Abstract
Growth hormone (GH) transgenic Atlantic salmon (Salmo salar) is one of the first transgenic animals being considered for commercial farming, yet ecological and genetic concerns remain should they enter the wild and interact reproductively with wild fish. Here, we provide the first empirical data reporting on the breeding performance of GH transgenic Atlantic salmon males, including that of an alternative male reproductive phenotype (i.e. small, precocially mature parr), in pair-wise competitive trials within a naturalised stream mesocosm. Wild anadromous (i.e. large, migratory) males outperformed captively reared transgenic counterparts in terms of nest fidelity, quivering frequency and spawn participation. Similarly, despite displaying less aggression, captively reared nontransgenic mature parr were superior competitors to their transgenic counterparts in terms of nest fidelity and spawn participation. Moreover, nontransgenic parr had higher overall fertilisation success than transgenic parr, and their offspring were represented in more spawning trials. Although transgenic males displayed reduced breeding performance relative to nontransgenics, both male reproductive phenotypes demonstrated the ability to participate in natural spawning events and thus have the potential to contribute genes to subsequent generations.
Keywords: alternative reproductive tactics, genetically modified organisms, growth hormone transgenesis, introgression, precocially mature male parr, reproduction
Introduction
Growth-enhancing transgenic biotechnologies have attracted considerable interest from the global aquaculture industry, particularly with regard to Atlantic salmon. However, similar to domesticated strains, concerns have been raised regarding the ecological and genetic effects that may arise if these organisms were to enter the wild (Kapuscinski and Hallerman 1991; Devlin et al. 2006; Kapuscinski et al. 2007). A principal concern involves the potential genetic impacts of fertile transgenic organisms interbreeding with wild populations into which their genes may introgress. For example, risk models indicate that Trojan gene effects may occur, whereby the transgene spreads by enhanced mating advantage but the resulting offspring have reduced viability, which leads to the eventual extinction of populations (Muir and Howard 1999, 2002; Howard et al. 2004). However, there has yet to be any empirical research documenting the ability of growth hormone (GH) transgenic Atlantic salmon to breed naturally and introgress with wild populations. Moreover, there is little understanding of the role that alternative reproductive phenotypes may play in such introgression.
The breeding system of Atlantic salmon exhibits two alternative male reproductive phenotypes, large anadromous adults that have migrated to sea and returned to their natal streams, and small precocial parr that have matured in freshwater, having never been to sea. Anadromous males develop specialised secondary sexual characters to fight other males and court for access to ovipositing females, while precocial parr mature at a fraction of the size of the anadromous phenotype and use their small size and cryptic colouration to sneak fertilisations (reviewed in Fleming 1996). Both male reproductive phenotypes may form dominance hierarchies among themselves for access to spawning females through aggressive behavioural interactions. While the fertilisation success of anadromous males is typically greater than that of mature parr, reports of precocial parr fertilisation rates have ranged from 11% to 65% of the available eggs (reviewed in Fleming and Reynolds 2004). Thus, both male reproductive phenotypes can contribute substantially to the next generation and represent potential routes for the introduction of transgenes into wild populations.
The extent of transgene introgression into wild populations would depend on the fitness of transgenic individuals in the receiving environment, which may vary along a continuum featuring high fitness, leading to the fixation of the transgene, at one end and low fitness, leading to its elimination within a few generations, at the other (Muir and Howard 1999, 2002). Perhaps more commonly, however, the fitness of transgenic organisms would lie between these poles and create, for example, an outbreeding depression scenario where transgene-induced maladaptive traits pose a threat to the viability of the entire receiving population (Hedrick 2001).
This outbreeding depression scenario is representative of the concerns associated with wild salmonid populations exposed to strains that have experienced domestication selection (McGinnity et al. 2003; Tymchuk et al. 2007; Fraser et al. 2008). In Atlantic salmon, anadromous adults from aquaculture strains (farmed) exhibit atypical spawning behaviour, including reductions in aggressive displays towards other males, quivering and nest fidelity, which may contribute to observations of reduced reproductive success (Fleming 1996; Fleming et al. 2000; Weir et al. 2004). In contrast, studies exploring the relative reproductive behaviour and success of mature farmed and wild parr have found that farmed parr perform similarly to or better than wild parr (Garant et al. 2003; Weir et al. 2005). Regardless of the relative spawning success of farmed and wild males, both reproductive phenotypes have demonstrated the potential for the introgression of farmed genes into wild populations and the disruption of locally adapted phenotypic traits (Hindar et al. 2006; Garcia de Leaniz et al. 2007; Fraser et al. 2010).
Comparisons of reproductive performance between GH transgenic and nontransgenic salmonids are limited. Similar to observations with farmed adults, previous efforts have reported reduced reproductive performance in hatchery-reared transgenic relative to wild coho salmon (Oncorhynchus kisutch; Bessey et al. 2004; Fitzpatrick et al. 2011). While these results represent the expectations of a first-generation transgenic escapee scenario, GH transgenic Atlantic and coho salmon represent two species carrying two unique transgene constructs with two distinct life histories (e.g. rarely do coho salmon mature precocially as parr; Fleming 1998). For example, previous work has demonstrated differences in the onset of transgene-induced phenotypic expression between the two species, which may have important implications for early survival (Sundstrom et al. 2004, 2005; Lohmus et al. 2010; Moreau et al. 2011). Potentially more important are the distinct differences in reproductive phenotypes that may have implications for introgression (Valosaari et al. 2008), as seen in the reproductive performance differences between anadromous and mature parr Atlantic salmon males of farmed origin (Fleming 1996; Fleming et al. 2000; Garant et al. 2003; Weir et al. 2005).
The aim of this study was to compare the breeding performance of GH transgenic and nontransgenic Atlantic salmon males of both alternative reproductive phenotypes to test for the potential of the transgene to introgress into wild populations. We conducted two separate experiments in a naturalised stream mesocosm. First, to assess the ability of first-generation, farmed transgenic males to contribute reproductively, the breeding behaviour and participation of captively reared, anadromous transgenic males (approximating farmed fish) were observed in pair-wise competitive trials with wild males, as well as alone with wild females. Second, to assess the ability of transgenic fish to contribute reproductively as precocial parr, the breeding behaviour, performance and reproductive success of captively reared, transgenic and nontransgenic precocial parr were compared in pair-wise competitive trials.
Methods
Experimental fish
In 1989, a transgene construct consisting of GH cDNA from Chinook salmon, Oncorhynchus tshawytscha (Walbaum), and an ocean pout, Macrozoarces americanus L., antifreeze protein gene promoter (opAFP-GHc2), was inserted into the genome of wild Atlantic salmon collected from the Exploits and Colinet Rivers, Newfoundland, Canada (Du et al. 1992). A stable transgenic line (EO-1α; Yaskowiak et al. 2006) was created and has since been cultured at the Ocean Sciences Centre (OSC), Memorial University of Newfoundland. The competitive breeding trials between transgenic and wild anadromous salmon were conducted in 2006 and involved fifth- and sixth-generation anadromous males from this captive transgenic line. Wild anadromous males and females for these trials were collected from the Exploits River (48°55′N, 55°40′W), Newfoundland, Canada, in September of that year and transferred to the OSC. Parr, both mature and immature individuals, were also included in the 2006 trials to simulate the natural structure of the breeding system. They were derived from eight single pair crosses produced in the fall of 2004 that involved wild, Exploits River salmon, with the subsequent offspring captively reared to the parr stage at the OSC.
The competitive breeding trials to assess the ability of transgenic relative to nontransgenic fish to contribute reproductively as precocial parr were undertaken in 2007. The mature transgenic parr were age 0+, having been produced in the fall of 2006 by eight single pair crosses between St. John River (aquaculture strain) males, hemizygous for the EO-1α transgene, and wild Exploits River females. True to Mendelian inheritance patterns, crosses of hemizygous to wild-type individuals result in ca. half the offspring inheriting the GH transgene (Shears et al. 1992). Because of the tremendous growth induced by transgenesis, it is difficult to compare size- and age-matched transgenic and nontransgenic individuals. Therefore, to reduce these potential sources of variation, half of the mature nontransgenic parr used in the trials were 0+ offspring from the above 2006 crosses and the other half were 1+ offspring from five single pair crosses of wild, Exploits River parents, produced in 2005. To facilitate natural breeding and competitive interactions, anadromous females and males collected from the Exploits River during September 2007 were transferred to the OSC and used in the trials.
Prior to both the anadromous and parr competition experiments, all animals were housed in fibreglass tanks under a natural photoperiod and fed a standard salmonid dry feed (Corey Feed Mills, Fredericton, NB, Canada) ad libitum, 3–5 times weekly. Feeding of the anadromous transgenic fish ceased in early fall, preceding the breeding season (wild anadromous fish captured in early fall were not fed). Parr continued to be fed until they were introduced into the breeding trials. Prior to experimentation, all potential transgenic individuals were screened using the polymerase chain reaction (PCR) amplification protocol described in Deitch et al. (2006). To facilitate night behavioural observations in the breeding trials, the fish were exposed to a low-light regime with standard facility light installations. All measurement and tagging procedures were performed under mild anaesthesia (MS-222; Western Chemical Inc., Ferndale, WA, USA), and fish were treated in accordance with the guidelines provided by the Canadian Council on Animal Care and with the approval of Memorial University's Institutional Animal Care Committee.
Experimental design
A fully contained stream mesocosm was constructed out of a large, indoor concrete raceway and used for the competitive breeding trials (Fig. 1). To divide the mesocosm into two replicate breeding channels (1.25 × 7.8 × 0.25 m), a fibreglass partition was placed along the centre of the mesocosm and screens of plastic mesh fencing, framed with PVC pipe, were installed at each end. Two external pumps (1.5 hp, Dynamo®; Pentair Water Pool and Spa, Inc., Sanford, NC, USA) were placed at opposite ends of the mesocosm to generate a unidirectional, circulating current (range: 8–98 cm/s; mean ± SE: 22.3 ± 0.24 cm/s). The bottom of the mesocosm was covered with cobble (∼5–10 cm diameter; ∼40 cm deep) and large rocks (20–30 cm diameter) to naturalise the breeding channels and provide the salmon with nest substrate.
Figure 1.
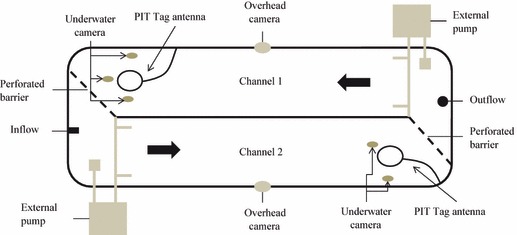
An illustration of the naturalised stream mesocosm (1.25 × 7.8 × 0.25 m per channel), which was divided into two channels and used to compare the reproductive performance of growth hormone transgenic and nontransgenic Atlantic salmon (Salmo salar) males, both as anadromous fish and as precocial parr. Behavioual data were collected using a combination of video observation and passive integrated transponder tag detection, with the respective underwater cameras and antenna moved in response to the location of female nesting activity. Thick arrows indicate the direction of water flow.
Anadromous male experiments
The behaviour of anadromous transgenic and nontransgenic males was compared during pair-wise competitive breeding trials between 18 November and 16 December 2006. Each trial consisted of a single female, a focal pair of anadromous males and a complement of parr (5 mature males and 10 immature) to naturalise the mesocosm with respect to the Atlantic salmon breeding system. Six weeks prior to the onset of experimentation, fork length (cm) and mass (g) measurements were recorded for all anadromous males and females. It was not possible to size-match competing anadromous males because of substantial size differences between the transgenic and nontransgenic fish (Table 1). To allow for individual identification, anadromous fish were marked with uniquely coded Petersen disc tags (3.4 cm diameter; Floy Tag & Manufacturing Inc., Seattle, WA, USA) just below the dorsal fin.
Table 1.
Mean fork length (cm; ±SE) and mass (g; ±SE) of the mature Atlantic salmon used in competitive breeding experiments comparing growth hormone transgenic and nontransgenic alternative reproductive phenotypes. In 2007, six trials compared age 0+ transgenic (T) versus 1+ nontransgenic (NT) parr and five trials compared 0+ transgenic versus 0+ nontransgenic parr; the size of parr involved are reported separately for each age comparison below. The N for each fish type is provided in parentheses
| Length (cm) | Mass (g) | ||||
|---|---|---|---|---|---|
| Year | Fish type | Transgenic | Nontransgenic | Transgenic | Nontransgenic |
| 2006 | Anadromous female | – | 54.26 ± 1.20 (11) | – | 1620.3 ± 150.8 (11) |
| Anadromous male* | 65.45 ± 0.83 (11) | 55.74 ± 1.61 (11) | 2862.2 ± 134.2 (11) | 1604.6 ± 180.1 (11) | |
| Mature parr | – | 14.39 ± 1.14 (30) | – | 37.99 ± 3.18 (30) | |
| Anadromous female | – | 59.36 ± 1.83 (9) | – | 1925.6 ± 140.4 (9) | |
| Anadromous male | – | 62.13 ± 1.49 (7) | – | 2202.3 ± 287.5 (7) | |
| 2007 | Mature parr (T 0+ vs NT 1+) | 15.22 ± 0.56 (6) | 14.14 ± 0.28 (6) | 36.10 ± 3.52 (6) | 30.9 ± 1.91 (6) |
| Mature parr* (T 0+ vs NT 0+) | 9.33 ± 0.70 (5) | 7.84 ± 0.39 (5) | 20.6 ± 2.45 (5) | 12.85 ± 1.40 (5) | |
Instances where transgenic males were larger than nontransgenic males in length and mass.
Each breeding trial (n = 11) consisted of two phases: the competitive and noncompetitive phases. The competitive phase included both the anadromous transgenic (n = 11) and nontransgenic (n = 11) males competing directly for breeding opportunities with the female. To separate the effects of courting and mate choice from intersexual competition on breeding performance, the noncompetitive phase involved providing each of the transgenic (n = 8) and nontransgenic (n = 6) males sole access to the female. The order by which each of the two males had sole access to the female was alternated among trials. Each trial phase consisted of 1–5 spawning events (a female will spawn 3–8 times typically, depending on her size; Fleming 1996). However, to standardise among trials, a maximum of two spawns per phase were included in the behavioural analyses. The duration of each phase was dependent on the spawning behaviour of the individuals, with a phase being terminated following two confirmed spawning events. In cases where no spawning occurred (n = 4; all transgenic males in the absence of competition), a maximum duration of 36 h was applied to each phase.
Precocious male parr experiments
The behaviour of transgenic and nontransgenic precocious male parr was compared in pair-wise competitive breeding trials (n = 11) between 15 November and 22 December 2007. Each trial consisted of an anadromous male and female pair, a focal pair of mature male parr and four immature parr (2 transgenic and 2 nontransgenic). In most cases, it was not possible to size-match competing mature parr because of substantial size differences between transgenic and nontransgenic parr (Table 1). Each breeding trial consisted of 1–4 spawning events; however, a maximum of the first two spawns per trial (referred to subsequently as spawn A or spawn B) were included in the behavioural analyses. Similar to the 2006 experiments, the anadromous fish were measured for fork length (cm) and mass (g) and tagged with uniquely coded Petersen disc tags, all of which was completed 3 weeks prior to the experiments. The parr to be used in the experiments were either tagged with a passive integrated transponder (PIT, model RI-TRP-WRHP, 23.1 × 3.9 mm and 0.6 g; Texas Instruments Inc., Dallas, TX, USA) or marked using visible implant elastomer (Northwest Marine Technology Inc., Shaw Island, WA, USA) 6 weeks prior to the experiment. PIT tags were inserted into the body cavity through a small, ventral incision made anterior to the pelvic girdle, which was closed with a single suture using surgical thread. For parr deemed too small for a PIT tag (i.e. <10 cm), elastomer was injected ventrally, just under the skin with a fine needle to provide a small, unique mark. Just prior to the beginning of each trial, the fork length (cm) and mass (g) of the experimental parr were measured. Adipose fin clips were collected following the trials for all fish involved and placed in 1.5-mL microcentrifuge tubes containing 99% ethanol and stored in a −20°C freezer.
Between 1 and 3 h following each spawning event, the trial was temporarily disrupted to collect the eggs laid for subsequent parentage analyses. Eggs were excavated from the gravel with the aid of a suction system based on the venturi effect and then counted, transferred into spawn-specific plastic mesh baskets and reared in Heath incubation trays. The effect of these disruptions appeared to be limited to the latency of breeding behaviour resumption, which ranged from 15 min to 3 h.
Behavioural observations
For the anadromous male experiments, breeding behaviour was monitored 24 h per day using a combination of live and recorded video observations. The video monitoring system included two overhead surveillance cameras, equipped with remote pan, tilt and zoom capabilities that recorded directly to a computer, and underwater cameras (SEA-CAM; Borel Manufacturing Inc., Alameda, CA, USA) positioned near female nest sites that recorded directly to individual HDD/DVD recorders. Each spawn was monitored with one overhead and 2–3 underwater cameras, simultaneously.
During the anadromous male experiments, behavioural data were collected for 60 min before (prespawn) and 30 min after (postspawn) each spawning event. For trial phases where no spawning event occurred, observations were conducted for 5-min intervals every 30 min for the duration of the phase (i.e. a total of 360 min of observation time). The behaviours recorded included, nest fidelity, anadromous male–male aggression, quivering and spawn participation (Table 2).
Table 2.
An ethogram describing the spawning behaviours measured during paired competitive trials between transgenic and nontransgenic Atlantic salmon males of both the anadromous and the parr reproductive phenotypes
| Behaviour | Description | Unit of measure |
|---|---|---|
| Nest Fidelity | The time the focal male spends with a nesting female. | Proportion of time the focal male attends the nest with the female present. |
| Overt Aggression | Male–male overt aggressive actions including chasing, charging, biting and fighting (Fleming 1996). | Frequency of all overt aggressive behaviours performed by the focal male. |
| Quivering | A courting behaviour, where the focal male vibrates its body while aligned in parallel with the female. | Frequency of all quivers performed by the focal male. |
| Spawn Participation | The active participation of the focal male during a spawning event. | Presence or absence of active participation during a spawning event. |
For the precocious male parr experiments, breeding behaviour was also monitored 24 h per day. A PIT tag detection system was used in addition to live and recorded video observations from 3 to 4 underwater cameras stationed around the nest site. The PIT tag detection system monitored the presence/absence and time data on parr around the nest site (Armstrong et al. 2001) and was designed in a manner similar to that detailed in Roussel et al. (2000). Each unit (n = 2) consisted of a double-gate loop antenna (100 cm diameter) that was positioned so as to encircle an individual nest site of a spawning female. The antennae was connected to a PIT tag reader (model Series 2000; RI-CTL-MB2A; Texas Instruments Inc) powered by a 12-V battery. Data were input into a palmtop computer (Dell™ Axim™ X51; Round Rock, TX, USA) with a custom-designed software program (Roussel et al. 2000). Both cameras and PIT tag systems were positioned at nest sites shortly following female nest site selection (as indicated by the female's consistent digging at a focal site).
Based on observations conducted during the anadromous male experiments, parr behavioural data collection and analyses were adjusted to capture perceived differences between the two reproductive phenotypes. As such, behavioural data were collected over a continuous 75-min period, 52.5 min before and 22.5 min after each spawning event. For analysis, these data were segregated into three time periods including the spawn period (12.5 min before and after the spawning event), the prespawn period (40 min prior to the spawn period) and the postspawn period (10 min immediately after the spawn period). Behaviours recorded included, nest fidelity, parr–parr aggression and spawn participation (Table 2).
Parentage analyses
Parentage analyses were conducted exclusively for the mature parr experiments because the behavioural results from the anadromous male experiments made it unnecessary to assess breeding success at the genetic level. Shortly following hatching, a subsample of offspring from each spawn was placed in 1.5-mL microcentrifuge tubes with 99% ethanol and stored in a −20°C freezer. A total of 32 alevins were sampled from each spawn, unless fewer had survived. Parentage analyses were conducted on individuals from all 11 trials, with representation ranging from 1 to 4 spawns per trial, 27–119 eggs per trial and 13–32 eggs per spawn for a total of 715 eggs.
Microsatellite analyses were conducted at three highly polymorphic, tetranucleotide loci using primer sequences developed specifically for Atlantic salmon (Ssa202, O'Reilly et al. 1996; SSsp2215, SSsp2216, Paterson et al. 2004). The DNA of potential parents and offspring were extracted and purified using the Wizard® SV 96 Genomic DNA Purification System (Promega Corp., Madison, WI, USA), following the protocol provided by the manufacturer. PCR amplifications were performed in 10 μL solutions, containing 2–10 ng of sample DNA template, 0.2 mm of each dNTP, 0.5 μm of each of the labelled and unlabelled primers, 1* KCl buffer (10 mm Tris–HCl, pH 8.3), 2.5 mm MgCl2 and 0.5 U of Taq DNA polymerase. Thermal cyclers (model 2720; Applied Biosystems™, Foster City, CA, USA) were programmed under the following regime: (94°C for 2 min)*1, (94°C for 45 s, 58°C for 45 s, 72°C for 1 min)*35, (72°C for 15 min)*1 and finished with a 4°C hold. Subsequent to DNA amplification, the PCR products representing different primer sets from like samples were combined and purified using the MiniElute® 96 UF PCR Purification method (Qiagen Inc., Hilden, NRW, Germany), following the manufacturer's protocol. Microsatellite fragments were then separated and visualised with an Applied Biosystems™ 3130 Genetic Analyzer and the accompanying GeneMapper® 4.0 software (Applied Biosystems™). Two known reference samples were used as standards and run on each plate to monitor for allele size shifts and function as an internal plate indicator.
Given that each spawn involved a single female and three potential males, we used an allele exclusion-based approach to assign parentage, where potential parents are eliminated on the basis of Mendelian inheritance patterns at primer loci (O'Reilly et al. 1998). Specifically, offspring genotypes were compared to all potential parental genotype combinations from all breeding trials, using a custom-designed Microsoft® Excel exclusion macro. In cases where multiple parental crosses shared the most complete genotypic match (allelic match at two or three loci) to an offspring, assignment was assumed to the parental cross representing the particular trial and spawn corresponding to that offspring. In no circumstance did two parental crosses from the same trial and spawn share the most complete genotypic match. Moreover, all offspring were successfully assigned to a parental cross corresponding to the trial and spawn from which they were collected. All exclusion-based assignments were corroborated with the likelihood-based assignments produced using Cervus 3.0.3 (Field Genetics Ltd., London, UK).
Statistical analyses
For the anadromous male experiment, nest fidelity was modelled as a binomial logistic regression (LRb) with trial and genotype (transgenic or nontransgenic) as explanatory variables. Prespawn and spawn periods were analysed separately for the competitive phase; however, all periods were summed during the noncompetitive phase to allow for the comparison of the two genotypes because half of the transgenic males failed to spawn. Spawn participation was also modelled as a binomial LR with explanatory variables that included genotype and phase. Quivering count data from prespawn and spawn periods were summed, as there were no differences between the periods, and a LR with Poisson error (LRp) was fit, where genotype, phase and trial served as explanatory variables. For similar reasons, overt aggression count data were summed across spawn periods and phases and analysed with the Wilcoxon signed rank test with continuity correction. In cases where data were available for multiple spawns within a phase, the mean value of the behavioural measure was used for analyses. All observations were standardised with respect to observation time.
Similar statistical models to those used for the competitive phase of the anadromous male experiments were used for analogous behavioural data in the mature parr experiments. Spawn identity (spawn A or B) was used in an analogous fashion to experimental phase in the anadromous male experiments. For analysis of male fertilisation success in the parr experiments, the number of eggs fathered by either the anadromous male, transgenic parr or nontransgenic parr from each trial was summed across spawns and tested using two approaches. First, a series of Wilcoxon signed rank tests were used to compare the relative fertilisation success between all three male types. Second, the overall proportions of offspring fertilised by transgenic and nontransgenic parr across all trials were compared by a two-sample test of binomial proportions.
Any over-dispersed data were accounted for by applying an empirical scale parameter by specifying either quasi-likelihood binomial or Poisson errors in the model. All data were analysed using the R statistical software application (version: R-2.10.1.; http://www.r-project.org) following a hypothesis testing approach. Statistical significance was measured at a 5% alpha level of type I error.
Results
Anadromous males
The captive-reared, transgenic males were significantly larger than the wild, nontransgenic males in terms of both mass (Table 1; paired t-test; t1, 10 = 6.03, P < 0.001) and length (paired t-test; t1, 10 = 5.14, P < 0.001). Despite a clear size advantage for transgenic males, there were no differences in the frequency of overt aggressive behaviours relative to nontransgenic males (Fig. 2A; Wilcoxon signed rank test: V1, 10 = 34.20, P = 0.057). However, nontransgenic males demonstrated a competitive advantage over transgenic males in all other breeding behaviours measured. In the presence of competition, nontransgenic males spent significantly more time at the nest with the females (nest fidelity) than did transgenic males during both the prespawn and postspawn periods (Table 3). Nontransgenic males also had higher nest fidelity than transgenic males in the absence of competition. Moreover, unlike both the prespawn (LRb; χ2 = 0.40, P = 1) and postspawn (LRb; χ2 = 6.79, P = 0.731) periods of direct competition, there was a significant trial effect on nest fidelity (LRb; χ2 = 20.56, P < 0.001) in the absence of competition, indicative of the high variation in behaviour observed. The quivering frequency of nontransgenic males was greater than that of transgenic males (LR; χ2 = 41.456, P < 0.001), with no effect of competition (Fig. 2; LRp; χ2 = 1.00, P = 0.606) or trial (LRp; χ2 = 15.63, P = 0.111). Furthermore, nontransgenic males participated in more spawning events than transgenic males regardless of the presence or absence of competition (Fig. 3; LRp; χ2 = 22.60, P < 0.001).
Figure 2.
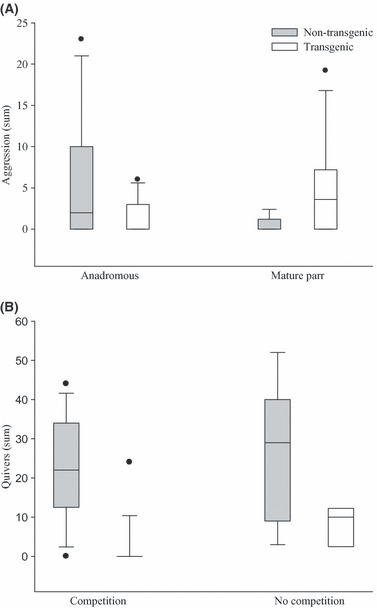
Standard box plot frequencies of (A) overt aggressive behaviours by transgenic and nontransgenic anadromous and parr males during paired competitive breeding trials and (B) quivering by transgenic and nontransgenic anadromous males during the competitive and noncompetitive phases. For graphical purposes, these data were standardised to a 90-min observation period. The top and bottom of each box represent the upper (75%) and lower (25%) quantiles, respectively. The horizontal line within each box indicates the median. The vertical lines (whiskers) extending from the upper and lower quantiles represent the maximum and minimum values of the distribution, excluding the outliers. The outliers are represented by the dots located beyond the maximum and minimum whiskers.
Table 3.
Nest fidelity (proportion of time spent with nesting female) of anadromous growth hormone transgenic and nontransgenic Atlantic salmon males during paired competitive breeding trials. Each breeding trial included phases of competition and no competition. During competition, both the transgenic and the nontransgenic males competed directly for breeding opportunities with the female. During no competition, males had sole access to a spawning female. Data from each trial were analysed 60 min before (prespawn) and 30 min after (postspawn) each spawning event. In trials with no spawning (n = 4; all transgenic males in the absence of competition), analyses were based on observations conducted for 5-min intervals every 30 min for the duration of the phase (i.e. a total of 360 min of observation time). For statistical analyses, nest fidelity during the no competition phase was not segregated into periods
| Phase | Period | Genotype | N | Median | 0.25 Quantile | 0.75 Quantile | Range | Statistics |
|---|---|---|---|---|---|---|---|---|
| Competition | Prespawn | Transgenic | 11 | 0 | 0 | 0.06 | 0–0.98 | χ2 = 19.33, P < 0.001 |
| Nontransgenic* | 11 | 1 | 0.89 | 1 | 0–1 | |||
| Postspawn | Transgenic | 11 | 0.07 | 0 | 0.52 | 0–0.88 | χ2 = 14.85, P < 0.001 | |
| Nontransgenic* | 11 | 0.83 | 0.58 | 1 | 0–1 | |||
| No Competition | – | Transgenic | 8 | 0.91 | 0.64 | 0.97 | 0.02–1 | χ2 = 7.09, P < 0.01 |
| Nontransgenic* | 6 | 0.96 | 0.94 | 0.98 | 0.89–1 |
The genotype with greater nest fidelity for each comparison.
Figure 3.
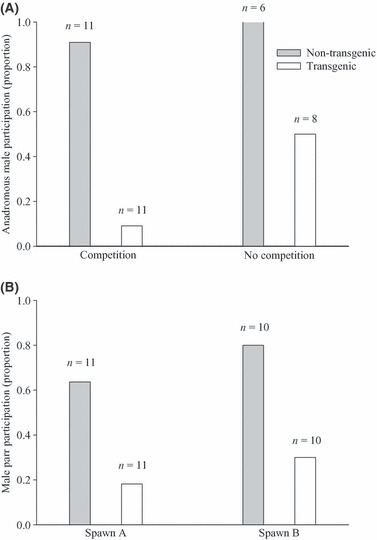
The spawn participation (presence/absence during a spawning event) of growth hormone transgenic and nontransgenic Atlantic salmon (Salmo salar) males during paired competitive breeding trials. Spawning behaviour and success were measured between transgenic and nontransgenic males of both the anadromous (A) and the parr (B) reproductive phenotypes.
Precocious male parr
In trials involving 1+ nontransgenic and 0+ transgenic parr, there were no significant differences in mass (paired t-test; t1, 5 = −1.37, P = 0.231) and length (paired t-test; t1, 5 = −1.63, P = 0.163) between the two groups (Table 1). However, in trials where both parr types were age 0+, the transgenic parr were significantly larger than the nontransgenic parr in terms of both mass (paired t-test; t1, 4 = −5.325, P = 0.006) and length (paired t-test; t1, 4 = −3.47, P = 0.026). Similarly, when age is ignored and the aforementioned data are analysed collectively, the transgenic parr were significantly larger than the nontransgenic parr in terms of both mass (paired t-test; t1, 10 = −3.42, P < 0.001) and length (paired t-test; t1, 10 = −3.26, P < 0.001). There were no significant differences in behaviour between trials involving 0+ and 1+ nontransgenic parr; thus, these were combined for subsequent analyses. Transgenic parr performed more overt aggressive behaviours than nontransgenic parr (Fig. 2; Wilcoxon signed rank test; V1, 10 = 26.5, P = 0.042). However, nontransgenic parr demonstrated greater nest fidelity than transgenic parr during all the comparisons save one; nest fidelity was similar during the postspawn period of spawn A (Table 4). There were no trial effects observed on nest fidelity. Greater nest fidelity was accompanied by greater spawn participation by nontransgenic relative to transgenic parr (Fig. 3; LRb; χ2 = 11.20, P < 0.001), and the levels of participation were similar across spawns (LRp; χ2 = 0.13, P = 0.72).
Table 4.
Nest fidelity (proportion of time spent with nesting female) during paired competitive breeding trials of mature male parr that were growth hormone transgenic and nontransgenic. The first (A) and second (B) spawns from each trial were analysed for the period 52.5–12.5 min before the spawn (prespawn), 12.5 min on either side of the spawn (spawn), and 12.5–22.5 after the spawn (postspawn)
| Spawn | Period | Genotype | N | Median | 0.25 Quantile | 0.75 Quantile | Range | Statistics |
|---|---|---|---|---|---|---|---|---|
| A | Prespawn | Transgenic | 11 | 0 | 0 | 0.04 | 0–0.88 | χ2 = 5.27 |
| Nontransgenic* | 11 | 0.87 | 0.09 | 1 | 0–1 | P = 0.022 | ||
| Spawn | Transgenic | 11 | 0 | 0 | 0.04 | 0–1 | χ2 = 4.70 | |
| Nontransgenic* | 11 | 0.98 | 0.64 | 1 | 0.01–1 | P = 0.030 | ||
| Postspawn | Transgenic | 11 | 0 | 0 | 0.38 | 0–1 | χ2 = 1.58 | |
| Nontransgenic | 11 | 0.77 | 0.11 | 1 | 0–1 | P = 0.209 | ||
| B | Prespawn | Transgenic | 10 | 0.02 | 0 | 0.72 | 0–1 | χ2 = 4.51 |
| Nontransgenic* | 10 | 0.98 | 0.52 | 1 | 0–1 | P = 0.034 | ||
| Spawn | Transgenic | 10 | 0.02 | 0 | 0.40 | 0–1 | χ2 = 5.19 | |
| Nontransgenic* | 10 | 1 | 0.55 | 1 | 0.05–1 | P = 0.023 | ||
| Postspawn | Transgenic | 10 | 1 | 0 | 0.39 | 0–1 | χ2 = 5.89 | |
| Nontransgenic* | 10 | 1 | 1 | 1 | 0–1 | P = 0.015 |
The genotype with greater nest fidelity.
The fertilisation success of both transgenic and nontransgenic parr was low (Table 5). Wilcoxon signed ranked tests confirmed that anadromous males dominated both transgenic (V1, 10 = 66.0, P < 0.001) and nontransgenic (V1, 10 = 66.0, P < 0.001) parr in fertilisation success across breeding trials. Furthermore, transgenic and nontransgenic parr fertilisation success did not differ significantly across trials (Wilcoxon signed rank test; V1, 10 = 16.0, P = 0.295). The overall (trial ignored) fertilisation success of nontransgenic parr, however, was significantly higher than that of transgenic parr (binomial test; χ2 = 15.98, P < 0.001), and offspring fathered by nontransgenic parr were represented in more trials.
Table 5.
The fertilisation success (proportion of eggs fertilised) of wild anadromous males and growth hormone transgenic and nontransgenic mature male parr during 11 pair-wise competitive breeding trials. Representation indicates the number of trials where successful fertilisation was observed by a male type
| Male type | Median | 0.25 Quantile | 0.75 Quantile | Range | Representation |
|---|---|---|---|---|---|
| Anadromous male* | 0.98 | 0.92 | 1 | 0.59–1 | 11 |
| Transgenic parr | 0 | 0 | 0.06 | 0–0.22 | 1 |
| Nontransgenic parr | 0 | 0 | 0 | 0–0.41 | 5 |
The anadromous males fertilised significantly more offspring than either parr genotype.
Discussion
This study provides the first empirical observation on the breeding of and potential for transgene introgression by GH transgenic male Atlantic salmon, including that of alternative reproductive phenotypes. Transgenic anadromous males (i.e. large, fighter males), reared to maturity in captivity, were behaviourally outcompeted by their wild counterparts in terms of nest fidelity, quivering frequency and spawn participation. Similarly, despite having similar rearing histories and displaying more aggression, transgenic male parr (i.e. precocially mature, sneaker males) were inferior competitors to wild-type parr in terms of nest fidelity and spawn participation. Moreover, wild-type parr had higher overall fertilisation success than transgenic parr, and their offspring were represented in more spawning trials. Although transgenic males displayed reduced breeding performance relative to nontransgenics, both male reproductive phenotypes demonstrated the ability to participate in natural spawning events and thus have the potential to contribute genes to subsequent generations.
The reduced reproductive performance of captively reared, anadromous transgenic males relative to wild males parallels the results of similar studies comparing captively reared salmon to wild salmon. Varying degrees of exposure to captive environments and domestication selection have been shown to affect the breeding behaviour and success of adult salmonids negatively (Fleming and Gross 1993; Fleming et al. 1997; Berejikian et al. 2001a; Weir et al. 2004). Moreover, Bessey et al. (2004) observed that wild-exposed coho salmon males outcompeted captively reared transgenic males in terms of spawn participation, courtship and aggressive behaviours. Bessey et al. (2004) also observed that when transgenic and nontransgenic males were both reared in the laboratory, performance was poor irrespective of transgenesis (see also Fitzpatrick et al. 2011). Thus, the captive rearing environment appears to diminish the competitive and reproductive performance of the anadromous salmonid phenotype, irrespective of genetic background (Berejikian et al. 1997, 2001a,b). The current study can, therefore, not eliminate the possibility that the poor performance of the anadromous transgenic males has more to do with rearing environment than transgenesis because these variables were confounded. Nevertheless, comparisons of captively reared transgenic and wild andromous males mimic the environmental differences that represent an initial transgenic escapee invasion scenario and are thus valuable for predicting the probability of first-generation intraspecific hybridisation.
Reproductively isolated populations are predicted to genetically diverge because of adaptive and/or nonadaptive evolutionary pressures, such as selection to environmental variation, genetic drift, gene flow and chance mutations (Frankham et al. 2002; Allendorf and Luikart 2007; Garcia de Leaniz et al. 2007; Carlson and Seamons 2008). This evolutionary theory provides some perspective on two elements of the current study. First, the captively reared, anadromous transgenic males did not have an identical genetic background to the wild anadromous males with which they were compared. Specifically, the genetic background of the transgenic males consisted of two wild populations, one of which was the same as that of the wild males. Thus, in addition to captive rearing, intraspecific population differences may also have contributed to observations of reduced reproductive performance in transgenic relative to nontransgenic males. Second, evolutionary divergence among wild Atlantic salmon populations can potentially influence their relative reproductive performance when competing against transgenic invaders (Devlin et al. 2006; Kapuscinski et al. 2007; Hutchings and Fraser 2008). This study correctly mimics a likely invasion scenario, where the genetic background of the transgenic population differs from that of the wild population. However, contextualising these results with the general concerns of GH transgene introgression into wild populations must be performed with caution. It remains uncertain how the reproductive performance of this GH transgenic population would compare with other wild populations. Similarly, it is uncertain how the reproductive performance of this wild population would compare with other GH transgenic populations.
Previous studies comparing the reproductive behaviour and success of farmed and wild-type mature male parr have suggested that this alternative male reproductive phenotype may facilitate the interbreeding and introgression of farmed genes into wild populations (Garant et al. 2003; Weir et al. 2005). This rationale is based on observations of equal or greater breeding performance among farmed parr relative to wild-type parr coupled with the fact that maturation prior to anadromy increases the probability of survival to maturity and reduces generation time. In the current study, we found that the breeding behaviour and success of transgenic parr was inferior to that of wild-type parr, despite transgenic parr displaying more aggression. Moreover, transgenic parr sired fewer eggs than nontransgenic parr. When the data were paired by trial, however, no differences were observed in fertilisation success between the two groups, which may be due largely to a number of spawns where there was no parr contribution and the associated low statistical power. Interpretations based on the entirety of the behavioural and fertilisation findings suggest that the nontransgenic parr marginally outcompeted transgenic parr during spawning. Nevertheless, transgenic parr demonstrated a behavioural interest in spawning and contributed gametes to the next generation. Thus, the alternative male reproductive phenotype of early maturation in Atlantic salmon may facilitate the introgression of transgenes into wild populations in a similar manner to that observed with farmed strains.
In an effort to limit size differences between transgenic and nontransgenic parr during the paired behavioural trials, age differences existed between competing parr in some of the trials. There was no significant difference in transgenic performance, whether competing with 0+ (n = 5) or 1+ (n = 6) nontransgenic parr, although we acknowledge the statistical limitations associated with the low sample sizes. Moreover, despite holding a significant body size advantage, irrespective of nontransgenic parr age, and exhibiting increased overt aggressive behaviour, the reproductive success of transgenic parr was less than that of nontransgenic parr. While there is evidence both for (Thomaz et al. 1997; Koseki and Maekawa 2000) and against (Jones and Hutchings 2001, 2002) parr body size influencing spawning success, it has been suggested that large body size may be a stronger predictor of dominance under scenarios with few competing parr (Hutchings and Myers 1994; Jones and Hutchings 2001). However, in the present study, the breeding performance of transgenic parr appears to be inferior independent of size.
The reduced breeding performance of transgenic parr may be due, in part, to behavioural changes associated with GH transgenesis. Juvenile salmonids have shown distinct shifts in behavioural phenotypes in response to GH transgenesis, including increased foraging-induced aggression and reduced antipredator behaviour (Abrahams and Sutterlin 1999; Sundstrom et al. 2003, 2004). The reduced nest fidelity and spawn participation by mature transgenic parr relative to nontransgenic parr may be driven by transgene-induced hormonal changes. Gonadotropin-releasing hormone (GnRH) is thought to increase the expression of reproductive behaviours in many species (Maney et al. 1997; Yamamoto et al. 1997; Johnson et al. 2007; Munakata and Kobayashi 2010), including salmonids (Berejikian et al. 2003). For example, studies with the dwarf gourami (Colisa lalia) have indicated that male nest building behaviour is reduced when GnRH function is disrupted (Yamamoto et al. 1997; Munakata and Kobayashi 2010). Moreover, there is an existent, but poorly understood, association between the GH-IGF-I axis and the GnRH-gonadotropin-sex steroid axis (Holloway and Leatherland 1997a,b; Mercure et al. 2001; Bjornsson et al. 2002). Thus, GH transgenesis may influence the interactions between these two hormonal axes such that the breeding behaviour of mature male parr is negatively affected. However, empirical investigations are required to explore the effects of GH on reproductive hormones and behaviour.
A common method for conducting environmental risk assessments involves the use of quantitative models that estimate a defined measure of risk. For genetically modified organisms, the prospect of gene flow from transgenic escapees into wild populations is a key issue because of the potential influences the transgene may have on fitness. In response, models have been developed to estimate the fitness outcome of transgene introgression into wild populations (Muir and Howard 1999, 2001; Aikio et al. 2008; Valosaari et al. 2008; Ahrens and Devlin 2010). Frequently, the model parameters consist of empirical measurements of fitness-related life history traits such as growth, survival and reproductive probabilities, age at sexual maturity, female fecundity and male fertility (Muir and Howard 2002). The current study provides data on the relative breeding success of male salmon that are applicable to such predictive quantitative models. Specifically, we contribute to observations indicating captive-reared GH transgenic and farmed adult male salmon have a mating disadvantage relative to wild individuals, a gene flow scenario indicative of an initial invasion. Moreover, captive-reared nontransgenic precocial male parr demonstrated a modest mating advantage over transgenic individuals, a gene flow scenario comparable to subsequent generations following an invasion. Similar to the Japanese medaka (Oryzias latipes) work of Pennington et al. (2010), these findings are inconsistent with the assumption of a transgenic male mating advantage used in previous quantitative models (Hedrick 2001; Aikio et al. 2008; Valosaari et al. 2008), but see Howard et al. (2004), and emphasise the importance of basing parameter values on empirical data.
The present study, however, only provides an estimate of breeding success under a single set of physical and demographic environmental conditions consisting of paired males competing for single females. In the wild, male salmon will typically have access to multiple females simultaneously and have to contend with multiple competitors (Fleming 1996; Fleming and Reynolds 2004). Moreover, should transgenic animals get exposure to the wild environment prior to breeding (i.e. escape prior maturation), this may well alter their reproductive performance in a similar way, but opposite, to the effects captive rearing has on wild fish (e.g. Berejikian et al. 1997, 2001a; Bessey et al. 2004). As pointed out by Devlin et al. (2006), there are limitations and difficulties associated with collecting the breadth of empirical data required to accurately represent the full range of genotype by environment interactions affecting fitness-related life history traits in the wild. The findings of this study are valuable with respect to a first-generation invasion scenario; but beyond that, reproductive performance is difficult to predict and is, therefore, an unavoidable source of epistemic uncertainty for both quantitative and qualitative invasion models. Further work is thus required to compare the breeding performance of transgenic and nontransgenic salmon in a range of ecologically relevant scenarios.
Acknowledgments
The authors thank Dr Patrick T. O'Reilly for providing technical advice for the microsatellite analyses, Carolyn Harvie for providing the parentage exclusion macro, Curtis J. Pennell for providing the PIT tag detection system, Corinne Conway, Danny Ings and Madonna King for assistance with data collection and Aqua Bounty Farms Inc. for providing transgenic gametes. The authors also thank our reviewers for their valued input into earlier versions of this manuscript. Support was kindly provided by a collaborative grant led by Dr Eric M. Hallerman and funded by the USDA Biotechnology Risk Assessment Research Grants Programme.
References
- Abrahams MV, Sutterlin AM. The foraging and antipredator behaviour of growth-enhanced transgenic Atlantic salmon. Animal Behaviour. 1999;58:933–952. doi: 10.1006/anbe.1999.1229. [DOI] [PubMed] [Google Scholar]
- Ahrens RNM, Devlin RH. Standing genetic variation and compensatory evolution in transgenic organisms: a growth-enhanced salmon simulation. Transgenic Research. 2010;20:583–597. doi: 10.1007/s11248-010-9443-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aikio S, Valosaari KR, Kaitala V. Mating preference in the invasion of growth enhanced fish. Oikos. 2008;117:406–414. [Google Scholar]
- Allendorf FW, Luikart G. Conservation and the Genetics of Populations. Oxford, UK: Blackwell Publishing; 2007. [Google Scholar]
- Armstrong JD, Einum S, Fleming IA, Rycroft P. A method for tracking the behaviour of mature and immature salmon parr around nests during spawning. Journal of Fish Biology. 2001;59:1023–1032. [Google Scholar]
- Berejikian BA, Tezak EP, Schroder SL, Knudsen CM, Hard JJ. Reproductive behavioral interactions between wild and captively reared coho salmon (Oncorhynchus kisutch. Ices Journal of Marine Science. 1997;54:1040–1050. [Google Scholar]
- Berejikian BA, Tezak EP, Park L, LaHood E, Schroder SL, Beall E. Male competition and breeding success in captively reared and wild coho salmon (Oncorhynchus kisutch. Canadian Journal of Fisheries and Aquatic Sciences. 2001a;58:804–810. [Google Scholar]
- Berejikian BA, Tezak EP, Schroder SL. Reproductive behavior and breeding success of captively reared chinook salmon. North American Journal of Fisheries Management. 2001b;21:255–260. [Google Scholar]
- Berejikian BA, Fairgrieve WT, Swanson P, Tezak EP. Current velocity and injection of GnRHa affect reproductive behavior and body composition of captively reared offspring of wild chinook salmon (Oncorhynchus tshawytscha. Canadian Journal of Fisheries and Aquatic Sciences. 2003;60:690–699. [Google Scholar]
- Bessey C, Devlin RH, Liley NR, Biagi CA. Reproductive performance of growth-enhanced transgenic Coho Salmon. Transactions of the American Fisheries Society. 2004;133:1205–1220. [Google Scholar]
- Bjornsson BT, Johansson V, Benedet S, Einarsdottir IE, Hildahl J, Agustsson T, Jonsson E. Growth hormone endocrinology of salmonids: regulatory mechanisms and mode of action. Fish Physiology and Biochemistry. 2002;27:227–242. [Google Scholar]
- Carlson SM, Seamons TR. A review of fitness and its quantitative genetic components in salmonids: implications for adaptation to future change. Evolutionary Applications. 2008;1:222–238. doi: 10.1111/j.1752-4571.2008.00025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitch EJ, Fletcher GL, Petersen LH, Costa IASF, Shears MA, Driedzic WR, Gamperl AK. Cardiorespiratory modifications, and limitations, in post-smolt growth hormone transgenic Atlantic salmon Salmo salar. Journal of Experimental Biology. 2006;209:1310–1325. doi: 10.1242/jeb.02105. [DOI] [PubMed] [Google Scholar]
- Devlin RH, Sundstrom LF, Muir WM. Interface of biotechnology and ecology for environmental risk assessments of transgenic fish. Trends in Biotechnology. 2006;24:89–97. doi: 10.1016/j.tibtech.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Du SJ, Gong ZY, Fletcher GL, Shears MA, King MJ, Idler DR, Hew CL. Growth enhancement in transgenic Atlantic salmon by the use of an all fish chimeric growth-hormone gene construct. Bio-Technology. 1992;10:176–181. doi: 10.1038/nbt0292-176. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick JL, Akbarashandiz H, Sakhrani D, Biagi CB, Pitcher TB, Devlin H. Cultured growth hormone transgenic salmon are reproductively out-competed by wild-reared salmon in semi-natural mating arenas. Aquaculture. 2011;312:185–191. [Google Scholar]
- Fleming IA. Reproductive strategies of Atlantic salmon: ecology and evolution. Reviews in Fish Biology and Fisheries. 1996;6:379–416. [Google Scholar]
- Fleming IA. Pattern and variability in the breeding system of Atlantic salmon (Salmo salar), with comparisons to other salmonids. Canadian Journal of Fisheries and Aquatic Sciences. 1998;55:59–76. [Google Scholar]
- Fleming IA, Gross MR. Breeding success of hatchery and wild coho salmon (Oncorhynchus-kisutch) in competition. Ecological Applications. 1993;3:230–245. doi: 10.2307/1941826. [DOI] [PubMed] [Google Scholar]
- Fleming IA, Reynolds JD. Salmonid breeding systems. In: Hendry AP, Stearns SC, editors. Evolution Illuminated, Salmon and Their Relatives. Oxford: Oxford University Press; 2004. pp. 264–294. [Google Scholar]
- Fleming IA, Lamberg A, Jonsson B. Effects of early experience on the reproductive performance of Atlantic salmon. Behavioral Ecology. 1997;8:470–480. [Google Scholar]
- Fleming IA, Hindar K, Mjoelneroed IB, Jonsson B, Balstad T, Lamberg A. Lifetime success and interactions of farm salmon invading a native population. Proceedings of the Royal Society of London, Series B: Biological Sciences. 2000;267:1517–1523. doi: 10.1098/rspb.2000.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R, Ballou JD, Briscoe DA. Introduction to Conservation Genetics. Cambridge, UK: Cambridge University Press; 2002. [Google Scholar]
- Fraser DJ, Cook AM, Eddington JD, Bentzen P, Hutchings JA. Mixed evidence for reduced local adaptation in wild salmon resulting from interbreeding with escaped farmed salmon: complexities in hybrid fitness. Evolutionary Applications. 2008;1:501–512. doi: 10.1111/j.1752-4571.2008.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser DJ, Houde ALS, Debes PV, O'Reilly P, Eddington JD, Hutchings JA. Consequences of farmed-wild hybridization across divergent wild populations and multiple traits in salmon. Ecological Applications. 2010;20:935–953. doi: 10.1890/09-0694.1. [DOI] [PubMed] [Google Scholar]
- Garant D, Fleming IA, Einum S, Bernatchez L. Alternative male life-history tactics as potential vehicles for speeding introgression of farm salmon traits into wild populations. Ecology Letters. 2003;6:541–549. [Google Scholar]
- Garcia de Leaniz C, Fleming IA, Einum S, Verspoor E, Jordan WC, Consuegra S, Aubin-Horth N, et al. A critical review of adaptive genetic variation in Atlantic salmon: implications for conservation. Biological Reviews. 2007;82:173–211. doi: 10.1111/j.1469-185X.2006.00004.x. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. Invasion of transgenes from salmon or other genetically modified organisms into natural populations. Canadian Journal of Fisheries and Aquatic Sciences. 2001;58:841–844. [Google Scholar]
- Hindar K, Fleming IA, McGinnity P, Diserud A. Genetic and ecological effects of salmon farming on wild salmon: modelling from experimental results. Ices Journal of Marine Science. 2006;63:1234–1247. [Google Scholar]
- Holloway AC, Leatherland JF. Effects of N-methyl-D,L-aspartate (NMA) on growth hormone and thyroid hormone levels in steroid-primed immature rainbow trout (Oncorhynchus mykiss. Journal of Experimental Zoology. 1997a;279:126–132. [Google Scholar]
- Holloway AC, Leatherland JF. The effects of N-methyl-D,L-aspartate and gonadotropin-releasing hormone on in vitro growth-hormone release in steroid-primed immature rainbow trout, Oncorhynchus mykiss. General and Comparative Endocrinology. 1997b;107:32–43. doi: 10.1006/gcen.1997.6907. [DOI] [PubMed] [Google Scholar]
- Howard RD, DeWoody JA, Muir WM. Transgenic male mating advantage provides opportunity for Trojan gene effect in a fish. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2934–2938. doi: 10.1073/pnas.0306285101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings JA, Fraser DJ. The nature of fisheries- and farming-induced evolution. Molecular Ecology. 2008;17:294–313. doi: 10.1111/j.1365-294X.2007.03485.x. [DOI] [PubMed] [Google Scholar]
- Hutchings JA, Myers RA. The evolution of alternative mating strategies in variable environments. Evolutionary Ecology. 1994;8:256–268. [Google Scholar]
- Johnson MA, Tsutsui K, Fraley GS. Rat RFamide-related peptide-3 stimulates GH secretion, inhibits LH secretion, and has variable effects on sex behavior in the adult male rat. Hormones and Behavior. 2007;51:171–180. doi: 10.1016/j.yhbeh.2006.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MW, Hutchings JA. The influence of male parr body size and mate competition on fertilization success and effective population size in Atlantic salmon. Heredity. 2001;86:675–684. doi: 10.1046/j.1365-2540.2001.00880.x. [DOI] [PubMed] [Google Scholar]
- Jones MW, Hutchings JA. Individual variation in Atlantic salmon fertilization success: implications for effective population size. Ecological Applications. 2002;12:184–193. [Google Scholar]
- Kapuscinski AR, Hallerman EM. Implications of introduction of transgenic fish into natural ecosystems. Canadian Journal of Fisheries and Aquatic Sciences. 1991;48:99–107. [Google Scholar]
- Kapuscinski AR, Hayes KR, Li S, Dana G. Environmental Risk Assessment of Genetically Modified Organisms, Volume 3: Methodologies for Transgenic Fish. Oxfordshire, UK: CABI Publishing; 2007. [Google Scholar]
- Koseki Y, Maekawa K. Sexual selection on mature male parr of masu salmon (Oncorhynchus masou): does sneaking behavior favor small body size and less-developed sexual characters? Behavioral Ecology and Sociobiology. 2000;48:211–217. [Google Scholar]
- Lohmus M, Sundstrom LF, Bjorklund M, Devlin RH. Genotype-temperature interaction in the regulation of development, growth, and morphometrics in wild-type, and growth-hormone transgenic Coho Salmon. PLoS ONE. 2010;5:1–11. doi: 10.1371/journal.pone.0009980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney DL, Richardson RD, Wingfield JC. Central administration of chicken gonadotropin-releasing hormone-II enhances courtship behavior in a female sparrow. Hormones and Behavior. 1997;32:11–18. doi: 10.1006/hbeh.1997.1399. [DOI] [PubMed] [Google Scholar]
- McGinnity P, Prodoehl P, Ferguson A, Hynes R, Maoileidigh NO, Baker N, Cotter D, et al. Fitness reduction and potential extinction of wild populations of Atlantic salmon, Salmo salar, as a result of interactions with escaped farm salmon. Proceedings of the Royal Society of London, Series B: Biological Sciences. 2003;270:2443–2450. doi: 10.1098/rspb.2003.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercure F, Holloway AC, Tocher DR, Sheridan MA, Van der Kraak G, Leatherland JF. Influence of plasma lipid changes in response to 17 beta-oestradiol stimulation on plasma growth hormone, somatostatin, and thyroid hormone levels in immature rainbow trout. Journal of Fish Biology. 2001;59:605–615. [Google Scholar]
- Moreau DTR, Fleming IA, Fletcher GL, Brown JA. Growth hormone transgenesis does not influence territorial dominance or growth and survival of first-feeding Atlantic salmon Salmo salar L. in food-limited stream microcosms. Journal of Fish Biology. 2011;78:726–740. doi: 10.1111/j.1095-8649.2010.02888.x. [DOI] [PubMed] [Google Scholar]
- Muir WM, Howard RD. Possible ecological risks of transgenic organism release when transgenes affect mating success: sexual selection and the Trojan gene hypothesis. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:13853–13856. doi: 10.1073/pnas.96.24.13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir WM, Howard RD. Fitness components and ecological risk of transgenic release: a model using Japanese medaka (Oryzias latipes. American Naturalist. 2001;158:1–16. doi: 10.1086/320860. [DOI] [PubMed] [Google Scholar]
- Muir WM, Howard RD. Assessment of possible ecological risks and hazards of transgenic fish with implications for other sexually reproducing organisms. Transgenic Research. 2002;11:101–114. doi: 10.1023/a:1015203812200. [DOI] [PubMed] [Google Scholar]
- Munakata A, Kobayashi M. Endocrine control of sexual behavior in teleost fish. General and Comparative Endocrinology. 2010;165:456–468. doi: 10.1016/j.ygcen.2009.04.011. [DOI] [PubMed] [Google Scholar]
- O'Reilly PT, Hamilton LC, McConnell SK, Wright JM. Rapid analysis of genetic variation in Atlantic salmon (Salmo salar) by PCR multiplexing of dinucleotide and tetranucleotide microsatellites. Canadian Journal of Fisheries and Aquatic Sciences. 1996;53:2292–2298. [Google Scholar]
- O'Reilly PT, Herbinger C, Wright JM. Analysis of parentage determination in Atlantic salmon (Salmo salar) using microsatellites. Animal Genetics. 1998;29:363–370. [Google Scholar]
- Paterson S, Piertney SB, Knox D, Gilbey J, Verspoor E. Characterization and PCR multiplexing of novel highly variable tetranucleotide Atlantic salmon (Salmo salar L.) microsatellites. Molecular Ecology Notes. 2004;4:160–162. [Google Scholar]
- Pennington KM, Kapuscinski AR, Morton MS, Cooper AM, Miller LM. Full life-cycle assessment of gene flow consistent with fitness differences in transgenic and wild-type Japanese medaka fish (Oryzias latipes. Environmental Biosafety Research. 2010;9:41–57. doi: 10.1051/ebr/2010005. [DOI] [PubMed] [Google Scholar]
- Roussel JM, Haro A, Cunjak RA. Field test of a new method for tracking small fishes in shallow rivers using passive integrated transponder (PIT) technology. Canadian Journal of Fisheries and Aquatic Sciences. 2000;57:1326–1329. [Google Scholar]
- Shears MA, King MJ, Goddard SV, Fletcher GL. Gene transfer in salmonids by injection through the micropyle. In: Hew CL, Fletcher GL, editors. Transgenic Fish. Singapore: World Scientific; 1992. pp. 44–60. [Google Scholar]
- Sundstrom LF, Devlin RH, Johnsson JI, Biagi CA. Vertical position reflects increased feeding motivation in growth hormone transgenic coho salmon (Oncorhynchus kisutch. Ethology. 2003;109:701–712. [Google Scholar]
- Sundstrom LF, Lohmus M, Johnsson JI, Devlin RH. Growth hormone transgenic salmon pay for growth potential with increased predation mortality. Proceedings of the Royal Society of London, Series B: Biological Sciences. 2004;271:S350–S352. doi: 10.1098/rsbl.2004.0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundstrom LF, Lohmus M, Devlin RH. Selection on increased intrinsic growth rates in coho salmon, Oncorhynchus kisutch. Evolution. 2005;59:1560–1569. [PubMed] [Google Scholar]
- Thomaz D, Beall E, Burke T. Alternative reproductive tactics in Atlantic salmon: factors affecting mature parr success. Proceedings of the Royal Society of London Series B-Biological Sciences. 1997;264:219–226. [Google Scholar]
- Tymchuk WE, Sundstrom LF, Devlin RH. Growth and survival trade-offs and outbreeding depression in rainbow trout (Oncorhynchus mykiss. Evolution. 2007;61:1225–1237. doi: 10.1111/j.1558-5646.2007.00102.x. [DOI] [PubMed] [Google Scholar]
- Valosaari KR, Aikio S, Kaitala V. Male mating strategy and the introgression of a growth hormone transgene. Evolutionary Applications. 2008;1:608–619. doi: 10.1111/j.1752-4571.2008.00046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir LK, Hutchings JA, Fleming IA, Einum S. Dominance relationships and behavioural correlates of individual spawning success in farmed and wild male Atlantic salmon, Salmo salar. Journal of Animal Ecology. 2004;73:1069–1079. [Google Scholar]
- Weir LK, Hutchings JA, Fleming IA, Einum S. Spawning behaviour and success of mature male atlantic salmon (Salmo salar) parr of farmed and wild origin. Canadian Journal of Fisheries and Aquatic Sciences. 2005;62:1153–1160. [Google Scholar]
- Yamamoto N, Oka Y, Kawashima S. Lesions of gonadotropin-releasing hormone-immunoreactive terminal nerve cells: effects on the reproductive behavior of male dwarf gouramis. Neuroendocrinology. 1997;65:403–412. doi: 10.1159/000127203. [DOI] [PubMed] [Google Scholar]
- Yaskowiak ES, Shears MA, Agarwal-Mawal A, Fletcher GL. Characterization and multi-generational stability of the growth hormone transgene (EO-1 alpha) responsible for enhanced growth rates in Atlantic salmon. Transgenic Research. 2006;15:465–480. doi: 10.1007/s11248-006-0020-5. [DOI] [PubMed] [Google Scholar]


