Abstract
It is not clear which forms of plasticity in fitness-related traits are associated with invasive species. On one hand, it may be better to have a robust performance across environments. On the other, it may be beneficial to take advantage of limited favorable conditions. We chose to study a worldwide invasive species, Potamopyrgus antipodarum, and compare the plasticity of life-history traits of a sample of invasive genotypes to a sample of ancestral-range genotypes. We examined the responses to salinity in this freshwater snail because it varies spatially and temporally in the introduced range and contributes to variation in fitness in our system. We used a recently developed statistical method that quantifies aspects of differences in the shape among reaction norms. We found that the invasive lineages survived and reproduced with an increased probability at the higher salinities, and were superior to ancestral-range lineages in only two traits related to reproduction. Moreover, we found that in terms of traits related to growth, the invasive lineages have a performance optimum that is shifted to higher salinities than the ancestral-range lineages as well as having a narrower niche breadth. Contrary to the prediction of the general purpose genotype hypothesis, we found that invasive lineages tended to be opportunistic specialists.
Keywords: adaptation, invasive species, phenotypic plasticity
Introduction
When species expand their range or encounter dramatic environmental change, rapid adaptive phenotypic responses can prevent extinction and lead to demographic success. The capacity to produce adaptive phenotypic changes by phenotypic plasticity might be required to enhance the chances of persistence (Lande 2009). Dramatic environmental and range changes are often associated with species invasions. In fact, it has long been hypothesized that phenotypic plasticity increases invasion success (Baker 1965). However, we do not fully understand which forms of plasticity are associated with invasive species. Baker (1965) proposed the general purpose genotype hypothesis where a genotype exhibits an ability to produce different phenotypes across a range of environmental conditions that result in the maintenance of high fitness regardless of habitat. This hypothesis details just one kind of pattern of plasticity where the fitness of the genotype has little sensitivity to the environment. The pattern of environmental sensitivity of a genotype is commonly referred to as a reaction norm (Falconer and Mackay 1996). While it may be best if an organism can have a maximum fitness across all environments and little environmental sensitivity, physiological or morphological constraints may make this impossible. Given the particular conditions that are encountered, an organism may exhibit trade-offs between maximizing fitness in a particular environmental condition and maintaining a high performance across a range of conditions. This trade-off can be characterized as a contrast between the importance of robustness and opportunism (Richards et al. 2006).
Richards et al. (2006) identify different patterns of fitness plasticity that may be beneficial for invasion (Fig. 1A–C). These reaction norms or patterns of plasticity highlight a trade-off between robustness and opportunism. First, the fitness of an invasive genotype may vary little across environmental conditions but in a particular environmental condition the fitness may be lower than other non-invasive genotypes, therefore sacrificing opportunism for robustness (Fig. 1A: Jack-of-all-trades). In this case, invasive types have a broader niche breadth. Second, an invasive genotype may opportunistically take advantage of a favorable environmental condition, sacrificing robustness and leading to a narrower niche breadth (Fig. 1B: Master-of-some). Finally, it may be possible to relax this trade-off. In this case the genotype has both a robust fitness across harsh environmental conditions and can opportunistically take advantage of some conditions with a high fitness (Fig. 1C: Jack-and-Master). Here, the reaction norm of invasive types is shifted vertically relative to non-invasive types. It would be appropriate to consider this pattern to be produced by a general purpose genotype.
Figure 1.
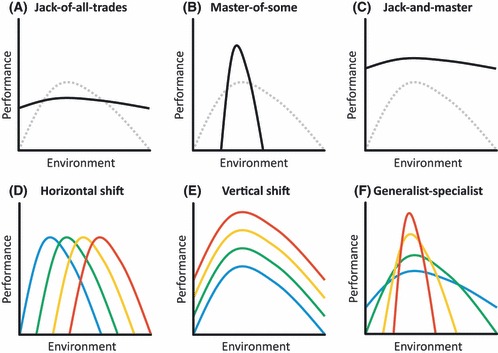
(A–C) Hypotheses of reaction norm variation contrasting the ancestral genotype (dashed gray lines) and the invasive genotype (solid black lines). (A) A Jack-of-all-trades invasive genotype may be robust to an environmental gradient and be able to maintain similar performance across conditions, leading to greater reaction norm breadth. (B) A Master-of-some invasive genotype may opportunistically take advantage of particular conditions and have high performance across a limited range of conditions, leading to narrower reaction norm breadth. (C) A Jack-and-Master invasive genotype relaxes this trade-off and is able to be robust to the environmental conditions and have a high performance, leading to a vertically shifted reaction norm. (D–F) Three modes of variation decomposed by TMV analysis. Shown are hypothetical reaction norms to emphasize differences in the shift. Colors represent four hypothetical genotypes. (D) Horizontal shifts are shown where the reaction norms differ only in the environment in which optimal performance occurs. (E) Vertical shifts are shown where the reaction norms differ in the mean performance across the environment. (F) Generalist-Specialist trade-off where the reaction norms differ in the breadth of high performance as well as differences between maximum and minimum performance.
Experimental studies of reaction norms often examine too few environmental conditions to fully distinguish between relevant differences in reaction norms, hence it is often difficult to objectively compare experimental data to these patterns (Izem and Kingsolver 2005). One way to empirically examine differences in reaction norms is to consider changes to a common underlying shape partitioned into three separate dimensions (Huey and Kingsolver 1989; Izem and Kingsolver 2005; Knies et al. 2006). Imagine a generalized shape of a reaction norm or performance curve where the fitness of a genotype increases across some environmental gradient to a point of maximum performance. Beyond this environmental condition, performance decreases across the gradient. An example of a trait that may display such a generalized shape would be thermal performance. The performance of a genotype might increase with increasing temperature up to a point at which the temperature degrades enzyme performance and results in a decline in performance (Knies et al. 2006). With this underlying model of a unimodal reaction norm, it is possible to describe three changes to this underlying shape.
Differences between the reaction norms of genotypes can be partitioned along three dimensions: horizontal shifts, vertical shifts, and generalist-specialist trade-offs (Izem and Kingsolver 2005). Horizontal shifts are differences in the environment in which optimal performance occurs (Fig. 1D). A horizontal shift means that some genotypes are able to exhibit their optimal fitness at a lower or higher value of an environmental gradient (Fig. 1D: contrast blue vs. red lines). Vertical shifts describe differences between genotypes in the mean performance across the environmental gradient (Fig. 1E). This means that some genotypes do better on average across the entire environmental gradient than other genotypes (Fig. 1E: contrast blue vs. red lines). Generalist-specialist trade-offs describe differences in the breadth of high performance as well as differences between maximum and minimum performance (Fig. 1F). This final trade-off describes how sharply the performance changes across favorable and unfavorable conditions of the environmental gradient. A generalist genotype will show a small difference between maximum and minimum performance and a nearly flat reaction norm across the environmental gradient (Fig. 1F: blue line). In contrast, a specialist will have a large difference of performance at the optimum environmental condition compared to the unfavorable conditions (Fig. 1F: red line). The performance will decline rapidly as the conditions change from the optimal or favorable condition. It is now possible to partition the variation among genotypes into these three shifts using a recently developed statistical method (Izem and Kingsolver 2005).
To determine which reaction norm shift is associated with successful invasive species, we chose to study Potamopyrgus antipodarum, an aquatic snail natively occurring in lakes and streams in New Zealand but a worldwide invader. Females are either obligately sexual or parthenogenetic, and clonal diversity is abundant and strongly structured geographically (Dybdahl and Lively 1995). Previous studies have characterized P. antipodarum in the ancestral-range as habitat specialists (Fox et al. 1996; Jokela et al. 1997). However, this species has also been introduced across the globe (e.g. Australia, western United States, and Europe) and become a highly successful invader across wide geographic ranges in the introduced locations. Studies suggest that some of these events may represent the independent colonizations of different genotypes from the ancestral-range of New Zealand (Zaranko et al. 1997; Schreiber et al. 1998; Richards et al. 2001; Stadler et al. 2005). We chose to examine responses to salinity because salinity varies spatially and temporally in the introduced range in the western United States and Europe and contributes to variation in fitness in our system (Jacobsen and Forbes 1997; Dybdahl and Kane. 2005). In addition, responses to salinity (i.e. an evolved response in plasticity) might have been important during dispersal and colonization that may have occurred via ballast water transfer (Zaranko et al. 1997). Studying a group of successful invasive genotypes and genotypes from the ancestral-range allows us the opportunity to look for general patterns of plasticity in invasive genotypes.
In this study, our goal is to understand how the reaction norms of fitness-related traits might differ between invasive genotypes and genotypes in the ancestral range across an environmental gradient of salinity. Are there differences in the invasive genotypes of P. antipodarum compared to the genotypes present in the ancestral-range of New Zealand? Specifically, we want to quantify differences in the form of phenotypic plasticity among these two samples: genotypes from the ancestral-range and the invaded range. We use the method described by Izem and Kingsolver (2005) to identify quantitative differences in the form of fitness plasticity of an invasive species. We then relate these quantitative shifts to the previously proposed reaction norm patterns of successful invasive species (Baker 1965; Richards et al. 2006).
Methods
Experimental setup
To compare fitness-related traits of invasive-range and ancestral-range genotypes, we obtained juvenile snails from lineages that had been isolated from natural populations in freshwater. The original laboratory populations were maintained in freshwater. The invasive-range lineage type was comprised of four clonal lineages: from two populations in the western US (Columbia River, WA and Snake River, ID), one from a population in Lake Ontario, NY, and one from a population from the Rhine River, Germany, referred to as clone A in Ponder (1988). The ancestral-range lineage type was comprised of clonal lineages that were obtained from lake populations on New Zealand's South Island: one from Lake Poerua, two from Lake Alexandrina, and one from Lake Mapourika (P32, AAI1, A14.1, and M2.3.29, respectively). Two ancestral-range sexual lineages were isolated: one from Lake Alexandrina (Alex stock SA) and one from Lake Ianthe (IAF4).
Juvenile snails were raised at five different salinity levels, ranging from freshwater to marine (0, 5, 10, 15, and 30 parts per thousand or ppt) for a maximum of 230 days. A total of 25 juvenile snails (1.0–1.5 mm shell length) were collected from each lineage, and five snails from each lineage were assigned to each of five salinity treatments. Juveniles were placed individually into plastic cups with freshwater (0 ppt). Snails in the higher salinity treatments were slowly acclimated over 2 days by transferring them every 6 h to successively higher salinities (+5 ppt) until their assigned treatment salinity were reached. Snails were fed Spirulina powder and the water from each cup was changed three times per week. Due to high mortality at 30 ppt for most lineage types, we were unable to use this treatment in comparative analyses of survival, growth, and reproduction.
Growth and reproduction measurement
Shell length was measured at the initiation of the experiment and every 2 weeks subsequently to determine growth rate. After 10 weeks, we collected measurements weekly and counted the number of offspring produced each week by each individual. For the sexual lineages, each female was provided with one male from the same population to mate with for the period of 2 weeks. Snails from the sexual lineages that were discovered to be males were removed from the analysis (n = 5). Because sexual females may have been limited by access to males, we excluded the ancestral-range sexual lineages when examining traits associated with reproduction. We terminated a snail from observation after 8 weeks of reproduction. We calculated the probability of survival of each type of snail (invasive clones, ancestral-range clones, and ancestral-range sexuals) per salinity treatment as the percent survival to the termination from observation.
We fit logistic curves to the growth data for each snail using the Nonlinear Least Squares method in Matlab (The Mathworks, Natick, MA, USA) following the methods in Dybdahl and Kane (2005). We used a logistic model because P. antipodarum stops growing and switches to reproduction at maturity (Jokela et al. 2003). We calculated individual growth rate as the change in length per day over the first 8 weeks of the experiment for all individuals that survived at least this long. We obtained estimates of asymptotic size (shell length) and time to asymptotic size (how long it took to reach that shell length) by the following method. We noted the shell length at the time of removal from the experiment from the logistic growth curve estimate. We then selected the earliest time point where the individual was within 0.2 mm of this shell length. The shell length and elapsed days at this point became the asymptotic size and time to asymptotic size.
For each lineage at each salinity level, we calculated the reproduction probability as the percent of individuals that both survived and reproduced for each type at each treatment level. Additionally, for only those individuals that reproduced, we calculated the time to first reproduction as the experimental day where the first offspring was observed. The shell length of the individual on that day was also recorded as the size at first reproduction. The total offspring production was calculated as the sum of offspring over the next 8 weeks regardless of whether or not there was continuous offspring output.
We estimated individual fitness (λ) as the age dependent population growth rate of an individual (McGraw and Caswell 1996; Caswell 2001). This multivariate method estimates the long term growth rate of a population as if it were composed of individuals with the characteristics measured. For each snail in each treatment we constructed an individual transition matrix of weekly fecundity and survival probabilities (McGraw and Caswell 1996). The first row consists of weekly fecundity rates. The remainder of the matrix is a diagonal representing the survival probability on a weekly basis. We calculated the individual fitness (λ) as the dominant eigenvalue of the matrix in Matlab (The Mathworks).
Statistical analysis
Our primary question was to identify differences among lineage types in the phenotypic plasticity of fitness-related traits. Hence, we considered individual genotypes as a random sample within each lineage type (invasive-range, ancestral-range). Salinity and lineage type were treated as fixed effects. We compared lineage types for the following set of traits: survival probability, probability of reproducing, growth rate, time to asymptotic shell size, time of first reproduction, shell size of first reproduction, and individual fitness.
To compare the survival probability among salinity treatments (0, 5, 10, 15 ppt) and between lineage types (invasive, ancestral-range clones, and ancestral-range sexual), we used a proportional hazard test (Cox regression) in SPSS 15 (SPSS Inc., Chicago, IL, USA). We used forward stepwise addition using likelihood ratio tests to compare models.
For traits related to reproduction, statistical analyses were computed with JMP® 8.0.1 (SAS Institute Inc, Cary, NC, USA). For probability of reproduction, defined as the fraction of individuals in the lineage type that survived and reproduced, we compared effects of salinity and lineage type using a generalized linear model with binomial error distribution and a logit fit. First, we calculated the fit of a model including all main effects and interactions (full model). We used likelihood ratio tests to find insignificant terms and selected the reduced model with the lowest corrected Akaike information criterion (AICc) score for final analysis.
For time of first reproduction and size of reproduction, we analyzed only the invasive and ancestral-range clonal lineage types across the lower salinities (range: 0–10 ppt) because the vast majority of individuals failed to reproduce at salinities higher than 10 ppt. We compared the effects of salinity and lineage type using a generalized linear model assuming a normal error distribution. We followed the same model selection procedure as previously described.
For our analysis of individual fitness as determined from our age-specific project matrices, we followed the advice of Caswell (2001) and used a bootstrap method to estimate 95% confidence intervals (Kalisz and McPeek 1992) as implemented in Matlab (The Mathworks). For each of 10 000 bootstrap samples, we drew individuals with replacement from a specific type and treatment level while maintaining sample size with the individual as the unit sampled. We then calculated 95% confidence using the bias corrected and accelerated percentile method (Efron 1987). Statistical comparisons were computed by permutation tests (Levin et al. 1996; Caswell 2001). To test for differences between type, we randomly permuted individuals between type but maintained salinity level. At each salinity level, we computed the squared difference in mean λ. We summed this squared difference across the four treatment levels (0–15 ppt). We created 10 000 permutation samples. We used this statistic to test the hypothesis that our measured difference was greater than what we observe by chance. To compare differences between type at specific salinity levels, we used a similar method. Here, individuals were randomly permuted between type at the specific salinity level. We computed the difference in mean λ of each new sample. We created 10 000 permutation samples. We used this statistic to test the hypothesis that our measured difference was greater than what we observe by chance.
Reaction norm decomposition
To characterize differences in the shape of the reaction norms among the different types (invasive vs. ancestral-range), we used a recently developed method called template mode variation (TMV) (Izem and Kingsolver 2005). This method works by fitting a common template or curve among all of the reaction norms. Next, the variation between types is decomposed or partitioned into three different changes of this common template. It is important to note that each change is given independent of the other changes. One of these changes is a difference in which environment produces optimal or maximum performance between the types (horizontal shift). Difference in the mean performance among types is a second kind of change (vertical shift). Thirdly, by adjusting the width of the common template while maintaining the area underneath (squeezing or stretching the curve) changes in niche breath can be estimated. This final variation has often been characterized as a generalist-specialist trade-off because it captures a trade-off between the maximum performance and the average performance. The TMV method allows one to partition the source of variance of reaction norms among types into these three separate categories (plus error): horizontal shifts, vertical shifts, and generalist-specialist trade-offs. In addition to this, for each type, one is able to obtain estimates of the magnitude of each shift independent of the other shifts. Horizontal shifts are measured in units of the environmental gradient (e.g. ppt salinity). Vertical shifts are defined in terms of the response measured (e.g. growth rate). Generalist-specialist trade-offs are a unitless measure (Izem and Kingsolver 2005).
We were able to use the TMV analysis to compare the growth rate (over first 8 weeks) and time to asymptotic size for two lineage types: invasive and ancestral-range (combined sexual and clonal). We combined the ancestral-range sexual with ancestral-range clonal lineages to increase the power of the statistical test. We transformed the time to asymptotic size by subtracting it from 230 days (maximum length of experiment) to produce reaction norms with a maximum value within the range of measured salinities which is an assumption of the TMV method (Izem and Kingsolver 2005). We fit third order polynomials to our reaction norms to conduct the TMV analysis. We were limited to this order by the number of salinity treatments (4). The TMV analysis was performed in Matlab (The Mathworks) using modified code obtained from Rima Izem (personal communication) that was developed for a previous publication (Izem and Kingsolver 2005).
Once we quantified each of the differences between the invasive and ancestral-range lineages, we related the results to the strategies of successful invasive genotypes proposed by Richards et al. (2006). If invasive types fit Jack-of-all-trades, we expect most of the variation in reaction norms to be explained by differences in niche breadth, with broader reaction norms for the invasives (Fig. 1A,F). If invasive types are Master-of-some, we expect most of the variation in reaction norms also to be explained by differences in niche breadth, with narrower reaction norms for invasives (Fig. 1B,F). If invasive types fit Jack-and-Master, we expect most of the variation in reaction norms to be explained by differences in vertical shift, with higher reaction norms for invasives (Fig. 1C,E). Other combinations of reaction norm shifts are possible, including horizontal shifts which suggest a change in the environmental conditions for optimal performance.
Results
Survival
Survival was significantly different among treatments and between types. We found that a fully saturated model of main effects and interactions was most appropriate (Table 1). From Fig. 2A, it is clear that low (0 ppt) and high salinities (15 ppt) are stressful conditions compared to the intermediate levels (5 and 10 ppt). The highest salinity had the most negative effect on survival across all types. The ancestral-range types, both clonal and sexual, performed worse at high salinity, compared to invasive types, although invasive types performed worse at the lowest salinity (Fig. 2A). While there was a significant interaction among the effects, we found it was due to differences between the invasive clones vs. ancestral types at only the lowest salinity (0 ppt) (P = 0.004).
Table 1.
Survival analysis
| Variables in models | Overall | Change from previous | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Salinity | Type | Salinity*Type | −LL | χ2 | d.f. | P-value | χ2 | d.f. | P-value |
| (A) Stepwise analysis of sources of variation for model | |||||||||
| + | − | − | 463.899 | 33.971 | 3 | <0.001 | 32.866 | 3 | 0.000 |
| + | + | − | 454.606 | 44.006 | 5 | <0.001 | 9.292 | 2 | 0.010 |
| + | + | + | 434.111 | 84.141 | 11 | <0.001 | 20.495 | 6 | 0.002 |
| Source | d.f. | P-value |
|---|---|---|
| (B) Full model breakdown of variation | ||
| Salinity | 3 | 0.002 |
| Type | 2 | 0.001 |
| Salinity*Type | 6 | 0.014 |
Salinity includes four treatment levels (0, 5, 10, and 15 ppt), Type includes three levels (Invasive clones, Ancestral clones, and Ancestral sexuals) and included a total of 195 observations. −LL is the −2*likelihood ratio and d.f. refers to the degrees of freedom. The null model −LL = 496.765.
Figure 2.
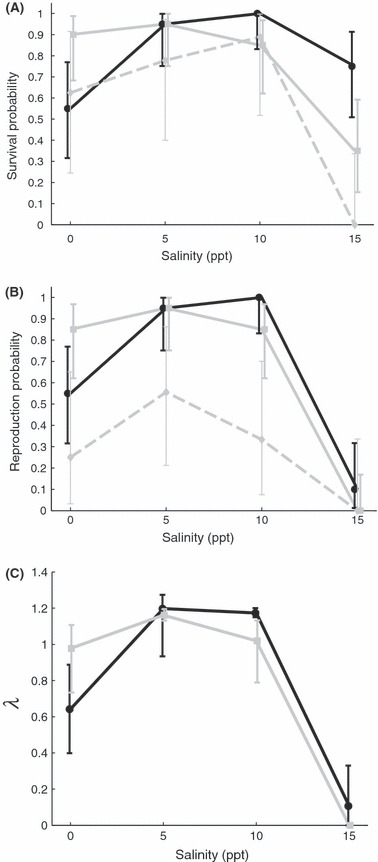
Analysis of survival and reproduction probability. Invasive type includes all four invasive lineages (solid black line and circles). Ancestral clones include all four ancestral clone lineages (solid grey line and squares) and ancestral sexuals includes both ancestral sexual lineages (dashed grey line and diamonds). Error bars show 95% confidence interval. (A) Plotted are the mean values of survival probability for each snail type in each salinity treatment. There was a significant interaction between type and salinity (P = 0.014). There was also a significant effect of type (P < 0.001) with the ancestral sexuals having a lower survival probability than the other types (invasive and ancestral clones). Salinity had a significant effect (P < 0.002) with the lowest (0 ppt) and highest (15 ppt) salinities having a lower survival probability than the intermediate salinities (5 and 10 ppt). (B) Plotted are the mean values of reproduction probability for each snail type in each salinity treatment. There was also a significant effect of type (P < 0.001) with the ancestral sexuals having a lower probability of reproduction than the other types (invasive and ancestral clones). There was a significant effect of salinity (P < 0.0001). Low salinity (0 ppt) had a negative effect of variable strength. The highest salinity (15 ppt) had a large negative effect on reproduction. (C) Plotted are the mean values of individual fitness (λ) for each snail type in each salinity treatment. There was a significant effect of type (P < 0.0248). Permutation tests revealed that the ancestral clones have a higher fitness at the lowest salinity (0 ppt) (P = 0.0351), but the invasive clones have a higher fitness at the intermediate salinity (10 ppt) (P = 0.0026).
Reproduction
For this analysis, we compared all types across the full salinity range (0–15 ppt). Our analysis on the probability of reproduction showed a significant effect of salinity (Table 2A). There was a sharp decline in the ability to reproduce occurring at the highest salinity (Fig. 2B). We found that ancestral-range sexuals had much lower probability of reproduction compared to both invasive and ancestral-range clonal lineages (Fig. 2B).
Table 2.
Analysis of reproductive traits
| Trait | Source | AICc | L-R χ2 | d.f. | P-value |
|---|---|---|---|---|---|
| (A) Probability of reproduction among all types | Full model | 152.1183 | 137.6289 | 11 | <0.0001 |
| Reduced model | 148.4101 | 128.0537 | 5 | <0.0001 | |
| Type | – | 26.5168 | 3 | <0.0001 | |
| Salinity | – | 107.2981 | <0.0001 | ||
| (B) Time of first reproduction among clonal types | Full model | 1037.0961 | 28.0800 | 5 | <0.0001 |
| Reduced model | 1033.8643 | 22.3948 | 1 | <0.0001 | |
| Type | – | 22.3948 | 1 | <0.0001 | |
| (C) Size at first reproduction among clonal types | Full model | 127.8856 | 47.2282 | 5 | <0.0001 |
| Reduced model | 123.3982 | 47.1671 | 3 | <0.0001 | |
| Type | – | 21.0314 | 1 | <0.0001 | |
| Salinity | – | 26.4824 | 2 | <0.0001 |
Full model is main effects plus interactions. Reduced model only contains significant effects which are listed immediately below. L-R χ2 is the likelihood ratio χ2 value and d.f. refers to the degrees of freedom. (A) Salinity includes four treatment levels (0, 5, 10, and 15 ppt), Type includes three levels (Invasive clones, Ancestral clones, and Ancestral sexuals) and includes a total of 195 observations. (B) Salinity includes three treatment levels (0, 5, and 10 ppt), Type includes two levels (Invasive clones and Ancestral clones) and included a total of 105 observations. (C) Salinity includes three treatment levels (0, 5, and 10 ppt), Type includes two levels (Invasive clones and Ancestral clones) and included a total of 105 observations.
When we compared the timing and size of first reproduction of the clonal types across the lowest salinity (0–10ppt), there was a significant difference between the invasive and ancestral-range clonal types (Table 2B,C). The invasive type reproduced earlier (Fig. 3A) and at a smaller size (Fig. 3B) across all salinities compared (0–10 ppt).
Figure 3.
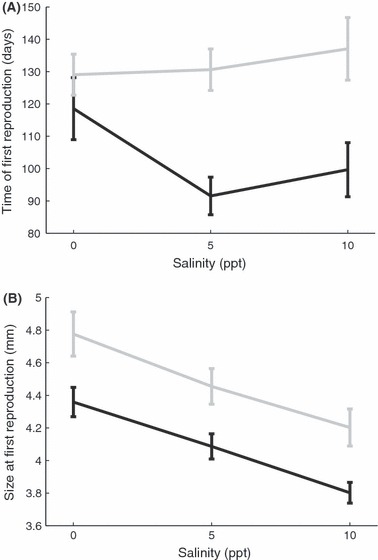
Analysis of specific traits related to reproduction. Invasive type includes all four invasive lineages (black lines and circles). Ancestral type includes all four clonal ancestral lineages (grey lines and squares). Error bars are ±1SE (A and B) or 95% confidence intervals (C). (A) Plotted are the mean values of time of first reproduction for each snail type in each salinity treatment. There was only a significant effect of type (P < 0.0001) with the invasive clones reproducing earlier across all salinities. (B) Plotted are the mean values of size at first reproduction for each snail type in each salinity treatment. There was a significant effect of type (P < 0.0001) and salinity (P < 0.0001) with the invasive clones reproducing at a smaller size across all salinities and snails reproducing at a smaller size as the salinity increased in concentration.
The analysis of individual fitness (λ) indicates the detrimental effect of high salinity (Fig. 2C), but we also found there is a significant difference between snail type (P = 0.0248) when tested with a permutation test. When we examined each salinity level individually, we found that at the lowest salinity (0 ppt) the ancestral clones had a higher individual fitness than the invasive lines (P = 0.0351). This relationship was reversed at the intermediate salinity (10 ppt), the invasive type had a higher individual fitness (P = 0.0026). At the other two salinity treatments, the types did not significantly differ (5 ppt: P = 0.3982; 15 ppt: P = 0.2394).
Growth
There are clear differences in the growth rate reaction norms of the invasive and ancestral types (Fig. 4A). The TMV analysis of the growth rate reaction norms suggests that most of the variation (68.65%) between the ancestral and invasive snail lineages is represented by a horizontal shift in optimum performance. Looking at the individual parameter estimates, one can see that the invasive types have a maximum growth rate at a higher salinity (∼7.5 ppt) compared to the ancestral types (∼3.9 ppt). This difference is evident in the reaction norm in Fig. 4A but is also clearly visualized in the reaction norm decomposition presented in Fig. 4C. Most of the remaining variation between the types was partitioned as a specialist-generalist trade-off (16.38%). The niche width, a unitless measure, of the specialist invasive lineages was smaller (0.94) compared to the generalist ancestral (1.05) (Fig. 4D). While the raw data does not make it obvious (Fig. 4A), very little of the variation (2.37%) is partitioned as a vertical shift or difference in mean performance (Fig. 4B).
Figure 4.
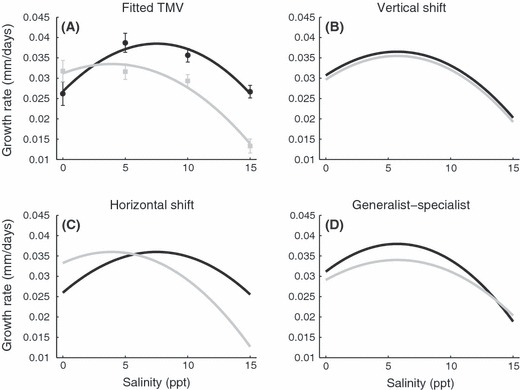
Common template fit (TMV) of growth rate. (A) Plotted are the mean values of growth rate ± 1SE in mm/day for each snail type in each salinity treatment (points) and fitted TMV curves (thick lines). Invasive type includes all four invasive lineages (black lines and circles). Ancestral type includes all four clonal ancestral lineages and both sexual lineages (grey lines and squares). (B–D) TMV decomposition of growth rate. (B) Differences in average performance (vertical shifts) accounted for 2.37% of the variation in reaction norms. The small difference is visualized with the invasive type having higher values (0.001 mm/day) than the ancestral type. (C) Differences in optimal salinity (horizontal shifts) accounted for 68.65% of the variation. The invasive type had a maximum growth rate at a higher salinity (7.5223 ppt) than the ancestral type (3.9136 ppt). (D) Differences in niche width (generalist-specialist trade-offs) accounted for 16.38% for the variation. The TMV analysis suggests that the invasive type is a specialist with a relative niche width of 0.9440 compared to the ancestral type with a niche width of 1.0536. The remaining 12.61% of the variation in reaction norms represented the error of the estimate.
Differences between the ancestral and invasive types in the time to asymptotic size were almost completely partitioned (88.04%) as a generalist-specialist trade-off by the TMV analysis (Fig. 5). As in the growth rate analysis, time to asymptotic size in the invasive type had a narrower niche width and their optimal salinity was higher than the ancestral type (Fig. 5B–D). Again, less than 1% of the variation was partitioned into a vertical shift in performance.
Figure 5.
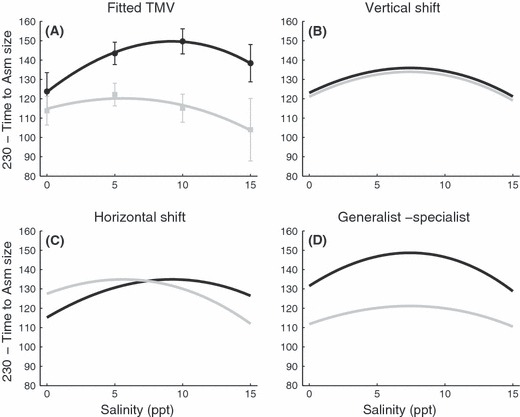
Common template fit (TMV) of time to asymptotic size. (A) Plotted are the mean values ± 1SE in days to reach asymptotic size subtracted from 230 for each snail type in each salinity treatment (points) and fitted TMV curves (thick lines). Invasive type includes all four invasive lineages (black lines and circles). Ancestral type includes all four clonal ancestral lineages and both sexual lineages (grey lines and squares). (B–D) TMV decomposition of time to asymptotic size. (B) Differences in average performance (vertical shifts) accounted for 0.77% of the variation in reaction norms. The small difference is visualized with the invasive type having higher values (2 days) than the ancestral type. (C) Differences in optimal salinity where asymptotic size occurs (horizontal shifts) accounted for 10.27% of the variation. The invasive type had a maximum at a higher salinity (9.2272 ppt) than the ancestral type (5.6059 ppt). (D) Differences in niche width (generalist-specialist trade-offs) accounted for 88.04% for the variation. The TMV analysis suggests that the invasive type is a specialist with a relative niche width of 1.4848 compared to the ancestral type with a niche width of 1.8215. The remaining 0.92% of the variation in reaction norms represented the error of the estimate.
Discussion
With the goal of understanding how the form of plasticity differs between invasive genotypes as compared to ancestral-range genotypes, we have characterized the reaction norms of survival, reproduction and growth across a salinity environmental gradient that is important to invasive potential. We found that the highest and lowest salinity environments were generally stressful, but performance depended on type. For survival probability, reproduction probability, and overall fitness (λ), invasive types tended to perform better at the higher salinities than ancestral range types, both sexual and clonal. Invasive types had a shorter time and smaller size to first reproduction at all salinities. TMV analysis for two measures of growth rate confirmed the reaction norm of invasive types tended to be narrower and shifted horizontally with maximum fitness at higher salinities. Overall, there was little support for the general purpose genotype hypothesis (vertical shift).
We quantitatively characterized differences in the reaction norms between invasive genotypes and genotypes in the ancestral-range. Richards et al. (2006) described three hypotheses or characterizations of reaction norms beneficial to invasion success: (i) Jack-of-all-trades, (ii) Master-of-some, and (iii) Jack-and-Master. The Jack-and-Master, is consistent with Baker's (1965) ideal invasive genotype, the general purpose genotype, because it relaxes this trade-off and produces high fitness in both stressful and favorable environmental conditions. We interpreted differences in the reaction norms of fitness-related traits between the invasive and ancestral types as three possible changes in performance (vertical shifts, horizontal shifts, or changes in niche breadth reflecting a generalist-specialist trade-off) (Izem and Kingsolver 2005). While different traits exhibited different combinations of shifts (e.g. horizontal and specialism), none were consistent with the Master-of-some or Jack-of-all-trades strategies. We saw no combination of the invasive type having a superior performance under favorable conditions and poor performance under stressful conditions (i.e. Master-of-some). We also did not observe the invasive type with a poor performance in favorable conditions and robust to the environmental condition (i.e. Jack-of-all-trades).
When species encounter novel environments during the course of a range expansion, plastic responses of the phenotype which maintains high fitness across environmental conditions would enhance the chance of persistence. Invasive species provide natural experiments to understand how reaction norms may be shaped during range expansion. The general purpose genotype hypothesis predicts that invasive genotypes should have a reaction norm that is robust across environmental conditions (Baker 1965), yet we uncovered little evidence to support this hypothesis in invasive lineages of P. antipodarum. Time to asymptotic size (i.e. time to reproductive maturity) was the only trait for which we observed most of the variation between types as a generalist-specialist trade-off (88%) using the recently developed methods of reaction norm decomposition (Izem and Kingsolver 2005). Contrary to the prediction, our evidence suggests that invasive lineages are specialized for high performance on a narrow range of salinities as compared to the wider reaction norm of the ancestral lineages. The same trend of the invasive type being a specialist was also observed in the growth rate data.
In general, we found that invasive types tended to display horizontally shifted reaction norms compared to ancestral types. Only two related traits, time and size at first reproduction, showed superiority across the environmental gradient. The invasive lineages were vertically shifted compared to the ancestral lineages. While not being completely robust to environmental conditions, they exhibited a similar robustness as the ancestral lineages as evident in the lack of significant interaction terms. The invasive type reproduces at smaller shell length and earlier in their lifespan compared to the ancestral clones in the timing and size of first reproduction. The invasive snails that could reproduce were able to take advantage of the conditions, both favorable and harsh, and were superior. Richards et al. (2006) suggests that this ability could be referred to as Jack-and-Master or a combination of both robustness and opportunism.
Shorter maturation time and smaller maturation size combined with a faster growth rate could represent a significant fitness benefit for the invasive type. Potamopyrgus antipodarum typically stops growing once it has reached reproductive size. If that size is smaller, it could reach this asymptotic size more quickly, even with a similar growth rate. These general qualities would be advantageous in invasive species. However, timing and size at first reproduction were only measured for snails that reproduced during the course of the experiment, representing only a small portion of the total sample. Other measures of fitness (λ), which did not show a vertical shift, contrast how populations might compare relative to one another. These specific traits of first reproduction only show how some individuals of the invasive type are superior to the ancestral type.
Among the clonal invasive-range and ancestral-range types, we observed an obvious difference in the level of salinity for which maximum survival occurred. The invasive lineages survive poorly at the lowest salinity where the ancestral-range clones do well. However, the invasive lineages were robust to the environmental stress of high salinity. We interpret the invasive-range type as showing a horizontal shift to a higher salinity optimum. The higher tolerance of the invasive types to increased salinity was also evident across other measures of performance. Our measure of individual fitness (λ) indicated that high fitness was maintained at a high salinity in the invasive type compared to the ancestral type. However, high fitness was not maintained in the invasive type at low salinity. The TMV analysis of growth rate attributed most of the variation between types as a horizontal shift (69%) and indicates a shift of maximum growth rate to a higher salinity environment. Beyond this, the reproductive ability was compromised in the ancestral types at the higher salinity levels, with the invasive types being the only lines successful at reproduction at the two highest levels (15 and 30 ppt).
We have shown that clones of P. antipodarum that have encountered new geographic range as invaders do not demonstrate characteristics of a general purpose genotype. In contrast, the invasive lineage shifts we measured are consistent with increased performance in stressful conditions, and we found limited evidence for increased performance across conditions. Our results are inconsistent with the hypothesis that invasive lineages are general purpose genotypes. Rather than having broad and flat reaction norms, our evidence suggests, at least with respect to this one environmental gradient, that invasive lineages are specialized as compared to the wide reaction norm of the ancestral lineages. Thus our results are broadly consistent with previous studies that suggest that clones of P. antipodarum are habitat specialists rather than general purpose genotypes (Dybdahl and Lively 1995; Fox et al. 1996; Jokela et al. 1997). This continues to raise the question as to how specialists genotypes have become so geographically widespread.
While some of the forms of plasticity we observed did not fit well with the previously established hypotheses, they are still potentially beneficial to invasion success. We observed shifts in the optimal salinity for survival, reproduction and growth. While these snails naturally occur in fresh water, it is possible that alternative environments would be encountered during the invasion process particularly early on or during transport (Zaranko et al. 1997). This high salinity environment could create strong selection pressures during the invasion event and result in lineages with the highest salinity tolerance surviving to colonize the invaded range. Here opportunism rather than robustness might be important if this high salin-ity environmental condition was the only one being encountered. Once in the invaded range, individuals may encounter a wide range of spatial heterogeneity in environmental conditions. General trends of increased performance in traits related to reproduction of the invasive type would give them an advantage across environment conditions that may be encountered (Jacobsen and Forbes 1997). Here robustness in fitness would be important during the course of their expansion across the invaded-range.
The goal of this paper was to quantify differences in plasticity among genotypes from two sources (ancestral and invaded range), which is the first step to understanding the potential origin of plasticity of an invasive species. The ability to compare invasive and ancestral-range genotypes within a species increased our ability to discern reaction norm differences while controlling for a number of potential differences that might have confounded our interpretation (e.g. phylogeny or evolutionary history). Sometimes comparisons of reaction norms are made between invasive species and other species in the invaded range. This kind of comparison specifically addresses how the shape of plasticity on the invasive species provides a competitive benefit over the local native species in the context of that particular invasion event. A comparison of a target invasive species and closely related non-invasive exotic species limits the influence of phylogenetic differences confounding plasticity differences. The comparison, ancestral-range vs. invaded-range, we used identified differences in the genotypes that may allow them to be more successful invaders whether it is the result of recent natural selection or existing variation already present in the ancestral-range. Detecting an evolved response in this way can be confounded by differences in genetic variation caused by founder effects.
The question remains is the pattern of phenotypic plasticity that we observe in an invasive genotype the direct rapid response to selection in the invaded range? If there is evidence of differences between ancestral-range genotypes and invasive ones, then it is possible the plasticity evolved in the invaded range by specific adaptations of genotypes (Richards et al. 2006). Alternatively, if there is no difference between the genotypes of the two origins, then non-evolutionary explanations may be important for explaining the potential success or failure of an invasive species (Facon et al. 2006). There could be increases of dispersal into the invaded range or the invaded range environmental may be modified allowing successful invasion. The design used in this paper was a comparison of invasive and ancestral-range genotypes. Other studies have used this kind of comparison to understand the source of plasticity in an invasive genotype (Kaufman and Smouse 2001; DeWalt et al. 2004). We were able to sample multiple genotypes from globally distributed invasive ranges, but to understand the origin of plasticity it would be ideal to sample genotypes in the ancestral-range from the proposed source of the invasive genotypes. It is possible that the invasive clones already had the observed plasticity in the ancestral-range. Even if this was the case, our results highlight key qualities of the invasive lineages that make them superior invaders. It remains an open question as to why these invasive lineages would have not expanded to become common across the ancestral-range.
Conclusions
In this paper, we provided evidence of a widely distributed invasive aquatic invertebrate that shows clear evidence of being an opportunistic specialist. In contrast to predictions that invasive lineages may represent general purpose genotypes, our analyses of several fitness-related traits across a salinity gradient indicate that invasive lineages are adapted to a higher salinity compared to lineages from the ancestral-range. In addition, the invasive lineages were marked by a narrowing of breadth in the reaction norms.
Acknowledgments
We wish to thank Jeremiah Busch, Richard Gomulkiewicz, Emily Jones, Ursel Schütte, and three anonymous reviewers for comments on the manuscript. We thank Brandon Dalton and Tiffany Smith for help in with snail care and growth measurement data collection. The National Science Foundation provided financial support to MFD, EPL, and DMD. EPL was supported by a Research Development Grant from Penn State Altoona. The IGERT Program in Evolutionary Modeling provided additional financial support to DMD.
Literature cited
- Baker HG. Characteristics and modes of origins of weeds. In: Baker HG, Stebbins GL, editors. The Genetics of Colonizing Species. New York, NY, USA: Academic Press; 1965. pp. 147–172. [Google Scholar]
- Caswell H. Matrix Population Models: Construction, Analysis, and Interpretation. 2nd edn. Sunderland, MA: Sianauer Associates; 2001. [Google Scholar]
- DeWalt SJ, Denslow JS, Hamrick JL. Biomass allocation, growth, and photosynthesis of genotypes from native and introduced ranges of the tropical shrub Clidemia hirta. Oecologia. 2004;138:521–531. doi: 10.1007/s00442-003-1462-6. [DOI] [PubMed] [Google Scholar]
- Dybdahl MF, Kane SL. Adaptation vs. phenotypic plasticity in the success of a clonal invader. Ecology. 2005;86:1592–1601. [Google Scholar]
- Dybdahl MF, Lively CM. Diverse, endemic and polyphyletic clones in mixed populations of a fresh-water snail (Potamopyrgus antipodarum. Journal of Evolutionary Biology. 1995;8:385–398. [Google Scholar]
- Efron Bradley. Better bootstrap confidence intervals. Journal of the American Statistical Association. 1987;82:171–185. [Google Scholar]
- Facon B, Genton BJ, Shykoff J, Jarne P, Estoup A, David P. A general eco-evolutionary framework for understanding bioinvasions. Trends in Ecology & Evolution. 2006;21:130–135. doi: 10.1016/j.tree.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. Essex: Longman Group Limited; 1996. [Google Scholar]
- Fox JA, Dybdahl MF, Jokela J, Lively CM. Genetic structure of coexisting sexual and clonal subpopulations in a freshwater snail (Potamopyrgus antipodarum) Evolution. 1996;50:1541–1548. doi: 10.1111/j.1558-5646.1996.tb03926.x. [DOI] [PubMed] [Google Scholar]
- Huey RB, Kingsolver JG. Evolution of thermal sensitivity of ectotherm performance. Trends in Ecology & Evolution. 1989;4:131–135. doi: 10.1016/0169-5347(89)90211-5. [DOI] [PubMed] [Google Scholar]
- Izem R, Kingsolver JG. Variation in continuous reaction norms: quantifying directions of biological interest. American Naturalist. 2005;166:277–289. doi: 10.1086/431314. [DOI] [PubMed] [Google Scholar]
- Jacobsen R, Forbes VE. Clonal variation in life-history traits and feeding rates in the gastropod, Potamopyrgus antipodarum: performance across a salinity gradient. Functional Ecology. 1997;11:260–267. [Google Scholar]
- Jokela J, Lively CM, Fox JA, Dybdahl MF. Flat reaction norms and ‘‘frozen’’ phenotypic variation in clonal snails (Potamopyrgus antipodarum) Evolution. 1997;51:1120–1129. doi: 10.1111/j.1558-5646.1997.tb03959.x. [DOI] [PubMed] [Google Scholar]
- Jokela J, Lively CM, Dybdahl MF, Fox JA. Genetic variation in sexual and clonal lineages of a freshwater snail. Biological Journal of the Linnean Society. 2003;79:165–181. [Google Scholar]
- Kalisz S, McPeek MA. Demography of an age-structured annual: resampled projection matrices, elasticity analyses, and seed bank effects. Ecology. 1992;73:1082–1093. [Google Scholar]
- Kaufman SR, Smouse PE. Comparing indigenous and introduced populations of Melaleuca quinquenervia (Cav.) Blake: response of seedlings to water and pH levels. Oecologia. 2001;127:487–494. doi: 10.1007/s004420000621. [DOI] [PubMed] [Google Scholar]
- Knies JL, Izem R, Supler KL, Kingsolver JG, Burch CL. The genetic basis of thermal reaction norm evolution in lab and natural phage populations. PLoS Biology. 2006;4:1257–1264. doi: 10.1371/journal.pbio.0040201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. Adaptation to an extraordinary environment by evolution of phenotypic plasticity and genetic assimilation. Journal of Evolutionary Biology. 2009;22:1435–1446. doi: 10.1111/j.1420-9101.2009.01754.x. [DOI] [PubMed] [Google Scholar]
- Levin L, Caswell H, Bridges T, DiBacco C, Cabrera D, Plaia G. Demographic responses of estuarine polychaetes to pollutants: life table response experiments. Ecological Applications. 1996;6:1295–1313. [Google Scholar]
- McGraw JB, Caswell H. Estimation of individual fitness from life-history data. American Naturalist. 1996;147:47–64. [Google Scholar]
- Ponder WF. Potamopyrgus antipodarum, a molluscan colonizer of Europe and Australia. Journal of Molluscan Studies. 1988;54:271–286. [Google Scholar]
- Richards DC, Cazier LD, Lester GT. Spatial distribution of three snail species, including the invader Potamopyrgus antipodarum, in a freshwater spring. Western North American Naturalist. 2001;61:375–380. [Google Scholar]
- Richards CL, Bossdorf O, Muth NZ, Gurevitch J, Pigliucci M. Jack of all trades, master of some? On the role of phenotypic plasticity in plant invasions. Ecology Letters. 2006;9:981–993. doi: 10.1111/j.1461-0248.2006.00950.x. [DOI] [PubMed] [Google Scholar]
- Schreiber ESG, Glaister A, Quinn GP, Lake PS. Life history and population dynamics of the exotic snail Potamopyrgus antipodarum (Prosobranchia : Hydrobiidae) in Lake Purrumbete, Victoria, Australia. Marine and Freshwater Research. 1998;49:73–78. [Google Scholar]
- Stadler T, Frye M, Neiman M, Lively CM. Mitochondrial haplotypes and the New Zealand origin of clonal European Potamopyrgus, an invasive aquatic snail. Molecular Ecology. 2005;14:2465–2473. doi: 10.1111/j.1365-294X.2005.02603.x. [DOI] [PubMed] [Google Scholar]
- Zaranko DT, Farara DG, Thompson FG. Another exotic mollusc in the Laurentian Great Lakes: The New Zealand native Potamopyrgus antipodarum (Gray 1843) (Gastropoda, Hydrobiidae) Canadian Journal of Fisheries and Aquatic Sciences. 1997;54:809–814. [Google Scholar]


