizes (i.e., allelic richness, AR; HP-RARE 1.0, Kalinowski 2005). Differences in the response variables (number of alleles, allelic richness, observed or expected heterozygosity) were then separately compared among time periods (i.e., before versus after the disturbance), and across all microsatellite loci using paired-sample t-tests. Heterozygosity from before versus after the disturbance was also compared in GENEPOP (Raymond and Rousset 1995) by calculating pairwise FST between time periods. Finally, data were log-transformed where appropriate to ensure normality; false rejection rate was set at α = 0.05 throughout, unless otherwise noted.
Quantitative genetic variation
The estimation of quantitative genetic parameters for PCL and body mass was based on pooled samples (i.e., all individuals from 1995 to 2000, n = 583, versus all individuals from 2001 to 2007, n = 768) because the smaller sample sizes of year-specific models prevented convergence. For similar reasons, growth rate could not be analyzed for either period. Analyses were performed with multivariate mixed model REML estimation in ASReml V.2 (for general details see Kruuk 2004; Garant and Kruuk 2005). We previously tested different mixed models in our population (see DiBattista et al. 2009), and so we here only consider estimates from the most-likely model. This model included additive genetic variance and maternal identity (i.e.,‘dam’) as random effects, plus age, sex, and cohort as fixed effects. The ‘dam’ term accounts for maternal effects (for review see Räsänen and Kruuk 2007), which appear strong in this particular system (DiBattista et al. 2009), and reflect both nongenetic maternal and genetic dominance effects; for simplicity, however, we hereafter refer to these jointly as only maternal effects. The form of the mixed model was as follows:
where y is a vector of phenotypic values; b is a vector of fixed effects; a is a vector of random effects of the additive genetic merit of each individual; u is a vector containing other random effects included in the model (i.e., maternal identity); e is a vector of residual values; and X, Z1, and Z2 are design matrices relating phenotypic observations to their corresponding fixed or random effects (Lynch and Walsh 1998). For each relevant trait, the total phenotypic variance (VP) was partitioned into the additive genetic (VA), maternal (VM + D), and residual variance (VR). Narrow-sense heritability (h2) was estimated as the ratio of additive genetic variance to total phenotypic variance (h2 = VA/VP). Statistical significance was assessed with likelihood ratio tests that compared the full model to a reduced model that lacked the parameter in question. Recent sensitivity analyses showed that quantitative genetic parameters are quite robust to pedigree error in this system (DiBattista et al. 2009). Parameters from before to after the disturbance were compared using bivariate models where trait values from the before and after disturbance periods were coded as being different traits; their genetic (or maternal, or residual) variance was either constrained to be equal or allowed to be estimated separately for each period. A significant improvement of the model likelihood under unconstrained conditions, when compared to the constrained model, would be taken as evidence for a difference in variance components among periods.
Selection analyses
We used standard methods (Lande and Arnold 1983; Schluter 1988; Brodie et al. 1995; Janzen and Stern 1998) to assess the strength of natural selection acting on juvenile body size and growth rate at Bimini. Analyses for the cohorts from 1995 to 2000 were reported earlier (DiBattista et al. 2007), and we here apply identical methods to cohorts from 2001 to 2005. The last cohort we analyzed was 2005 because 3 years of postbirth data are best for confirming whether individuals live or die before leaving the nursery site (DiBattista et al. 2007). Selection on PCL and mass was estimated by relating these traits for individuals at the start of an interval (year i) to whether or not these individuals survived to the end of that interval (year i + 1). Selection on growth was estimated by relating the change in length between year i and year i + 1 (here age-0 to age-1) to survival between year i + 1 and year i + 2 (here age-1 to age-2). Because growth rate data were only available for a subset of the fish, estimates of selection on the other traits excluded data for growth rate (to maximize N).
Before analysis, trait values were standardized to a mean of zero and a standard deviation of unity (Lande and Arnold 1983) within each combination of cohort and age class. Any tagged sharks captured at the end of a given year-long interval, or in any subsequent year, were known to have survived through that interval (absolute fitness = 1). Any tagged sharks not recaptured at the end of a given interval, or in any subsequent year, were assumed to have died (absolute fitness = 0). This assumption was shown to be valid in our previous analysis of selection (DiBattista et al. 2007). Absolute fitness was standardized to relative fitness for each shark by dividing its absolute fitness (0 or 1) over an interval, by the mean fitness of all individuals for that combination of cohort/age/interval.
Logistic regressions (Janzen and Stern 1998) of relative fitness on each standardized trait value were then used to estimate selection differentials (i). Similarly, multiple logistic regressions were used to estimate selection gradients (β), which account for correlations among the measured traits. Body mass was excluded when calculating gradients because it was too highly correlated (r = 0.99) with body length (see discussion in Mitchell-Olds and Shaw 1987). Coefficients presented here were converted to their linear equivalents to facilitate comparison with other studies (Kingsolver et al. 2001). Pooled selection coefficients were then compared from before (1995–2000) to after (2001–2005) the disturbance for each trait (i.e., PCL, mass, and growth) with Student's t-tests. Finally, we used univariate cubic splines (Schluter 1988; glmsWIN1.0 spline program, Schluter 2000) to visualize the form of selection acting on each trait for each combination of cohort and age. To facilitate interpretation, we here used raw trait data and absolute fitness rather than standardized values.
Results
Number of breeding adults and juvenile mortality
We identified 117 unique mothers and 487 unique fathers over the course of the study. The average number of mothers that gave birth at Bimini each year was 16.08 ± 1.19 SEM (range: 9–23) and the average number of fathers was 38.31 ± 3.34 SEM (range: 20–62) (see Fig. 4). The number of fathers increased following the disturbance (t = −3.17, df = 11, P = 0.009), as did the number of mothers, although the latter was marginally nonsignificant (t = −1.95, df = 11, P = 0.078). Moreover, ‘year’ was positively correlated with the number of fathers (β = −4491.43, r2 = 0.54, P = 0.004) and mothers (β = −1303.26, r2 = 0.49, P = 0.03). In short, more adult lemon sharks contributed to the juvenile population after the disturbance than before it. Such an increase in the number of assigned parents over time does not however appear to be an artifact of including more potential parents in later years for pedigree reconstruction analyses. Indeed, we had a similar proportion of offspring not assigned to parents before versus after the disturbance (data not shown).
Figure 4.
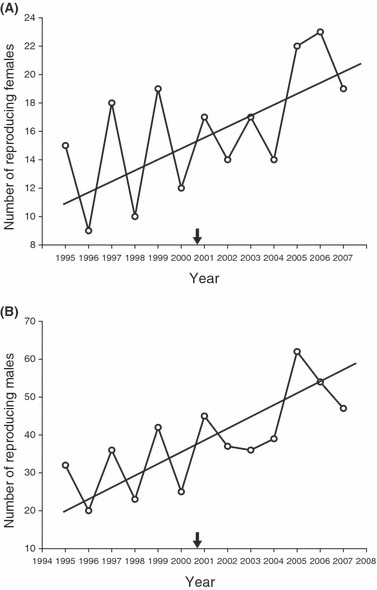
Number of reproducing males (A) and females (B) using the Bimini nursery site each year from 1995 to 2007. It should be noted that the majority of these individuals (92% of females and 99% of males) were genetically inferred and never physically captured. The black arrow indicates the approximate onset of disturbance on the x-axis.
The proportion of age-0 lemon sharks that did not survive their first year also appeared to increase following the disturbance (t = −3.20, df = 9, P = 0.011), although this was not the case for mortality between age-1 and age-2 (t = −0.27, df = 8, P = 0.79; Fig. 3). Similarly, ‘year’ was positively correlated with age-0 mortality (β = 0.018, r2 = 0.51, P = 0.009), but not mortality between age-1 and age-2 (β = 0.003, r2 = 0.012, P = 0.76), which suggests that the disturbance had only a modest effect on juvenile mortality rates.
Neutral genetic variation
We also found consistent increases in measures of neutral genetic variation from before to after the disturbance (see Table 1 and Fig. 5). This difference among time periods was supported by comparisons of genetic variation for some (mean number of alleles: t = −4.28, df = 10, P = 0.002; allelic richness: t = −3.36, df = 10, P = 0.007), but not all microsatellite metrics (i.e., observed heterozygosity: t = −1.95, df = 10, P = 0.08; expected heterozygosity: t = −2.14, df = 10, P = 0.06). Genotypes from before versus after the disturbance were significantly different, however, when considering FST (P < 0.001); results were also similar when step-wise mutation model estimators were considered (RST), and so only FST is presented here. Moreover, trends for an increase in genetic variation after the disturbance was supported by the number of alleles, but not heterozygosity, when data from each year was considered separately (see Fig. 5). Such changes could not be explained by an increase in the number of related individuals after the disturbance either; average pairwise relatedness (r) estimated using MARK v. 3.0 (Ritland 2004) was no different between time periods (before disturbance: r = 0.003 ± 0.006; after disturbance: r = 0.004 ± 0.008; see DiBattista et al. 2009).
Table 1.
Mean number of alleles per locus (A), allelic richness (AR), the observed (HO) and expected (HE) heterozygosity, as well as FIS and FST for each loci in all lemon sharks captured in the North Bimini lagoon from 1995 to 2000 (n = 449) versus from 2001 to 2007 (n = 682)
| 1995–2000 | 2001–2007 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Microsatellite ID | A | AR | HO | HE | A | AR | HO | HE | FIS | FST |
| LS22 | 18 | 14.042 | 0.88 | 0.90 | 20 | 14.5 | 0.89 | 0.90 | 0.015 | 0.002 |
| LS30 | 14 | 9.8 | 0.67 | 0.71 | 17 | 11.87 | 0.77 | 0.79 | 0.049 | 0.009 |
| LS48 | 25 | 20.86 | 0.95 | 0.94 | 26 | 21.19 | 0.94 | 0.94 | 0.002 | 0.003 |
| LS54 | 5 | 4.089 | 0.53 | 0.54 | 5 | 4.073 | 0.59 | 0.58 | 0.003 | 0.001 |
| LS75 | 5 | 4.55 | 0.66 | 0.70 | 6 | 4.71 | 0.72 | 0.72 | 0.028 | 0.003 |
| LS52 | 37 | 25.96 | 0.94 | 0.95 | 41 | 26.72 | 0.94 | 0.94 | 0.008 | 0.003 |
| LS572 | 7 | 5.56 | 0.74 | 0.73 | 8 | 6.19 | 0.71 | 0.72 | 0.015 | 0.001 |
| LS542 | 10 | 6.5 | 0.66 | 0.63 | 11 | 7.9 | 0.68 | 0.69 | −0.009 | 0.004 |
| LS596 | 12 | 10.43 | 0.83 | 0.87 | 13 | 10.71 | 0.90 | 0.86 | −0.007 | 0.004 |
| LS801 | 22 | 13.55 | 0.83 | 0.80 | 23 | 15.98 | 0.84 | 0.84 | −0.006 | 0.001 |
| LS560 | 9 | 8.21 | 0.88 | 0.84 | 10 | 8.67 | 0.86 | 0.85 | −0.027 | 0.002 |
| Average | 14.91* | 11.23 | 0.78 | 0.78 | 16.36 | 12.05 | 0.80 | 0.80 | 0.006 | 0.003† |
| SE | 0.96 | 0.13 | 0.007 | 0.005 | 0.32 | 0.38 | 0.007 | 0.004 | 0.0018 | 0.0007 |
These time periods correspond to before and after the beginning of the large-scale development project at Bimini.
Numbers in bold are significantly different before versus after the disturbance (paired sample t-test, P < 0.05).
FST significantly different when comparing offspring genotypes from before versus after the disturbance (P < 0.001).
Figure 5.
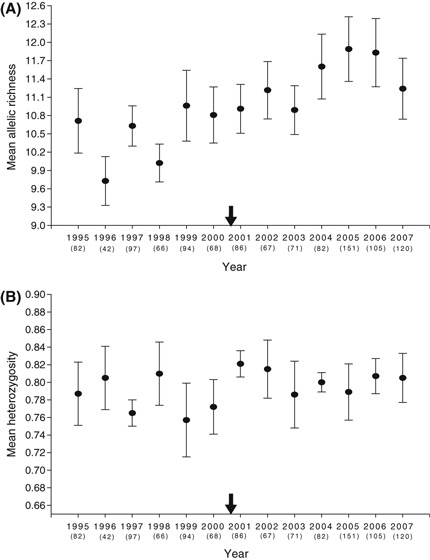
Mean allelic richness (A) and expected heterozygosity (B) for juvenile lemon sharks captured at Bimini, Bahamas from 1995 to 2007 (n = 1131). Values are means ± 1 SEM. The black arrow indicates the approximate onset of disturbance on the x-axis.
Quantitative genetic variation
Several changes were evident from before to after the disturbance (Table 2). First, additive genetic variance was significantly higher after the disturbance for PCL (χ2 = 3.90, df = 1, P = 0.048) and mass (χ2 = 7.91, df = 1, P = 0.005). Second, residual variance was significantly lower for PCL (χ2 = 5.16, df = 1, P = 0.023) and also for mass (χ2 = 18.04, df = 1, P < 0.001). Third, maternal effects, already strong before the disturbance (i.e., 29% and 20% of the phenotypic variance for PCL and mass) were even stronger afterward (PCL, χ2 = 6.30, df = 1, P = 0.012; Mass, χ2 = 16.25, df = 1, P < 0.001).
Table 2.
Estimates of variance components (VA, additive genetic variance; VM + D, nongenetic maternal and genetic dominance variance; VR, residual variance; VP, phenotypic variance) and heritability (h2) with their standard error, as well as coefficients of variation for morphological traits (PCL, precaudal length; Mass) in a natural lemon shark population based on the ‘animal model’
| Traits/models* | n | VA | VM + D | VR | VP | h2 | CVA |
|---|---|---|---|---|---|---|---|
| Animal/dam random effects model, age, sex, and cohort as fixed effects | |||||||
| Before | |||||||
| PCL | 583 | 1.92 ± 0.98† | 1.94 ± 0.75†‡ (0.29) | 2.83 ± 0.64† (0.42) | 6.68 ± 0.73† | 0.29 ± 0.15 | 2.86 |
| Mass | 583 | 0.015 ± 0.013† | 0.019 ± 0.008† (0.20) | 0.063 ± 0.009† (0.65) | 0.097 ± 0.008† | 0.15 ± 0.13 | 9.21 |
| After | |||||||
| PCL | 768 | 5.51 ± 0.89 | 4.91 ± 1.29 (0.42) | 1.14 ± 0.51 (0.10) | 11.56 ± 1.26 | 0.48 ± 0.09 | 4.87 |
| Mass | 768 | 0.055 ± 0.014 | 0.072 ± 0.018 (0.45) | 0.032 ± 0.009 (0.20) | 0.159 ± 0.017 | 0.34 ± 0.094 | 19.54 |
Variance components are estimated from pooled juvenile shark samples caught prior to (1995–2000, n = 583), or following (2001–2007, n = 768) an on-going anthropogenic disturbance at Bimini. All values are mean ± SE. Numbers in parentheses represent quantitative genetic parameters (i.e., VM+D and VR) expressed as a ratio of VP.
Each model includes ‘dam’ as a random factor to account for possible maternal effects present in the population.
Significantly different from variance components estimated in the ‘after’ time period based on likelihood ratio tests, P < 0.05.
Parameter estimates in bold are significantly different from 0 (i.e., chi-square analysis, P < 0.05 in all cases).
Natural selection
In the following, linear selection differentials and gradients are referred to jointly as ‘selection coefficients’ when they showed similar trends. Before the disturbance (1995–2000), age-0 selection coefficients for length varied in sign (Fig. 6A,B), but selection for mass was negative in five of six cohorts (and significant in one: 1996, P = 0.043; Fig. 6E). After the disturbance (2001–2005), age-0 selection coefficients for length and mass were negative in every cohort except 2005 (although none were significant, Fig. 6A,B,E). Overall, mean coefficients did not differ significantly from before versus after the disturbance for age-0 sharks (Student's t-test; differentials: PCL, t = −0.60, df = 9, P = 0.56; Mass, t = −0.16, df = 9, P = 0.88; gradients: PCL, t = −0.47, df = 9, P = 0.65). In general, then, selection on age-0 size traits was variable through time and not consistently different between time periods, a result confirmed by visual inspection of cubic splines (Fig. 7; also see DiBattista et al. 2007 for comparison).
Figure 6.
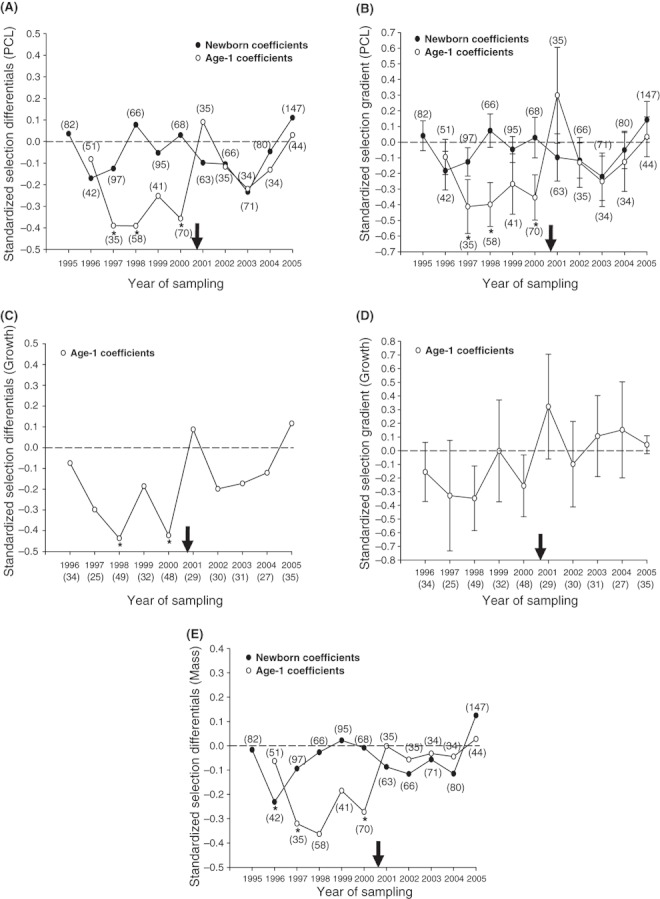
Linear (directional) selection coefficients acting on the length (A,B), mass (E), and growth rate (C,D) of newborn (i.e., age-0) and age-1 juvenile lemon sharks. Values are means ± 1 SEM. The black arrow indicates the approximate onset of disturbance on the x-axis.
Figure 7.
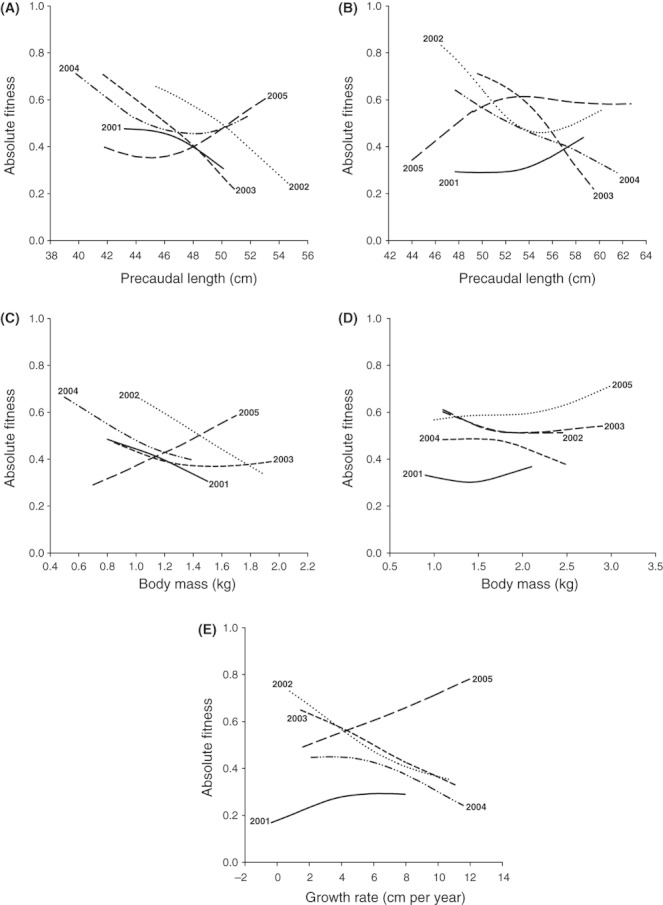
Relationship between initial precaudal length (A,B), body mass (C,D), or growth rate (E) and an individual's absolute fitness for each cohort of age-0 (A,C) and age-1 (B,D,E) juvenile lemon sharks. The lines are univariate cubic splines (see Schluter 1988). Growth was calculated for the interval preceding that (i.e., age-0 to age-1) over which selection was estimated (i.e., age-1 to age-2) and thus only available for age-1 juveniles.
Before the disturbance, age-1 selection coefficients were similar in sign to those for age-0 sharks, but much stronger. Coefficients for length, mass, and growth rate were negative in all cohorts from 1996 to 2000, and 10 of the 25 estimates were significant (Fig. 6). After the disturbance, only three out of five coefficients were negative for each PCL and mass (Fig. 6A,B,E), and only one out of five coefficients was negative for growth (Fig. 6D); none were statistically significant here (Fig. 6). Moreover, mean coefficients from before the disturbance were significantly higher for all traits (and thus stronger) than after the disturbance (Student's t-test; differentials: PCL, t = −2.75, df = 8, P = 0.025; Mass, t = −3.95, df = 8, P = 0.004; Growth, t = −2.35, df = 8, P = 0.047; gradients: PCL, t = −2.43, df = 8, P = 0.041; Growth, t = −3.45, df = 8, P = 0.009). In short, selection on size and growth was less negative and much weaker in age-1 sharks after the disturbance; cubic spline visualizations confirm these interpretations (Fig. 7; also see DiBattista et al. 2007 for comparison).
Discussion
Relative to before habitat loss at our main study site (North Sound, Bimini, Bahamas), samples afterward showed (i) an increase in the number of adult sharks successfully breeding, (ii) a modest transient increase in juvenile mortality, (iii) an increase in neutral genetic variation in the juvenile population, (iv) an increase in additive genetic and maternal effect variation for key juvenile life history traits, and (v) a dramatic change in selection on the same life history traits. Most of these results were not generally expected after habitat loss (Frankham 1995; Young et al. 1996; Cushman 2006; McClure et al. 2008), and so we now examine each in more detail and provide possible explanations. Although these explanations are necessarily speculative, we can at least eliminate some of them, leaving the remainder as viable hypotheses to help motivate and guide future work.
Number of breeders and juvenile mortality
Habitat loss is generally expected to cause a decrease in population size (Carvajal-Rodríguez et al. 2005; Johansson et al. 2007; McClure et al. 2008). In contrast to this straightforward expectation, we found that the number of breeding individuals has actually increased at Bimini after the disturbance (Fig. 4). To interpret this result, we must first recognize that previous expectations are generally based on situations where the segment of the population being considered is the same that which would be sensitive to the habitat loss, which is not the case here. We are in fact considering successful adult breeders that only use the site for parturition. Habitat loss would therefore not be expected to negatively impact the survival of these breeders. Instead, it might influence their choice of breeding site.
We can first see several possible explanations for why breeding population size has not decreased following habitat loss. The first hypothesis implies a constraint imposed by philopatry. That is, adult lemon sharks might continue to use nursery sites after a human-caused disturbance simply because past selection in formerly stable environments has led to the evolution of strong philopatry, even though the current disturbance would actually disfavor reproduction at that site (see Pärt 1994; Travis and Dytham 1999; Hendry and Stearns 2004). The second hypothesis is that the nursery site might not actually be degraded for juvenile lemon sharks or, at the very least, these individuals are initially resilient to the effects of disturbance. Indeed, juvenile mortality rates increased only slightly after the disturbance and have since largely recovered to predisturbance levels (Fig. 3). Juveniles have also been tracked within the most damaged areas of the nursery (S. Gruber unpublished data), with some individuals spending much of their time in those locations; high juvenile site fidelity coupled with high survival therefore suggests resilience to changing conditions.
We can also offer several hypotheses for why the size of the breeding population has actually increased. One possibility is that lemon sharks from other sites are increasingly attracted to Bimini. This implies that the disturbance increased carrying capacity or created new opportunities for mating, which seems unlikely in this species. Another possibility is that local recruitment to the breeding population has increased simply because of a run of good conditions 12–13 years earlier when the current adults were juveniles, although this cannot be tested because most current breeding adults would have been juveniles before our study period. And, of course, these possibilities may act in combination. For instance, a slight increase in local recruitment of philopatric females to the breeding population might attract an increasing number of nonphilopatric males from other sites (since mating occurs after parturition). Finally, the same number of adults might be returning to the nursery site but more of them might be successful in producing offspring. Regardless of the reason, however, an increase in the number of adults that successfully managed to reproduce at Bimini is expected to have consequences for genetic variation in this population.
Neutral genetic variation
Previous theoretical and empirical work has suggested that anthropogenic disturbances, particularly habitat loss or fragmentation, are often associated with reductions in neutral genetic variation (Lowe et al. 2005; DiBattista 2008). This was not the case in our study, wherein neutral genetic variation actually increased (Table 1 and Fig. 5). Moreover, opportunistic sampling at nearby undisturbed sites did not show a similar increase in neutral genetic variation over the same time period. In particular, we were able to catch and tag 189 newborn sharks at the south Bimini nursery between 1995 and 2007. This nursery site is six kilometers away from the North Sound disturbance area, directly across a wide sand flat that impedes water exchange, and the southern site is largely free from mangrove removal or reductions in seagrass (see Jennings et al. 2008). In contrast to the North Sound, we found no temporal change at the southern site in the number of alleles (mean number of alleles: t = −1.35, df = 10, P = 0.21; allelic richness: t = −0.21, df = 10, P = 0.84) or heterozygosity (i.e., observed heterozygosity: t = −0.55, df = 10, P = 0.60; expected heterozygosity: t = −0.52, df = 10, P = 0.62). In addition, FST (or FIS) was no different between time periods for this control site (see Appendix A). These finding suggest that factors specific to the North Bimini site caused the increase in genetic variation.
The simplest and most direct explanation is the aforementioned increase in breeding population size; declines in genetic variation are really only expected with declines in population size. We therefore here extend those previous discussions to a more explicit consideration of neutral genetic variation. First, it might simply take more time to observe decreases in neutral genetic variation (Lowe et al. 2005) – especially in long-lived philopatric adults that are not directly killed by the disturbance. This is certainly possible given the long generation time of lemon sharks (∼20 years, Schultz et al. 2008) and their late age-at-maturity (12–13 years, Brown and Gruber 1988). The potential buffering effect of a long generation time on changes in genetic variation has been inferred in other vertebrate taxa (Hailer et al. 2006; Lippé et al. 2006; Bishop et al. 2009), and so we favor this possibility. Of course, we also expect that adult breeding population size will have to decrease before appreciable losses in genetic variation are observed in the juvenile population. That said, juvenile lemon sharks clearly benefit from mangroves and seagrass beds (Feldheim and Edren 2002; Franks 2007; Wiley and Simpfendorfer 2007), and these habitats have clearly declined at the nursery site; we therefore cannot rule out future problems for the Bimini population (e.g.,Fahrig 2001; Wiegand et al. 2005).
Second, high levels of gene flow among nursery sites might buffer against losses in neutral genetic variation. In theory, even relatively low levels of gene flow between remnant populations can have this effect (Lowe et al. 2005). Although female lemon sharks seem philopatric, as discussed above, males are probably not – as seen in both the lack of genetic differentiation among nursery sites (Feldheim et al. 2001) and the fact that few males sire pups at Bimini more than once (Feldheim et al. 2004; DiBattista et al. 2009). Indeed, increasing male dispersal provides a reasonable explanation for why genetic variation increased through time at Bimini (Fig. 5), which could be investigated in future with additional exhaustive sampling at proximal nursery sites.
Additive genetic variance and maternal effects
Although we might have expected a decrease in additive genetic variance (VA) owing to disturbance (see Introduction), we instead observed an increase. Several potential explanations exist and we start by quickly listing those that are not particularly likely. First, genetic bottlenecks can sometimes convert epistatic variation into additive genetic variation (Bryant and Meffert 1990; Goodnight 1995; Merilä and Sheldon 1999). In our population, however, a bottleneck has not been evident. Second, VA can in theory increase under stressful conditions (e.g., food shortage), owing to increased mutation, selection, or the expression of cryptic genetic variation (Hoffman and Merilä 1999; McGuigan and Sgrò 2009). Field tests, however, have typically found that the VA of morphological traits is lower in poor environments (Charmantier and Garant 2005) and unfavourable conditions (Merilä 1997; Merilä and Sheldon 2001; Hendfickx et al. 2008). Third, different sample sizes (n = 583 before vs 768 after, see Table 2) might have caused lower statistical power before the disturbance, but this does not explain why the effect size (amount of change) is so large (Table 2). Fourth, VA might have increased simply because VP increased. Indeed, VP increased by 73% for PCL and 67% for mass, perhaps because of the increasing number of adults or increasing environmental variation. This does not provide the full answer, however, because the increase in VP was much smaller than the increase in VA (187% and 267% for PCL and mass, respectively).
What then is a reasonable explanation for the increase in VA, both in absolute terms and in proportion to other variance components? As above, an obvious possibility is the increasing number of adults contributing offspring to the Bimini nursery (see Fig. 4). This increase might have at least two effects. First, it might increase the total phenotypic variance in the parent pool (Table 2). Second, it might increase the relative contribution of VA because the new males might be immigrants originating from other nursery sites. Indeed, we have previously described dramatic variation in juvenile size and growth among lemon shark populations, suggesting the possibility of adaptive divergence in these traits (DiBattista et al. 2007, 2009). Here, then, is a possible example from nature where migration among populations in different selection regimes might increase additive genetic variation (also see Alleaume-Beharira et al. 2006; Lopez et al. 2008).
We now discuss the interesting increase in maternal effects, which again cannot be explained solely by the increase in phenotypic variation (Table 2). We previously showed that maternal effects represent an important source of phenotypic variation for early life-history traits in the lemon shark (DiBattista et al. 2009), which is not surprising given the year-long gestation period. One potential explanation for the increase in maternal effects variance after the disturbance is based on this close association between a mother and her offspring prior to birth. In resource limited or degraded environments, offspring might be more dependent on nutrition received from their mother during gestation (also see Charmantier and Garant 2005), particularly for species, such as lemon sharks, that do not show postnatal parental care (Pratt and Casey 1990). Indeed, a strong association between mother and offspring may act to reduce mismatches between traits and fitness in rapidly changing environments (Galloway 2005). Another explanation is that with the possibility of more adult individuals pairing after the disturbance, more dominance effects might be captured by VM + D (in addition to VA, since dominance was not modeled explicitly here; see Kozielska et al. 2003).
Natural selection
Anthropogenic disturbances can profoundly influence natural or sexual selection in wild populations (Stockwell et al. 2003; Hendry et al. 2006, 2008; Seehausen 2006; Darimont et al. 2009). These changes might then cause maladaptation that leads to population declines (e.g.,Both et al. 2006) and possible extirpations. To date, however, no studies have directly examined selection in natural populations both before and after habitat loss.
Before the onset of habitat loss at Bimini, natural (viability) selection favored small size and slow growth, particularly in the shark's second year of life; the main driver of this effect is thought to be predation (DiBattista et al. 2007). Indeed, foraging by juvenile lemon sharks typically takes place near the mangrove roots, which affords some protection from the pronounced inter- and intra-specific predation at this site (Morrissey and Gruber 1993; B. Franks unpublished data). Large and fast-growing juveniles, however, probably forage more frequently and in riskier situations (i.e., away from mangrove cover) to satisfy their greater metabolic requirements. This behavior should increase predation risk and therefore decrease their survival relative to small, slow growing individuals (likely predators at Bimini are not gape-limited). More generally, a number of studies have suggested that faster growing individuals are more susceptible to predators (Brown and Braithwaite 2004; Biro et al. 2004; Carlson et al. 2008).
After the onset of habitat loss, natural selection weakened considerably and no longer favored small size and slow growth (Figs 6 and 7). In fact, the last year in which selection was estimated (2005), coefficients were positive for both size and growth. This change is unlikely to be the result of random fluctuations because the difference was quite consistent across multiple years. We hypothesize that the most likely explanation is that habitat loss altered size-specific predation pressure. Given that some of the mangrove cover at Bimini has now been removed (Jennings et al. 2008) more individuals may need to forage away from the mangroves, thereby potentially ‘levelling the playing field’ with respect to size- and growth-related predation rates. Indeed, all the changes noted here are consistent with the scenario of facing a novel environment. Regardless of the specific mechanism, our study provides a clear example of how habitat loss can alter the fitness landscape experienced by natural populations. The consequences of this alternation remain to be seen.
Summary and conclusion
We have shown a human-induced shift in the fitness landscape for juvenile lemon sharks at Bimini — habitat loss has changed the pattern of natural selection. The long-term consequences of this change are not yet known, but they certainly warrant further investigation. In addition to possible changes in means and variances for the specific traits, the altered selection might influence demographic and evolutionary parameters.
To date, however, habitat loss appears not to have negatively affected the number of breeding adults, juvenile mortality rates, neutral genetic variation, or additive genetic variation for the studied traits. We have suggested a number of hypotheses, which warrant future investigation, for these initially unexpected results. It is also true that negative effects may only become evident with increasing time, as has been the case in other natural systems (Kuo and Janzen 2004; Goossens et al. 2005; Lippé et al. 2006), but they may also not appear at all.
A remaining question is if and how particular populations might adapt under the accelerated changes brought about by humans activities. This potential for evolutionary rescue depends on a host of factors that include population connectivity, initial population size, mortality rates, adaptive plasticity (or maternal effects), genetic variation, and the strength and form of selection (Bürger and Lynch 1995; Gomulkiewicz and Holt 1995; Boulding and Hay 2001; Price et al. 2003; Räsänen and Kruuk 2007; Orr and Unckless 2008; Bell and Gonzalez 2009). We considered many of these factors in our study population, which revealed several interesting patterns. These patterns raised new questions that might be profitable for integrating into future research efforts: (i) how does philopatry in long-lived species buffer or exacerbate the effects of environmental change at different temporal scales, and (ii) how does dispersal among populations maintain genetic variation that might otherwise be lost? Although these questions have been considered before, we suggest that they might be better incorporated into the study of evolutionary responses to environmental change.
Acknowledgments
This research was supported by the National Science Foundation Biological Oceanography Program under grant OCE-0623283 to S.G. and K.F. This study was also funded in part by a Natural Sciences and Engineering Research Council of Canada postgraduate fellowship to J.D.D., as well as by grants from the Company of Biologists, the Canadian Society of Zoology, the Bimini Biological Field Station, the Field Museum, the National Fish and Wildlife Foundation, Florida Sea Grant, National Geographic Society, University of Illinois at Chicago Research Foundation, Québec-Océans, and PADI's Project AWARE. We also thank Rose Mann and Lacey Hoover for their kind efforts to secure private funding, and we are indebted to the Hoover Foundation, and Drs. Tadashi and Toshi Fujino, for generous private support. We gratefully acknowledge the following corporate support: Mario Aiello, Davey Marine; Mercury Division, Brunswick Corporation; The Carolina Skiff and Sundance Boat corporations; Digital Angel Corporation (Destron), especially Sean Casey; Andrea Obrian, Bimini Island Air; and Cathy Bosch of Pelican Products. This research was carried out under a permit from the Department of Fisheries of the Commonwealth of the Bahamas (Michael Braynan, director). Thanks to the numerous students and volunteers who have diligently sampled over many years at Bimini, sometimes in inclement conditions. Thanks also to S. Kessel and D. Jennings for the photos of North Bimini. Genetic work was carried out in the Field Museum's Pritzker Laboratory for Molecular Systematics and Evolution, operated with support from the Pritzker Foundation. We finally thank two anonymous reviewers for their insightful comments, which greatly improved the quality of this work.
Appendix A
Mean number of alleles per locus (A), allelic richness (AR), the observed (HO) and expected (HE) heterozygosity, as well as FIS and FST for each loci in all newborn lemon sharks captured opportunistically at the South Bimini nursery from 1995 to 2000 (n = 75) versus from 2001 to 2007 (n = 113).
| 1995–2000 | 2001–2007 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Microsatellite ID | A | AR | HO | HE | A | AR | HO | HE | FIS | FST | |
| LS22 | 11 | 10.36 | 0.80 | 0.84 | 16 | 14.10 | 0.85 | 0.88 | 0.008 | 0.0003 | |
| LS30 | 11 | 9.39 | 0.76 | 0.68 | 11 | 9.50 | 0.73 | 0.69 | 0.039 | 0.004 | |
| LS48 | 20 | 18.85 | 0.96 | 0.92 | 22 | 19.66 | 0.91 | 0.92 | −0.001 | 0.0005 | |
| LS54 | 5 | 4.83 | 0.61 | 0.62 | 4 | 3.92 | 0.57 | 0.55 | 0.003 | 0.002 | |
| LS75 | 4 | 4 | 0.75 | 0.70 | 5 | 4.40 | 0.65 | 0.65 | 0.029 | 0.002 | |
| LS52 | 26 | 22.94 | 0.95 | 0.93 | 26 | 21.03 | 0.97 | 0.93 | −0.003 | 0.001 | |
| LS572 | 7 | 6.64 | 0.79 | 0.74 | 6 | 5.39 | 0.79 | 0.73 | 0.007 | 0.003 | |
| LS542 | 6 | 5.96 | 0.61 | 0.61 | 7 | 6.28 | 0.75 | 0.69 | −0.019 | 0.0005 | |
| LS596 | 12 | 11.19 | 0.89 | 0.86 | 12 | 11.56 | 0.86 | 0.86 | −0.009 | 0.0007 | |
| LS801 | 15 | 13.17 | 0.83 | 0.78 | 17 | 14 | 0.90 | 0.85 | −0.003 | 0.0001 | |
| LS560 | 9 | 8.81 | 0.83 | 0.81 | 8 | 7.39 | 0.93 | 0.82 | −0.007 | 0.001 | |
| Average | 11.45 | 10.56 | 0.80 | 0.77 | 12.18 | 10.66 | 0.81 | 0.79 | 0.004 | 0.001 | |
| SEM | 3.45 | 3.18 | 0.24 | 0.23 | 3.67 | 3.21 | 0.24 | 0.23 | 0.001 | 0.0005 | |
This sampling site is far removed from the development project (i.e., 6 km) and largely free from mangrove removal or reductions in seagrass (see Jennings et al. 2008); we therefore treat this as a control site.
Literature cited
- Alleaume-Beharira M, Pen IR, Ronce O. Geographical patterns of adaptation within a species’ range: interactions between drift and gene flow. Journal of Evolutionary Biology. 2006;19:203–215. doi: 10.1111/j.1420-9101.2005.00976.x. [DOI] [PubMed] [Google Scholar]
- Bell G, Gonzalez A. Evolutionary rescue can prevent extinction following environmental change. Ecology Letters. 2009;12:942–948. doi: 10.1111/j.1461-0248.2009.01350.x. [DOI] [PubMed] [Google Scholar]
- Biro PA, Abrahams MV, Post JR, Parkinson EA. Predators select against high growth rates and risk-taking behaviour in domestic trout populations. Proceedings of the Royal Society of London B. 2004;271:2233–2237. doi: 10.1098/rspb.2004.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop JM, Leslie AJ, Bourquin SL, O'Ryan C. Reduced effective population size in an overexploited population of the Nile crocodile (Crocodylus niloticus. Biological Conservation. 2009;142:2335–2341. [Google Scholar]
- Both C, Bouwhuis S, Lessells CM, Visser ME. Climate change and population declines in a long-distance migratory bird. Nature. 2006;441:81–83. doi: 10.1038/nature04539. [DOI] [PubMed] [Google Scholar]
- Boulding EG, Hay T. Genetic and demographic parameters determining population persistence after a discrete change in the environment. Heredity. 2001;86:313–324. doi: 10.1046/j.1365-2540.2001.00829.x. [DOI] [PubMed] [Google Scholar]
- Branstetter S. Early life history implications of selected Carcharinoid and Lamnoid sharks of the Northwest Atlantic. In: Pratt HL, Gruber SH, Taniuchi T, editors. Elasmobranchs as Living Resources. Washington, US: NOAA Technical Report 90; 1990. [Google Scholar]
- Brodie ED, Moore AJ, Janzen FJ. Visualizing and quantifying natural selection. Trends in Ecology and Evolution. 1995;10:313–318. doi: 10.1016/s0169-5347(00)89117-x. [DOI] [PubMed] [Google Scholar]
- Brown C, Braithwaite VA. Size matters: a test of boldness in eight populations of the poeciliid Brachyraphis episcopi. Animal Behavior. 2004;68:1325–1329. [Google Scholar]
- Brown CA, Gruber SH. Age assessment of the lemon shark, Negaprion brevirostris, using tetracycline validated vertebral centra. Copeia. 1988;1998:747–753. [Google Scholar]
- Bryant EH, Meffert LM. Multivariate phenotypic differentiation among bottleneck lines of the housefly. Evolution. 1990;44:660–668. doi: 10.1111/j.1558-5646.1990.tb05946.x. [DOI] [PubMed] [Google Scholar]
- Bürger R, Lynch M. Evolution and extinction in a changing environment: a quantitative-genetic analysis. Evolution. 1995;49:151–163. doi: 10.1111/j.1558-5646.1995.tb05967.x. [DOI] [PubMed] [Google Scholar]
- Caizergues A, Rätti O, Helle P, Rotelli L, Ellison L, Rasplus J. Population genetic structure of male black grouse (Tetrao tetrix L.) in fragmented vs. continuous landscapes. Molecular Ecology. 2003;12:2297–2305. doi: 10.1046/j.1365-294x.2003.01903.x. [DOI] [PubMed] [Google Scholar]
- Carlson SM, Olsen EM, Vøllestad LA. Seasonal mortality and the effect of body size: a review and an empirical test using individual data on brown trout. Functional Ecology. 2008;22:663–673. [Google Scholar]
- Carroll SP. Facing change: forms and foundations of contemporary adaptation to biotic invasions. Molecular Ecology. 2008;17:361–372. doi: 10.1111/j.1365-294X.2007.03484.x. [DOI] [PubMed] [Google Scholar]
- Carvajal-Rodríguez A, Rolán-Alvarez E, Caballero A. Quantitative variation as a tool for detecting human-induced impacts on genetic diversity. Biological Conservation. 2005;124:1–13. [Google Scholar]
- Chapman DD, Babcock EA, Gruber SH, DiBattista JD, Franks BR, Kessel SA, Guttridge T, et al. Long-term natal site-fidelity by immature lemon sharks (Negaprion brevirostris) at a subtropical island. Molecular Ecology. 2009;18:3500–3507. doi: 10.1111/j.1365-294X.2009.04289.x. [DOI] [PubMed] [Google Scholar]
- Charmantier A, Garant D. Environmental quality and evolutionary potential: lessons from wild populations. Proceedings of the Royal Society of London B. 2005;272:1415–1425. doi: 10.1098/rspb.2005.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagno LJV. FAO Species Catalogue: Sharks of the World. Rome, Italy: Food and Agriculture Organization of the United Nations; 1984. [Google Scholar]
- Crnokrak P, Roff DA. Inbreeding depression in the wild. Heredity. 1999;8:260–270. doi: 10.1038/sj.hdy.6885530. [DOI] [PubMed] [Google Scholar]
- Cushman SA. Effects of habitat loss and fragmentation on amphibians: a review and prospectus. Biological Conservation. 2006;128:231–240. [Google Scholar]
- Darimont CT, Carlson SM, Kinnison MT, Paquet PC, Reimchen TE, Wilmers CC. Human predators outpace other agents of trait change in the wild. Proceedings of the National Academy of Sciences. 2009;106:952–954. doi: 10.1073/pnas.0809235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBattista JD. Patterns of genetic variation in anthropogenically impacted populations. Conservation Genetics. 2008;9:141–156. [Google Scholar]
- DiBattista JD, Feldheim KA, Gruber SH, Hendry AP. When bigger is not better: selection against large size, high condition, and fast growth in juvenile lemon sharks. Journal of Evolutionary Biology. 2007;20:201–212. doi: 10.1111/j.1420-9101.2006.01210.x. [DOI] [PubMed] [Google Scholar]
- DiBattista JD, Feldheim KA, Thibert-Plante X, Gruber SH, Hendry AP. A genetic assessment of polyandry and breeding site fidelity in lemon sharks. Molecular Ecology. 2008a;17:3337–3351. doi: 10.1111/j.1365-294X.2008.03833.x. [DOI] [PubMed] [Google Scholar]
- DiBattista JD, Feldheim KA, Gruber SH, Hendry AP. Are indirect genetic benefits associated with polyandry? A test in a natural population of lemon sharks. Molecular Ecology. 2008b;17:783–795. doi: 10.1111/j.1365-294X.2007.03623.x. [DOI] [PubMed] [Google Scholar]
- DiBattista JD, Feldheim KA, Garant D, Gruber SH, Hendry AP. Evolutionary potential of a large marine vertebrate: quantitative genetic parameters in a wild population. Evolution. 2009;63:1051–1067. doi: 10.1111/j.1558-5646.2008.00605.x. [DOI] [PubMed] [Google Scholar]
- England PR, Osler GHR, Woodworth LM, Montgomery ME, Briscoe DA, Frankham R. Effects of intense versus diffuse population bottlenecks on microsatellite genetic diversity and evolutionary potential. Conservation Genetics. 2003;4:595–604. [Google Scholar]
- Fahrig L. How much habitat is enough? Biological Conservation. 2001;100:65–74. [Google Scholar]
- Feldheim KA, Edren SMC. Impacts of dredging on marine-communities — The Bimini lemon shark. Bahamas Journal of Science. 2002;9:28–35. [Google Scholar]
- Feldheim KA, Gruber SH, Ashley MV. Population genetic structure of the lemon shark (Negaprion brevirostris) in the western Atlantic: DNA microsatellite variation. Molecular Ecology. 2001;10:295–303. doi: 10.1046/j.1365-294x.2001.01182.x. [DOI] [PubMed] [Google Scholar]
- Feldheim KA, Gruber SH, Ashley MV. The breeding biology of lemon sharks at a tropical nursery lagoon. Proceedings of the Royal Society of London B. 2002;269:1655–1661. doi: 10.1098/rspb.2002.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldheim KA, Gruber SH, Ashley MV. Reconstruction of parental microsatellite genotypes reveals female polyandry and philopatry in the lemon shark, Negaprion brevirostris. Evolution. 2004;10:2332–2342. doi: 10.1111/j.0014-3820.2004.tb01607.x. [DOI] [PubMed] [Google Scholar]
- Frankham R. Conservation genetics. Annual Review of Genetics. 1995;29:305–327. doi: 10.1146/annurev.ge.29.120195.001513. [DOI] [PubMed] [Google Scholar]
- Frankham R. Relationship of genetic variation to population size in wildlife. Conservation Biology. 1996;10:1500–1508. [Google Scholar]
- Frankham R. Genetics and extinction. Biological Conservation. 2005;126:131–140. [Google Scholar]
- Franks BR. US: Drexel Univeristy; 2007. The spatial ecology and resource selection of juvenile lemon sharks (Negaprion brevirostris) in their primary nursery areas. PhD thesis. [Google Scholar]
- Fraser DJ, Hansen MM, Østergaard S, Tessier N, Legault M, Bernatchez L. Comparative estimation of effective population sizes and temporal gene flow in two contrasting population systems. Molecular Ecology. 2007;16:3866–3889. doi: 10.1111/j.1365-294X.2007.03453.x. [DOI] [PubMed] [Google Scholar]
- Galloway LF. Maternal effects provide phenotypic adaptation to local environmental conditions. New Phytologist. 2005;166:93–100. doi: 10.1111/j.1469-8137.2004.01314.x. [DOI] [PubMed] [Google Scholar]
- Garant D, Kruuk LEB. How to use molecular marker data to measure evolutionary parameters in wild populations. Molecular Ecology. 2005;14:1843–1859. doi: 10.1111/j.1365-294X.2005.02561.x. [DOI] [PubMed] [Google Scholar]
- Godt MJ, Johnson BR, Hamrick JL. Genetic diversity and population size in four southern Appalachian plant species. Conservation Biology. 1996;10:796–805. [Google Scholar]
- Gomulkiewicz R, Holt RD. When does evolution by natural selection prevent extinction? Evolution. 1995;49:201–207. doi: 10.1111/j.1558-5646.1995.tb05971.x. [DOI] [PubMed] [Google Scholar]
- Goodnight CJ. Epistasis and the increase in additive genetic variance: implications for phase 1 of Wright's shifting balance process. Evolution. 1995;49:502–511. doi: 10.1111/j.1558-5646.1995.tb02282.x. [DOI] [PubMed] [Google Scholar]
- Goossens B, Chikhi L, Jalil M, Acrenaz M, Lackman-Acrenaz I, Mohamed M, Andau P, et al. Patterns of genetic diversity and migration in increasingly fragmented and declining orang-utan (Pongo pygmaeus) populations from Sabah, Malaysia. Molecular Ecology. 2005;14:441–456. doi: 10.1111/j.1365-294X.2004.02421.x. [DOI] [PubMed] [Google Scholar]
- Gordon SP, Reznick DN, Kinnison MT, Bryant MJ, Weese DJ, Räsänen K, Millar NP, et al. Adaptive changes in life history and survival following a new guppy introduction. The American Naturalist. 2009;174:34–45. doi: 10.1086/599300. [DOI] [PubMed] [Google Scholar]
- Gruber SH, Parks W. Mega-resort development on Bimini: sound economics or environmental disaster. Bahamas Journal of Science. 2002;9:2–18. [Google Scholar]
- Gruber SH, De Marignac JRC, Hoenig JM. Survival of juvenile lemon sharks at Bimini, Bahamas, estimated by mark-depletion experiments. Transactions of the American Fisheries Society. 2001;130:376–384. [Google Scholar]
- Hailer F, Helander B, Folkestad AO, Ganusevich SA, Garstad S, Hauff P, Koren C, et al. Bottlenecked but long-lived: high genetic diversity retained in white-tailed eagles upon recovery from population decline. Biology Letters. 2006;2:316–319. doi: 10.1098/rsbl.2006.0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanski I, Saccheri I. Molecular-level variation affects population growth in a butterfly metapopulation. PLOS Biology. 2006;4:719–726. doi: 10.1371/journal.pbio.0040129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendfickx F, Maelfait JP, Lens L. Effect of metal stress on life history divergence and quantitative genetic architecture in a wolf spider. Journal of Evolutionary Biology. 2008;21:183–193. doi: 10.1111/j.1420-9101.2007.01452.x. [DOI] [PubMed] [Google Scholar]
- Hendry AP, Kinnison MT. Perspective: the pace of modern life: measuring rates of contemporary microevolution. Evolution. 1999;53:1637–1653. doi: 10.1111/j.1558-5646.1999.tb04550.x. [DOI] [PubMed] [Google Scholar]
- Hendry AP, Stearns SC, editors. Evolution Illuminated: Salmon and Their Relatives. New York: Oxford University Press; 2004. [Google Scholar]
- Hendry AP, Grant PR, Grant BR, Ford HA, Brewer MJ, Podos J. Possible human impacts on adaptive radiation: beak size bimodality in Darwin's finches. Proceedings of the Royal Society of London B. 2006;273:1887–1894. doi: 10.1098/rspb.2006.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry AP, Farrugia TJ, Kinnison MT. Human influences on rates of phenotypic change in wild animal populations. Molecular Ecology. 2008;17:20–29. doi: 10.1111/j.1365-294X.2007.03428.x. [DOI] [PubMed] [Google Scholar]
- Hoffman AA, Merilä J. Heritable variation and evolution under favourable and unfavourable conditions. Trends in Ecology and Evolution. 1999;14:96–102. doi: 10.1016/s0169-5347(99)01595-5. [DOI] [PubMed] [Google Scholar]
- Janzen FJ, Stern HS. Logistic regression for empirical studies of multivariate selection. Evolution. 1998;52:1564–1571. doi: 10.1111/j.1558-5646.1998.tb02237.x. [DOI] [PubMed] [Google Scholar]
- Jennings DE, Gruber SH, Franks BR, Kessel ST, Robertson AL. Effects of large-scale anthropogenic development on juvenile lemon shark (Negaprion brevirostris) populations of Bimini, Bahamas. Environmental Biology of Fishes. 2008;83:369–377. [Google Scholar]
- Johansson M, Primmer CR, Merilä J. Does habitat fragmentation reduce fitness and adaptability? A case study of the common frog (Rana temporaria) Molecular Ecology. 2007;16:2693–2700. doi: 10.1111/j.1365-294X.2007.03357.x. [DOI] [PubMed] [Google Scholar]
- Kalinowski ST. HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Molecular Ecology Notes. 2005;5:187–189. [Google Scholar]
- Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- Kingsolver JG, Hoekstra HE, Hoekstra JM, Berrigan D, Vignieri SN, Hill CE, Hoang A, et al. The strength of phenotypic selection in natural populations. The American Naturalist. 2001;157:245–261. doi: 10.1086/319193. [DOI] [PubMed] [Google Scholar]
- Kinnison MT, Hairston NG., Jr Eco-evolutionary conservation biology: contemporary evolution and the dynamics of persistence. Functional Ecology. 2007;21:444–454. [Google Scholar]
- Kinnison MT, Unwin MJ, Quinn TP. Eco-evolutionary vs. habitat contributions to invasion in salmon: experimental evaluation in the wild. Molecular Ecology. 2007;17:405–414. doi: 10.1111/j.1365-294X.2007.03495.x. [DOI] [PubMed] [Google Scholar]
- Kozielska M, Krzemińska A, Radwan J. Good genes and the maternal effects of polyandry on offspring reproductive success in the bulb mite. Proceedings of the Royal Society of London B. 2003;271:165–170. doi: 10.1098/rspb.2003.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruuk LEB. Estimating genetic parameters in natural populations using the “animal model”. Philosophical Transactions of the Royal Society of London B. 2004;359:873–890. doi: 10.1098/rstb.2003.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C, Janzen F. Genetic effects of a persistent bottleneck on a natural population of ornate box turtles (Terrapene ornata. Conservation Genetics. 2004;5:425–437. [Google Scholar]
- Lande R, Arnold SJ. The measurement of selection on correlated characters. Evolution. 1983;37:1210–1226. doi: 10.1111/j.1558-5646.1983.tb00236.x. [DOI] [PubMed] [Google Scholar]
- Leimu R, Mutikainen P, Koricheva J, Fischer M. How general are positive relationships between plant population size, fitness and genetic variation. Journal of Ecology. 2006;94:942–952. [Google Scholar]
- Lippé C, Dumont P, Bernatchez L. High genetic diversity and no inbreeding in the endangered copper redhorse, Moxostoma hubbsi (Catostomidae, Pisces): the positive sides of a long generation time. Molecular Ecology. 2006;15:1769–1780. doi: 10.1111/j.1365-294X.2006.02902.x. [DOI] [PubMed] [Google Scholar]
- Lopez S, Rousset F, Shaw FH, Shaw RG, Ronce O. Migration load in plants: role of pollen and seed dispersal in heterogeneous landscapes. Journal of Evolutionary Biology. 2008;21:294–309. doi: 10.1111/j.1420-9101.2007.01442.x. [DOI] [PubMed] [Google Scholar]
- Lowe AJ, Boshier D, Ward M, Bacles CFE, Navarro C. Genetic resource impacts of habitat loss and degradation: reconciling empirical evidence and predicted theory for neotropical trees. Heredity. 2005;95:255–273. doi: 10.1038/sj.hdy.6800725. [DOI] [PubMed] [Google Scholar]
- Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland: Sinauer; 1998. [Google Scholar]
- Lynch M, Conery J, Bürger R. Mutation accumulation and the extinction of small populations. The American Naturalist. 1995;146:489–518. [Google Scholar]
- Manire CH, Gruber SH. Effect of M-type dart tags on field growth of juvenile lemon sharks. Transactions of the American Fisheries Society. 1991;120:776–780. [Google Scholar]
- Marshall TC, Slate J, Kruuk LEB, Pemberton JM. Statistical confidence for likelihood based paternity inference in natural populations. Molecular Ecology. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- McClure MM, Carlson SM, Beechie TJ, Pess GR, Jorgensen JC, Sogard SM, Sultan SE, et al. Evolutionary consequences of habitat loss for Pacific anadromous salmonids. Evolutionary Applications. 2008;1:300–318. doi: 10.1111/j.1752-4571.2008.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuigan K, Sgrò CM. Evolutionary consequences of cryptic genetic variation. Trends in Ecology and Evolution. 2009;24:305–311. doi: 10.1016/j.tree.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Merilä J. Quantitative trait and allozyme divergence in the Greenfinch (Carduelis chloris, Aves: Fringillidae) Biological Journal of the Linnean Society. 1997;61:243–266. [Google Scholar]
- Merilä J, Sheldon BC. Genetic architecture of fitness and non-fitness traits — empirical patterns and development of ideas. Heredity. 1999;83:103–109. doi: 10.1046/j.1365-2540.1999.00585.x. [DOI] [PubMed] [Google Scholar]
- Merilä J, Sheldon BC. Avian quantitative genetics. Current Ornithology. 2001;9:179–255. [Google Scholar]
- Mitchell-Olds T, Shaw RG. Regression analysis of natural selection: statistical inference and biological interpretation. Evolution. 1987;41:1149–1161. doi: 10.1111/j.1558-5646.1987.tb02457.x. [DOI] [PubMed] [Google Scholar]
- Morrissey JF, Gruber SH. Habitat selection by juvenile lemon sharks, Negaprion brevirostris. Environmental Biology of Fishes. 1993;38:311–319. [Google Scholar]
- Orr HA, Unckless RL. Population extinction and the genetics of adaptation. The American Naturalist. 2008;172:160–169. doi: 10.1086/589460. [DOI] [PubMed] [Google Scholar]
- Pärt T. Male philopatry confers a mating advantage in the migratory collared flycatcher, Ficedula albicollis. Animal Behavior. 1994;48:401–409. [Google Scholar]
- Pelletier F, Clutton-Brock T, Pemberton J, Tuljapurkar S, Coulson T. The evolutionary demography of ecological change: linking trait variation and population growth. Science. 2007;315:1571–1574. doi: 10.1126/science.1139024. [DOI] [PubMed] [Google Scholar]
- Pratt HL, Jr, Casey JG. Shark reproductive strategies as a limiting factor in directed fisheries, with a review of Holden's method of estimating growth- parameters. In: Pratt HL, Gruber SH, Taniuchi Y, editors. Elasmobranchs as Living Resources. Washington, US: NOAA Technical Report 90; 1990. [Google Scholar]
- Price TD, Qvarnström A, Irwin DE. The role of phenotypic plasticity in driving genetic evolution. Proceedings of the Royal Society of London B. 2003;270:1433–1440. doi: 10.1098/rspb.2003.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Räsänen K, Kruuk LEB. Maternal effects and evolution at ecological time-scales. Functional Ecology. 2007;21:408–421. [Google Scholar]
- Raymond M, Rousset F. GENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Reznick DN, Ghalambor CK. The population ecology of contemporary adaptations: what empirical studies reveal about the conditions that promote adaptive evolution. Genetica. 2001;112-113:183–198. [PubMed] [Google Scholar]
- Ritland K. MARK—Genetic Marker Analysis Program. Vancouver, BC, Canada: University of British Columbia; 2004. [Google Scholar]
- Rountree RA, Able KW. Seasonal abundance, growth, and foraging habits of juvenile smooth dogfish, Mustelus canis, in a New Jersey estuary. US Fisheries Bulletin. 1996;94:533–534. [Google Scholar]
- Schluter D. Estimating the form of natural selection on a quantitative trait. Evolution. 1988;42:849–861. doi: 10.1111/j.1558-5646.1988.tb02507.x. [DOI] [PubMed] [Google Scholar]
- Schluter D. 2000. http://www.zoology.ubc.ca/~schluter/splines.html.
- Schultz JK, Feldheim KA, Gruber SH, Ashley MV, McGovern TM, Bowen BW. Global phylogeography and seascape genetics of the lemon shark (genus Negaprion. Molecular Ecology. 2008;17:5336–5348. doi: 10.1111/j.1365-294X.2008.04000.x. [DOI] [PubMed] [Google Scholar]
- Seehausen O. Conservation: losing biodiversity by reverse speciation. Current Biology. 2006;16:R334–R337. doi: 10.1016/j.cub.2006.03.080. [DOI] [PubMed] [Google Scholar]
- Stockwell CA, Hendry AP, Kinnison MT. Contemporary evolution meets conservation biology. Trends in ecology and evolution. 2003;18:94–102. [Google Scholar]
- Sunnucks P, Hales DF. Numerous transposed sequences of mitochondrial cytochrome oxidase I-II in aphids of the genus Sitobion (Hemiptera: Aphididae) Molecular Biology and Evolution. 1996;13:510–524. doi: 10.1093/oxfordjournals.molbev.a025612. [DOI] [PubMed] [Google Scholar]
- Travis JMJ, Dytham C. Habitat persistence, habitat availability and the evolution of dispersal. Proceedings of the Royal Society of London B. 1999;266:723–728. [Google Scholar]
- Wang J. Sibship reconstruction from genetic data with typing errors. Genetics. 2004;166:1963–1979. doi: 10.1534/genetics.166.4.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples RS. Genetic estimates of contemporary effective population size: to what time periods do the estimates apply? Molecular Ecology. 2005;14:3335–3352. doi: 10.1111/j.1365-294X.2005.02673.x. [DOI] [PubMed] [Google Scholar]
- White GC, Burnham KP. Program MARK: survival estimation form populations of marked animals. Bird Study. 1999;46:120–138. [Google Scholar]
- Wiegand T, Revilla E, Moloney KA. Effects of habitat loss and fragmentation on population dynamics. Conservation Biology. 2005;19:108–121. [Google Scholar]
- Wiley TR, Simpfendorfer CA. The ecology of elasmobranchs occurring in the everglades national park, Florida: implications for conservation and management. Bulletin of Marine Science. 2007;80:171–189. [Google Scholar]
- Young A, Boyle T, Brown T. The population genetic consequences of habitat fragmentation for plants. Trends in Ecology and Evolution. 1996;11:413–418. doi: 10.1016/0169-5347(96)10045-8. [DOI] [PubMed] [Google Scholar]


