Abstract
Gene flow between domesticated species and their wild relatives is receiving growing attention. This study addressed introgression between wheat and natural populations of its wild relatives (Aegilops species). The sampling included 472 individuals, collected from 32 Mediterranean populations of three widespread Aegilops species (Aegilops geniculata, Ae. neglecta and Ae. triuncialis) and compared wheat field borders to areas isolated from agriculture. Individuals were characterized with amplified fragment length polymorphism fingerprinting, analysed through two computational approaches (i.e. Bayesian estimations of admixture and fuzzy clustering), and sequences marking wheat-specific insertions of transposable elements. With this combined approach, we detected substantial gene flow between wheat and Aegilops species. Specifically, Ae. neglecta and Ae. triuncialis showed significantly more admixed individuals close to wheat fields than in locations isolated from agriculture. In contrast, little evidence of gene flow was found in Ae. geniculata. Our results indicated that reproductive barriers have been regularly bypassed during the long history of sympatry between wheat and Aegilops.
Keywords: admixture, amplified fragment length polymorphism, crop-to-wild gene flow, introgression, transposable element-based markers
Introduction
Many domesticated plants hybridize and backcross spontaneously with their wild relatives, and the potential for gene exchanges involving crop species is receiving growing attention since the last decade (Ellstrand et al. 1999; Felber et al. 2007). Effective transfer of crop genes to wild relatives has been reported in at least 27 cases for European agro-ecosystems, encompassing many plant families (Felber et al. 2007). Crop-to-wild gene flow has important evolutionary consequences for local relatives, as it can promote the origin of highly competitive genotypes that can develop into aggressive weed populations (e.g. Beta vulgaris × B. maritima hybrids, Ellstrand et al. 1999; Amaranthus tuberculatus × A. hybridus, Trucco et al. 2009).
The wheat–Aegilops species complex provides an excellent model to document and assess the impact of crop-to-wild gene flow. Wheat is a crop in which ancestors (i.e. Aegilops species) frequently grow in sympatry across the Mediterranean area, in northern Europe and in the United States of America. More specifically, durum wheat (Triticum turgidum) is an allotetraploid crop species (2n = 4x = 28; genome BA, see van Slageren 1994 for genome nomenclature), with seven sets of chromosomes originating from Triticum urartu (A genome) and Ae. speltoides (B genome) (Waines and Barnhart 1992). It is mainly grown under semi-arid climates in western Asia, southern Europe and North America for pasta and semolina production. However, 95% of the worldwide production is bread wheat (T. aestivum), an allohexaploid species (2n = 6x = 42; genome BAD), resulting from the hybridization of durum wheat (BA genome) with Ae. tauschii, a wild relative that contributed the D genome. Aegilops is a Mediterranean–Western Asiatic genus that has been regularly used by breeders for wheat improvement programmes (Schneider et al. 2008). Aegilops species occur in open habitats, such as cultivation edges, roadsides, pastures, orchards or olives groves, and show flowering overlap with Triticum (Zaharieva and Monneveux 2006). Natural F1 hybrids have been regularly reported under natural conditions (van Slageren 1994; Guadagnuolo et al. 2001; Morrison et al. 2002; David et al. 2004; Karagöz 2006; Loureiro et al. 2006, 2007a, 2009). For instance, hybridization rates ranging from 0.20% to 0.45% have been measured between Ae. geniculata and wheat when grown together at less than 1 m (Loureiro et al. 2006). In southern Europe, wheat is thus particularly likely to hybridize with the three most common Aegilops species (van Slageren 1994): Ae. geniculata (2n = 4x = 28, genome MU), Ae. triuncialis (2n = 4x = 28, genome CU) and Ae. neglecta (2n = 4x = 28 and 2n = 6x = 42, genomes MU and MUN, respectively).
Introgression between wheat and Aegilops species is however presumed to be limited because meiosis in F1 hybrids generally fails, resulting in male sterility and low female fertility (ranging between 1% and 4%; Zaharieva and Monneveux 2006). Nevertheless, backcrosses have been reported from experimental settings, suggesting that wheat DNA may readily introgress into Aegilops populations (Schoenenberger et al. 2005, 2006; Loureiro et al. 2007a, 2009). Furthermore, backcrossing in Ae. cylindrica generally restores male fertility (Schoenenberger et al. 2006), with values as high as 15.1% and 37.4% for BC1 and BC2 generations (Zaharieva and Monneveux 2006). Recombination between wheat and Aegilops homoeologous chromosomes has been commonly reported in F1 hybrids (Wang et al. 2000; David et al. 2004; Cifuentes et al. 2006) and seems to depend on the occurrence of pairing control genes (Sears 1976) or on the arrangement of homologous transposable elements (TEs) (Chantret et al. 2005; Parisod et al. 2010). Such recombination events may promote the introgression of wheat genes into Aegilops genomes. In the three studied species (i.e. U, M, N and C genomes), homoeologous pairing seems to preferentially involve wheat A and D chromosomes (Fernández-Calvín 1992; Cifuentes et al. 2006; Zaharieva and Monneveux 2006).
The introgression of wheat DNA into Aegilops genomes represents a likely scenario that has been rarely examined in European natural populations (but see Weissmann et al. 2005). This study aims to document gene flow between tetraploid or hexaploid wheat and three common Mediterranean polyploid Aegilops species: Ae. geniculata, Ae. triuncialis and Ae. neglecta. It relies on the detection of wheat genetic markers in naturally occurring Aegilops populations and contrasts populations growing along wheat fields from geographically isolated populations. Here, introgression of wheat DNA into Aegilops genomes is highlighted with genome-wide amplified fragment length polymorphism (AFLP) fingerprinting and fast-evolving, wheat-specific TE insertion markers.
Material and methods
Sampling and plant material
Three Aegilops species were investigated in this study (Table 1): Ae. geniculata (247 individuals, 20 populations), Ae. neglecta (92 individuals, 11 populations) and Ae. triuncialis (133 individuals, 15 populations), totalling 472 Aegilops individuals. Seeds from distinct individuals were collected in naturally occurring populations in Spain, France, Italy, Croatia and Turkey and were subsequently grown at the Botanical Garden of the University and City of Neuchâtel. Because Ae. neglecta includes tetraploid and hexaploid cytotypes, chromosome numbers were checked in the surveyed populations (two individuals per population), using aceto-carmine staining as in Schoenenberger et al. (2006). All Ae. neglecta populations surveyed here were hexaploid (data not shown). Based on morphological inspection following van Slageren (1994), one spontaneous F1 hybrid between Ae. geniculata and T. aestivum was detected in the progeny of a Spanish population that grew along a wheat field (population SP6, Castille e Leon, near Valdesimonte).
Table 1.
Aegilops populations included in the survey with the number of sampled individuals for Aegilops geniculata (Ae. gen.), Ae. neglecta (Ae. negl.) and Ae. triuncialis (Ae. triun.) indicated
| Population | NDD* | EDD* | Distance to wheat cultivation† | Ae. gen | Ae. negl | Ae. triun | |
|---|---|---|---|---|---|---|---|
| Croatia | |||||||
| HR1 | 43.51 | 16.09 | C | 10 m | 2 | 10 | |
| HR2 | 43.47 | 16.69 | C | 30 m | 8 | 9 | |
| France | |||||||
| FR1 | 43.68 | 3.91 | C | 1 m | 17 | ||
| FR2 | 44.05 | 6.06 | C | 50 m | 11 | ||
| FR3 | 43.39 | 5.13 | D | 600 m | 11 | ||
| FR4 | 43.71 | 3.68 | D | 1.3 km | 16 | ||
| Italy | |||||||
| IT1 | 43.59 | 11.57 | C | 1 m | 9 | ||
| IT2 | 43.18 | 11.14 | C | 1 m | 15 | ||
| IT3 | 43.07 | 10.91 | C | 1 m | 11 | 1 | |
| IT4 | 44.69 | 9.99 | D | 100 m | 12 | ||
| IT5 | 43.15 | 11.18 | D | 120 m | 17 | ||
| IT6 | 43.18 | 11.37 | D | 400 m | 4 | 1 | |
| IT7 | 43.54 | 11.42 | D | 1.2 km | 12 | ||
| IT8 | 42.47 | 11.34 | D | >15 km | 12 | ||
| Spain | |||||||
| SP1 | 40.01 | −2.96 | C | 1 m | 23 | 4 | |
| SP2 | 39.98 | −2.09 | C | 1 m | 15 | 6 | |
| SP3 | 38.26 | −5.70 | C | 1 m | 5 | 13 | |
| SP4 | 39.31 | −5.72 | C | 1 m | 11 | ||
| SP5 | 39.46 | −5.17 | C | 1 m | 7 | ||
| SP6 | 41.22 | −3.88 | C | 1 m | 22 | 9 | |
| SP7 | 39.22 | −1.94 | C | 10 m | 5 | 9 | |
| SP8 | 37.54 | −4.34 | C | 10 m | 7 | ||
| SP9 | 39.97 | −4.71 | C | 20 m | 11 | 7 | |
| SP10 | 40.75 | −4.66 | C | 20 m | 14 | ||
| SP11 | 39.17 | −6.25 | D | 200 m | 14 | 7 | |
| SP12 | 36.77 | −5.27 | D | 800 m | 3 | 12 | 8 |
| SP13 | 39.85 | −2.02 | D | 2.4 km | 25 | ||
| SP14 | 37.46 | −4.32 | D | 2.8 km | 14 | ||
| SP15 | 38.15 | −3.02 | D | >5 km | 13 | 7 | |
| SP16 | 37.57 | −4.29 | D | >5 km | 5 | ||
| SP17 | 36.76 | −5.39 | D | 10.4 km | 7 | ||
| Turkey | |||||||
| TU1 | 38.43 | 43.26 | D | >15 km | 11 | ||
Geographical coordinates (decimal degrees): NDD, latitude; EDD, longitude.
Classified according to distance to wheat cultivation: C, close to wheat (closer than 50 m) or D, distant from wheat (farther than 50 m).
Because Aegilops and wheat species share a recent common evolutionary history, involving frequent polyploidy events, it is inherently difficult to distinguish introgression from incomplete lineage sorting. Our sampling design aims to circumvent this limitation by comparing Aegilops populations collected within or further away than 50 m from cultivated fields (i.e. ‘close to wheat’ and ‘distant from wheat’ populations, respectively). This distance is assumed conservative, because the majority of pollen-mediated gene flow between wheat fields was reported within 30 m (Matus-Cadiz et al. 2004; but see Matus-Cadiz et al. 2007 reporting traces of gene flow at 2.75 km). We considered all populations located along cultivated fields, irrespectively of the crop being grown, as the presence of wheat could not be excluded during the preceding years because of culture rotations. The data set was completed with 70 wheat reference accessions collected during field sampling or provided by the agronomic research station ‘Agroscope Changins-Wädenswil’. Wheat reference individuals included 28 lines of tetraploid (BA genomes, including T. durum and T. turgidum) and 42 lines of hexaploid wheat (BAD genomes, including T. aestivum and T. spelta), representing a significant fraction of the wheat genetic diversity cultivated throughout this area (Table S1, Miller 1987). This data set was organized in three subsets by pooling the individuals of each Aegilops species with the wheat references (i.e. Ae. geniculata, Ae. neglecta and Ae. triuncialis subsets).
Amplified fragment length polymorphism fingerprinting
DNA was extracted from 10 mg of silica-dried leaves using a CTAB protocol (Chen and Ronald 1999). DNA concentrations were standardized at 10 ng/ml. We followed the AFLP protocol developed by Gugerli et al. (2008). Reactions were conducted with a PTC-100 (MJ Research) thermocycler, with samples randomly distributed in 96-well plates. Two selective PCR primer pairs were used (EACT-MCTG and EAGT-MCAT), with FAM-labelled EcoRI primers. PCR products were analysed with an ABI 3730XL capillary sequencer (Applied Biosystems, Foster City, CA, USA; service provided by Macrogen Inc. Seoul, Korea). Raw electropherograms were analysed with PEAKSCANNER V1.0 (ABI, using default peak detection parameters except a light smoothing and 100 rfu as fluorescence threshold) to detect and calculate the size of AFLP bands. The scoring was performed on the complete set of individuals (i.e. 472 Aegilops and the 70 wheat individuals) using RawGeno, an automated scoring R CRAN package (Arrigo et al. 2009). The library was settled as follows: scoring range = 100–400 bp, minimum bin width = 0 bp, maximum bin width = 1 bp. AFLP reactions were independently replicated, with five individuals and one blank distributed on each 96-well plate to check for plate variability and 11 chosen randomly individuals from each plate (i.e. representing 12% of the final data set) to eliminate nonreproducible bands. As recommended by Vekemans et al. (2002), the correlation between AFLP band size and frequency among individuals was assessed, but no significant correlation was observed, suggesting a limited impact of homoplasy in our results.
Detection of wheat introgression in Aegilops sp. genomes
Amplified fragment length polymorphism results were displayed with principal co-ordinates analyses, computed on a Jaccard distance matrix between individuals, as implemented in the ‘vegan’ R CRAN package. Furthermore, two statistical approaches were combined to address the question of wheat introgression into wild relatives: (i) a model-based Bayesian clustering analysis and (ii) a nonmodel-based fuzzy clustering analysis. We tested for an association between admixture proportions in each Aegilops subset using Spearman's rank correlations. Finally, generalized linear mixed models, implemented in the ‘glmmPQL’ R CRAN package, were used to test whether admixture proportions of ‘close to wheat’ and ‘distant from wheat’ individuals differed statistically. The population origin of individuals was declared as a covariate to remove its effect on admixture proportions. Computations were performed separately for each Aegilops data set. Models were fitted using a binomial distribution with logit link.
Bayesian clustering analyses
Bayesian inference methods (STRUCTURE 2.2, Falush et al. 2007), working without a priori information on the grouping of individuals, were used to estimate admixture proportions in Aegilops–Triticum individuals. Based on the AFLP data set, the admixture model of STRUCTURE computes the probability that a given allele has its ancestry in each of K groups (with K being user-defined). The probabilities are then averaged over all loci to compute an admixture proportion for each genotype included in the analysis. As wheat and Aegilops species shared a common ancestor, allelic frequencies were possibly correlated among the K groups and we used the admixture model with correlated allele frequencies and a dominant coding of data (the remaining parameters were left with default settings). The algorithm uses a Markov Chain Monte Carlo (MCMC) framework to explore a parameter space considering individual admixture proportions, locus-specific ancestries, population allele frequencies and the expected admixture of the data set. Each set of parameter values is then evaluated by computing the probability of the model predictions given the AFLP data. This probability computation assumes Hardy–Weinberg equilibrium within the K groups. The MCMC algorithm was set up for 500 000 initiation steps (i.e. ‘burn-in’ phase, without results recording), followed by 1 000 000 steps for data acquisition. Each STRUCTURE analysis was replicated ten times, and only runs with the highest maximum-likelihood values were kept for further analyses.
Admixture estimations were performed for each Aegilops subset separately with K = 2 (i.e. the focal Aegilops sp. and wheats). Accordingly, we obtained PAegilops and Pwheat, the respective admixture proportions to the Aegilops sp. and the wheat genetic groups. The distribution of Pwheat values was first compared using generalized linear mixed models within each Aegilops subset, according to the occurrence of individuals as ‘close to wheat’ versus ‘distant from wheat’ (the population origin of individuals was declared as a random covariate to remove its effects). Finally, Pwheat values were displayed on geographical maps and compared between populations within each Aegilops species (this information is also reported in Table S2).
We also used InStruct (Gao et al. 2007), an adapted version of the STRUCTURE algorithm, to consistently address potential departures from Hardy–Weinberg equilibrium, because of self-fertilization being reported in our study organisms (Hammer 1980). InStruct explicitly incorporates the probability of inbreeding when grouping individuals and estimating admixture proportions. The algorithm uses a MCMC framework to explore a parameter space considering the allelic frequencies and the selfing rate in each population, the number of generations since the last selfing event within each simulated lineage, the population assignment of each individual and the respective admixture proportions of each individual within each population. Maximizing the likelihood of the model, given the empirical data, allows estimating admixture proportions for each individual included in the analysis. Computations were performed on servers provided by the Computational Biology Service Unit (CBSU), Cornell University, (http://cbsuapps.tc.cornell.edu), using default parameters except assuming allopolyploidy. Results obtained with STRUCTURE and InStruct were highly congruent (see Appendix S1 for further details). To limit the inclusion of false-positives, we report here admixture estimations of the former program because they were the most conservative.
Although STRUCTURE and InStruct provided consistent and congruent results, both could potentially lead to biased admixture estimations. More specifically, it remained unclear how both programs addressed dominant data in a polyploid context, given that AFLP bands can potentially originate from multiple homoeologous chromosomes. Because of this limitation, we also used a nonmodel-based approach for estimating admixture proportions (see the following paragraph).
Fuzzy clustering analysis (c-means)
We used the fuzzy c-means clustering (hereafter ‘FCM’), an extension of the k-means algorithm working without biological assumptions (Bezdek 1981). Such multidimensional partitioning methods were already successfully applied to AFLPs to detect differentiated polyploid lineages (Burnier et al. 2009; Arrigo et al. 2010). The FCM algorithm follows an iterative process to (i) allocate individuals within a predefined number of groups (K) in a way to minimize the intragroup variance and (ii) calculate memberships of individuals to the K groups. The membership is related to the inverse of the Euclidean distance computed between a focal individual and the centroid of the K groups, thus assuming that each individual has a degree of belonging to each cluster. Being normalized so that the sum equals one, membership can be considered as an admixture measure (e.g. see Kuehn et al. 2004; Gompert et al. 2010). Membership computation relies on a fuzzification parameter (r, ranging between one and infinity) acting on the stringency of clustering. In an admixture detection context, r must be understood as the ‘sensitivity’ of the analysis and asks for optimization: using r ≍ 1 produces nonfuzzy clusters (i.e. memberships take either 1 or 0 as values), while larger r values increase the fuzziness of clustering until all individuals are equally assigned to all genetic groups. As the choice of r is subjective to a certain extent, we tested r values ranging between 1.25 and 2 (as commonly reported in ecological studies, see Gompert et al. 2010). All trials provided results highly congruent with those of STRUCTURE (see Appendix S2), and we presented admixture measures obtained with r = 1.5.
Fuzzy c-means clustering admixture proportions were computed for each Aegilops individuals in comparison with the 70 wheat reference individuals. Analyses were performed considering each Aegilops subset separately with K = 2 groups. A total of 1000 runs were performed per Aegilops subset, and results from runs minimizing the intragroup variance were reported (we used methods implemented in the ‘e1071’ R CRAN package). As explained above, FCM admixture proportions were compared using generalized linear mixed models within each Aegilops subset, according to the occurrence of individuals as ‘close to wheat’ versus ‘distant from wheat’ (the population origin of individuals was declared as a random covariable to remove its effects).
Sequences marking wheat-specific transposable element insertions
Based on extensive sequencing of the chromosome 3B of wheat, Charles et al. (2008) developed markers amplifying specific TE insertions (i.e. encompassing the terminus of a given TE insertion and its flanking genomic region). Given the high sequence turnover of TE fractions in plant genomes (e.g. Parisod et al. 2010 and references therein), TE insertions offer fast-evolving and highly specific markers that are expected to show low homoplasy, helping to confidently document introgression events into Aegilops genomes. Furthermore, TE insertions can be reliably dated by comparing their long terminal repeats (SanMiguel et al. 1996). Here, four TE insertions (loci 11, 70, 81 and 89) were selected and amplified according to Charles et al. (2008). These TE insertions were located on two different BACs (TA3B63B7 for loci 11 and 81; TA3B95C9 for loci 70 and 89) of the long arm of chromosome 3B of wheat. Loci 11 and 81 were separated by 36.5 kb, while loci 70 and 89 were located at 52 kb from each other. TE insertions surveyed by loci 11, 89 and 81 were dated at 3.7 ± SE 0.54 million years ago (hereafter MYA), 2.5 ± SE 0.31 MYA and 1.5 ± SE 0.12 MYA, respectively (Charles et al. 2008), and thus happened after the divergence of the wheat B genome from the U, M, N and C Aegilops genomes (i.e. estimated at 3.5 ± SE 0.5 MYA; Dvorak and Akhunov 2005; Salse et al. 2008). Whether the locus 70 was exclusive to the 3B wheat chromosome is less clear, as this TE insertion was dated at 5.04 ± SE 0.2 MYA (Charles et al. 2008). The four TE-based markers wheat markers were successfully amplified in all of the 54 Triticum individuals tested (i.e. 41 individuals of T. aestivum and 13 T. turgidum) and were further screened in all Ae. triuncialis and Ae. neglecta individuals and in 111 Ae. geniculata individuals (i.e. five individuals per population). Fragments resulting from positive amplifications were sequenced (service provided by Macrogen Inc.) to confirm homology with wheat sequences. The percentage of diverging base pairs between Aegilops and wheat sequences was computed using MEGA (Kumar et al. 2008), and only sequences having more than 95% identity were conservatively considered as homologous.
Results
The AFLP analysis provided a total of 191 fully reproducible bands (113 and 78 bands for EACT-MCTG and EAGT-MCAT, respectively), with an average of 50 bands per individual. The data set included several species-specific loci that clearly discriminated Ae. geniculata, Ae. neglecta, Ae. triuncialis, tetraploid and hexaploid wheats based on principal co-ordinates analysis (Fig. 1).
Figure 1.
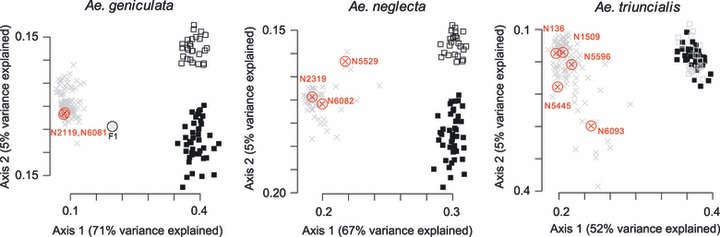
Principal co-ordinates analyses of the three amplified fragment length polymorphism subsets computed using the Jaccard distance (i.e. 247, 92 and 133 individuals of Aegilops geniculata, Ae. neglecta and Ae. triuncialis were, respectively, compared with 70 wheat reference individuals). The two-first axes and their contribution in explaining the global variance are displayed. Aegilops (x) and wheat (□– tetraploid, BA genome and ▪ – hexaploid, BAD genome) individuals are labelled according to their taxonomic status. Aegilops individuals with positive TE-based marker amplifications are highlighted, and their corresponding accession numbers are provided. The Ae. geniculata x Triticum aestivum hybrid is highlighted (F1).
Bayesian clustering analysis
STRUCTURE consistently assigned individuals to genetic groups representing the taxonomic species (i.e. the focal Aegilops sp. and wheats). Individuals were considered as admixed when their wheat admixture proportions (hereafter Pwheat) exceeded 0.1 (see Appendix S1 for further details). STRUCTURE outlined different results between Aegilops species (Fig. 2A). For instance, the majority of Ae. geniculata individuals obtained low wheat assignment probabilities, with only seven individuals (2.8% of the sampling) showing wheat admixture proportions greater than 0.1 (the highest probability, Pwheat = 0.385, was observed for the F1 hybrid between Ae. geniculata and T. aestivum). No significant Pwheat difference was measured between ‘close to wheat’ and ‘distant from wheat’ individuals for this species (Fig. 2A).
Figure 2.
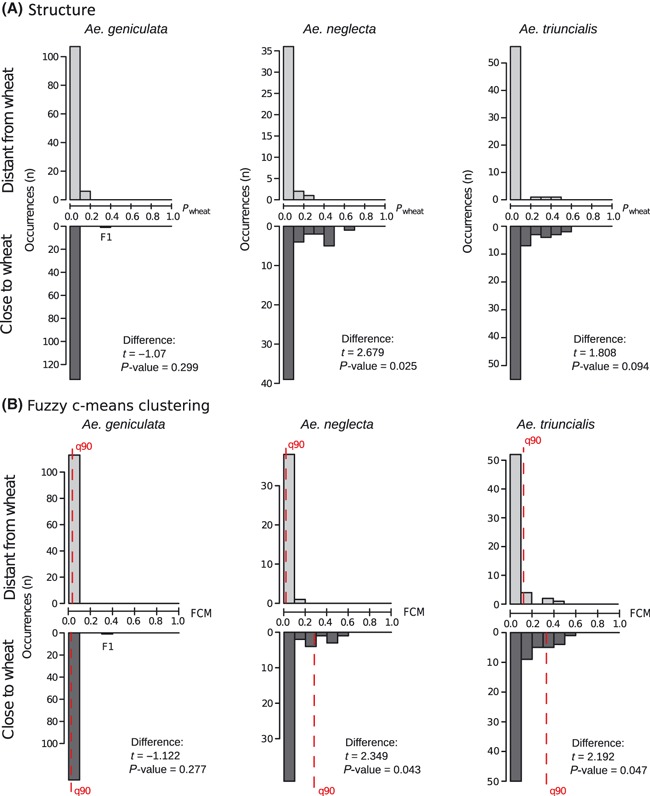
Distribution of admixture proportions for the surveyed Aegilops individuals, based on (A) the admixture proportions of fuzzy c-means clustering (FCM) and (B) wheat assignment probabilities (Pwheat, computed using STRUCTURE), according to the proximity of Aegilops individuals to wheat cultivations. Statistical differences between individuals collected close to wheat fields (i.e. closer than 50 m from cultivation) and those collected in wheat distant areas (i.e. located at more than 50 m from cultivation) were tested using generalized linear mixed models of which results and P-value are reported (binomial distribution, logit link, the effect of population origin was removed by declaring it as a covariable). The Aegilops geniculata × Triticum aestivum F1 hybrid detected on morphological grounds is highlighted. For FCM results, the 90% quantile of admixture proportions (q90) is indicated.
In marked contrast, the Ae. neglecta and Ae. triuncialis subsets presented numerous intermediate genotypes. In particular, 17 individuals of Ae. neglecta (24% of the subset) and 22 individuals of Ae. triuncialis (16% of the subset) were admixed (i.e. Pwheat > 0.1). In addition, most admixed individuals were collected close to wheat cultivation (Fig. 2A), as indicated by significant differences of Pwheat values between ‘close to wheat’ and ‘distant from wheat’ individuals (t = 2.349, P = 0.043 and t = 2.192, P = 0.047, for Ae. neglecta and Ae. triuncialis, respectively). At the population scale (Fig. 3 and Table S2), admixed individuals could represent a large proportion of the sampling effort. For instance, more than 25% of the sampled individuals were admixed in seven of the populations (i.e. HR1, HR2, IT1, SP5 and HR1, SP3, SP12, in Ae. neglecta and Ae. triuncialis subsets, respectively). For both species, the few admixed individuals collected ‘distant from wheat’ occurred mostly in population SP12, a pasture located at 800 m distance from current crop cultivations. Finally, one admixed individual was detected at more than 15 km from cultivations (IT8 population).
Figure 3.
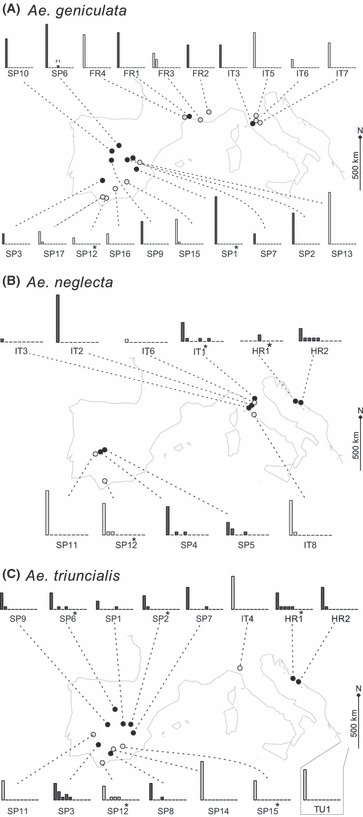
Detailed geographical distribution of the surveyed populations, for (A) Aegilops geniculata, (B) Ae. neglecta and (C) Ae. triuncialis. The distributions of admixtures proportions, as estimated using STRUCTURE, are displayed for each population using histograms [x-axis: admixture proportions ranging from 0 (pure Aegilops) to 1 (pure wheat), split into 10 equivalent admixture classes, y-axis: number of individuals per admixture class]. Populations collected close to wheat fields (i.e. closer than 50 m from cultivation) and those collected in wheat distant areas (i.e. located at more than 50 m from cultivation) are displayed in dark and light grey, respectively. Populations where TE-based markers were detected are highlighted with a star (*). Note that the TU1 population (Ae. triuncialis) is located outside of the mapped area.
Fuzzy clustering analysis
The FCM algorithm consistently assigned Aegilops sp. and wheat individuals to distinct genetic groups and allowed for computing admixture proportions. The obtained results were highly congruent with those obtained with STRUCTURE (Appendix S2). For all three Aegilops species, FCM admixture proportions followed exponential-shaped distributions and outlined a considerable amount of Aegilops individuals with admixed genotypes (Fig. 2B). Results varied between Aegilops species, as revealed by 90% quantile of admixture proportion distributions (hereafter ‘q90’) indicating that Ae. geniculata presented globally the lowest admixture proportions (q90 = 0.03), while Ae. neglecta and Ae. triuncialis had higher values (with q90 = 0.15 and 0.26, respectively). In addition, Ae. neglecta individuals collected close to wheat fields showed significantly higher admixture proportions than individuals collected far away from cultivations (Fig. 2B, = 0.11 and 0.33, t = 2.679 and P-value = 0.025). Similar trends were reported for Ae. triuncialis, although the difference was not significant (q90 = 0.02 and 0.28, t = 1.808 and P-value = 0.094).
Cross-check of introgression detection methods
Admixture estimations obtained with STRUCTURE and the nonmodel-based FCM algorithm were revealed by Spearman's rank correlation tests as strongly and significantly congruent (Table 2). Associations however varied between the three Aegilops subsets. The best associations were observed in the Ae. neglecta and Ae. triuncialis data sets, while Ae. geniculata showed lower correlations.
Table 2.
Correlations among admixture estimations used to highlight gene flow between wheat and Aegilops with amplified fragment length polymorphisms
| Subset | FCM –Pwheat† |
|---|---|
| Ae. geniculata | 0.65*** |
| Ae. neglecta | 0.83*** |
| Ae. triuncialis | 0.95*** |
†Spearman's rank correlation (ρ, 1000 permutations, ***P-value < 0.001).
Validation from wheat TE insertions
The screening of four loci marking wheat-specific TE insertions resulted in a total of 18 successful amplifications in Aegilops individuals. Among those fragments, 13 sequences diverged between 0% and 2.6% from corresponding wheat sequences and were considered as homologous (GenBank accessions numbers HQ659715 to HQ659741). Those 13 fragments were observed in a total of 10 individuals (3% of the sampling) from all three Aegilops species (Table 3). In particular, two, three and eight wheat-specific TE insertions were successfully amplified in Ae. geniculata, Ae. neglecta and Ae. triuncialis, respectively. The co-occurrence of multiple TE-based markers was observed in only two individuals (N5596 and N6063), which presented two and three wheat-specific TE insertions, respectively. In addition, wheat-specific TE insertions were detected in individuals already highlighted as admixed by AFLPs (e.g. N5529, N6063). These wheat sequences were detected in seven distinct populations of Aegilops, mostly located ‘close to wheat’ fields. As a notable exception, a wheat-specific TE insertion was detected in an Ae. triuncialis individual collected in SP15, a population located at more than 5 km from current wheat cultivation. While most of these populations contained only one positive individual, two populations (HR1 and SP12) showed, respectively, two and three individuals with wheat-specific TE insertions.
Table 3.
Introgression events evidenced with wheat-specific transposable element (TE)-based markers
| TE-based markers† | ||||||
|---|---|---|---|---|---|---|
| Individual (Pop.) | Distance to wheat cultivation* | 11 | 70 | 81 | 89 | |
| Ae. geniculata | ||||||
| N2119 (SP1) | C | 1 m | 1 | |||
| N6081 (SP12) | D | 800 m | 1 | |||
| Ae. neglecta | ||||||
| N5529 (HR1) | C | 10 m | 1 | |||
| N6082 (SP12) | D | 800 m | 1 | |||
| N2319 (IT1) | C | 1 m | 1 | |||
| Ae. triuncialis | ||||||
| N5445 (HR1) | C | 10 m | 1 | |||
| N136 (SP2) | C | 1 m | 1 | |||
| N1509 (SP6) | C | 1 m | 1 | |||
| N6063 (SP12) | D | 800 m | 1 | 1 | 1 | |
| N5596 (SP15) | D | >5 km | 1 | 1 | ||
Classified according to distance to wheat cultivation: C, close to wheat (closer than 50 m) or D, distant from wheat (farther than 50 m).
Loci 11, 70, 81 and 89, see Material and methods for details.
Discussion
Evidence of gene flow between wheat and Aegilops species
Genome-wide AFLP markers have proved reliable to detect hybridization and introgression in a range of species (O'Hanlon et al. 1999; Monte et al. 2001; Pester et al. 2003; Bonin et al. 2007). Our study compared populations collected close and distant from wheat cultivations, using two molecular marker systems as well as two distinct statistical approaches (i.e. model-based STRUCTURE and nonmodel-based FCM). As a whole, results offered congruent insights (Figs 2 and 3) by showing admixed Aegilops individuals to occur more frequently close to wheat cultivations. Although homoplasy in our AFLP data set cannot be formally excluded, we think that this potential bias had only slightly affected our results. First, homoplasy is expected to introduce misleading homologies between nonrelated individuals, which would have decreased the global discrimination power of our AFLPs by making wheat and Aegilops genotypes appear more similar. Furthermore, if it occurred, homoplasy would probably have affected admixture estimations on a random basis. In contrast, we found here a significant effect of cultivation proximity on admixture proportions of Aegilops individuals, supporting the robustness of the insights offered here by AFLPs.
Amplified fragment length polymorphisms clearly distinguished the different Aegilops species and highlighted a large number of individuals showing introgression from wheat, representing more than 25% of the sampling effort in several populations (Fig. 3, Table S2). AFLPs most likely detected recent introgression events (i.e. ranging from F1s to early generation backcrosses), because few wheat-specific bands were expected to be retained in advanced backcrossed individuals, thus providing marginal contributions to STRUCTURE and FCM admixture proportions. Accordingly, ancient introgression events likely remained underestimated with this approach.
On the other hand, indirect observations gathered with AFLPs were largely confirmed by fast-evolving markers based on TE insertions, with 10 Aegilops individuals successfully amplifying up to 13 TE insertion sites specific to wheat (Table 3). For instance, individuals issued from recent introgression were revealed with the co-occurrence of several TE-based markers (e.g. N5596 or N6063 that were already detected with AFLPs). Ancient introgression events might also have been detected with TE-based markers, because Aegilops individuals showing low admixture levels with AFLPs were amplifying wheat-specific TE insertions (e.g. N2119, N6081, N2319, N1509). However, TE-based markers used here (i.e. loci 11, 70, 81 and 89) were linked on the chromosome 3B of wheat, and only admixed individuals having retained the corresponding wheat genomic regions could be detected. Therefore, individuals highlighted as admixed with AFLPs but not amplifying any wheat-specific TE insertions probably represented introgressed individuals having retained other parts of the wheat genome and TE-based markers also certainly underestimated introgression. This reliable approach should be extended to genome-wide markers of TE insertions to document the magnitude of such gene flow, both in terms of extent and integrity of wheat chromosomes retained within admixed Aegilops genomes.
Variation in gene flow according to mating system and remoteness of populations
Our approach revealed only limited gene flow from wheat to Ae. geniculata, while Ae. neglecta and Ae. triuncialis showed substantial evidence of introgression (Fig. 1). Both wheat and Aegilops produce mostly cleistogamous flowers and are considered largely autogamous (Hammer 1980). The observed pattern of gene flow might however be associated with mating system differences between the three Aegilops species. Aegilops neglecta and Ae. triuncialis have a well-developed stigma and produce abundant pollen, while Ae. geniculata has a short stigma and produces limited amounts of pollen. This suggests that the former species may be more allogamous (Hammer 1980) and thus more likely to hybridize with wheat.
The majority of admixed Aegilops sp. individuals occurred within 50 m from wheat fields (i.e. ‘close to wheat’ populations, Figs 2 and 3). Accordingly, the majority of Aegilops × Triticum F1 hybrids detected in natural conditions occurred close to wheat cultivations (Zaharieva and Monneveux 2006). As pollen-mediated gene flow from wheat is expected to be more important along wheat fields than in remote locations (i.e. 20–30 m pollen is a commonly reported range for pollen movements, Matus-Cadiz et al. 2004; Hanson et al. 2005; Loureiro et al. 2007b), this observation is not surprising and indicates that wheat to Aegilops gene flow might be a local-scale phenomenon. Nevertheless, a several admixed individuals were observed in populations distant from wheat cultivations (i.e. the SP12, SP15 and IT8 populations, collected at 800 m, >5 km and >15 km from cultivations, respectively, Fig. 3). Because long-distance pollen dispersal of wheat has been reported (De Vries 1971; Zaharieva and Monneveux 2006; Matus-Cadiz et al. 2007), it might partly explain our results. In addition, cattle or human activities could also promote long-distance seed dispersal (van Slageren 1994), which could explain the presence of introgressed individuals in populations remotely connected to present wheat cultivation (e.g. the SP15 or the IT8 populations). Finally, long-term persistence of wheat alleles in Aegilops populations should also be considered. For instance, Weissmann et al. (2005) reported traces of ancient gene flow from wheat in individuals of Ae. peregrina collected in fields abandoned for more than 30 years. This situation could for instance explain the presence of admixed individuals into the three surveyed species, in SP12 a population standing at 800 m from cultivations. Finally, the individual N5596 (population SP15), which showed wheat-specific TE insertions (Table 3), but a limited signature of hybridization based on AFLPs (Fig. 3), could also represent an ancient hybridization event.
Containment of wheat genes
As a whole, our study offered a qualitative survey of natural populations questioning the commonly admitted strength of reproductive barriers limiting the introgression of wheat genes into Aegilops (i.e. autogamy and sterility of F1 hybrids). Accordingly, it questions wheat gene containment strategies relying strictly on allopatry and suggests that the release of commercial transgenic wheat in southern Europe should take into account the possible introgression of transgenes into wild Aegilops populations. These conclusions might also be extended to north-western America, where Ae. triuncialis was recently introduced and now invades the pastures of California. In practical terms, these results outline gene flow from wheat to Aegilops as a potential issue that has to be explicitly addressed during risk assessment studies associated with the release of genetically modified wheat cultivars. More specifically, escape routes of transgenes (i.e. pollen- versus seed-mediated gene flow), dispersal abilities (i.e. cattle-mediated seed movements) and consequences on the fitness of admixed individuals should be systematically investigated (see also Willenborg and Van Acker 2008). In conclusion, suitable strategies limiting the potential spread and persistence of transgenes in natural populations should be developed if transgenic wheats were to be released in European or American agrosystems.
Acknowledgments
We are grateful to the team of the Botanical Garden of the University and City of Neuchâtel for their help in plants cultivation. We thank Boulos Chalhoub, Harry Belcram and Jérémy Just for providing assistance regarding TE-based markers. We are also grateful to two anonymous reviewers for valuable comments on earlier versions of the manuscript. Nadir Alvarez, Anouk Sarr, Celia Bueno, Jérôme Goudet, Graham Chapman, Jacques David, Geert Kleier, Philippe Küpfer, Mélanie Thobor and Ilhan Kaya provided useful comments and technical assistance. This project was funded by the National Centre of Competence in Research (NCCR) Plant Survival and the PNR-59, two research programmes of the Swiss National Science Foundation.
Data archiving statement
Raw data for this paper are archived in Dryad. http://dx.doi.org/10.5061/dryad.8955
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. Wheat references included in the analysis.
Table S2. Distribution of structure admixture proportions at the population level.
Appendix S1. Comparing STRUCTURE and InStruct.
Appendix S2. Optimizing the sensitivity of fuzzy c-means clustering.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Arrigo N, Tuszynski JW, Ehrich D, Gerdes T, Alvarez N. Evaluating the impact of scoring parameters on the structure of intra-specific genetic variation using RawGeno, an R package for automating AFLP scoring. BMC Bioinformatics. 2009;10:33. doi: 10.1186/1471-2105-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigo N, Felber F, Parisod C, Buerki S, Alvarez N, David J, Guadagnuolo R. Origin and expansion of the allotetraploid Aegilops geniculata, a wild relative of wheat. New Phytologist. 2010;187:1170–1180. doi: 10.1111/j.1469-8137.2010.03328.x. [DOI] [PubMed] [Google Scholar]
- Bezdek J. Pattern Recognition with Fuzzy Objective Function Algoritms. New York: Plenum Press; 1981. [Google Scholar]
- Bonin A, Ehrich D, Manel S. Statistical analysis of amplified fragment length polymorphism data: a toolbox for molecular ecologists and evolutionists. Molecular Ecology. 2007;16:3737–3758. doi: 10.1111/j.1365-294X.2007.03435.x. [DOI] [PubMed] [Google Scholar]
- Burnier J, Buerki S, Arrigo N, Kupfer P, Alvarez N. Genetic structure and evolution of Alpine polyploid complexes: Ranunculus kuepferi (Ranunculaceae) as a case study. Molecular Ecology. 2009;18:3730–3744. doi: 10.1111/j.1365-294X.2009.04281.x. [DOI] [PubMed] [Google Scholar]
- Chantret N, Salse J, Sabot F, Rahman S, Bellec A, Laubin B, Dubois I, et al. Molecular basis of evolutionary events that shaped the hardness locus in diploid and polyploid wheat species (Triticum and Aegilops. Plant Cell. 2005;17:1033–1045. doi: 10.1105/tpc.104.029181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles M, Belcram H, Just J, Huneau C, Viollet A, Couloux A, Segurens B, et al. Dynamics and differential proliferation of transposable elements during the evolution of the B and A genomes of wheat. Genetics. 2008;180:1071–1086. doi: 10.1534/genetics.108.092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DH, Ronald PC. A rapid DNA minipreparation method suitable for AFLP and other PCR applications. Plant Molecular Biology Reporter. 1999;17:53–57. [Google Scholar]
- Cifuentes M, Blein M, Benavente E. A cytomolecular approach to assess the potential of gene transfer from a crop (Triticum turgidum L.) to a wide relative (Aegilops geniculata Roth.) Theoretical and Applied Genetics. 2006;112:657–664. doi: 10.1007/s00122-005-0168-z. [DOI] [PubMed] [Google Scholar]
- David JL, Benavente E, Bres-Patry C, Dusautoir JC, Echaide M. Are neopolyploids a likely route for a transgene walk to the wild? The Aegilops ovata × Triticum turgidum durum case. Biological Journal of the Linnean Society. 2004;82:503–510. [Google Scholar]
- De Vries AP. Flowering biology of wheat, particularly in view of hybrid seed production - a review. Euphytica. 1971;20:152–170. [Google Scholar]
- Dvorak J, Akhunov ED. Tempos of gene locus deletions and duplications and their relationship to recombination rate during diploid and polyploid evolution in the Aegilops-Triticum alliance. Genetics. 2005;171:323–332. doi: 10.1534/genetics.105.041632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellstrand NC, Prentice HC, Hancock JF. Gene flow and introgression from domesticated plants into their wild relatives. Annual Review of Ecology and Systematics. 1999;30:539–563. [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felber F, Kozlowski G, Arrigo N, Guadagnuolo R. Genetic and ecological consequences of transgene flow to the wild flora. In: Fiechter A, Suatter C, editors. Advances in Biochemical Engineering/Biotechnology. Berlin: Springer-Verlag; 2007. pp. 173–205. [DOI] [PubMed] [Google Scholar]
- Fernández-Calvín B, Orellan J. Relationship between pairing frequencies and genome affinity estimations in Aegilops ovata ×Triticum aestivum hybrid plants. Heredity. 1992;68:165–172. doi: 10.1038/hdy.1992.25. [DOI] [PubMed] [Google Scholar]
- Gao H, Williamson S, Bustamante C. An MCMC approach for joint inference of population structure and inbreeding rates from multi-locus genotype data. Genetics. 2007;176:1635–1651. doi: 10.1534/genetics.107.072371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompert Z, Lucas LK, Fordyce JA, Forister ML, Nice CC. Secondary contact between Lycaeides idas and L. melissa in the Rocky Mountains: extensive admixture and a patchy hybrid zone. Molecular Ecology. 2010;19:3171–3192. doi: 10.1111/j.1365-294X.2010.04727.x. [DOI] [PubMed] [Google Scholar]
- Guadagnuolo R, Savova-Bianchi D, Felber F. Gene flow from wheat (Triticum aestivum L.) to jointed goatgrass (Aegilops cylindrica Host.), as revealed by RAPD and microsatellite markers. Theoretical And Applied Genetics. 2001;103:1–8. [Google Scholar]
- Gugerli F, Englisch T, Niklfeld H, Tribsch A, Mirek Z, Ronikier M, Zimmermann NE, et al. Relationships among levels of biodiversity and the relevance of intraspecific diversity in conservation - a project synopsis. Perspectives in Plant Ecology, Evolution and Systematics. 2008;10:259–281. [Google Scholar]
- Hammer K. Vorarbeiten zur Monographischen Darstellung von Wildpflanzensortimenten: Aegilops L. Kulturpflanze. 1980;28:33–180. [Google Scholar]
- Hanson BD, Mallory-Smith CA, Shafii B, Thill DC, Zemetra R. Pollen-mediated gene flow from blue aleurone wheat to other wheat cultivars. Crop Science. 2005;45:1610–1617. [Google Scholar]
- Karagöz A. Hybridization in Turkish Aegilops L. species. Pakistan Journal of Biological Science. 2006;9:2243–2248. [Google Scholar]
- Kuehn R, Haller H, Schroeder W, Rottmann O. Genetic Roots of the Red Deer (Cervus elaphus) Population in Eastern Switzerland. Journal of Heredity. 2004;95:136–143. doi: 10.1093/jhered/esh019. [DOI] [PubMed] [Google Scholar]
- Kumar S, Dudley J, Nei M, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro I, Escorial MC, Garcia-Baudin JM, Chueca MC. Evidence of natural hybridization between Aegilops geniculata and wheat under field conditions in central Spain. Environmental Biosafety Research. 2006;5:105–109. doi: 10.1051/ebr:2006020. [DOI] [PubMed] [Google Scholar]
- Loureiro I, Escorial MC, Garcia-Baudin JM, Chueca MC. Hybridization between wheat (Triticum aestivum) and the wild species Aegilops geniculata and A. biuncialis under experimental field conditions. Agriculture Ecosystems & Environment. 2007a;120:384–390. [Google Scholar]
- Loureiro I, Escorial MC, Gonzáles-Anduja JL, Garcia-Baudin JM, Chueca MC. Wheat pollen dispersal under semiarid field conditions: potential outcrossing with Triticum aestivum and Triticum turgidum. Euphytica. 2007b;156:25–37. [Google Scholar]
- Loureiro I, Escorial MC, Garcia-Baudin JM, Chueca MC. Hybridization, fertility and herbicide resistance of hybrids between wheat and Ae. biuncialis. Agronomy for Sustainable Development. 2009;29:237–245. [Google Scholar]
- Matus-Cadiz MA, Hucl P, Horak MJ, Blomquist LK. Gene flow in wheat at the field scale. Crop Science. 2004;44:718–727. [Google Scholar]
- Matus-Cadiz MA, Hucl P, Dupuis B. Pollen-mediated gene flow in wheat at the commercial scale. Crop Science. 2007;47:573–581. [Google Scholar]
- Miller TE. Systematics and Evolution. In: Lupton EGH, editor. Wheat breeding – its scientific basis. London, New York: Chapman & Hall Ltd; 1987. pp. 1–28. [Google Scholar]
- Monte JV, De Nova PJG, Soler C. AFLP-based analysis to study genetic variability and relationships in the Spanish species of the genus Aegilops. Hereditas. 2001;135:233–238. doi: 10.1111/j.1601-5223.2001.00233.x. [DOI] [PubMed] [Google Scholar]
- Morrison LA, Riera-Liazrazu O, Cremieux L, Mallory-Smith CA. Jointed goatgrass (Aegilops cylindrica host) × wheat (Triticum aestivum L.) hybrids: hybridization dynamics in Oregon wheat fields. Crop Science. 2002;42:1863–1872. [Google Scholar]
- O'Hanlon PC, Peakall R, Briese DT. Amplified fragment length polymorphism (AFLP) reveals introgression in weedy Onopordum thistles: hybridization and invasion. Molecular Ecology. 1999;8:1239–1246. doi: 10.1046/j.1365-294x.1999.00646.x. [DOI] [PubMed] [Google Scholar]
- Parisod C, Alix K, Just J, Petit M, Sarilar V, Mhiri C, Ainouche M, et al. Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytologist. 2010;186:37–45. doi: 10.1111/j.1469-8137.2009.03096.x. [DOI] [PubMed] [Google Scholar]
- Pester TA, Ward SM, Fenwick AL, Westra P, Nissen SJ. Genetic diversity of jointed goatgrass (Aegilops cylindrica) determined with RAPD and AFLP markers. Weed Science. 2003;51:287–293. [Google Scholar]
- Salse J, Chagué V, Bolot S, Magdelenat G, Huneau C, Pont C, Belcram H, et al. New insights into the origin of the B genome of hexaploid wheat: evolutionary relationships at the SPA genomic region with the S genome of the diploid relative Aegilops speltoides. BMC Genomics. 2008;9:555. doi: 10.1186/1471-2164-9-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- Schneider A, Molnar I, Molnár-Láng MM. Utilisation of Aegilops (goatgrass) species to widen the genetic diversity of cultivated wheat. Euphytica. 2008;163:1–19. [Google Scholar]
- Schoenenberger N, Felber F, Savova-Bianchi D D, Guadagnuolo R. Introgression of wheat DNA markers from A, B and D genomes in early generation progeny of Aegilops cylindrica Host x Triticum aestivum L. hybrids. Theoretical and Applied Genetics. 2005;111:1338–1346. doi: 10.1007/s00122-005-0063-7. [DOI] [PubMed] [Google Scholar]
- Schoenenberger N, Guadagnuolo R, Savova-Bianchi D, Kupfer P, Felber F. Molecular analysis, cytogenetics and fertility of introgression lines from transgenic wheat to Aegilops cylindrica host. Genetics. 2006;174:2061–2070. doi: 10.1534/genetics.106.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears ER. Genetic-control of chromosome-pairing in wheat. Annual Review of Genetics. 1976;10:31–51. doi: 10.1146/annurev.ge.10.120176.000335. [DOI] [PubMed] [Google Scholar]
- van Slageren MW. Wild wheats: A monograph of Aegilops L. and Ambylopyrum (Jaub and Spach) Eig (Poaceae) Wageningen, The Netherlandsand Int. Cent. for Agric. Res. in Dry Areas, Aleppo, Syria. Wageningen: Agricultural Univ; 1994. [Google Scholar]
- Trucco F, Tatum T, Rayburn AL, Tranel P. Out of the swamp: unidirectional hybridization with weedy species may explain the prevalence of Amaranthus tuberculatus as a weed. New Phytologist. 2009;184:819–827. doi: 10.1111/j.1469-8137.2009.02979.x. [DOI] [PubMed] [Google Scholar]
- Vekemans X, Beauwens T, Lemaire M, Roldan-Ruiz I. Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Molecular Ecology. 2002;11:139–151. doi: 10.1046/j.0962-1083.2001.01415.x. [DOI] [PubMed] [Google Scholar]
- Waines JG, Barnhart D. Biosystematic research in Aegilops and Triticum. Hereditas. 1992;116:207–212. [Google Scholar]
- Wang ZN, Hang A, Hansen J, Burton C, Mallory-Smith CA, Zemetra RS. Visualization of A- and B-genome chromosomes in wheat (Triticum aestivum L.) x jointed goatgrass (Aegilops cylindrica Host) backcross progenies. Genome. 2000;43:1038–1044. doi: 10.1139/g00-080. [DOI] [PubMed] [Google Scholar]
- Weissmann S, Feldman M, Gressel J. Sequence evidence for sporadic intergeneric DNA introgression from wheat to a wild Aegilops species. Molecular Biology and Evolution. 2005;22:2055–2062. doi: 10.1093/molbev/msi196. [DOI] [PubMed] [Google Scholar]
- Willenborg CJ, Van Acker RC. The biology and ecology of hexaploid wheat (Triticum aestivum L.) and its implications for trait confinement. Canadian Journal of Plant Science. 2008;88:997–1013. [Google Scholar]
- Zaharieva M, Monneveux P. Spontaneous hybridization between bread wheat (Triticum aestivum L.) and its wild relatives in Europe. Crop Science. 2006;46:512–527. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


