Abstract
Biological invasions have significant ecological, evolutionary and economic consequences. Ants are exemplary invaders and their invasion success is frequently attributed to a shift in social structure between native and introduced populations. Here, we use a multidisciplinary approach to determine the social structure, origin and expansion of the invasive Argentine ant, Linepithema humile, in Australia by linking behavioural and genetic studies with indicators of dispersal pathways and propagule pressure. Behavioural assays revealed a complete absence of aggression within and between three cities – Melbourne, Adelaide and Perth – spanning 2700 km across Australia. Microsatellite analyses showed intracity genetic homogeneity and limited but significant intercity genetic differentiation. Exceptions were two Perth nests that likely represent independent translocations from Adelaide. These patterns suggest efficient local gene flow with more limited jump dispersal via transport corridors between cities. Microsatellite analyses of L. humile from potential source regions, combined with data from port interceptions, trade pathways and the timeline of spread within Australia, implicate the main European supercolony as the source of L. humile in Melbourne. Such an introduction probably then redistributed across Australia and spread to New Zealand to form an expansive Australasian supercolony.
Keywords: biological invasions, intraspecific aggression, invasion history, invasive ants, Linepithema humile, microsatellites, source populations
Introduction
Biological invasions, a key component of global change, have significant ecological, evolutionary and economic consequences (Vitousek et al. 1997). The biology of native and introduced populations of invasive species can differ as a result of genetic changes associated with introduction events, affecting invasive success and impact (Sakai et al. 2001). Molecular techniques and behavioural studies play an important role in providing insight into the introduction history of invasive species, taking advantages of changes after introduction to estimate social structure, source populations, phylogeographic relationships between populations, and genetic diversity within and between populations that can reflect the severity of founder effects (Holway and Suarez 1999; Lee 2002). Integrating these studies with economic indicators of invader movement (e.g., port interception records for unintentional introductions; pathways and volumes of continental and international trade) can facilitate reconstruction of invasion histories (Puth and Post 2005; Hulme 2009).
Among invertebrates, ants are exemplary invaders (Holway et al. 2002a). Ecological success and impact in their introduced ranges is frequently attributed to a shift in social structure between the native and introduced ranges. Most invasive ants studied to date are polygnous (i.e. multiple-queened), polydomous (i.e. multiple nests), unicolonial and able to form supercolonies, colonies that are so large that direct cooperative interactions between workers from separated nests become impossible (Helantera et al. 2009). The formation of these supercolonies may reduce costs linked with territoriality and result in high worker densities, thus promoting ecological dominance and impact in the introduced ranges (Holway et al. 1998), most notably reductions in native ant fauna diversity and abundance, impacts on some nonant invertebrates and vertebrates, and disruption to ecosystem processes (Lach and Hooper-Bui 2010).
The Argentine ant (Linepithema humile) exemplifies a successful ant invader. Native to the Paraná River drainage system in southern South America, L. humile has been introduced throughout the world via human trade and established on six continents and many islands (Suarez et al. 2001; Wild 2004). Native populations are characterized by small mutually antagonistic colonies but sometimes form highly localized supercolonies tens to hundreds of metres in size (Tsutsui et al. 2000; Heller 2004). These native colonies are typically genetically differentiated from each other (Tsutsui et al. 2000; Pedersen et al. 2006) and form closed breeding units, each possessing a single mitochondrial DNA haplotype (Vogel et al. 2009).
In contrast, in the introduced ranges, L. humile forms unicolonial populations or supercolonies that are several magnitudes of order larger than those in the native range, which lack territorial behaviour and aggression, sometimes extending across hundreds to thousands of kilometres (e.g., California: Suarez et al. 1999; Europe: Giraud et al. 2002). In some regions, localized supercolonies are also present, probably the result of further primary introductions from the native range or secondary introductions from other introduced ranges (California: Suarez et al. 2002; Catalonia: Giraud et al. 2002; Japan: Sunamura et al. 2009b; south-eastern United States: Buczkowski et al. 2004). Introduced populations typically display limited genetic differentiation and possess a single haplotype across much larger areas than that seen in native populations (Tsutsui and Case 2001; Giraud et al. 2002; Vogel et al. 2010).
In Australia, the first verified discovery of L. humile in Australia was made in 1939 (and maybe as early as 1931 –Wetterer et al. 2009) in the eastern suburbs of Melbourne, Victoria (Pasfield 1968). Thereafter, its range expanded rapidly across southern Australia [Western Australia (1941), New South Wales (1950), Tasmania (1951), South Australia (1979) and Queensland (2002)] (Suhr et al. 2009). Across the continent, L. humile displaces native ant species (Victoria –Rowles and O'Dowd 2007, 2009a; South Australia –Walters 2006; Western Australia –Heterick et al. 2000; Thomas and Holway 2005), which can disrupt native seed dispersal mutualisms and facilitate dispersal of some invasive plant species (Rowles and O'Dowd 2009b).
In this study, we use intraspecific aggression assays and 11 microsatellite markers to determine the behaviour and genetic structure of L. humile within and between three cities – Melbourne, Adelaide and Perth – spanning 2700 km across southern Australia. We integrate this with genetic data of 11 potential source populations from the native and other introduced ranges, 20 years of Australian port interception records of the Argentine ant, and Australian and international trade pathway information. This multidisciplinary approach helped us to make inferences about the origin and expansion of the L. humile invasion in Australia, understand patterns in social structure of introduced populations of L. humile around the globe and reconstruct worldwide movement patterns of this important invader beyond what could be inferred from behaviour and genetic structure alone (e.g., Sunamura et al. 2009a; van Wilgenburg et al. 2010; Vogel et al. 2010).
Materials and methods
Sampling
Linepithema humile workers were collected across southern Australia in 2004 and 2005. Fifteen workers from each of eight nests were sampled in each of three cities: Melbourne (nests ME1 to ME8, all between 4–72 km apart), Adelaide (AD1–8, 1–8 km apart) and Perth (PE1–8, 4–51 km apart). Two nests were sampled from each of the northern, eastern, southern and western urban quadrants. Sampling sites included sidewalks, residential gardens, nature reserves, industrial buildings, university grounds and plant nurseries.
Fifteen workers were also obtained from each of two nests in the native range in Argentina – Ita Ibate (II) and Costanera Sur (CS), and nine nests from other introduced ranges in Spain from Girona (EU) and Barcelona (CT), part of the dominant main European and smaller and restricted Catalonian supercolonies, respectively (Giraud et al. 2002; C Gómez pers. comm.), in the United States two from California, La Jolla (CA1) and Davis (CA2) – both part of the large Californian supercolony (Tsutsui et al. 2000; ND Tsutsui pers. comm.), and one from North Carolina in Raleigh (NC), two in New Zealand from Auckland (NZ1) and Wellington (NZ2) – both part of the New Zealand supercolony (Corin et al. 2007a), one in South Africa from Brackenfell in the Cape region (SA), and one from the island of Saint Helena in the mid-Atlantic Ocean (SH).
Intraspecific aggression
Intraspecific aggression in L. humile across southern Australia was assessed using a standard live 1-1 aggression assay (Holway et al. 1998). Assays were performed from 1 day to within a week of collection between all pairwise combinations of eight nests within each city and four randomly selected nests between each city. For both intracity and intercity comparisons, we placed single workers from queen-right nest pairs into an 8-mL (1 cm diameter × 5 cm tall) Fluon™-coated (Northern Products, Inc., Woonsocket, RI, USA) glass vial for 5 min. Interactions were scored at 30-s intervals where 0 = ignore (no interaction), 1 = touch (antennation), 2 = avoid (antennation and one or both retreat in opposite direction), 3 = aggression (raising of gaster) and 4 = fighting (extended aggression including biting, pulling and using chemical defence compounds). Five trials were carried out for each nest pair, and workers were only used once. A score of three or higher was considered aggressive behaviour.
Microsatellite analysis
To quantify genetic structure in L. humile from each of 24 Australian nests and 11 nests from potential source populations, we genotyped 15 ants per nest at 11 polymorphic microsatellite loci: Lhum-11, Lhum-13, Lhum-14, Lhum-19, Lhum-28, Lhum-33, Lhum-35, Lhum-39, Lhum-52, Lihu-H and Lihu-T1 (Krieger and Keller 1999; Suarez et al. 1999; Ingram and Palumbi 2002). Workers were placed in 100% EtOH and stored at −80°C following collection. DNA was extracted individually using the chelex extraction method (Ingram and Gordon 2003) and stored at −20°C.
Polymerase chain reaction (PCR) amplifications were performed in 25 μL reactions with 2.5 μL reaction buffer (1×), 2.5 μL MgCl2 (2 mm), 2 μL dNTPs (0.2 mm), 0.05 μL labelled IR-dye forward primer (1 pmol), 0.45 μL unlabelled forward primer (10 pmol), 0.5 μL unlabelled reverse primer (10 pmol), 14.5 μL ddH20, 0.5 μL Taq DNA polymerase (1 μg/μL) and 2 μL DNA template in the Applied Biosystems PCR Gene Systems 2700 (Life Technologies, Inc., Carlsbad, CA, USA). PCR cycle parameters were 95°C (2 min), 36 cycles of denaturation of 95°C (30 s), annealing (Lhum-11: 55°C, Lhum-13, Lhum-14 and Lhum-19: 53°C, Lhum-39 and Lihu-T1: 58°C, and Lhum-28, Lhum-33, Lhum-35, Lhum-52 and Lihu-H: 60°C) (1 min), 72°C (3 min) and a final extension step 72°C (2 min). PCR products were diluted between 1:1 and 1:3 with ddH20, run on a LI-COR® 4200 Global Edition IR2 system (LI-COR Biosciences, Inc., Lincoln, NE, USA) and analysed using SAGA 2.1 software (LI-COR Biosciences, Inc., Lincoln, NE, USA).
Genetic diversity and structure
The number of alleles (A) and expected heterozygosities (HE) were calculated for all nests in Australia and from potential source populations (n = 35) using Genepop 3.4 (Raymond and Rousset 1995). Lhum-33 was monomorphic in all Australian nests and subsequently excluded from intra-Australia analyses. We report the mean HE values for each city population and Australia overall by averaging HE values across nests within cities and across Australia. All nests and loci were examined for departures from Hardy–Weinberg proportions and linkage equilibrium. Given the large number of tests performed, a few departures from Hardy–Weinberg proportions were expected but linkage disequilibrium was not detected. Measures of allelic richness were standardized for sample size using a rarefaction method implemented in HP-Rare (Kalinowski 2005).
Across Australia and globally, pairwise genetic differentiation between Australian nests and nest groups and potential source populations were estimated using FST (Wright 1965), the significance of the differentiation being tested using 10 000 permutations (Arlequin 3.01: Excoffier et al. 2005). Australian nests were pooled into five nest groups corresponding to genetic substructure revealed in intra-Australia analyses: Melbourne (number of nests = 8), Adelaide (n = 8), Perth (n = 6), PE2 (n = 1) and PE8 (n = 1). Spatial hierarchical analyses of molecular variance (AMOVA) among workers within nests (intranest), nests within cities (intracity) and nests between cities (intercity) across Australia were carried out based on FST (Weir and Cockerham 1984) in Arlequin 3.01. Statistical significances of variance components were assessed based upon 10 000 permutations.
Principal component analysis (PCA) of allelic frequency data was performed to visualize genetic relationships among Australian nests alone, and between Australian nest groups and potential source populations using PCAgen (developed by J. Goudet, http://www2.unil.ch/popgen/softwares/pcagen.htm). The percentage of inertia of each PCA axis and the significance of each principal component were assessed from 10 000 randomizations. The Bayesian model-based clustering method (BAPS 5.2: Corander et al. 2004, 2003) was used to estimate genetic substructure among L. humile nests in Australia and potential source populations. Using the group-level option, workers within nests were merged into single samples. The maximum number of clusters (K) was set to the number of nests sampled, 24 for the Australian analysis and 35 for all Australian and potential source population nests. In both cases, the analysis was repeated ten times to ensure consistent and robust results.
Unrooted consensus trees (additive tree model) based on Cavalli-Sforza's chord measure of genetic distance (Cavalli-Sforza and Edwards 1967) were constructed in PHYLIP (v3.68; Felsenstein 2008) to depict the relationships among Australian nests, and between Australian nest groups and potential source populations. For each tree, 2000 bootstrapped allele frequency datasets were constructed (using SEQBOOT), genetic distances calculated (using GENDIST) and trees constructed (using FITCH). The program CONSENSE generated the final consensus tree using the majority rule criterion (>50% bootstrap values).
A Bayesian assignment test implemented in GENECLASS2 (Piry et al. 2004) was run to assess the probability of Australian nest groups belonging to one of the potential source populations (the ‘reference’ populations). We used the Rannala and Mountain (1997) criterion method of assignment based on group allele frequencies and set the threshold to 0.001 (i.e. the sample group fits the reference population with greater than 99% likelihood). As Australia has been identified as the source of the New Zealand supercolony (Corin et al. 2007b), the test was run with and without the two New Zealand populations.
Interception records and trade pathways
We analysed detailed port interception records for L. humile made by the Australian Quarantine and Inspection Service between 1988 and 2007. Analysis was restricted to records (n = 62) with Melbourne, Adelaide or Perth as the port of entry (POE) and only records originating from countries where L. humile is known to occur were included. We used volumes of nonbulk freight movements between Melbourne, Adelaide and Perth to help infer likely movement pathways of L. humile across southern Australia (DOTARS 2007). We also used percentages of the total import value ($) for main international trading partners of Australia to infer likely source regions for the introduction of L. humile over three periods of time – 1937–1939, 1966–1968 and 2006–2007 (Meredith and Dyster 1999; ABS 2007).
Results
Intraspecific aggression
None of the intraspecific aggression trials among L. humile workers from intracity (n = 540 trials) or intercity (n = 240 trials) nest pairs across southern Australia resulted in aggression. The highest level of interaction observed was 1 (touch, nonaggressive). Consequently, aggression scores showed no correlation with geographical distance between nest pairs even across large distances (Fig. 1).
Figure 1.
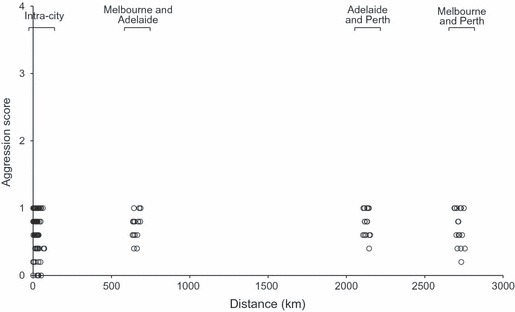
Mean level of intraspecific aggression between Linepithema humile workers for intracity (1–72 km apart) and intercity (634–2757 km apart) comparisons of paired nests (n = 5 assays for each pairwise comparison).
Genetic diversity and structure
A total of 59 alleles were found across 10 microsatellite loci when genotypes were assessed from 24 L. humile nests (15 ants per nest) across southern Australia. Similar numbers of alleles were detected in each city (42, 49 and 46 alleles in Melbourne, Adelaide and Perth, respectively). The total number of alleles per locus varied from 2 to 15 (Table 1). Most alleles were shared across the continent, with intracity nest pairs sharing on average 72% of alleles and intercity nest pairs sharing 61% of alleles. When intracity nests were pooled into population samples, 13 private alleles were identified (Melbourne = 5 alleles, Adelaide = 6, Perth = 2) and over half of these originated from a single locus, Lhum-35. For the whole Australian dataset, expected heterozygosities (HE, taking the Wahlund effect into account) ranged across loci from 0.017 to 0.660, with the average locus having a HE of 0.384. While the number of workers sampled per nest was small, and allowing for the large number of trials, observed heterozygosities did not deviate significantly from expected. PE2 and PE8 shared three alleles, one from each of three loci, exclusively with some Adelaide nests – the di-allele Lhum-14 at the second allele (AD5 and AD8), Lhum-28 (AD4 and AD8) and Lihu-T1 (AD7).
Table 1.
The number of alleles (A) and expected heterozygosity (HE) at 10 microsatellite loci for Linepithema humile populations in Melbourne, Adelaide, Perth and the whole Australian dataset
| Melbourne | Adelaide | Perth | Australia | |||||
|---|---|---|---|---|---|---|---|---|
| Locus | A | HE | A | HE | A | HE | A | HE |
| Lhum-11 | 4 | 0.333 | 4 | 0.609 | 5 | 0.451 | 5 | 0.465 |
| Lhum-13 | 6 | 0.649 | 7 | 0.634 | 5 | 0.547 | 8 | 0.610 |
| Lhum-14 | 1 | 0.000 | 2 | 0.019 | 2 | 0.033 | 2 | 0.017 |
| Lhum-19 | 5 | 0.659 | 5 | 0.705 | 5 | 0.337 | 6 | 0.567 |
| Lhum-28 | 2 | 0.010 | 6 | 0.383 | 5 | 0.239 | 6 | 0.211 |
| Lhum-35 | 10 | 0.383 | 8 | 0.535 | 8 | 0.574 | 15 | 0.497 |
| Lhum-39 | 5 | 0.590 | 6 | 0.224 | 6 | 0.339 | 6 | 0.384 |
| Lhum-52 | 2 | 0.152 | 2 | 0.496 | 2 | 0.472 | 2 | 0.374 |
| Lihu-H | 4 | 0.686 | 5 | 0.638 | 5 | 0.657 | 5 | 0.660 |
| Lihu-T1 | 3 | 0.080 | 4 | 0.050 | 3 | 0.026 | 4 | 0.052 |
| All | 42 | 49 | 46 | 59 | ||||
| Mean | 0.354 | 0.429 | 0.368 | 0.384 | ||||
Considerable insight into the genetic structure of L. humile in Australia came from FST estimates and AMOVA analyses (Tables 2 and 3). Within cities, there was little genetic divergence among nests (Table 2), with two of the eight Perth nests providing an exception. Adelaide formed the most genetically homogenous group of nests with an average FST of only 0.001. Only one of 28 pairwise nest comparisons differed significantly from zero (P < 0.05). Two-level hierarchical AMOVA (Table 3) supported this, revealing 99.7% of genetic differentiation occurred within nests. Melbourne nests were slightly more heterogenous with an average FST of 0.009. Eight of 28 pairwise comparisons were significantly different from zero (P < 0.05) and only 2.2% of the variation occurred among nests. Perth was the most genetically heterogenous group of nests (average intracity FST = 0.111). Of 28 pairwise nest comparisons, 26 were significantly different from zero (P < 0.05) and this was reflected as a higher level of internest genetic variation at 12.1% for Perth (intranest variation being 87.9% of the city total). Each of the loci considered separately exhibited this relatively high proportion of internest variation (Table 3). Closer examination of the Perth population revealed two nests that were responsible for this relatively greater heterogeneity compared to Adelaide or Melbourne – Perth nests two and eight (PE2 and PE8, respectively). Differentiation of PE2 and PE8 from the other Perth nests was indicated by higher average FST values of 0.109 and 0.242, respectively (Table 2). The average Perth pairwise nest FST decreased from 0.111 to 0.061 when PE2 and PE8 were excluded.
Table 2.
Comparison of FST estimates (means with range in parentheses) between Linepithema humile nests and nest groups from Australia and potential source populations based on 11 microsatellite markers. The number of nests in comparison between locations is given in brackets
| Location | Melbourne [8] | Adelaide [8] | Perth [8] | Perth [6] | PE2 | PE8 |
|---|---|---|---|---|---|---|
| Australia | ||||||
| Melbourne [8] | 0.009 (−0.032 to 0.097) | |||||
| Adelaide [8] | 0.116 (0.034 to 0.209) | 0.001 (−0.015 to 0.033) | ||||
| Perth [8] | 0.185 (0.063 to 0.303) | 0.138 (−0.001 to 0.234) | 0.111 (0.010 to 0.266) | |||
| Perth [6] | 0.187 (0.096 to 0.291) | 0.161 (0.098 to 0.234) | 0.073 (0.010 to 0.128) | 0.061 (0.010 to 0.106) | ||
| PE2 | 0.105 (0.063 to 0.139) | 0.016 (−0.001 to 0.034) | 0.105 (0.078 to 0.128) | 0.109 (0.080 to 0.128) | ||
| PE8 | 0.255 (0.211 to 0.303) | 0.119 (0.105 to 0.144) | 0.218 (0.078 to 0.266) | 0.242 (0.210 to 0.266) | 0.078 | |
| Native and other introduced ranges | ||||||
| Ita Ibate, Argentina (II) | 0.494 (0.460 to 0.520) | 0.460 (0.422 to 0.492) | 0.474 (0.409 to 0.514) | 0.487 (0.467 to 0.514) | 0.409 | 0.459 |
| Costanera Sur, Argentina (CS) | 0.449 (0.427 to 0.474) | 0.427 (0.402 to 0.448) | 0.446 (0.397 to 0.484) | 0.455 (0.430 to 0.484) | 0.397 | 0.442 |
| Catalonia (CT) | 0.484 (0.451 to 0.513) | 0.431 (0.384 to 0.462) | 0.461 (0.378 to 0.522) | 0.482 (0.467 to 0.522) | 0.378 | 0.418 |
| Europe Main (EU) | 0.068 (0.004 to 0.101) | 0.053 (0.030 to 0.083) | 0.087 (0.030 to 0.150) | 0.091 (0.062 to 0.150) | 0.030 | 0.121 |
| La Jolla, California (CA1) | 0.116 (0.081 to 0.148) | 0.084 (0.051 to 0.125) | 0.148 (0.062 to 0.190) | 0.155 (0.090 to 0.176) | 0.062 | 0.190 |
| Davis, California (CA2) | 0.157 (0.131 to 0.182) | 0.089 (0.076 to 0.108) | 0.196 (0.091 to 0.230) | 0.213 (0.162 to 0.230) | 0.091 | 0.195 |
| North Carolina (NC) | 0.258 (0.185 to 0.303) | 0.131 (0.091 to 0.178) | 0.252 (0.137 to 0.308) | 0.269 (0.253 to 0.308) | 0.137 | 0.261 |
| Auckland, New Zealand (NZ1) | 0.046 (0.014 to 0.087) | 0.181 (0.137 to 0.231) | 0.218 (0.150 to 0.308) | 0.216 (0.150 to 0.308) | 0.161 | 0.288 |
| Wellington, New Zealand (NZ2) | 0.016 (−0.005 to 0.054) | 0.148 (0.096 to 0.214) | 0.192 (0.129 to 0.278) | 0.187 (0.129 to 0.272) | 0.136 | 0.278 |
| South Africa (SA) | 0.466 (0.439 to 0.502) | 0.457 (0.422 to 0.490) | 0.502 (0.430 to 0.556) | 0.523 (0.498 to 0.556) | 0.430 | 0.451 |
| Saint Helena (SH) | 0.465 (0.442 to 0.499) | 0.455 (0.419 to 0.489) | 0.495 (0.422 to 0.549) | 0.516 (0.493 to 0.549) | 0.422 | 0.445 |
*Perth is considered in several ways: all eight nests, six nest excluding nests 2 (PE2) and 8 (PE8) that diverged from other Perth nests, and PE2 and PE8 separately.
Table 3.
Two-way hierarchical AMOVA for Linepithema humile nests in Melbourne, Adelaide and Perth, and three-way AMOVA for the whole Australian dataset. The percentage of genetic variance explained by each hierarchical level is given for each and over all 10 microsatellite loci
| Melbourne | Adelaide | Perth | Australia | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Intranest | Internest | Intranest | Internest | Intranest | Internest | Intranest | Intracity | Intercity | |
| Lhum-11 | 100.73 | −0.73 | 98.17 | 1.83 | 93.45*** | 6.55*** | 85.31** | 2.49*** | 12.20*** |
| Lhum-13 | – | – | 100.29 | −0.29 | 85.81*** | 14.19*** | 81.88*** | 4.87*** | 13.25*** |
| Lhum-14 | 97.67 | 2.33 | 100.27 | −0.27 | 97.42 | 2.58 | 98.08 | 1.66* | 0.26 |
| Lhum-19 | 97.65* | 2.35* | 100.59 | −0.59 | 69.36*** | 30.64*** | 79.62*** | 7.70*** | 12.69*** |
| Lhum-28 | 99.26 | 0.74 | 97.80 | 2.20 | 81.21*** | 18.79*** | 84.17*** | 8.40*** | 7.43*** |
| Lhum-35 | 98.63 | 1.37 | 101.76 | −1.76 | 89.85*** | 10.15*** | 89.44*** | 3.76*** | 6.80*** |
| Lhum-39 | 95.75** | 4.25** | 98.80 | 1.20 | 95.50** | 4.50** | 85.83*** | 3.33*** | 10.84*** |
| Lhum-52 | 99.17 | 0.83 | 98.78 | 1.22 | 93.90** | 6.10** | 75.17* | 2.56*** | 22.28*** |
| Lihu-H | 98.91 | 1.09 | 100.22 | −0.22 | 90.84*** | 9.16*** | 94.76*** | 3.39*** | 1.84** |
| Lihu-T1 | 89.11*** | 10.89*** | 101.03 | −1.03 | 99.16 | 0.84 | 94.13*** | 5.76*** | 0.11 |
| All | 97.82*** | 2.18*** | 99.73 | 0.27 | 87.95*** | 12.05*** | 84.86*** | 4.45*** | 10.69*** |
P < 0.05
P < 0.01
P < 0.00.
Significant genetic heterogeneity occurred across Australia (Tables 2 and 3). The global FST (i.e. averaged over all Australian nests) was 0.147 (P < 0.0005). Three-level hierarchical AMOVA revealed that 10.7% of overall genetic variation continentwide was attributed to intercity nest differentiation. Again, most variation was within nests (84.9%), while the remaining 4.4% of variation occurred among nests within cities. Melbourne and Perth nests were the most genetically differentiated from each other (average FST = 0.185), whereas Melbourne and Adelaide were the most genetically similar (average FST = 0.116) and Adelaide and Perth nests were moderately differentiated (average FST = 0.138). All pairwise nest FST comparisons between Melbourne and Adelaide, and Melbourne and Perth were significant (P < 0.05), while only 59 of 64 comparisons were significantly differentiated for Adelaide and Perth nests. The remaining five nonsignificant comparisons were between PE2 and Adelaide nests (average FST = 0.016; i.e. PE2 was no more genetically distant to any Adelaide nest as any of the Adelaide nests were to each other despite a separation distance of 2115 km). PE8 on the other hand was distinctive to all other Australian nests (except PE2; FST 0.078, P < 0.05, Table 2), but was more similar to Adelaide (average FST = 0.119) than to Melbourne or other Perth nests (average FST value = 0.255 and 0.242, respectively; Table 2).
Three additional analyses complemented FST and AMOVA results. First, the 24 nests largely clustered into three ‘city’ groups – Melbourne, Perth and Adelaide – when subjected to PCA (Fig. 2A). PCA explained 68% of the total variation in genetic relationships among Australian nests and identified two significant axes (P = 0.0005 for both). Principal component 1 (PC1) explained 36% of the variation and PC2, 32%. However, PE2 and PE8 grouped with the Adelaide nests. Second, phylogenetic analysis (Fig. 2B) revealed three well-supported clades of genetically similar nests, each clade largely corresponding to a city population (bootstrap value 89%). Within the Adelaide clade, PE2 and PE8 branched off to form their own clade (bootstrap value 79%– with PE8 being the most divergent). In each major clade, additional minor clades of closely related nests were indicated, two in each of the Melbourne and Perth clades and one in the Adelaide clade. Genetic similarity between nest pairs did not depend upon geographical proximity. Third, Bayesian analysis identified four clusters of genetically similar L. humile nests in Australia (Fig. 3; Bayesian probability, P = 1 for all runs). All Adelaide and Melbourne nests each corresponded to their own cluster. Excluding PE2 and PE8, the remaining Perth nests formed the third cluster. PE2 grouped with the Adelaide cluster, and PE8 formed the fourth cluster alone. Thus, with two exceptions in Perth, intracity nests show a high level of genetic similarity.
Figure 2.
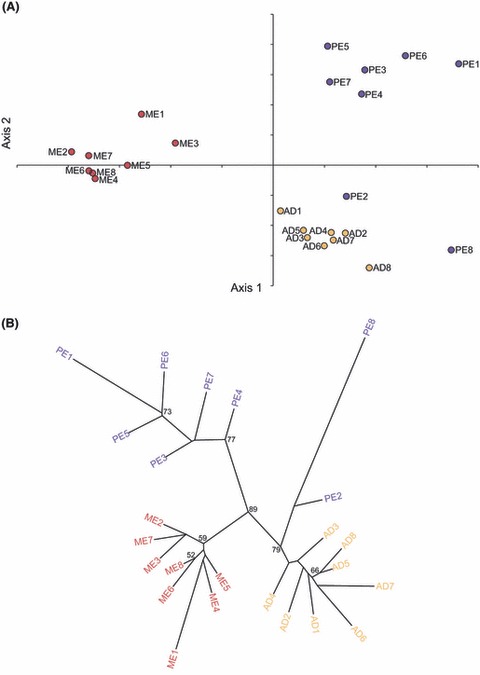
(A) Principal component analysis based on allelic frequency data at 10 microsatellite loci for Linepithema humile nests in Melbourne, Adelaide and Perth. The proportion of inertia for both axes are significant (axes 1 and 2: P = 0.0005) and explain 35.61% and 32.41% of the variance, respectively. Levels of significance were derived from 2000 iterations. (B) Consensus of 2000 additive trees based on Cavalli-Sforza chord measures to depict relationships among Linepithema humile nests in Melbourne, Adelaide and Perth (nodal values are the percentage of bootstraps >50% in which the relevant grouping occurred).
Figure 3.
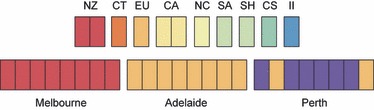
Genetic structure analysis of all Linepithema humile nests (n = 35) using BAPS, with Australian nests being extracted to the lower row. Nests include those from Australia (Melbourne, Adelaide and Perth) and from the native (II = Ita Ibate, CS = Costanera Sur) and other introduced ranges (NZ = New Zealand, CA = California, NC = North Carolina, CT = Catalonia, EU = Europe, SA = South Africa, SH = Saint Helena). Each bar represents a nest and colours correspond to BAPS groups.
Genetic diversity and structure and potential source populations
When genetic data from the Australian dataset and potential source populations were combined, 114 alleles were found in total across 11 microsatellite loci (Table 4). The number of alleles and allelic richness per potential source population varied from 26 to 59 and 2.21 to 3.31, respectively. The allelic richness for the Australian dataset was 2.63. Expected heterozygosities ranged from 0.269 to 0.531 and did not differ significantly from observed heterozygosities. Conspicuous were the higher levels of allelic richness and expected heterozygosity in the two native range populations. Private alleles in these two populations accounted for over half of the total (61%).
Table 4.
The number of alleles (A), allelic richness (AR), private alleles (AP), frequency of private alleles (AF) and expected heterozygosity (HE) over 11 microsatellite loci for potential source populations of Linepithema humile from the native and other introduced ranges (n = 15 workers per population)
| Potential source population | A | AR | AP | AF | HE |
|---|---|---|---|---|---|
| Native range | |||||
| Ita Ibate, Argentina (II) | 47 | 3.62 | 23 | 0.174 | 0.516 |
| Costanera Sur, Argentina (CS) | 37 | 3.02 | 7 | 0.159 | 0.533 |
| Other introduced ranges | |||||
| Catalonia (CT) | 30 | 2.39 | 5 | 0.301 | 0.408 |
| Europe Main (EU) | 31 | 2.50 | – | – | 0.402 |
| La Jolla, California (CA1) | 31 | 2.50 | – | – | 0.382 |
| Davis, California (CA2) | 30 | 2.44 | 1 | 0.033 | 0.374 |
| North Carolina (NC) | 26 | 2.21 | 1 | 0.083 | 0.314 |
| Auckland, New Zealand (NZ1) | 27 | 2.12 | – | – | 0.267 |
| Wellington, New Zealand (NZ2) | 31 | 2.26 | 1 | 0.067 | 0.314 |
| South Africa (SA) | 28 | 2.28 | – | – | 0.361 |
| Saint Helena (SH) | 26 | 2.24 | – | – | 0.392 |
Linepithema humile nests in Australia were less genetically differentiated from each other relative to their differentiation from potential source populations (Table 2). The average global pairwise combination FST was 0.213 (P < 0.001) compared to the Australian average of 0.147. Australian nests were most genetically similar to CA1, CA2, EU, NC, NZ1 and NZ2 (average FST = 0.016–0.288). FST comparisons between Australia and New Zealand were not significantly different from zero. Australian nests were considerably more divergent from CT, CS, II, SA and SH [average FST = 0.378–0.523 (Table 2)].
Bayesian analysis detected nine genetically distinct clusters of L. humile nests across Australia and potential source populations (Fig. 3; Bayesian probability, P > 0.99 for all runs). Three of these clusters largely corresponded to the three Australian city populations sampled in this study – the Adelaide group included EU, PE2 and PE8. The Melbourne group included NZ1 and NZ2, and the Perth group contained only the remaining six Perth nests. PE8, which was distinct from the other Australian nests in the Australia-wide analysis, was indistinguishable from Adelaide and PE2 in this global analysis.
PCA and phylogenetic analysis provided very similar patterns of association when the Australian nest groups of L. humile identified by Bayesian analysis were assessed together with the 11 potential source populations (Fig. 4). From the phylogenetic tree, five divergent branches were obvious, three leading to single potential source populations (CS, CT and II), one to two sub-branches of a potential source population each (SA and SH) and one to eleven genetically similar sub-branches of Australian nest groups and potential source populations (bootstrap value 100% for all 2000 comparisons) (Fig. 4B). In the largest branch, all five Australian nest groups grouped with CA1, CA2, EU, NC, NZ1 and NZ2. NZ1 and NZ2 were consistently paired (bootstrap value 99%) and together grouped with the Melbourne nest group (bootstrap value 92%). The two Perth nest outliers, PE2 and PE8, usually grouped with each other (bootstrap value 52%) and together most frequently grouped with the Adelaide nest group (bootstrap value 65%). The Perth nest group and EU did not group with other populations but were in the sub-branch that included the other Australian nest groups and New Zealand populations.
Figure 4.
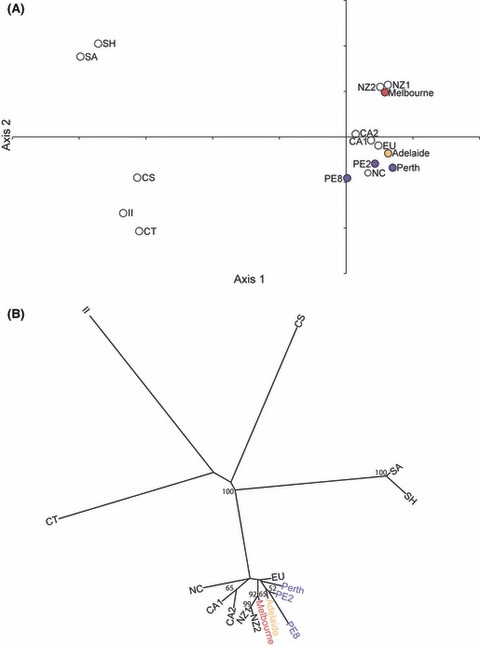
(A) Principal component analysis based on allelic frequency data at 11 microsatellite loci for Linepithema humile nest groups in Australia and populations from the native range and other introduced ranges. Perth nests were separated (Perth, PE2 and PE8) based on BAPS. The proportion of inertia of the first axis is significant (P = 0.0035) and explains 36.76% of the variance in the data. The second principal component (16.30%) is not significant (P = 0.490). Levels of significance were derived from 2000 iterations. (B) Consensus of 2000 additive trees based on Cavalli-Sforza chord measures to depict genetic relationships between Linepithema humile nest groups in Australia and populations from the native range and other introduced ranges (nodal values are the percentage of bootstraps >50% in which the relevant grouping occurred).
Genetic assignment tests for the five Australian L. humile nest groups using GENECLASS2 (threshold P < 0.001) assigned them all to EU with scores of 100% when both New Zealand populations were appropriately omitted from the analyses.
Interception records and introduction pathways
A total of 62 interceptions of L. humile were recorded at ports of entry in Melbourne, Adelaide and Perth between 1988 and 2007 (Fig. 5). The source regions of these interceptions were diverse and included Asia, Europe, New Zealand, North America and South America. However, 71% of interceptions were sourced from the United States (Fig. 5). Interception records were few in Adelaide and Perth but sourced from Europe and Japan, as well as the United States. The one L. humile interception record from South America was from Chile, outside its native range (Argentina, Paraguay or Uruguay).
Figure 5.
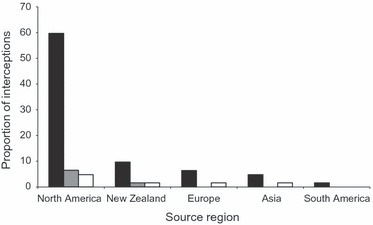
Proportion of port of entry (POE) interception records for Linepithema humile in Australia from 1988 to 2007 [Melbourne (solid bars, n = 51), Adelaide (shaded bars, n = 5) and Perth (open bars, n = 6)] from source regions (North America, Europe, Asia and South America).
Discussion
An Australian supercolony
The complete absence of intraspecific aggression within and between three city populations of L. humile sampled across southern Australia strongly suggests that these populations behave as a single disjunct supercolony spread over 2700 km (i.e. Melbourne to Perth). Previous inferences about the behaviour and genetic structure of Argentine ants in Australia have been made from either single nests (e.g., Vogel et al. 2010; van Wilgenburg et al. 2010) or from a restricted distribution of its entire introduced range (Suhr et al. 2009). This supercolony is likely to extend beyond these three population centres to regional cities and towns in Victoria (Björkman-Chiswell et al. 2008) and other Australian states (e.g., Brisbane, Queensland and Hobart, Tasmania, Suhr et al. 2009). The expansive Australian supercolony mirrors massive supercolonies elsewhere worldwide that lack aggression across hundreds to thousands of kilometres (Suarez et al. 1999; Giraud et al. 2002). While these populations of L. humile across Australia act socially as a single supercolony, we note they do not function ecologically as one, but rather as mosaics of smaller, distinct colonies consisting of groups of interacting nests as suggested by Heller et al. (2008). Spread of L. humile appears limited by abiotic conditions, especially water availability (Holway et al. 2002b). In Australia, large stretches of unsuitable habitat (e.g., arid and semi-arid natural areas, dryland pastures) between urban centres are likely to limit the distribution and expansion of L. humile (Walters and Mackay 2003, 2004).
The genetic structure of L. humile in Australia was consistent with low connectivity of L. humile between urban centres. It also suggested that homogeneity within cities is a result of effective local gene flow; by in large, each city possesses a characteristic set of allele frequencies. This pattern appears most similar to L. humile from the main European supercolony, where moderate genetic differentiation and higher allele diversity were found across Europe but still accompanied by the absence of intraspecific aggression (Giraud et al. 2002). Similarly, we found no relationship between genetic similarity and intraspecific aggression in L. humile across Australia. Nest pairs showed no aggression irrespective of whether as few as 45% or as many as 87% alleles were shared. In contrast, Tsutsui et al. (2000) showed that nest pairs in Argentina sharing 17–63% alleles were aggressive, whereas those in California sharing >75% of alleles were not. Nest pairs in Australia shared on average fewer alleles (64%) than California (75%) but more than the south-east United States (57%), where workers from nests within the region nevertheless showed strong intraspecific aggression (Buczkowski et al. 2004).
Our results are consistent with the pattern of low-to-nil genetic differentiation within L. humile that has been found in almost all other introduced populations at local and rangewide scales (local: Suhr et al. 2009; Ingram and Gordon 2003; rangewide: Tsutsui and Case 2001; Buczkowski et al. 2004; Corin et al. 2007a). At the local scale, this homogeneity is best explained by colony budding where queens disperse on foot, resulting in the homogenization of allele frequencies. Because the maximum yearly rate of spread is ∼150 m, local-scale dispersal is unlikely to account completely for low-to-nil intracity genetic differentiation and cannot account for it at the rangewide scale (Suarez et al. 2001). The average distance of human-assisted jump dispersal in L. humile has been estimated at >150 km (Suarez et al. 2001). Human-assisted dispersal within and between cities is likely to explain low genetic differentiation at these broader spatial scales. Rail (58%) followed by road transport (24%) is the predominant modes of movement of nonbulk freight in Australia annually (DOTARS 2007), highlighting the importance of transport corridors in facilitating intercity movement of L. humile.
Genetic heterogeneity was greatest in the Perth population of L. humile, largely attributable to the PE2 and PE8 nests. The exclusive sharing of alleles between PE2 and PE8 with some Adelaide nests contributed to their dissimilarity to Perth nests and similarity to Adelaide nests. Additionally, PE2 grouped with Adelaide in Bayesian and phylogenetic analyses and PCA. This suggests that PE2 is a recent intracontinental translocation from Adelaide to Perth, although we cannot exclude the possibility of an intercontinental introduction from an Adelaide-like population. PE8, on the other hand, stood alone in the Australian Bayesian analysis and PCA, but like PE2, branched with Adelaide in both phylogenetic trees and clustered with the Adelaide group in the global PCA and Bayesian analysis. This shows PE8 was slightly more genetically distinct than PE2 to the other Adelaide nests, despite its genetic affinity to PE2. This suggests that PE8 is a separate recent introduction into Perth, possibly also from Adelaide although several worldwide sources are possible.
Spread in Australia
Within Australia, the initial colonization of L. humile probably occurred on the eastern coast in Melbourne in 1939, with an intracontinental pathway of establishment proceeding from Melbourne to Adelaide and Perth. Linepithema humile is easily moved (Holway et al. 2002a), suggesting intracontinental spread is more prevalent than intercontinental invasions. Once established in Melbourne, L. humile likely spread locally by colony budding and then rapidly expanded geographically at the city- and continentwide scales through human-mediated jump dispersal. The establishment of the Argentine Ant Act in Western Australia in 1954 illustrates an early appreciation of the potential intracontinental movement of these ants (Jenkins and Forte 1973). Although recorded for the first time in Perth in 1950 and in Adelaide in 1979, it is highly probable that it established in Adelaide long before since it had already spread to 69 suburbs across Adelaide by then (Madge 1979). Colonies of L. humile from Melbourne were intercepted in potted plants from Melbourne at the South Australian border in 1975 (Madge and Caon 1987), consistent with an east-to-west spread, as is the asymmetric trade flow between Melbourne, Adelaide and Perth, with 33% more nonbulk freight moving from east to west (DOTARS 2007). Given this, we might have expected reduced genetic diversity between Melbourne and the more western cities (Nei et al. 1975). However, the total number of alleles and expected heterozygosities per loci hardly differed among cities, and in fact, the allelic richness was similar for the Australian supercolony and the Main European population (2.63 and 2.50, respectively). The most parsimonious explanation of these data would seem to be an early establishment on the eastern coast in Melbourne, perhaps by a large founding colony or several smaller independent introductions, followed by ongoing western intracontinental human-assisted dispersal.
Arrival and spread from potential source populations
Integrating behavioural and genetic data with information from interception records and historical trade pathways allowed us to begin to reconstruct the likely introduction history of L. humile into Australia. It is highly unlikely the Australian supercolony resulted from primary introduction events from the native range as there are no records of L. humile interceptions in Melbourne, Adelaide or Perth that came from its native range, although a single interception in Sydney originated from Argentina (E. L. Suhr and D. J. O'Dowd, unpublished data). Furthermore, the direct source of most non-native populations is usually other introduced populations (Suarez et al. 2008).
We propose a Europe-to-Australia introduction pathway as the most likely source of the Australian supercolony. Genetic differentiation (FST) and phylogenetic analyses showed Australian nests and nest groups were more genetically similar to the introduced Californian, Main European, New Zealand and North Carolina populations (FST = −0.005–0.308; clade bootstrap value = 100%). Bayesian analysis then confirmed the Main European population as genetically similar to the Adelaide, PE2 and PE8 nests. Finally, a single assignment test assigned all Australian nest groups to the Main European population, implicating the main European supercolony as the most likely source of L. humile in Australia. This conclusion is in agreement with the view that the main European supercolony descended from the earliest recorded introduction of L. humile on Madeira before 1858 (Wetterer and Wetterer 2006) and that many supercolonies throughout the introduced ranges likely descended from Madeira or from the same ancestral population (Wetterer and Wetterer 2006; van Wilgenburg et al. 2010).
Recent POE interception records of L. humile in Australia predominantly reflected interceptions from the United States; few were sourced from Europe ports. However, trade flows to Australia have changed markedly as Argentine ants were first reported in Australia over 70 years ago. Trade volume records indicate that the primary potential source region for introduction of L. humile into Australia has shifted over time from Europe (1937–1939) to the United States (1968–1969), which was later surpassed by Asia (2006–2007) (Table 5). In the early 20th century, Australia's largest trading partners were the United Kingdom and Europe, which reflected Australia's historical links with these regions. In the 1960s, trade between Britain and Australia declined as Australia's trading focus shifted to the United States. Another shift occurred in 1989 as Asian nations gradually overtook the United States as Australia's top trading partners. It is interesting, for example, that interceptions of L. humile from Japan, where it was first detected in 1993 (Sugiyama 2000), have only been recorded since 2003. The interception record database used in our study covers the period from 1988 to 2007, which reflects the last two decades where Australia's main trading partners were the United States or in Asia. However, heavy trade flow from Europe at the time of first recording of the Argentine ant in Australia is consistent with the genetic similarity between L. humile from the Australian and main European supercolonies.
Table 5.
Percentage of total import value ($) for main trading partners of Australia from each import source region between 1937 and 2007
| Time period | |||
|---|---|---|---|
| Import source region | 1937–1939 | 1966–1969 | 2006–2007 |
| North America | 22.3 | 25.6 | 13.8 |
| New Zealand | 1.9 | 1.9 | 3.1 |
| Europe | 44.7 | 34.7 | 9.2 |
| Asia | 15.8 | 22.8 | 41.2 |
Sources: Meredith and Dyster 1999; ABS 2007. Each region includes some or all of the following countries: North America (USA, Canada), New Zealand, Europe (Belgium, France, Germany, Italy, Luxembourg and the Netherlands) and Asia (China, India, Indonesia, Japan, Malaysia, Papua New Guinea, Persian Gulf, Republic of Korea, Singapore, South and Southeast Asia and Thailand).
Australia is increasingly likely to provide a source of L. humile that helps facilitate a dynamic world supercolony of this species. For example, genetic differentiation (FST), and Bayesian and phylogenetic analyses showed L. humile nests in Melbourne and New Zealand were indistinguishable from each other. The Melbourne population was previously hypothesized as the source of the New Zealand supercolony based on matching mitochondrial DNA haplotypes (Corin et al. 2007b). Although Corin et al. (2007b) did not sample L. humile from other parts of its Australian range, our results using microsatellite markers support their contention. Of all our FST comparisons (except EU and ME3), only those between Melbourne and New Zealand nests did not differ significantly from zero. These results, along with the asymmetry in propagule pressure of L. humile from Melbourne into New Zealand (Suhr et al. 2009), support the Melbourne population as the source of the New Zealand supercolony. In addition, L. humile from the Australian and New Zealand supercolonies lack aggression towards one another (van Wilgenburg et al. 2010), suggesting that they behave as an Australasian supercolony.
This Australasian supercolony appears to be part of an intercontinental supercolony that includes parts of Europe, North America and north-east Asia (Sunamura et al. 2009a; Vogel et al. 2010; van Wilgenburg et al. 2010). Introduced L. humile from the largest supercolonies in California, Hawaii, Europe, Japan and Australasia are mutually nonantagonistic (Sunamura et al. 2009a; van Wilgenburg et al. 2010). Interestingly, L. humile in Melbourne, Australia share similar cuticular hydrocarbons to those from these supercolonies in California, Europe and Hawaii (Brandt et al. 2009). As overall hydrocarbon similarity and intraspecific aggression are negatively correlated (Suarez et al. 2002), L. humile from these supercolonies in New Zealand and Japan may also share similar cuticular hydrocarbons. Finally, members of the intercontinental supercolony share a single mtDNA haplotype (Vogel et al. 2010), suggesting they descend from a single source supercolony.
Acknowledgments
We thank P Davis, M Widmer and T Smith at the Department of Agriculture and Food, Western Australia for assistance with Argentine ant collections and intraspecific aggression assays in Perth; C Gómez, C Green, D Holway, P Lester, J Silverman, A Suarez and N Tsutsui for providing Argentine ant specimens from the native range and other introduced ranges; and A. Suarez, R. Crozier, B. Schlick-Steiner and F. Steiner for improving the manuscript. Market Access and Biosecurity (Department of Agriculture, Fisheries and Forestry) provided access to the Pest and Disease Information and INCIDENTS databases. We acknowledge support from the Department of Primary Industries, Victoria through the Nancy Millis Postgraduate Research Award and the Australian Research Council through their Special Research Centre Program. This is publication no. 206 from the Australian Centre for Biodiversity at Monash University.
Literature cited
- ABS. Catalogue 5368.0 – International Trade in Good and Services, Australia, December 2007. Canberra, ACT: Australian Bureau of Statistics; 2007. [Google Scholar]
- Björkman-Chiswell BT, van Wilgenburg E, Thomas ML, Swearer SE, Elgar MA. Absence of aggression but not nestmate recognition in an Australian population of the Argentine ant, Linepithema humile. Insectes Sociaux. 2008;55:207–212. [Google Scholar]
- Brandt M, van Wilgenburg E, Tsutsui ND. Global-scale analyses of chemical ecology and population genetics in the invasive Argentine ant. Molecular Ecology. 2009;18:997–1005. doi: 10.1111/j.1365-294X.2008.04056.x. [DOI] [PubMed] [Google Scholar]
- Buczkowski G, Vargo EL, Silverman J. The diminutive supercolony: the Argentine ants of the southeastern United States. Molecular Ecology. 2004;13:2235–2242. doi: 10.1111/j.1365-294X.2004.02261.x. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Edwards AWF. Phylogenetic analysis: models and estimation procedures. American Journal of Human Genetics. 1967;19:233–257. [PMC free article] [PubMed] [Google Scholar]
- Corander J, Waldmann P, Sillanpää MJ. Bayesian analysis of genetic differentiation between populations. Genetics. 2003;163:367–374. doi: 10.1093/genetics/163.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corander J, Waldmann P, Marttinen P, Sillanpää MJ. BAPS 2: enhanced possibilities for the analysis of genetic population structure. Bioinformatics. 2004;20:2363–2369. doi: 10.1093/bioinformatics/bth250. [DOI] [PubMed] [Google Scholar]
- Corin SE, Abbott KL, Ritchie PA, Lester PJ. Large-scale unicoloniality: the population and colony structure of the invasive Argentine ant (Linepithema humile) in New Zealand. Insectes Sociaux. 2007a;54:275–282. [Google Scholar]
- Corin SE, Lester PJ, Abbott KL, Ritchie PA. Inferring historical introduction pathways with mitochondrial DNA: the case of introduced Argentine ants (Linepithema humile) into New Zealand. Diversity and Distributions. 2007b;13:510–518. [Google Scholar]
- DOTARS. Perth-Adelaide Corridor Strategy. Canberra, ACT: Department of Transport and Regional Services; 2007. [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package), Version 3.68. Seattle, WA: Department of Genome Sciences, University of Washington; 2008. [Google Scholar]
- Giraud T, Pedersen JS, Keller L. Evolution of supercolonies: the Argentine ants of southern Europe. Proceedings of the National Academy of Science USA. 2002;99:6075–6079. doi: 10.1073/pnas.092694199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helantera H, Strassmann JE, Carrillo J, Queller DC. Unicolonial ants: where do they come from, what are they and where are they going? Trends in Ecology and Evolution. 2009;24:341–349. doi: 10.1016/j.tree.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Heller NE. Colony structure in introduced and native populations of the invasive Argentine ant, Linepithema humile. Insectes Sociaux. 2004;51:378–386. [Google Scholar]
- Heller NE, Ingram KK, Gordon DM. Nest connectivity and colony structure in unicolonial Argentine ants. Insectes Sociaux. 2008;55:397–403. [Google Scholar]
- Heterick BE, Casella J, Majer JD. Influence of Argentine ant and coastal brown ant (Hymenoptera: Formicidae) invasions on ant communities in Perth gardens, Western Australia. Urban Ecosystems. 2000;4:277–292. [Google Scholar]
- Holway DA, Suarez AV. Animal behavior: an essential component of invasion biology. Trends in Ecology and Evolution. 1999;14:328–331. doi: 10.1016/s0169-5347(99)01636-5. [DOI] [PubMed] [Google Scholar]
- Holway DA, Suarez AV, Case TJ. Loss of intraspecific aggression in the success of a widespread invasive social insect. Science. 1998;282:949–952. doi: 10.1126/science.282.5390.949. [DOI] [PubMed] [Google Scholar]
- Holway DA, Lach L, Suarez AV, Tsutsui ND, Case TJ. The causes and consequences of ant invasions. Annual Review of Ecology and Systematics. 2002a;33:181–233. [Google Scholar]
- Holway DA, Suarez AV, Case TJ. The role of abiotic factors in governing susceptibility to invasion: a test with a widespread invasive social insect. Ecology. 2002b;83:1610–1619. [Google Scholar]
- Hulme PE. Trade, transport and trouble: managing invasive species pathways in an era of globalization. The Journal of Applied Ecology. 2009;46:10–18. [Google Scholar]
- Ingram KK, Gordon DM. Genetic analysis of dispersal dynamics in an invading population of Argentine ants. Ecology. 2003;84:2832–2842. [Google Scholar]
- Ingram KK, Palumbi SR. Characterization of microsatellite loci for the Argentine ant, Linepithema humile, and their potential for analysis of colony structure in invading Hawaiin populations. Molecular Ecology Notes. 2002;2:94–95. [Google Scholar]
- Jenkins CFH, Forte PN. Chemicals for Argentine ant control. Journal of Agriculture Western Australia. 1973;14:195–196. [Google Scholar]
- Kalinowski ST. HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Molecular Ecology Notes. 2005;5:187–189. [Google Scholar]
- Krieger MJB, Keller L. Low polymorphism at 19 microsatellite loci in a French population of Argentine ants (Linepithema humile. Molecular Ecology. 1999;8:1078–1080. [Google Scholar]
- Lach L, Hooper-Bui LM. Consequences of ant invasions. In: Lach L, Parr CL, Abbott KL, editors. Ant Ecology. Oxford: Oxford University Press; 2010. pp. 261–286. [Google Scholar]
- Lee CE. Evolutionary genetics of invasive species. Trends in Ecology and Evolution. 2002;17:386–391. [Google Scholar]
- Madge PE. 1979. Argentine ant. South Australia Department of Agriculture Fact Sheet AGDEX 612.
- Madge PE, Caon G. 1987. Technical Report number 111: Argentine ant, Department of Agriculture South Australia.
- Meredith D, Dyster B. Australia in the Global Economy: Continuity and Change. Oakleigh: Cambridge University Press; 1999. [Google Scholar]
- Nei M, Maruyama T, Chakraborty R. The bottleneck effect and genetic variability in populations. Evolution. 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- Pasfield G. Argentine ants. Australian Natural History. 1968;16:12–15. [Google Scholar]
- Pedersen JS, Krieger MJB, Vogel V, Giraud T, Keller L. Native supercolonies of unrelated individuals in the invasive Argentine ant. Evolution. 2006;60:782–791. [PubMed] [Google Scholar]
- Piry S, Alapetite A, Cornuet J-M, Paetkau D, Baudouin L, Estoup A. GeneClass2: a software for genetic assignment and first-generation migration detection. Journal of Heredity. 2004;95:536–539. doi: 10.1093/jhered/esh074. [DOI] [PubMed] [Google Scholar]
- Puth LM, Post DM. Studying invasion: have we missed the boat. Ecology Letters. 2005;8:715–721. [Google Scholar]
- Rannala B, Mountain JL. Detecting immigration by using multilocus genotypes. Proceedings of the National Academy of Science USA. 1997;94:9197–9201. doi: 10.1073/pnas.94.17.9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Rowles AD, O'Dowd DJ. Interference competition by Argentine ants displaces native ants: implications for biotic resistance to invasion. Biological Invasions. 2007;9:73–85. [Google Scholar]
- Rowles AD, O'Dowd DJ. Impacts of the invasive Argentine ant on native ants and other invertebrates in coastal scrub in southeastern Australia. Austral Ecology. 2009a;34:239–248. [Google Scholar]
- Rowles AD, O'Dowd DJ. New mutualism for old: indirect disruption and direct facilitation of seed dispersal following Argentine ant invasion. Oecologia. 2009b;158:709–716. doi: 10.1007/s00442-008-1171-2. [DOI] [PubMed] [Google Scholar]
- Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, Baughman S, et al. The population biology of invasive species. Annual Review of Ecology and Systematics. 2001;32:305–332. [Google Scholar]
- Suarez AV, Tsutsui ND, Holway DA, Case TJ. Behavioral and genetic differentiation between native and introduced populations of the Argentine ant. Biological Invasions. 1999;1:43–53. [Google Scholar]
- Suarez AV, Holway DA, Case TJ. Patterns of spread in biological invasions dominated by long-distance jump dispersal: insights from Argentine ants. Proceedings of the National Academy of Science USA. 2001;98:1095–1100. doi: 10.1073/pnas.98.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez AV, Holway DA, Liang D, Tsutsui ND, Case TJ. Spatiotemporal patterns of intraspecific aggression in the invasive Argentine ant. Animal Behaviour. 2002;64:697–708. [Google Scholar]
- Suarez AV, Holway DA, Tsutsui ND. Genetics and behavior of a colonizing species: the invasive Argentine ant. The American Naturalist. 2008;172:S72–S84. doi: 10.1086/588638. [DOI] [PubMed] [Google Scholar]
- Sugiyama T. Invasion of Argentine ant, Linepithema humile, into Hiroshima Prefecture, Japan. Japanese Journal of Applied Entomology and Zoology. 2000;44:127–129. [Google Scholar]
- Suhr EL, McKechnie SW, O'Dowd DJ. Genetic and behavioural evidence for a city-wide supercolony of the invasive Argentine ant Linepithema humile (Mayr) (Hymenoptera: Formicidae) in southeastern Australia. Australian Journal of Entomology. 2009;48:78–82. [Google Scholar]
- Sunamura E, Espadaler X, Sakamoto H, Suzuki S, Terayama M, Tatsuki S. Intercontinental union of Argentine ants: behavioral relationships among introduced populations in Europe, North America, and Asia. Insectes Sociaux. 2009a;56:143–147. [Google Scholar]
- Sunamura E, Hatsumi S, Karino S, Nishisue K, Teraama M, Kitade O, Tatsuki S. Four mutually incompatible Argentine ant supercolonies in Japan: inferring invasion history of introduced Argentine ants from their social structure. Biological Invasions. 2009b;11:2329–2339. [Google Scholar]
- Thomas ML, Holway DA. Condition-specific competition between invasive Argentine ants and Australian Iridomyrmex. Journal of Animal Ecology. 2005;74:532–542. [Google Scholar]
- Tsutsui ND, Case TJ. Population genetics and colony structure of the Argentine ant (Linepithema humile) in its native and introduced ranges. Evolution. 2001;55:976–985. doi: 10.1554/0014-3820(2001)055[0976:pgacso]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Tsutsui ND, Suarez AV, Holway DA, Case TJ. Reduced genetic variation and the success of an invasive species. Proceedings of the National Academy of Science USA. 2000;97:5948–5953. doi: 10.1073/pnas.100110397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitousek PM, D'Antonio CM, Loope LL, Rejmanek M, Westbrooks R. Introduced species: a significant component of human-casued global change. New Zealand Journal of Ecology. 1997;21:1–16. [Google Scholar]
- Vogel V, Pedersen JS, d'Ettorre P, Lehmann L, Keller L. Dynamics and genetic structure of Argentine ant supercolonies in their native range. Evolution. 2009;63:1627–1639. doi: 10.1111/j.1558-5646.2009.00628.x. [DOI] [PubMed] [Google Scholar]
- Vogel V, Pedersen JS, Giraud T, Krieger MJB, Keller L. The worldwide expansion of the Argentine ant. Diversity and Distributions. 2010;16:170–186. [Google Scholar]
- Walters AC. Invasion of Argentine ants (Hymenoptera: Formicidae) in South Australia: impacts on community composition and abundance of invertebrates in urban parklands. Austral Ecology. 2006;31:567–576. [Google Scholar]
- Walters AC, Mackay DA. An experimental study of the relative humidity preference and survival of the Argentine ant, Linepithema humile (Hymenoptera: Formicidae): comparisons with a native Iridomyrmex species in South Australia. Insectes Sociaux. 2003;50:355–360. [Google Scholar]
- Walters AC, Mackay DA. Comparisons of upper thermal tolerances between the invasive Argentine ant (Hymenoptera: Formicidae) and two native Australian ant species. Annals of the Entomological Society of America. 2004;97:971–975. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wetterer JK, Wetterer AL. A disjunct Argentine ant metacolony in Macaronesia and southwestern Europe. Biological Invasions. 2006;8:1123–1129. [Google Scholar]
- Wetterer JK, Wild AL, Suarez AV, Roura-Pascual N, Espadaler X. Worldwide spread of the Argentine ant, Linepithema humile (Hymenoptera: Formicidae) Myrmecological News. 2009;12:187–194. [Google Scholar]
- Wild AL. Taxonomy and distribution of the Argentine ant, Linepithema humile. Annals of the Entomological Society of America. 2004;97:1204–1215. [Google Scholar]
- van Wilgenburg E, Torres CW, Tsutsui ND. The global expansion of a single ant supercolony. Evolutionary Applications. 2010;3:136–143. doi: 10.1111/j.1752-4571.2009.00114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The interpretation of population structure by F-statistics with special regards to systems of mating. Evolution. 1965;19:393–420. [Google Scholar]


