Abstract
Artificial breeding programs initiated to enhance the size of animal populations are often motivated by the desire to increase harvest opportunities. The introduction of non-native genotypes, however, can have negative evolutionary impacts. These may be direct, such as introgressive hybridization, or indirect via competition. Less is known about the effects of stocking with native genotypes. We assayed variation at nine microsatellite loci in 902 steelhead trout (Oncorhynchus mykiss) from five rivers in British Columbia, Canada. These samples were collected over 58 years, a time period that spanned the initiation of native steelhead trout broodstock hatchery supplementation in these rivers. We detected no changes in estimates of effective population size, genetic variation or temporal genetic structure within any population, nor of altered genetic structure among them. Genetic interactions with nonmigratory O. mykiss, the use of substantial numbers of primarily native broodstock with an approximate 1:1 male-to-female ratio, and/or poor survival and reproductive success of hatchery fish may have minimized potential genetic changes. Although no genetic changes were detected, ecological effects of hatchery programs still may influence wild population productivity and abundance. Their effects await the design and implementation of a more comprehensive evaluation program.
Keywords: effective population size, gene flow, genetic drift, historical DNA samples, microsatellite DNA, native broodstock, salmonid, temporal analysis
Introduction
Salmonid fishes have contributed much to fundamental studies in evolution (e.g., Aspinwall 1974; Hendry and Stearns 2004). In particular, the diversity of environments they inhabit and their propensity for isolation via reproductive homing (or philopatry) have proven invaluable in assessing the roles of local adaptation, genetic drift, and gene flow in driving the evolution of population structure (reviewed in Adkison 1995; Hendry and Stearns 2004; Fraser et al. 2011). This link between evolutionary theory and empirical work on salmonids has also contributed significantly to conservation genetics (Ford 2004; Hard 2004; Waples 2004).
One area in particular that has benefited from salmonid research concerns the effects of artificial breeding programs on the genetics of populations (Young 2003). Supportive breeding, whereby a fraction of the population is bred and raised in captivity before release into the wild, is commonly used in conservation in an effort to rebuild breeding populations and/or to increase harvest opportunities (Ryman and Laikre 1991; Frankham et al. 2010). The potential negative evolutionary impacts of such programs have received particular attention in the management of salmonid fishes where an extensive history of fish culture and translocation has often focused on improving fishing opportunities over rebuilding spawning abundance per se (Stickney 1996; Utter 1998). One important potential impact of supportive breeding is that it may produce changes in the effective population size of supported populations by inducing variability in family size, especially in long-term programs for organisms like salmonid fishes that are characterized by high fecundities (Ryman and Laikre 1991; Ryman et al. 1995; Van Doornik et al. 2011). Given that effective population size is a crucial parameter of populations that influences the retention of genetic variation across generations, supportive breeding programs can have great relevance to overall conservation goals of managed populations (Ryman and Laikre 1991; Frankham et al. 2010).
Non-native cultured fish can impact the genetics of natural populations either indirectly by reducing population size through competition or directly through introgression between artificially propagated fish and wild populations. This may result in partial or complete replacement of native populations, reduced population size, and loss of genetic diversity. Nevertheless, the extent of such effects can be complex and unpredictable (e.g., Utter 1998; Reisenbichler and Rubin 1999; Reisenbichler 2004). Although there is evidence that stocking of non-native fish has reduced or changed genetic diversity among some wild populations (e.g. Oncorhynchus spp.: Reisenbichler 2004; Williamson and May 2005; Salmo spp.; Garcia-Marin et al. 1999; Hansen 2002; Hansen et al. 2009; Salvelinus spp.: Englbrecht et al. 2002; Marie et al. 2010), natural selection may act against non-native individuals and hybrids between native and non-native individuals (e.g. Poteaux et al. 1998; Hansen et al. 2000b; Miller et al. 2004), and there may be little or no introgression between wild and hatchery fish (e.g. Englbrecht et al. 2002; Hansen 2002; Kostow et al. 2003; Ruzzante et al. 2004; Taylor et al. 2007).
So-called ‘conservation-based’ fish-hatchery programs attempt to counter potential changes in genetic diversity by using native broodstock (Brannon et al. 2004). Less is known about the genetic effects of these initiatives despite potential negative evolutionary impacts. Performance traits may be negatively affected in the native population via competition or introgression if broodstock is perpetuated from hatchery-reared fish that experience artificial selection regimes in captivity (Utter 1998; Reisenbichler and Rubin 1999; Araki et al. 2007a, 2009; Caroffino et al. 2008). The annual collection of native wild broodstock can help reduce such effects, but potential impacts will also depend on the number of progeny released and the proportion that are harvested upon return; large numbers of hatchery fish have the potential to overwhelm the native population demographically and genetically (Eldridge and Naish 2007). Annual releases of large numbers of hatchery fish can also bring about genetic homogenization by increasing gene flow among populations (Slatkin 1985; Reisenbichler and Phelps 1989; Adkison 1995; Eldridge and Naish 2007), especially as hatchery-produced fish sometimes show lower reproductive site fidelity (Quinn 1993; Jonsson et al. 2003).
Additionally, a small number of breeders, a skewed sex ratio, or high annual variance in the spawning population size may diminish the effective population size (Ryman and Laikre 1991; Hansen et al. 2000a; Wang and Ryman 2001) and thereby increase genetic drift. This can alter patterns of genetic diversity (decrease genetic diversity, increase temporal genetic instability within populations and increase differentiation among them; Tessier et al. 1997; Hansen et al. 2000a), potentially reducing or constraining local adaptation, especially under changing environments. The collection of sufficient broodstock with a 1:1 male-to-female ratio can minimize reductions in effective population size (Tessier et al. 1997; Wang and Ryman 2001; Eldridge and Killebrew 2008).
In order to better manage natural resources undergoing hatchery supplementation, it is important to obtain an understanding of the biological effects of management decisions that use native broodstock (Brannon et al. 2004; Reisenbichler 2004). Genetic monitoring of archived samples collected over time may provide a tool for quantifying the potential for hatchery programs to change the diversity and structure of indigenous gene pools and hence, guide subsequent management plans (Schwartz et al. 2007; Allendorf et al. 2008; Van Doornik et al. 2011). Archived scale samples have been used previously for comparative studies (e.g., Hansen 2002; Hansen et al. 2009), and opportunities exist in British Columbia (BC), Canada, where there is a long history of supplementation through ongoing hatchery programs that span decades and have involved many genetically variable steelhead trout (the migratory, anadromous form of rainbow trout Oncorhynchus mykiss Walbaum) populations (e.g., Beacham et al. 1999, 2000, 2004; Heath et al. 2001, 2002; Hendry et al. 2002).
Heggenes et al. (2006) utilized historical scale samples to assess possible changes in the genetic diversity of steelhead trout populations native to the Kitimat River of BC's central coast (Fig. 1). Hatchery steelhead trout smolts (young silver-colored salmonids that are preparing to leave freshwater for the sea) of known number and origin have been released in the Kitimat River on an annual basis since 1984. Despite 20 years of operation, the numbers of juvenile hatchery fish released (an average of 51 000 per year totaling about 1 million) had no discernible impact on heterozygosity or population structure (although a slight decrease in allelic richness was detected). This lack of genetic change was likely attributable at least in part to adequate numbers of native broodstock used each year, as well as the relatively large size of the native-spawning population compared with the number of hatchery fish (Heggenes et al. 2006).
Figure 1.
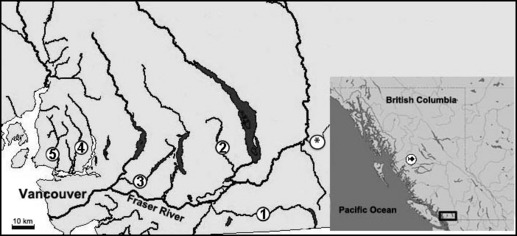
Map showing the geographic location of the rivers from which steelhead trout (Oncorhynchus mykiss) were sampled in southwestern British Columbia, Canada. 1, Chilliwack; 2, Chehalis; 3, Alouette; 4, Seymour; and 5, Capilano rivers. *Denotes the Coquihalla River, which is the source of some broodstock used in hatchery supplementation of the Chehalis River. The  symbol denotes the Kitimat River, study site for Heggenes et al. (2006).
symbol denotes the Kitimat River, study site for Heggenes et al. (2006).
To ascertain whether or not the results from the Kitimat River study by Heggenes et al. (2006) have any generality that may be used to inform broader management, our study extended this analysis to five other river systems, from southwestern BC. Given that many rivers throughout the southern mainland coast of BC have shown evidence of declines in the number of wild adult steelhead trout since about 1990 (Smith and Ward 2000; Ahrens 2004), our study serves to develop a baseline to monitor genetic change in rivers with declining numbers of wild spawners (cf. Van Doornik et al. 2011). In addition to riverine and marine environmental conditions, there is concern that perturbations by hatchery supplementation have contributed to this general decline, at least in rivers with hatchery fish present (Smith and Ward 2000). Using multilocus genotypes, the consequences of these supplementation programs on genetic diversity and population structure were evaluated in samples of steelhead trout collected over 58 years in five river systems. Ryman et al. (1995) suggested that supportive breeding programs should operate with the conservation genetic goal of inducing no more drift (and loss of diversity) in a supported population than would be observed had the population been left on its own. In conducting our study, our expectation for success of the genetic aspects of the steelhead trout hatchery program in BC was that while harvest opportunities increase with supplementation, there should be no change in genetic diversity or divergence among populations beyond what they would have experienced had they been left on their own without supplementation.
Materials and methods
Summary of steelhead trout fish culture and stocking in the study localities
Steelhead trout tissues were obtained from five relatively small river systems (approximately 6–60+ kilometers long) in southwestern BC, Canada, that support major recreational fisheries: the Chilliwack, Chehalis, and Alouette rivers (all of which drain into the lower Fraser River), and the Capilano and Seymour rivers (which drain into nearby Burrard Inlet, Fig. 1). These systems have a long history of recreational exploitation and all occur in watersheds that have been affected to varying degrees by urbanization, changes in water quality, impoundments, and flow diversions and controls, although habitat improvement projects have been undertaken, again, to varying degrees (Table 1; see also Lill 2002 and Ahrens 2004). Using salmon hatcheries located on each river, ongoing steelhead trout hatchery supplementation programs began on these rivers in the late 1970s and early 1980s (Table 1) as part of the federal Salmonid Enhancement Program initiated, in part, to mitigate the effects of increasing angling pressure, water use, and habitat degradation on steelhead trout abundance. The primary goal of these hatchery programs, however, remains to supply terminal recreational harvest opportunities and not to rebuild the wild populations in a demographic sense. Consequently, ‘success’ of these supplementation programs is defined as enhancing the angling opportunities for steelhead trout by increasing the numbers of harvestable, hatchery fish in each system with no to minimal negative ecological and genetic impacts on wild fish. Negative ecological impacts could include competition with wild fish for food in freshwater (as juveniles) or the ocean (as adults), and competition for spawning areas and mates as adults. Negative genetic impacts could include interbreeding of nonharvested hatchery fish with wild fish in nature, shifts in the genetic structure of populations through large releases of hatchery fish relative to wild fish production, and declines in effective population size of wild spawning fish by ‘mining’ the natural population for use as hatchery broodstock, or by increasing variance in family-specific survival (reviewed by Araki and Schmid 2010). Our study focuses on an evaluation of changes in neutral population genetic diversity and structure following the beginnings of hatchery supplementation.
Table 1.
Summary of important demographic and habitat characteristics of five steelhead trout (Oncorhynchus mykiss) populations sampled in the study. Length = length of river currently accessible from the sea for upstream migrating fish, MAD = mean annual discharge (cubic meters/s), Estimated census size = estimate of adult steelhead trout in the river during spawning period from snorkeling swim counts and professional opinion (the value to the right of the slash is the estimated capacity, at 13% marine survival, based on habitat availability), H:W = ratio of hatchery to wild smolts (wild estimated using biostandards/discharge models), Broodstock = average numbers of winter (w) and summer (s) run males and females used in hatchery program (all wild unless indicated), Major habitat perturbations = summary of major changes to river system since European settlement
| River | Chilliwack | Chehalis | Alouette | Seymour | Capilano | |
|---|---|---|---|---|---|---|
| Length (km) | 60+ | 20+ | 23 | 18.5 | 5.5 | |
| MAD (cms) | 67 | 41 | 6.2 | 16.1 | 19 | |
| Hatchery programs for each river using native broodstock* | Year of 1st release | 1978 | 1984 | 1979 | 1981 | 1973 |
| Year of 1st hatchery (clipped) capture | 1980 | 1985 | 1982 | 1982 | 1982 | |
| Approx. total hatchery release no. until most recent sampling year†: | Total | 2 890 000 | 1 600 000‡ | 550 000 | 780 000 | 230 000 |
| Annual mean | 116 000 | 66 000 | 23 000 | 31 000 | 23 000 | |
| Standard deviation | 12 000 | 18 000 | 9000 | 12 000 | 16 000 | |
| Smolt H:W | 1.1: 1 | 10.8: 1 | 7.7: 1 | Unknown | 6.1: 1 | |
| Estimated census size of wild steelhead trout§ | >2000/4000 | 200–500/700 | 200–500/600 | 100–200*/450 | <100*/300 | |
| Mean no. of annual broodstock (with standard deviation) and % hatchery fish | Female | 33 (11) 0% | Winter Run: 10 (4) 8% | 8 (3) 0% | 14 (5) 41%** | 16 (4.6) 53% |
| Summer Run: 9 (5) 93%, 0% | ||||||
| Male | 30 (10) 0% | Winter Run: 9 (4) 0% | 7 (2) 0% | 12 (5) 35%** | 13 (3.3) 51% | |
| Summer Run: 4(3) 36%, 38%¶ | ||||||
| Mean annual % of wild (unclipped) steelhead (with standard deviation) in total run between year of 1st hatchery (clipped) capture and most recent sampling year, based on | Steelhead Harvest Analysis†† | 68 (11) | 49 (12) | 62 (13) | 60 (14) | 57 (9) |
| Adult snorkel counts | 89 (3) | 65 (10) | 80 (8) | 59 (17) | 47 (4) | |
| Major habitat perturbations | Logging‡‡ Urban§§ | Logging‡‡ | Dam¶¶,*** Logging‡‡ Urban§§ | Dam¶¶,††† Logging‡‡ Urban§§ | Dam¶¶,††† Logging‡‡ Urban§§ | |
| Conservation status‡‡‡ | RMZ | RMZ | CC | ECC | ECC |
While not detailed, stocking records provide enough information to show that any stocking prior to these was comparatively sporadic and involved low numbers of fish (G. Wilson, unpublished data).
Total given to the nearest ten thousand; annual mean and standard deviation given to the nearest thousand.
Of the total, 77% of smolts originate from native, winter run adult broodstock and 23% is from non-native, summer-run broodstock.
Winter and summer runs combined (where applicable). Estimated census size = estimate of adult steelhead trout in the river during spawning period from snorkeling swim counts and professional opinion (the value to the right of the slash is the estimated capacity, at 13% marine survival, based on habitat availability, see Lill 2002). Escapement estimates have been made on multiple occasions for each stream throughout the 2000–2010 period and represent the typical escapement over this period.
Percentage of native hatchery fish listed, followed by percentage of non-native wild fish from the nearby Coquihalla River (see Fig. 1).
Ministry of Environment data on file from Steelhead Harvest Analysis results, see DeGisi (1999) for description.
Both winter and summer run broodstock are a mix of wild and hatchery origin adults
Historical, pre-forest practice code logging, hydrologic recovery thought to be achieved.
Lower most approximately 30% of river bank/watershed developed with associated dikes, channelization and estuary impacts.
Dam closure dates are 1928, 1954, 1927 for the Alouette, Capilano, and Seymour rivers, respectively. Regulated flow regimes and low summer flows.
Winter run migration barrier.
Summer run migration barrier, summer and winter runs share riverine habitat below dam since dam closure dates.
From Lill (2002). Conservation zones from Johnston et al. (2002) are extreme conservation concern (ECC): Populations believed to be at 15% or less of habitat capacity and subject to extinction; conservation concern (CC): Populations are estimated to be between 15% and 30% of capacity. Routine management zone (RMZ): populations between 30% and 100% of capacity and managed primarily in terms of recreational fishing opportunities and yield (see Johnston et al. 2002).
The hatcheries operate under various common guidelines following a policy that commits to the conservation of native wild populations (Ludwig 1995): (i) use of wild, native broodstock that are randomly collected annually within each river throughout the spawning run, (ii) employing a 1:1 mating design, (iii) marking (adipose fin clip) of all released smolts to allow for selective harvest (of freshwater-resident and adult returns of hatchery fish), (iv) release of smolts in the lower portions of rivers to reduce competition with wild juveniles, and (v) no transplants between rivers. Two of the rivers (Alouette and Chilliwack rivers) also contain substantial wild populations upstream of their hatcheries in areas that are closed to fishing (Nelson et al. 2005).
Although broadly adhered to, some deviations from these ‘conservation-based’ procedures have occurred (Table 1):
In attempts to attain target broodstock numbers in the face of dwindling numbers of wild fish, all but the Chilliwack and Alouette rivers have supplemented wild, native broodstock to a greater or lesser extent with wild-caught, hatchery-reared (fin-clipped) returns (Table 1).
When a 1:1 male-to-female broodstock ratio was not attained, individual males and/or females were occasionally used in more than one mating.
The exception to the use of native broodstock was the Chehalis hatchery, where a population of steelhead trout that return to spawn between June and October (‘summer-run’) was introduced in 1986 from the nearby Coquihalla River (Fig. 1, Table 1).
Mean proportions of wild (unclipped) steelhead trout in the total run since the first year of hatchery (fin-clipped) captures following the inception of supplementation using native broodstock are equal to or exceed 49% (Table 1). This indicates that the conservation goal of keeping the hatchery to wild fish ratio to <1 has been achieved, despite no annual adjustments of smolt release targets (Ludwig 1995). With the exception of Capilano River (whose posthatchery supplementation samples were collected after the first hatchery releases, but before the first recorded hatchery returns), hatchery fish comprised a significant proportion of the returning steelhead trout population in these rivers; estimates range from an annual mean of 51% of total run for the Chehalis River and 53% for the Capilano River (based on angler catch statistics) to between 11% (from adult snorkel counts) and 32% (based on angler catch statistics) from the Chilliwack River (Table 1).
Smolt release targets were determined by consideration of historical spawning run sizes, impact on wild populations by anglers, angler accessibility to water, proximity to human population centers, and carrying capacity with an attempt to balance wild and hatchery returns to a 1:1 ratio (Ludwig 1995). The annual mean smolt release number over the study period within each river was in the tens of thousands, reaching a hundred thousand for the Chilliwack River, and total releases over the study period's time scale of several decades range from many hundreds of thousands to nearly 3 million (Table 1).
Sample collection
Data from 902 adult fish samples were collected, with an average of 180.4 (SD ± 75, range 106–288) samples distributed across a mean of 4.0 (SD ± 1.2, range 3–6) time points for the five rivers that have a history of prehatchery scale collection and a hatchery program. At least one temporal point (range 1–3) from every river was sampled before the initiation of hatchery supplementation using native broodstock, and either two or three time points were comprised of samples collected afterward (Table 2). These will be referred to hereafter as pre- and posthatchery groups, respectively. Regrettably, there were no samples available for comparable time periods for rivers that had not undergone hatchery supplementation within the study area.
Table 2.
Steelhead (Oncorhynchus mykiss) sampled from five hatchery-supplemented rivers in southwestern British Columbia
| Wild/hatchery and winter/summer run composition of samples listed under ‘Sample size’ | |||||
|---|---|---|---|---|---|
| River | Population code* | Sampling time span | Sample size | Wild, unclipped (Wi) and hatchery, fin-clipped (H) returns | Winter (W) and summer (S) run returns |
| Chilliwack | CH48 | 1948–1949 | 40 | 40 Wi | 40 W |
| CH58 | 1958–1959 | 49 | 49 Wi | 49 W | |
| CH68 | 1968–1969 | 50 | 50 Wi | 50 W | |
| CH93 | 1993–1996 | 50 | 25 Wi: 25 H | 50 W | |
| CH98 | 1998–1999 | 49 | 49 Wi | 49 W | |
| CH04 | 2004–2005 | 50 | 50 Wi | 50 W | |
| Chehalis | CE51 | 1951–1954 | 53 | 53 Wi | 53 W |
| CE83 | 1983–1985 | 76 | 76 Wi | 76 W | |
| CE95 | 1995–1997 | 49 | 43 Wi: 6 H | 40 W: 9 S† | |
| CE99 | 1999–2001 | 35 | 35 Wi | 35 W | |
| Alouette | AL57 | 1957–1958 | 32 | 32 Wi | 32 W |
| AL83 | 1983–1985 | 43 | 31 Wi: 12 H | 43 W | |
| AL99 | 1999–2002 | 31 | 31 Wi | 31 W | |
| Seymour | SE50 | 1950–1959 | 41 | 41 Wi | 41 unknown |
| SE69 | 1969–1978 | 27 | 27 Wi | 27 unknown | |
| SE88 | 1988–1989 | 36 | 36 Wi | 1 W: 35 S | |
| SE01 | 2001–2006 | 78 | 78 unknown | 78 unknown | |
| Capilano | CA56 | 1956–1961 | 24 | 24 Wi | 24 unknown |
| CA77 | 1977–1978 | 40 | 40 Wi | 40 unknown | |
| CA79 | 1979–1982 | 49 | 49 Wi | 49 unknown | |
| Total | 902 (mean = 45 SD ± 14) | 781 Wi: 43 H: 78 unknown | 599 W: 44 S: 259 | ||
Population codes include the initial year of sampling and represent a range of years as indicated. Prehatchery supplementation samples are highlighted in boldface. Posthatchery supplementation samples refer to those collected after the first release and return of hatchery fish originating from native broodstock (refer to Table 1 for dates). The exception to this is Capilano River, where the posthatchery samples refer to samples collected after the first releases but before the first recorded hatchery returns. As such, this river's samples explore potential indirect effects of competition from hatchery releases while serving as a control for temporal change that may be associated with direct (introgression) and indirect (competition) impacts of returning adult hatchery fish. The wild/hatchery and winter/summer run composition of each sample size is also listed. For instance, the 40 samples from CH48 consist of 40 wild, winter run steelhead trout. The CE95 sample consists of 43 wild fish, 6 hatchery fish of which 40 were winter run and nine summer run. ‘Unknown’ means that the breakdown into wild and hatchery spawners was not determined.
Summer run steelhead were introduced by the Chehalis hatchery using broodstock from the nearby Coquihalla River (see Fig. 1).
Steelhead trout have variable migration run timing and are typically classified as either ‘summer run’ which enter rivers from the sea between May and September or ‘winter run’ which enter rivers from November to April (Withler 1966). This migration timing variation is thought to represent repeated, independent evolutionary responses to flow regimes in rivers, with fish entering rivers when flow regimes are suitable to facilitate upstream movements to spawning areas. Summer run steelhead trout typically have reduced gonad development and hold in areas of the river until they spawn the following spring (i.e., up to 1 year after they entered the river). Winter run fish typically enter streams with advanced gonad development and also spawn in the spring (i.e., within 6 months of entering the river). In some systems, significant genetic divergence between summer and winter steelhead trout from the same stream has been detected, but such differentiation is typically very low [e.g., 1% of total diversity (Nielsen and Fountain 1999; see also Chilcote et al. 1980)]. In our system, two of the study rivers have both summer and winter runs of steelhead trout (Seymour and Capilano rivers), and three have a native winter run only (Chilliwack, Alouette, and Chehalis rivers). We obtained separate samples of winter (N = 41) and summer run (N = 37) steelhead trout in the Seymour River for the 2001–2004 time period.
In order to generate adequate sample sizes for population genetic analyses (at least 30 individuals per time point wherever possible: mean N = 45, SD ± 14, Table 2), samples were pooled across consecutive years within rivers. Temporal analyses within rivers was enabled by a sampling design whereby each temporal grouping is composed of samples collected over a shorter period of time than that which separates it from other temporal groupings (Table 2). With 80% of sample points pooled over three or fewer years, most cases involved pooling over only 1 or 2 years. Low levels of repeat spawning (<10% spawn a second time, Hooten et al. 1987) and age at maturity of 4–6 years in steelhead trout (Maher and Larkin 1955; Withler 1966; Caverly 1978; G. Wilson, unpublished data) suggest that this is a reasonable strategy. In addition, we were unable to conduct analyses on all yearly samples because some localities have very low sample sizes within any given year (e.g., Alouette and Capilano rivers had only 1 and 2 years, respectively, with more than 30 fish sampled). Samples from two rivers involved more extensive pooling: the two prehatchery time points from the Seymour River spanned 9-year periods; and one posthatchery sample from the Seymour River and one prehatchery from the Capilano River spanned a 5-year period. These pooled temporal groupings within localities were separated by a decade or more, with only four exceptions (Table 2) where temporal groupings were separated by 1–5 years.
Microsatellite genotyping
Genomic DNA was extracted from up to 10 dried scales per individual, or from approximately 20 mg of adipose fin tissue stored in 95% ethanol. The Qiagen spin column-based DNA extraction procedures were used and samples were then stored at −20°C. Genetic variation was assayed at nine microsatellite loci (Omy77, Ssa456, Ssa197, Oneu14, Oneu8, Ots3, OkiA3, Ots103, Ssa85) chosen for their utility as population genetic markers for O. mykiss populations within BC (Heggenes et al. 2006; Tamkee et al. 2010). These loci were amplified from the DNA extractions by polymerase chain reaction (PCR) using flourescently labeled primers outlined in the procedures of Heggenes et al. (2006). Allelic variation was then assayed using a CEQ 8000 Genetic Analysis System (Beckman Coulter, Fullerton, CA, USA) with CEQ DNA Size Standard Kit-400 as the internal size standard.
Statistical analyses
Exploring factors that potentially confound analysis
Older tissue samples may yield a lower quantity of more degraded DNA; consequently, they are more susceptible to genotyping errors (Taberlet et al. 1996), such as short allele dominance and allelic dropout. Short allele dominance refers to preferential amplification of small alleles resulting in larger alleles specifically failing to amplify in heterozygotes (Wattier et al. 1998). It yields a specific pattern of deficiencies and excesses of particular genotypes that can readily be distinguished from Hardy–Weinberg deviations caused by nonrandom mating (van Oosterhout et al. 2004). We used MICROCHECKER version 2.2.3 (van Oosterhout et al. 2004) to detect any of these patterns in our dataset.
Allelic dropout refers to the failure of an allele that is present in very low copy number to amplify in any given PCR by chance, independent of allele size and locus. It can yield a pattern of heterozygote deficiencies that is similar to nonpanmixia (Taberlet et al. 1999). As a quantitative measure of the direction and extent of any population divergence from Hardy–Weinberg equilibrium (HWE), Weir and Cockerham's (1984) estimator f of the inbreeding coefficient, FIS, was estimated at each locus within each time point using FSTAT version 2.9.3 (Goudet 2001). A pattern of Hardy–Weinberg disequilibria associated with sample age could highlight potential allele dropout in our dataset.
To assess more subtle patterns of potential inbreeding within populations, we tested for the level of relatedness within populations against the null hypothesis of no relatedness. A high degree of relatedness could result from nonrandom mating, but it could also arise even when there is random mating within a very small population. Although not providing a direct measure of inbreeding per se (as in FIS calculations), relatedness analysis may provide a more sensitive test of inbreeding as it is based on pairwise comparisons among individuals. This was implemented by a permutation re-sampling test in IDENTIX version 1.1 (Belkhir et al. 2002). The observed distribution of both the mean and variance of pairwise relatedness coefficients within each sample time point (rxy, Queller and Goodnight 1989) were compared with a null distribution of 1000 multilocus genotypes expected under panmixia generated by random re-sampling of the original data (cf. Small et al. 2009). Even when mean rxy does not vary from the null expectation, indicating that individuals within a sample are no more genetically related than expected in a random mating population, a significantly higher variance in the observed rxy can indicate that the sample is composed of several independent groups of related individuals, where pairwise comparisons involve either related or unrelated individuals (Belkhir et al. 2002; Small et al. 2009).
The Fisher exact test assessed genotypic linkage disequilibrium (LD) among pairs of loci within each time point, as well as overall, using a Markov chain method in GENEPOP version 3.3 (Raymond and Rousset 2001).
Within-population analyses (effective population size, genetic variation, and genetic structure) in steelhead trout subject to hatchery supplementation using native broodstock
We examined potential changes in the size of the breeding population associated with hatchery operations in each of our samples. There are a number of methods available to infer effective number of breeders (Nb) and/or effective population size (Ne) from genetic data (e.g., see Waples 2005; Palstra and Ruzzante 2008; Palstra et al. 2009). Two general kinds of methods are those that estimate these parameters from a single time sample (e.g., methods based on LD or sibship assignments, Hill 1981; Wang 2009) and those that compare allele frequencies between two temporally spaced samples (i.e., so-called temporal methods, Waples 1989; Jorde and Ryman 1995). For our purposes, we wanted to be able to compare Nb or Ne between pre- and posthatchery samples which necessitated each estimate being independent of all others. Because this is not possible with the temporal methods, we used the sibship assignment method implemented by Colony version 2 (Wang 2009) to estimate the effective population size for each sample point. This method infers the contemporary effective population size from estimated sibship frequencies, drawing on the idea that a smaller population will result in a higher proportion of sibs in any given random sample. Importantly in our instance, the sibship procedure can be applied to subpopulations experiencing immigration as well as nonrandom mating, the latter of which is likely to be present in the posthatchery samples (Wang 2009). We ran two short runs in Colony (results were identical; between runs and longer runs on a subset of the sample points produced similar values), under a polygynous mating system with prior unknown allele frequencies.
Despite these strengths of the sibship-type analyses, our data offer a number of complications to the straightforward interpretation of estimates of Ne (see Discussion). For instance, our analyses estimate the effective number of breeders (Nb) not Ne itself because our samples consisted of adult fish with overlapping, not discrete, generations. In addition, all methods we employed assume closed populations, which is likely not strictly true for steelhead trout. Given the uncertainty surrounding genetic estimates of Nb and/or Ne, we also estimated Nb using LD methods (Waples and Do 2008), the standard temporal method of Waples (1989) both implemented in NeEstimator (Peel et al. 2004; Ovenden et al. 2007) as well as a modified temporal method implemented in SalmonNb (Waples et al. 2007).
Once estimates of Nb are obtained for single cohorts, they can be multiplied by the generation time to yield an estimate of Ne (e.g., Heath et al. 2002). Because steelhead trout mature at variable ages, our samples consist of multiple cohorts (age at maturity in our study area is composed of typically more than 90% of 4–6-year-olds, Maher and Larkin 1955; Withler 1966; Caverly 1978; G. Wilson, unpublished data). The time period between most samples, however, exceeded one generation, so our samples can be considered as single cohorts when comparing across time periods. The Seymour River's prehatchery samples, however, consisted of samples pooled across 9 years and several other samples were pooled across 2–5 years (Table 1). Pooling across years within a temporal time period will result in an upward bias in Nb estimates because multiple years are contributing variation to a single time period. To account for this bias, we multiplied the estimated Nb by 5/y where y = the number of years of pooling and 5 represents the typical generation time for steelhead trout in our system. This quantity provided our final Colony-based estimates of Ne that were used in all subsequent analyses.
Finally, sample size can affect both the accuracy and precision of estimates of Ne (e.g., Palstra and Ruzzante 2008; Wang 2009). Indeed, we observed a positive and significant correlation between our point estimates of Ne and sample size across all 20 temporal samples (r18 = 0.64, P = 0.002, Table S1), but average sample sizes for the pre- and posthatchery treatment groups within localities or pooled across localities were no different from each other (see Results). Consequently, differences between groupings of pre- and posthatchery time points were compared between pooled samples of pre- (N = 9) and posthatchery groups (N = 11) across rivers, using one-way analysis of variance (ANOVA) and Levene's test for homogeneity of variances using PAST, a general spreadsheet-based statistical package (Hammer et al. 2001).
To assess temporal changes associated with hatchery operations in genetic diversity, relatedness and structure within steelhead trout populations, we tested the significance of any difference between group averages of allelic richness (R, El Mousadik and Petit 1996), gene diversity (HE, Nei 1988), rxy and FST (θ, Weir and Cockerham 1984) using 15 000 permutations in FSTAT. For this, groupings of pre- and posthatchery time points were again compared within each river wherever possible, i.e., whenever there were at least two time points both pre- and posthatchery (Chilliwack, Chehalis and Seymour, but not Alouette and Capilano rivers), as well as between pooled samples of pre- and posthatchery groups across rivers. Analysis of variance (ANOVA) was used to test for differences between pre- and posthatchery groupings where there was only one time point available in at least one of the groups (Alouette and Capilano rivers). As allelic richness is independent of sample size, with rarefaction standardizing it to the smallest sample size, bias from unequal sample sizes in inter-sample comparisons is avoided, although a loss in statistical power can be expected (Leberg 2002). Sample size, however, did not vary significantly across these group comparisons (see Results) so differences in allele numbers (NA) were also assessed using ANOVA.
To examine temporal changes in genetic structure within each population over time, and the potential impact of hatchery supplementation on this, pairwise multilocus FST were estimated by θ (Weir and Cockerham 1984) between all samples and the significance of any genetic differentiation/distances was tested using a procedure implementing 10 000 permutations in ARLEQUIN version 3.01 (Excoffier et al. 2005). Similarities among samples were visualized using factorial correspondence analysis (FCA) to project time point means in microsatellite allele frequency space using GENETIX version 4.03 (Belkhir et al. 2001).
The contribution of temporal change to the total genetic variation observed in this system was estimated using the analysis of molecular variance approach (AMOVA: Excoffier et al. 1992) in ARLEQUIN, which partitioned the total genetic variance into covariance components associated with different levels of genetic structure: within individual time points across the entire study system, between time points within rivers and among rivers. Ten thousand permutations of individual genotypes among samples (either between or within groups), or whole samples among groups, tested the significance of each index of differentiation.
Among-population analyses (effective population size, genetic variation, and genetic structure) in steelhead trout populations subject to hatchery supplementation using native broodstock
To compare the extent of temporal changes within populations associated with hatchery operations, among-population analyses were also conducted. Differences among populations in estimated effective population size were investigated by testing the significance of any difference in population averages of Ne using ANOVA. The significance of any difference between group averages of R, HE, rxy and θ were again tested using 15 000 permutations in FSTAT. Once again, differences in allele numbers (NA) were also assessed using ANOVA, as sample size did not vary significantly across these group comparisons (see Results). Pairwise multilocus θ were calculated by grouping time points within each population as a single sample. The significance of any genetic differentiation was again tested by permutation in ARLEQUIN. FCA helped depict inter-population similarities.
Temporal changes in the degree of genetic structure among populations since the inception of hatchery supplementation using native broodstock were also explored; the significance of any difference between group θ averages was tested: one group composed the oldest samples from each river, and the other was comprised of the most recent ones (CH48, AL57, CE51, and SE50 versus CH04, AL99, CE99, and SE01, see Table 2 for population codes). We used 15 000 permutations in FSTAT to test the significance of any difference in θ between these two groups. Samples from the Capilano River, which did not have a sufficiently recent sample for comparison, were excluded (although its inclusion using the most recent sample did not alter the overall findings; data not shown).
We used AMOVA to assess the spatial genetic structure between populations prior to, and after, the initiation of hatchery supplementation using native broodstock by partitioning the total genetic variance into covariance components associated with different levels of genetic structure: within individual time points from these two groupings, between oldest and most recent time points within each river, and among rivers. Ten thousand permutations of individual genotypes among samples (either between or within groups), or whole samples among groups, tested the significance of each index of differentiation.
Finally, we used the model-based Bayesian clustering analysis within STRUCTURE (Pritchard et al. 2000) to assess population structure spatially, across time periods and with respect to the identity of samples as pre- or postsupplementation. Under our null hypothesis of no major effects of hatchery supplementation on microsatellite DNA variation, we expected to see genetic structure that was primarily organized spatially, with temporal samples (whether they were pre- or postsupplementation) clustering by river (e.g., all Chilliwack River samples would segregate within a single cluster separate from other rivers and there would be no distinct pre- and postsupplementation clusters). We conducted six separate analysis: all sample localities and time periods together in one analysis (i.e., 20 ‘population’ samples) and then five separate analyses treating each locality and their multiple time periods separately (i.e., six, four, three, three, and four ‘population’ samples for the Chilliwack, Chehalis, Alouette, Capilano, and Seymour rivers, respectively). For each analysis, we used the admixture model with a burn-in of 50 000 iterations followed by an additional 150 000 iterations, replicated five times. We ran simulations with hypothesized numbers of populations (K) ranging from K = 1–25 (5 more than total number of samples in the first analysis and for K values equal to double the number of population samples in each single locality analysis, Table 2). In the STRUCTURE analyses, we expected that if there was significant temporal variation in genetic structure within localities then the most likely number of genetic populations per river would be some value >1. We expected the same result if there were significant changes to population structure after hatchery supplementation began, with the additional expectation that the multiple genetic populations within localities would be primarily structured into pre- and postsupplementation genetic clusters.
For all analyses, significance criteria for each group of tests conducted were determined according to the sequential Bonferroni correction (Rice 1989).
Results
Hardy–Weinberg equilibrium, linkage disequilibrium, and polymorphism
Four of the 180 tests performed on individual loci in each time point showed significant deviations from genotypic frequencies expected under HWE (P < 0.05 for Oneu14 at CH48 and SE50, Omy77 at AL83, and Ssa85 at SE69), suggesting that allelic dropout was not a serious confounding factor in the majority of older samples. In addition, MICROCHECKER did not find any evidence of short allele dominance at any locus. FIS for each time point at each locus in HWE was estimated to range between −0.383 and 0.471, while FIS values estimated for those that deviated significantly from HWE ranged between 0.351 and 0.791. Three of these high positive FIS values, however, were found in some of our older samples (see above) which repeatedly amplified poorly at these loci (i.e., generally about half the samples amplified at these loci for these samples) suggesting that poor tissue quality influenced PCR amplification in a minority of cases.
Concordance with HWE within populations suggested no significant inbreeding. In addition, all of the mean pairwise relatedness values were negative (Fig. 2A) and not significantly different from what would be expected under the assumption of random mating (all P > 0.05). In four of five comparisons, however, the posthatchery samples had smaller negative relatedness values (Fig. 2A). In addition, the variance in rxy for all but one time point (ranging from 0.06 to 0.18) did not vary significantly from that expected under panmixia (all P > 0.05, Fig. 2B). The Seymour River 1988–1989 sample did show a significantly greater variance in rxy than expected by chance (P = 0.024, Fig. 2B).
Figure 2.
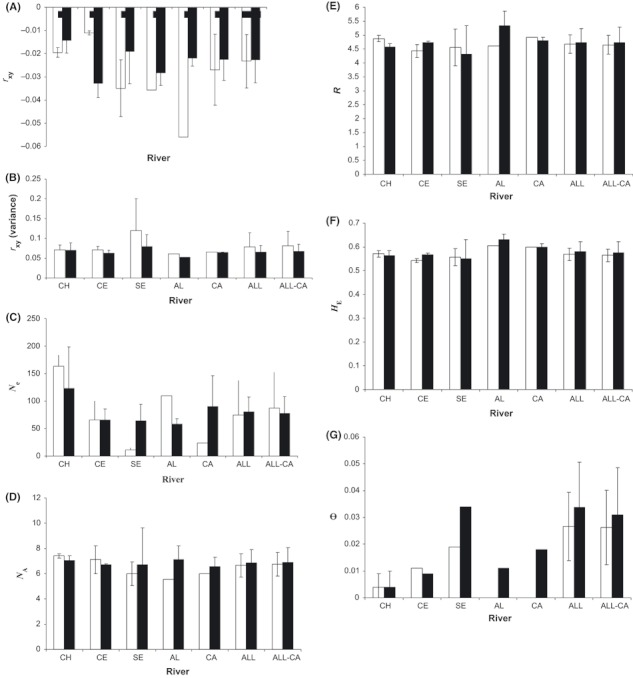
Average measures of population size, genetic variation, and structure in steelhead trout (Oncorhynchus mykiss) within rivers before (empty bars) and after (filled bars) the inception of hatchery supplementation using native broodstock. Based on variation of 902 samples that were genotyped at five or more of nine assayed microsatellite loci. Refer to Table 2 for population codes; ALL refers to all rivers combined; ALL-CA refers to all rivers except the Capilano River. Standard deviations given where applicable. (A) Mean relatedness (rxy); (B) Variance in rxy; (C) Effective population size (Ne); (D) Mean number of alleles (NA); (E) Allelic richness (R); (F) Gene diversity (HE); (G) FST (θ).
Pooled across time periods, there were no significant differences in the level of mean relatedness nor variance in rxy among all populations (minimum P = 0.25, Fig. S1A, B). No pair of loci was in LD, either within each time point or overall (P > 0.05). Temporal samples remained in HWE and linkage equilibrium regardless of their composition (wild : hatchery, winter : summer run, Table 2). Furthermore, FST (θ) between the summer and winter run steelhead trout from the Seymour River was low and nonsignificantly different from 0 (θ = 0.005, P > 0.1), so these samples were pooled for all subsequent analyses.
A high level of polymorphism was observed at all loci (four to 34 alleles per locus, mean = 16). There was a significant correlation between mean time point sample size (N = 45, SD ± 14) and mean number of alleles per locus within each time point (r18 = 0.61, P = 0.004), but there was no difference in mean sample size across populations, which ranged from 35 (SD ± 7) to 53 (SD ± 17; F4,15 = 1.01, P = 0.44). In addition, there was no difference in mean sample size between pre- and posthatchery samples, either overall (pre: N = 44, SD ± 16; post: N = 46, SD ± 13; F1,18 = 0.20, P = 0.66) or within each population: Chilliwack River (pre: N = 50, SD ± 6; post: N = 46, SD ± 0.6; F1,4 = 1.09, P = 0.36), Chehalis River (pre: N = 65, SD ± 16; post: N = 42, SD ± 10; F1,2 = 2.79, P = 0.24), Seymour River (pre: N = 34, SD ± 10; N = post: 57, SD ± 30; F1,2 = 1.08, P = 0.41), Alouette River (pre: N = 32; post: N = 37, SD ± 8; F1,1 = 0.23, P = 0.71), and Capilano River (pre: N = 24; post: N = 45, SD ± 6; F1,1 = 6.92, P = 0.23).
Effective population size
We used a number of methods to estimate effective population size from the genetic data. These alternative estimates are not reported here in detail (but see Table S1); in general, however, the absolute and relative values of Nb (or Ne– see below) were highly correlated among all methods (r ranged between 0.79 and 0.89 among Colony, LD, and SalmonNb methods, Table S1). Despite these broad similarities, absolute values varied considerably among the methods with Colony estimates generally producing the lowest and least variable values and SalmonNb the highest and most variable estimates (Table S1).
There was little evidence to support the hypothesis that hatchery practices have altered the Ne within populations. Our sibship-based estimates of Ne ranged from 9 (Seymour River 1950) to 185 (Chilliwack River 1968 and 2004), and the mean values were virtually identical between groupings of pre- or posthatchery time points across all rivers (all P > 0.05, Fig. 2C). Comparisons within rivers were more limited, but Ne estimates were comparable between pre- and posthatchery times for the Chilliwack and Chehalis rivers, higher in prehatchery times for the Alouette River and higher in the posthatchery times for the Seymour and Capilano rivers (Fig. 2C).
There was an overall significant difference in mean Ne among all of the populations (F4,15 = 4.38, P = 0.015, Fig. S1C). The only significant pairwise comparison, however, was between the Seymour (mean Ne: 37) and Chilliwack rivers (mean Ne: 143; F1,8 = 17.39, P = 0.029).
Genetic variation
For each time point, NA, R, and HE averaged over all loci ranged from 4.67 to 8.78, 3.59 to 5.71 and from 0.49 to 0.65, respectively, yielding relatively narrow ranges of mean values for each population (Fig. 2D–F). Indeed, there were no significant changes in mean NA, R or HE between pre- and posthatchery groups, neither within any of the rivers nor across rivers (all P > 0.05, Fig. 2D–F).
There was no significant difference in mean NA (F4,15 = 0.62, P = 0.65, Fig. S1D) or R (P = 0.27, Fig. S1E) among all the populations, but there was a significant difference in mean HE overall (P = 0.02, Fig. S1F). Pairwise comparisons revealed that the Alouette River had significantly higher HE than that found within three (Chilliwack, P = 0.01; Chehalis, P = 0.01; Seymour, P = 0.02) of the other four rivers.
Temporal genetic structure
Mean FST (θ) was unchanged between pre- or posthatchery time points within the Chilliwack, Chehalis and Seymour rivers, as well as across pooled samples of pre- and posthatchery groups from all of the rivers (all P > 0.05, Fig. 2G). Although the range of temporal mean θ values within the pre- and posthatchery groupings from each river was relatively low (0.004–0.034), FCA suggested some differences in the degree of temporal stability exhibited by the different populations (Fig. 3). For instance, the Chilliwack River time points cluster more closely together compared with the other populations, while those from Seymour River are the most scattered in FCA space (Fig. 3). The extent of any such differences was, however, not detected by a comparison among rivers of mean temporal genetic structure within each population (P = 0.66, Fig. S1G).
Figure 3.
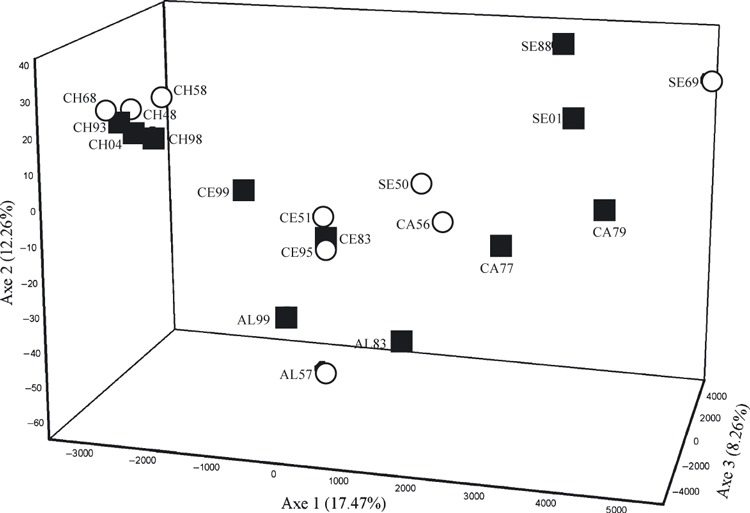
Plot of mean factorial correspondence scores along the first three axes for time point samples of 902 steelhead trout (Oncorhynchus mykiss) based on variation at five or more of nine assayed microsatellite loci. Refer to Table 2 for population codes. The status of time points highlighted as before (empty circles) or after (filled squares) the initiation of hatchery supplementation using native broodstock.
Pairwise values of θ indicated that there was, indeed, temporal stability within populations. Only one of 33 comparisons between time periods within populations was significantly differentiated (θ = 0.03, P < 0.001); this was the two posthatchery supplementation Seymour River time points. The hierarchical analysis of allele frequency variation also highlighted the temporal stability within populations; of the total genetic variation in the study system, there was no significant genetic variation among time points within populations (P = 1.00), 2.0% resided among populations (P < 0.001), with the vast majority of the variation (98.0%, P < 0.001) being found within individual temporal points.
The results from the STRUCTURE analyses were consistent with temporal stability within localities; in all cases and across all replicates, the most likely number of genetic populations for each locality across time periods was one (Table S2 and Figs S2).
Spatial genetic structure
In contrast to the consistent temporal stability in genetic structure within populations, there was significant overall spatial genetic structure among them (θ = 0.018, P < 0.001). All but one of the ten pairwise comparisons between populations showed a significant difference (θ ranged from 0.011 to 0.031, P > 0.05); the exception was the comparison between the Capilano and Seymour rivers (θ = 0.000, P = 0.64). FCA showed groupings of the river samples into two broad geographic clusters: those from the Chilliwack River and those from the remaining four rivers: Capilano, Seymour, Alouette and Chehalis rivers (Fig. 3).
There was, however, no evidence that the degree of spatial genetic structuring among populations had changed significantly with the onset of hatchery operations that use native broodstock; the hierarchical analysis of allele frequency variation highlights the congruence in θ (P = 0.61) between the oldest (θ = 0.022) and most recent (θ = 0.029) samples within hatchery supplemented rivers. Of the total genetic variation among these two groupings, there was no significant genetic variation between the oldest and most recent time points within populations (P = 1.00), with 2.8% residing among populations (P = 0.001). The vast majority of variation in microsatellite allele frequencies (97.8%, P < 0.001) was found within individual time intervals.
Again, the STRUCTURE analysis supported FST-based analyses. When all population samples (N = 20) were analyzed together, the most likely number of genetic populations was K = 7 and localities were clearly distinct from each other (Fig. S2, Table S2). Additional genetic structure beyond that of the five localities appeared to be associated with some temporal subdivision within the Capilano and Seymour rivers (Fig. S3). These same samples were also relatively distinct in the FCA (Fig. 3) and suggested some segregation between the two prehatchery Seymour River samples and between the pre- and posthatchery Capilano River samples.
Discussion
No discernible impact of hatchery supplementation using native broodstock
Our temporal analysis of steelhead trout from five rivers in southwestern British Columbia that have been subject to hatchery supplementation using native broodstock found no evidence of genetic changes associated with the onset and continuation of supplementation, neither within individual populations nor in the relationships among them, as measured using microsatellite DNA variation.
Temporal stability in genetic diversity within populations
Our data revealed no detectable changes in our estimates of effective population size or in measures of intra-population genetic variation since the onset of hatchery operations. Even given the limited statistical power of our analysis, consistent qualitative trends toward reduced variation in posthatchery samples were not observed. The level of polymorphism of the loci employed could potentially limit our ability to detect change in genetic diversity, yet even levels of gene diversity, which are less constrained than those of allelic richness, detected no changes in genetic diversity over time.
Although the variation in microsatellite loci and sample sizes analyzed warrants caution in inter-study comparisons of genetic diversity, one particular comparison is particularly relevant to our study. Using the same set of markers employed in the current study, a survey of steelhead trout from another native broodstock hatchery supplemented river in BC (Heggenes et al. 2006) reported values of mean gene diversity (0.57) and allelic richness (3.78) similar to those from our study (mean HE = 0.58, mean R = 4.7). Heggenes et al. (2006) also found little evidence for changes in genetic diversity associated with the onset of hatchery operations (no significant change in gene diversity, but a slight, significant, decline in allelic richness), supporting our conclusion that hatchery supplementation using native broodstock has had little detectable effect on neutral genetic variation within the steelhead trout populations that we assayed. Indeed, such temporal stability of genetic diversity is also characteristic of other, relatively unperturbed steelhead trout populations in BC (Heath et al. 2002; see also Van Doornik et al. 2011 for Chinook salmon, Oncorhynchus tshawytscha).
Temporal stability of spatial genetic structure
The overall differentiation between the five populations in this study (θ = 0.018) indicated a low but significant degree of population subdivision whose magnitude is consistent with that found in other studies over a comparable geographic scale (Beacham et al. 2000 [mean within watershed θ = 0.025]; Heath et al. 2002 [mean within watershed θ = 0.047]). Indeed, the partitioning of the majority of genetic variation within populations is typical of steelhead trout populations (e.g. Beacham et al. 1999, 2000, 2004; Heath et al. 2001, 2002), and anadromous salmonids more generally (Hendry et al. 2004).
Consistent with Heggenes et al. (2006), the temporal stability of this genetic structure, and consistently low levels of inbreeding over time periods spanning several decades indicate no discernible association between genetic differentiation and the hatchery operations that use native broodstock within these steelhead trout populations. Such temporal stability of genetic structure is characteristic of steelhead trout populations from other regions (e.g. Beacham et al. 1999, 2000, 2004; Heath et al. 2001; Hauser et al. 2006; Heggenes et al. 2006). Our results are also consistent with temporal studies using archived fish scales of other anadromous fish that have reported remarkable stability in genetic structure over several decades (Salmo salar: Nielsen et al. 1997, 1999; Tessier and Bernatchez 1999; Salmo trutta: Hansen et al. 2002; O. tshawytscha: Van Doornik et al. 2011). The Capilano and Seymour rivers, however, displayed the greatest tendency to some variability through time with respect to the degree of divergence among temporal samples. This is consistent with these populations experiencing some of the lowest estimated effective population sizes through time (thus promoting genetic drift of allele frequencies), especially in the Seymour River.
The sensitivity of our study to detect potential changes in genetic diversity and structure could, of course, be increased with more temporal sample points and greater numbers of individuals screened. Nevertheless, the lack of even qualitative temporal trends of change in our estimates of effective population size, genetic diversity and structure contrasts sharply with changes observed in the genetic structure in brown trout (S. trutta) that were associated with stocking of nonlocal strains of hatchery trout. These changes were detected despite fewer replicates and smaller average sample sizes (one pre- and poststocking sample for each of six rivers, with mean sample size of 33, SD ± 10, Hansen et al. 2009).
Conservation implications and recommendations
Hatchery management practices can influence the neutral genetic structure of indigenous populations, even when releases are from native broodstock, by effecting patterns of competition, introgression, genetic drift, and gene flow (Araki et al. 2007a; Eldridge and Naish 2007; Caroffino et al. 2008). In contrast to potential negative genetic impacts, however, our study corroborates and extends the findings of Heggenes et al. (2006) that supplementation using native broodstock has had negligible impact on the diversity and structure of the neutral genetic variation of at least some steelhead trout populations. Several factors likely contribute to this lack of a detectable impact.
First, wild fish that spawn in portions of our study rivers located upstream of the hatcheries may help to buffer potential negative effects from downstream releases of hatchery fish into the lower river reaches. Indeed, the location of hatcheries in the lower half of most of the rivers studied here likely separates many hatchery (which rarely swim upstream of the hatcheries) and wild spawners spatially. This feature is shared with the Kitimat River hatchery (Heggenes et al. 2006), and the importance to wild steelhead trout in these areas has been recognized in the Chilliwack River where such upstream refuges are closed to fishing (Nelson et al. 2005). By contrast, the Capilano and Seymour rivers, where hatcheries are located at dams that block access to upper reaches (believed to be used historically by summer run steelhead trout, Labelle 2007), do not benefit from such potential refuges.
Second, each of the rivers we studied contains some resident rainbow trout that probably interact to some unknown extent with steelhead trout. Some estimates of resident rainbow trout abundance are available for all rivers and are obtained during annual snorkel swims used to enumerate spawning fish. Resident rainbow trout are identified as those spawning fish that are between 20 and 45 cm total length, and they are usually found at relative abundances of 0.6–0.9 to that of adult steelhead trout (G. Wilson, unpublished data, N = 31 counts across seven southwestern BC streams between 2002 and 2010). Christie et al. (2011) demonstrated that up to 20% of alleles detected in anadromous steelhead trout may originate from matings with resident rainbow trout in the Hood River, Oregon. Consequently, resident wild rainbow trout may contribute substantially to the gene pool of sympatric steelhead trout and act as a buffer against homogenization from hatchery fish (Araki et al. 2007b; Christie et al. 2011).
Third, the lack of discernible genetic changes is also probably due, in part, to low survival of hatchery fish in the face of considerable release numbers. For instance, the annual escapement of wild steelhead trout into the Chilliwack River is about 4000 fish. Assuming a 50:50 sex ratio, a typical fecundity of 5000 eggs, an egg-to-fry survival rate of about 6.5% (Ward and Slaney 1993; van Dishoeck et al. 1998), and a fry-to-smolt (typically 2–3 years old, Maher and Larkin 1955; Withler 1966; Caverly 1978) survival rate of 12% (Ward and Slaney 1993) leads to an estimated wild smolt production of some 81 250 fish. This compares to over 100 000 hatchery smolts produced every year and larger hatchery : wild smolt ratios occur in the other rivers (Table 1). The larger number of hatchery smolts in many systems has the potential to significantly reduce the inbreeding effective population size of steelhead trout, especially in rivers where relatively few parents were used in the hatchery program (e.g., Chehalis River, Table 1). Hatchery-origin steelhead trout in southwestern BC rivers, however, suffer a higher mortality rate compared with their wild produced counterparts, with average marine returns at about 1% and 4%, respectively, over the past 30 years and following a dramatic decline from about 13% in the early-1990s (McCubbing and Ward 2008). Still, catch statistics and adult snorkel counts clearly indicate that hatchery fish may comprise a considerable proportion of the returning steelhead trout populations (Table 1). Given that many of these are released upon capture in recreational fisheries (G. Wilson, unpublished data), hatchery fish have the potential to have an impact upon the indigenous population. This is particularly true, given the modest population sizes of spawning adults (only the Chilliwack River typically has a relatively stable population size of more than 2000 wild adults) and the downward trends in estimated wild spawner abundance over much of the time period included in our study, especially for the Capilano and Seymour rivers (Ahrens 2004). Markedly lower reproductive success of non-native, naturally spawning hatchery steelhead trout compared with native, wild fish, however, has been documented in several areas (a decrease of at least two-thirds the number of smolts produced per individual, Kostow et al. 2003; McLean et al. 2004; reviewed in Araki et al. 2008). The poorer performance of these hatchery fish is likely influenced by artificial selection and/or generations of inadvertent domestication selection (Kostow et al. 2003; McLean et al. 2004; reviewed in Araki et al. 2008). Hatchery fish that are produced from wild, native broodstock generally perform better than non-native fish, although they, and even their descendants, often still fare worse than wild fish (reviewed in Araki et al. 2008, 2009). In summary, the use of native broodstock coupled with their relatively poor survival and reproductive success have likely minimized the opportunities for gene flow between wild and hatchery steelhead trout and minimized changes to effective population size and genetic diversity in our study system. A lack of change to effective population size (and resultant genetic diversity) is expected when the reproductive rate (i.e., the number of returning spawners and their reproductive success) is lower for hatchery-produced fish (Ryman et al. 1995).
Fourth, hatchery management practices may have contributed to limiting the potential impact of hatchery fish on the neutral genetic structure of indigenous populations. Using sufficient numbers of native, nonhatchery reared (unclipped) broodstock, as well as an approximate 1:1 male-to-female broodstock ratio whenever possible (Table 1) have likely contributed to the apparent genetic stability within and between our study's populations by minimizing large changes and fluctuations in Ne. Indeed, Ne estimates were relatively stable over time, which agrees with Araki et al.'s (2007b) finding that hatchery supplementation using native broodstock each generation did not negatively impact Nb in another steelhead trout system. On the other hand, Araki et al. (2007b) found that in more traditional hatchery programs, where non-native broodstock spent multiple generations in hatcheries, exhibited decreased mean, and increased variance, in reproductive success (Araki et al. 2007b).
Finally, perhaps the relatively small scale of hatchery releases of steelhead trout contributed to the lack of detectable changes. For instance, across a comparable geographic scale as in our study, the number of fish released from coho salmon (Oncorhynchus kisutch) native broodstock hatcheries in Puget Sound, Washington, averaged more than 69 million individuals per river (N = 11) over 52 years, and intensity of stocking was negatively correlated with the extent of population structure (Eldridge and Naish 2007). By contrast, in our study release numbers averaged 1 152 000 individuals per river over approximately 20 years (Table 1). In addition to relatively lower survival and reproductive success of hatchery fish, hatchery release numbers may have been sufficiently low to avoid genetic homogenization via increased gene flow between hatchery and wild fish (Slatkin 1985; Adkison 1995; Eldridge and Naish 2007).
Perhaps surprisingly, even the Chehalis River winter run population displayed no genetic change over time despite the introduction of a summer run from non-native broodstock. Potential reasons why no genetic impact was detected include: poor survival and/or reproductive success of these introduced hatchery fish (Kostow et al. 2003); life history differences acting as reproductive isolation barriers between summer and winter run returns (e.g. Leider et al. 1984); and lack of genetic distinction between the two rivers’ populations which have yet to be compared directly. While one of the two posthatchery time points from the Chehalis River was composed entirely of wild winter run returns in our analysis, interestingly, the other also included some summer run fish (18%, Table 2). This suggests that a lack of genetic distinction between the two populations contributes, at least in part, to this lack of detectable genetic change although a larger sample of summer run adults is needed to assess this idea robustly.
Despite the lack of any difference in neutral genetic variation associated with hatchery supplementation found in this study, we cannot discount that there have been no genetic changes of any kind in the steelhead trout populations studied here; for example, quantitative trait loci that are responsible for influencing adaptive characters such as growth rate, age at maturity, behavior, run-timing, morphology, etc. cannot be directly evaluated with our approach of assaying microsatellite loci that represent neutral genetic loci. There is considerable debate in the population and conservation genetic literature concerning whether or not neutral variation can be used as a proxy measure for genetic variation responsible for phenotypic traits (reviewed by Merilä and Crnokrak 2001; Frankham et al. 2010). The microsatellite DNA data, however, are relevant to inferring aspects of the demography of steelhead trout that may influence evolutionary processes. For instance, the response of populations to selection at quantitative trait loci within environments can be influenced by their level of genetic connectivity (gene flow) with individuals that have dispersed from alternative environments (Lenormond 2002). In addition, potential response to selection can be influenced by effective population size, i.e., changes in allele frequency are dominated by drift when the product of Ne and the selection coefficient is <1 (Li 1978). The lack of detectable changes in neutral genetic patterns (e.g., population structure and its links to genetic connectivity/gene flow; effective population size) over time in the populations that we studied, however, implies that the influence of these demographic parameters on evolutionary processes, such as natural selection, has also probably not changed appreciably. Consequently, this argues that steelhead trout should have retained the capacity to respond to changing environments, at least within the geographic areas that we examined.
Our estimates of Ne, however, are subject to a number of caveats. First, our sampling was not ideal for estimating Ne within any one time period owing to violations of some of the simplifying assumptions (e.g., single cohorts) and the pooling of samples across some years. Our Ne estimates, therefore, might best be considered as estimating some quantity between Nb and Ne, and each such estimate likely applies to some unknown range of years within each time period. Still, such estimates are probably useful for relative comparisons between time periods, especially because most comparisons involved intervals of time encompassing multiple steelhead trout generations, a situation which should minimize the effects of any sampling biases (e.g., Palstra et al. 2009). In addition, our estimates of Ne are broadly consistent with those in Heath et al. (2002) who reported Nes of between 92 and 560 for three rivers, across three time periods each, from the Skeena River system in northern BC. In addition, it is probably not unreasonable that the rivers in our study system that are the longest, have the greatest mean annual discharge (and hence inferred aquatic habitat area) and have likely been the least disturbed (i.e., the Chilliwack and Chehalis rivers), tended to have the highest and least variable Ne estimates (Fig. 2; Table S1). By contrast, the Seymour and Capilano rivers have probably been the most perturbed, have the smallest habitats (particularly after the dams on these systems were completed) had the lowest and most variable Ne, and exhibited the greatest fluctuations in genetic differentiation across time periods.
While it is encouraging from our study that Ne values in each river have not experienced consistent declines since the onset of hatchery augmentation programs some further caveats suggest caution in interpretation of these results. First, our power to test for a pre- versus posthatchery effect was limited by small sample sizes within rivers (N = 6 maximum) and any pooled pre- and posthatchery effects (N = 11 maximum) were probably confounded somewhat by among river effects (e.g., variation in habitat changes through time). Second, our study was unable to employ suitable control populations, i.e., those surveyed over the same time frame and that have experienced no hatchery supplementation. Such samples do not exist in British Columbia and the rivers that we studied were those that experienced the most intense recreational fishing and were selected for hatchery supplementation for that reason. Other, less targeted and less accessible systems that could have served as controls were not subject to scale sample or broodstock collection over the same or similar time frame. Consequently, without such controls, it is impossible to eliminate the possibility that fish produced in the hatchery from native broodstock, and that spawned successfully in the wild, actually helped to maintain existing variation and structure in the studied populations.
Third, our estimates of mean Ne were low relative to the value of about 500 that Waples (1990) suggested was the minimum for long term viability in Oncorhynchus. Even estimates of Ne generated using the temporal methods (Table S1). By contrast, the Seymour and Capilano rivers have probably been the most perturbed, have the smallest habitats (particularly after the dams on these systems were completed) had the lowest and most variable Ne, and exhibited the greatest fluctuations in genetic differentiation across time periods.
While it is encouraging from our study that Ne values in each river have not experienced consistent declines since the onset of hatchery augmentation programs some further caveats suggest caution in interpretation of these results. First, our power to test for a pre- versus posthatchery effect was limited by small sample sizes within rivers (N = 6 maximum) and any pooled pre- and posthatchery effects (N = 11 maximum) were probably confounded somewhat by among river effects (e.g., variation in habitat changes through time). Second, our study was unable to employ suitable control populations, i.e., those surveyed over the same time frame and that have experienced no hatchery supplementation. Such samples do not exist in British Columbia and the rivers that we studied were those that experienced the most intense recreational fishing and were selected for hatchery supplementation for that reason. Other, less targeted and less accessible systems that could have served as controls were not subject to scale sample or broodstock collection over the same or similar time frame. Consequently, without such controls, it is impossible to eliminate the possibility that fish produced in the hatchery from native broodstock, and that spawned successfully in the wild, actually helped to maintain existing variation and structure in the studied populations.
Third, our estimates of mean Ne were low relative to the value of about 500 that Waples (1990) suggested was the minimum for long term viability in Oncorhynchus. Even estimates of Ne generated using the temporal methods (Table S1). By contrast, the Seymour and Capilano rivers have probably been the most perturbed, have the smallest habitats (particularly after the dams on these systems were completed) had the lowest and most variable Ne, and exhibited the greatest fluctuations in genetic differentiation across time periods.
While it is encouraging from our study that Ne values in each river have not experienced consistent declines since the onset of hatchery augmentation programs some further caveats suggest caution in interpretation of these results. First, our power to test for a pre- versus posthatchery effect was limited by small sample sizes within rivers (N = 6 maximum) and any pooled pre- and posthatchery effects (N = 11 maximum) were probably confounded somewhat by among river effects (e.g., variation in habitat changes through time). Second, our study was unable to employ suitable control populations, i.e., those surveyed over the same time frame and that have experienced no hatchery supplementation. Such samples do not exist in British Columbia and the rivers that we studied were those that experienced the most intense recreational fishing and were selected for hatchery supplementation for that reason. Other, less targeted and less accessible systems that could have served as controls were not subject to scale sample or broodstock collection over the same or similar time frame. Consequently, without such controls, it is impossible to eliminate the possibility that fish produced in the hatchery from native broodstock, and that spawned successfully in the wild, actually helped to maintain existing variation and structure in the studied populations.
Third, our estimates of mean Ne were low relative to the value of about 500 that Waples (1990) suggested was the minimum for long term viability in Oncorhynchus. Even estimates of Ne generated using the temporal methods (Table S1). By contrast, the Seymour and Capilano rivers have probably been the most perturbed, have the smallest habitats (particularly after the dams on these systems were completed) had the lowest and most variable Ne, and exhibited the greatest fluctuations in genetic differentiation across time periods.
While it is encouraging from our study that Ne values in each river have not experienced consistent declines since the onset of hatchery augmentation programs some further caveats suggest caution in interpretation of these results. First, our power to test for a pre- versus posthatchery effect was limited by small sample sizes within rivers (N = 6 maximum) and any pooled pre- and posthatchery effects (N = 11 maximum) were probably confounded somewhat by among river effects (e.g., variation in habitat changes through time). Second, our study was unable to employ suitable control populations, i.e., those surveyed over the same time frame and that have experienced no hatchery supplementation. Such samples do not exist in British Columbia and the rivers that we studied were those that experienced the most intense recreational fishing and were selected for hatchery supplementation for that reason. Other, less targeted and less accessible systems that could have served as controls were not subject to scale sample or broodstock collection over the same or similar time frame. Consequently, without such controls, it is impossible to eliminate the possibility that fish produced in the hatchery from native broodstock, and that spawned successfully in the wild, actually helped to maintain existing variation and structure in the studied populations.
Third, our estimates of mean Ne were low relative to the value of about 500 that Waples (1990) suggested was the minimum for long term viability in Oncorhynchus. Even estimates of Ne generated using the temporal methods (Table S1) produced some values that exceeded 500 only for the Chilliwack and Chehalis rivers, which supports the idea that even relatively pristine populations of steelhead trout would appear to require cautious management from a perspective provided by Ne (cf. Heath et al. 2002). While some wild populations of anadromous salmon and trout may have persisted at low numbers for long periods of time, many populations in southern BC have been adversely impacted by human activities in recent history (Slaney et al. 1996; McCubbing and Ward 2008). In these instances, persistently depressed population sizes may constrain their evolutionary potential, increase their susceptibility to stochastic changes, and compromise their long-term ability to adapt to environmental change.
In conclusion, our analysis of historical samples up to 58 years old demonstrated that different rivers harbor genetically distinct steelhead trout populations, confirming the importance of implementing appropriate supportive breeding strategies to maintain their inter- as well as intra-population genetic integrity. Our findings reinforce and expand the conclusions of Heggenes et al. (2006) and Araki et al. (2007b) that when appropriate management strategies are employed, hatchery supplementation using native broodstock can operate without necessarily negatively impacting the neutral genetic diversity and structure of steelhead trout populations over a time frame of at least several decades. Consequently, it is reassuring to some extent that hatchery supplementation can occur to help increase harvest levels with little detectable influence on the genetic characteristics that we assayed. In this sense, the steelhead hatchery program in many parts of BC may be considered successful; i.e., harvest opportunities have increased with no detectable effect on the wild component of the populations. By contrast, ecological effects of hatchery programs may significantly reduce wild population productivity and abundance even when no genetic risks are apparent (reviewed by Kostow 2009 and Araki and Schmid 2010). Consequently, the answer to the question of whether hatchery supplementation using native broodstock can provide benefits to wild steelhead trout production remains equivocal and context specific. While there is evidence that native broodstock programs can be compatible with wild steelhead trout conservation in rivers where wild fish are relatively abundant and productive, and that offer areas of no exploitation upstream of hatcheries (Nelson et al. 2005; Heggenes et al. 2006), there is little evidence to support their value as a tool to rebuild wild populations through the spawning of hatchery returns in rivers. In fact, some evidence suggests that they may even cause harm (reviewed by Ward 2006; see also Araki and Schmid 2010). Our study represents the beginnings of a more inclusive and comprehensive assessment of the performance of hatchery programs for steelhead trout, but it is a modest one. A comprehensive biological assessment of hatchery programs requires integration of genetic, demographic, and ecologic components relevant to the biology of steelhead trout set against clear conservation objectives (cf. Araki and Schmid 2010).
Acknowledgments
We appreciate the assistance of Christy Juteau and Craig Wightman, and staff at various hatcheries for assisting with sample collection and organization. Hatchery staff are also thanked for their help determining brood stock numbers, and smolt release numbers. We thank Ted Down (BC Ministry of Environment) for helpful comments in the preparation of the manuscript. The project was funded by a Living Rivers Trust Fund Foundation (LRTF) grant and Natural Science and Engineering Council of Canada (NSERC) Discovery and Equipment grants awarded to EBT. We thank Ken Ashley and Al Martin for promoting the interaction between LRTF and the University of British Columbia.
Data archiving
Data for this study are available in Dryad: doi:10.5061/dryad.bq583
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Average measures of relatedness,effective population size, genetic variation and structure in steelhead trout (Oncorhynchus mykiss) within hatchery supplemented rivers.
Figure S2. Admixture coefficients (q) for 902 steelhead trout (Oncorhynchus mykiss) from five rivers assayed at nine microsatellite DNA loci for each of up to six time periods each.
Figure S3. Mean (+SD, N = 5 replicates) proportions (q) of each of seven genetic groups (1–7) identified by STRUCTURE (Pritchard et al. 2000, Genetics, 155:945–959) within temporal samples of steelhead trout (Oncorhynchus mykiss) based on variation across ninemicrosatellite DNA loci.
Table S1. Estimates of effective population size or effective number of breeders for steelhead trout based on microsatellite DNA variation and employing three analytical methods.
Table S2. Results of STRUCTURE (version 2.3) analyses on microsatellite DNA variation in steelhead trout.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Literature cited
- Adkison MD. Population differentiation in Pacific salmon: local adaptation, genetic drift, or the environment? Canadian Journal of Fisheries and Aquatic Sciences. 1995;52:2762–2777. [Google Scholar]
- Ahrens R. The Status of Steelhead Trout (Oncorhynchus mykiss) in British Columbia. Vancouver, BC: Draft report prepared for the Pacific Fisheries Resource Conservation Council; 2004. [Google Scholar]
- Allendorf FW, England PR, Luikart G, Ritchie PA, Ryman N. Genetic effects of harvest on wild animal populations. Trends in Ecology and Evolution. 2008;23:327–337. doi: 10.1016/j.tree.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Araki H, Schmid C. Is hatchery stocking a help or harm? Evidence, limitations and future directions in ecological and genetic surveys. Aquaculture. 2010;308:S2–S11. [Google Scholar]
- Araki H, Cooper B, Blouin MS. Genetic effects of captive breeding cause a rapid, cumulative fitness decline in the wild. Science. 2007a;318:100–103. doi: 10.1126/science.1145621. [DOI] [PubMed] [Google Scholar]
- Araki H, Waples RS, Ardren WR, Cooper B, Blouin MS. Effective population size of steelhead trout: influence of variance in reproductive success, hatchery programs, and genetic compensation between life-history forms. Molecular Ecology. 2007b;16:953–966. doi: 10.1111/j.1365-294X.2006.03206.x. [DOI] [PubMed] [Google Scholar]
- Araki H, Berejikian B, Ford M, Blouin MS. Fitness of hatchery-reared salmonids in the wild. Evolutionary Applications. 2008;1:342–355. doi: 10.1111/j.1752-4571.2008.00026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H, Cooper B, Blouin MS. Carry-over effect of captive breeding reduces reproductive fitness of wild-born descendants in the wild. Biology Letters. 2009;5:621–624. doi: 10.1098/rsbl.2009.0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspinwall N. Genetic analysis of North American populations of pink salmon, Oncorhynchus gorbuscha, possible evidence for neutral mutation-random drift hypothesis. Evolution. 1974;28:295–305. doi: 10.1111/j.1558-5646.1974.tb00749.x. [DOI] [PubMed] [Google Scholar]
- Beacham TD, Pollard S, Le KD. Population structure and stock identification of steelhead trout in southern British Columbia, Washington, and the Columbia River based on microsatellite DNA variation. Transactions of the American Fisheries Society. 1999;128:1068–1084. [Google Scholar]
- Beacham TD, Pollard S, Le KD. Microsatellite DNA population structure and stock identification of steelhead trout (Oncorhynchus mykiss) in the Nass and Skeena Rivers in Northern British Columbia. Marine Biotechnology. 2000;2:587–600. doi: 10.1007/s101260000045. [DOI] [PubMed] [Google Scholar]
- Beacham TD, Le KD, Candy JR. Population structure and stock identification of steelhead trout (Oncorhynchus mykiss) in British Columbia and the Columbia River based on microsatellite variation. Environmental Biology of Fishes. 2004;69:95–109. [Google Scholar]
- Belkhir K, Borsa P, Chikhi N, Raufaste N, Bonhomme F. GENETIX 4.03, logiciel sous indows TM pour la génétique des populations. Montpellier, France: Université de Montpellier II; 2001. In Laboratoire Génome, Populations, Interactions, CNRS UMR 5000. [Google Scholar]
- Belkhir K, Castric V, Bonhomme F. IDENTIX, a software to test for relatedness in a population using permutation methods. Molecular Ecology Notes. 2002;2:611–614. [Google Scholar]
- Brannon EL, Amend DF, Cronin MA, Lannan JE, LaPatra S, McNeil WJ, Noble RE, et al. The controversy about salmon hatcheries. Fisheries. 2004;29:12–31. [Google Scholar]
- Caroffino DC, Miller LM, Kapuscinski AR, Ostazeski JJ. Stocking success of local-origin fry and impact of hatchery ancestry: monitoring a new steelhead (Oncorhynchus mykiss) stocking program in a Minnesota tributary to Lake Superior. Canadian Journal of Fisheries and Aquatic Sciences. 2008;65:309–318. [Google Scholar]
- Caverly A. 1978. Life history of winter steelhead (Salmo gairdneri) in the Vedder-Chilliwack River – based on scale samples collected between 1948 and 1975. Unpublished report available from BC Ministry of Environment Fish and Wildlife Branch, Region 2, Surrey, BC.
- Chilcote MW, Crawford BA, Leider SA. A genetic cornparison of sympatric populations of summer and winter steelheads. Transactions of the American Fisheries Society. 1980;109:203–206. [Google Scholar]
- Christie MR, Marine ML, Blouin MS. Who are the missing parents? Grandparentage analysis identifies multiple sources of gene flow into a wild population. Molecular Ecology. 2011;20:1263–1276. doi: 10.1111/j.1365-294X.2010.04994.x. [DOI] [PubMed] [Google Scholar]
- DeGisi JS. Precision and bias of the British Columbia steelhead harvest analysis. Skeena Region, Smithers, BC: Skeena Fisheries Report SK122, British Columbia Ministry of Environment, Fisheries Branch; 1999. [Google Scholar]
- van Dishoeck P, Slaney T, Korman J. 1998. Wild steelhead conservation planning in the Lower Mainland. Chilliwack/Vedder River. Unpublished report to Ministry of Environment, Lands, and Parks, Fish and Wildlife Management, Surrey, BC.
- El Mousadik A, Petit RJ. High level of genetic differentiation for allelic richness among populations of the argan tree (Argania spinosa (L.) Skeels) endemic to Morocco. Theoretical and Applied Genetics. 1996;92:832–839. doi: 10.1007/BF00221895. [DOI] [PubMed] [Google Scholar]
- Eldridge WH, Killebrew K. Genetic diversity over multiple generation of supplementation: an example from Chinook salmon using microsatellite and demographic data. Conservation Genetics. 2008;9:13–28. [Google Scholar]
- Eldridge WH, Naish KA. Long-term effects of translocation and release numbers on fine-scale population structure among coho salmon (Oncorhynchus kisutch. Molecular Ecology. 2007;16:2407–2421. doi: 10.1111/j.1365-294X.2007.03271.x. [DOI] [PubMed] [Google Scholar]
- Englbrecht CC, Schliewen U, Tautz D. The impact of stocking on the genetic integrity of Arctic char (Salvelinus) populations from the Alpine region. Molecular Ecology. 2002;11:1017–1027. doi: 10.1046/j.1365-294x.2002.01498.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse P, Quattro J. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Ford MJ. Conservation units and preserving diversity. In: Hendry AP, Stearns SC, editors. Evolution Illuminated: Salmon and Their Relatives. Oxford: Oxford University Press; 2004. pp. 338–357. [Google Scholar]
- Frankham R, Ballou JD, Briscoe DA. Introduction to Conservation Genetics. 2nd edn. Cambridge: Cambridge University Press; 2010. [Google Scholar]
- Fraser DJ, Weir LK, Bernatchez L, Hansen MM, Taylor EB. Extent and scale of local adaptation in salmonid fishes: review and meta-analysis. Heredity. 2011;106:404–420. doi: 10.1038/hdy.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Marin JL, Sanz N, Pla C. Erosion of the native genetic resources of brown trout in Spain. Ecology of Freshwater Fish. 1999;8:151–158. [Google Scholar]
- Goudet J. 2001. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available from http://www2.unil.ch/popgen/softwares/fstat.htm (accessed on 1 July 2011)
- Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica. 2001;4:9. http://palaeo-electronica.org/2001_1/past/issue1_01.htm. [Google Scholar]
- Hansen MM. Estimating the long-term effects of stocking domesticated trout into wild brown trout (Salmo trutta) populations: an approach using microsatellite DNA analysis of historical and contemporary samples. Molecular Ecology. 2002;11:1003–1015. doi: 10.1046/j.1365-294x.2002.01495.x. [DOI] [PubMed] [Google Scholar]
- Hansen MM, Nielsen EE, Ruzzante DE, Bouza C, Mensberg KLD. Genetic monitoring of supportive breeding in brown trout (Salmo trutta L.), using microsatellite DNA markers. Canadian Journal of Fisheries and Aquatic Sciences. 2000a;57:2130–2139. [Google Scholar]
- Hansen MM, Ruzzante DE, Nielsen EE, Mensberg KLD. Microsatellite and mitochondrial DNA polymorphism reveals life-history dependent interbreeding between hatchery and wild brown trout (Salmo trutta L.) Molecular Ecology. 2000b;9:583–594. doi: 10.1046/j.1365-294x.2000.00898.x. [DOI] [PubMed] [Google Scholar]
- Hansen MM, Ruzzante DE, Nielsen EE, Bekkevold D, Mensberg KLD. Long-term effective population sizes, temporal stability of genetic composition and potential forlocal adaptation in adramous brown trout ((Salmo trutta) populations. Molecular Ecology. 2002;11:2523–2535. doi: 10.1046/j.1365-294x.2002.01634.x. [DOI] [PubMed] [Google Scholar]
- Hansen MM, Fraser DJ, Meier K, Mensberg KLD. Sixty years of anthropogenic pressure: a spatio-temporal genetic analysis of brown trout populations subject to stocking and population declines. Molecular Ecology. 2009;18:2549–2562. doi: 10.1111/j.1365-294X.2009.04198.x. [DOI] [PubMed] [Google Scholar]
- Hard JJ. Evolution of Chinook salmon life history under size-selective harvest. In: Hendry AP, Stearns SC, editors. Evolution Illuminated: Salmon and Their Relatives. Oxford: Oxford University Press; 2004. pp. 315–337. [Google Scholar]
- Hauser L, R Seamons T, Dauer M, Naish KA, Quinn TP. An empirical verification of population assignment methods by marking and parentage data: hatchery and wild steelhead (Oncorhynchus mykiss) in Forks Creek, Washington, USA. Molecular Ecology. 2006;15:3157–3173. doi: 10.1111/j.1365-294X.2006.03017.x. [DOI] [PubMed] [Google Scholar]
- Heath DD, Pollardà S, Herbinger C. Genetic structure and relationships among steelhead trout (Oncorhynchus mykiss) populations in British Columbia. Heredity. 2001;86:618–627. doi: 10.1046/j.1365-2540.2001.00867.x. [DOI] [PubMed] [Google Scholar]
- Heath DD, Busch C, Kelly J, Atagi DY. Temporal change in genetic structure and effective population size in steelhead trout (Oncorhynchus mykiss. Molecular Ecology. 2002;11:197–214. doi: 10.1046/j.1365-294x.2002.01434.x. [DOI] [PubMed] [Google Scholar]
- Heggenes J, Beere M, Tamkee P, Taylor EB. Temporal comparisons of genetic diversity in steelhead (Oncorhynchus mykiss) before and after conservation hatchery operation in a coastal, boreal river. Transactions of the American Fisheries Society. 2006;135:251–267. [Google Scholar]
- Hendry AP, Stearns SC. Evolution Illuminated: Salmon and Their Relatives. Oxford: Oxford University Press; 2004. [Google Scholar]
- Hendry MA, Wenburg JK, Myers KW, Hendry AP. Genetic and phenotypic variation through the migratory season provides evidence for multiple populations of wild steelhead in the Dean River, British Columbia. Transactions of the American Fisheries Society. 2002;31:418–434. [Google Scholar]
- Hendry AP, Castric V, Kinnison MT, Quinn TP. The evolution of philopatry and dispersal: homing versus straying in salmonids. In: Hendry AP, Stearns SC, editors. Evolution Illuminated: Salmon and Their Relatives. Oxford: Oxford University Press; 2004. pp. 52–91. [Google Scholar]
- Hill WG. Estimation of effective population size from data on linkage disequilibrium. Genetical Research. 1981;38:209–216. [Google Scholar]
- Hooten RS, Ward BR, Lewynsky VA, Lirette MG, Facchin AR. 1987. Age and growth of steelhead in Vancouver Island populations. Fisheries Technical Circular No 77. BC Ministry of Environment and Parks, Victoria.
- Johnston NT, Parkinson EA, Tautz AF, Ward BR. 2002. A conceptual framework for the management of steelhead, Oncorhyncus mykiss. BC Ministry of Environment Fisheries Project Report No. RD101.
- Jonsson B, Jonsson N, Hansen LP. Atantic salmon straying from the River Imsa. Journal of Fish Biology. 2003;62:641–657. [Google Scholar]
- Jorde PE, Ryman N. Temporal allele frequency change and estimate of effective population size. Genetics. 1995;139:1077–1090. doi: 10.1093/genetics/139.2.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostow KE. Factors that contribute to the ecological risks of salmon and steelhead hatchery programs and some mitigating strategies. Reviews of Fish Biology and Fisheries. 2009;19:9–31. [Google Scholar]
- Kostow KE, Marshal AR, Phelps SR. Naturally spawning hatchery steelhead contribute to smolt production but experience low reproductive success. Transactions of the American Fisheries Society. 2003;132:780–790. [Google Scholar]
- Labelle M. 2007. A preliminary assessment of the survival rates of coho salmon and steelhead trout from the Capilano River, 1998–2007. Report prepared for the Greater Vancouver Regional District.
- Leberg PL. Estimating allelic richness: effects of sample size and bottlenecks. Molecular Ecology. 2002;11:2445–2449. doi: 10.1046/j.1365-294x.2002.01612.x. [DOI] [PubMed] [Google Scholar]
- Leider SA, Chilcote MW, Loch JJ. Spawning characteristics of sympatric populations of steelhead trout (Salmo gairdneri): evidence for partial reproductive isolation. Canadian Journal of Fisheries and Aquatic Sciences. 1984;41:1454–1462. [Google Scholar]
- Lenormond T. Gene flow and the limits to natural selection. Trends in Ecology and Evolution. 2002;17:183–189. [Google Scholar]
- Li H-W. Maintenance of genetic variability under the joint effect of mutation, selection, and random drift. Genetics. 1978;90:349–382. doi: 10.1093/genetics/90.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill AF. Greater Georgia Basin Steelhead Recovery Action Plan. North Vancouver, BC: AF Lill and Associates Ltd; 2002. p. 107. Prepared for Pacific Salmon Foundation. [Google Scholar]
- Ludwig B. British Columbia's trout hatchery program and the stocking policies that guide it. American Fisheries Society Symposium. 1995;15:139–143. [Google Scholar]
- Maher FP, Larkin PA. Life history of the steelhead trout of the Chilliwack River, British Columbia. Transactions of the American Fisheries Society. 1955;84:27–38. [Google Scholar]
- Marie LD, Bernatchez L, Garant D. Loss of genetic integrity correlates with stocking intensity in brook charr (Salvelinus fontinalis. Molecular Ecology. 2010;19:2025–2037. doi: 10.1111/j.1365-294X.2010.04628.x. [DOI] [PubMed] [Google Scholar]
- McCubbing DJF, Ward BR. 2008. Adult steelhead trout and salmonid smolt migration at the Keogh River, B.C., during winter and spring 2008. Report prepared for the Habitat Conservation Trust Fund.
- McLean JE, Bentzen P, Quinn TP. Differential reprodtive success of sympatric, naturally spawning hatchery and wild steelhead, Oncorhynchus mykiss. Environmental Biology of Fishes. 2004;69:359–369. [Google Scholar]
- Merilä J, Crnokrak P. Comparison of genetic differentiation at marker loci and quantitative traits. Journal of Evolutionary Biology. 2001;4:892–903. [Google Scholar]
- Miller LM, Close T, Kapuscinski R. Lower fitness of hatchery and hybrid rainbow trout compared to naturalized populations in Lake Superior tributaries. Molecular Ecology. 2004;13:3379–3388. doi: 10.1111/j.1365-294X.2004.02347.x. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1988. [Google Scholar]
- Nelson TC, Rosenau ML, Johnston NT. Behavior and survival of wild and hatchery-origin winter steelhead spawners caught and released in a recreational fishery. North American Journal of Fisheries Management. 2005;25:931–943. [Google Scholar]
- Nielsen JL, Fountain MC. Microsatellite diversity in sympatric reproductive ecotypes of Pacific steelhead (Oncorhynchus mykiss) from the Middle Fork Eel River, California. Ecology of Freshwater Fish. 1999;8:159–168. [Google Scholar]
- Nielsen EE, Hansen MM, Loeschcke V. Analysis of microsatellite DNA from old scale samples of Atlantic salmon Salmo salar: a comparison of genetic composition over 60 years. Molecular Ecology. 1997;6:487–492. [Google Scholar]
- Nielsen EE, Hansen MM, Loeschcke V. Genetic variation in time and space: microsatellite analysis of extinct and extant populations of Atlantic salmon. Evolution. 1999;53:261–268. doi: 10.1111/j.1558-5646.1999.tb05351.x. [DOI] [PubMed] [Google Scholar]
- van Oosterhout C, Weetman D, Hutchinson WF. Estimation and adjustment of microsatellite null alleles in nonequilibrium populations. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- Ovenden J, Peel D, Street R, Courtney A, Hoyle S, Peel SL, Podlich H. The genetic effective and adult census size of an Australian population of tiger prawns (Penaeus esculentus) Molecular Ecology. 2007;16:127–138. doi: 10.1111/j.1365-294X.2006.03132.x. [DOI] [PubMed] [Google Scholar]
- Palstra FP, Ruzzante DE. Genetic estimates of contemporary effective population size: what can they tell us about the importance of genetic stochasticity for wild population persistence? Molecular Ecology. 2008;17:3428–3447. doi: 10.1111/j.1365-294x.2008.03842.x. [DOI] [PubMed] [Google Scholar]
- Palstra FP, O'Connell MF, Ruzzante DR. Age structure, changing demography and effective population size in Atlantic salmon (Salmo salar. Genetics. 2009;182:1233–1249. doi: 10.1534/genetics.109.101972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel D, Ovenden JR, Peel SL. 2004. NeEstimator: software for estimating effective population size, Version 1.3. Queensland Government, Department of Primary Industries and Fisheries.
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteaux C, Beaudou D, Berrebi P. Temporal variations of genetic introgression in stocked brown trout populations. Journal of Fish Biology. 1998;53:701–713. [Google Scholar]
- Queller DC, Goodnight KF. Estimating relatedness using genetic markers. Evolution. 1989;43:258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x. [DOI] [PubMed] [Google Scholar]
- Quinn TP. A review of homing and straying in wild and hatchery-produced salmon. Fisheries Research. 1993;18:29–44. [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 3.3): population genetics software for exact tests and ecumenicism. Updated from Raymond, M., and F. Rousset. 1995. Journal of Heredity. 2001;86:248–249. [Google Scholar]
- Reisenbichler RR. Uncertainty and research needs for supplementing wild populations of anadromous Pacific salmon. In: Nickum MJ, Mazik PM, Nickum JG, MacKinlay DD, Mudrak V, editors. Propagated Fish in Resource Management. Bethesda, MD: American Fisheries Society Symposium 44; 2004. pp. 263–276. [Google Scholar]
- Reisenbichler RR, Phelps SR. Genetic variation in steelhead Salmo gairdneri from the north coast of Washington. Canadian Journal of Fisheries and Aquatic Sciences. 1989;46:66–73. [Google Scholar]
- Reisenbichler RR, Rubin SP. Genetic changes from artificial propagation of Pacific salmon affect the productivity and viability of supplemented populations. ICES Journal of Marine Science. 1999;56:459–466. [Google Scholar]
- Rice W. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Ruzzante DE, Hansen MM, Meldrup D, Ebert KM. Stocking impact and migration pattern in an anadromous brown trout (Salmo trutta) complex: where have all the stocked spawning sea trout gone? Molecular Ecology. 2004;13:1433–1445. doi: 10.1111/j.1365-294X.2004.02162.x. [DOI] [PubMed] [Google Scholar]
- Ryman N, Laikre L. Effects of supportive breeding on the genetically effective population size. Conservation Biology. 1991;5:325–329. [Google Scholar]
- Ryman N, Jorde PE, Laikre L. Supportive breeding and variance effective population size. Conservation Biology. 1995;9:1619–1628. [Google Scholar]
- Schwartz MK, Luikart G, Waples RS. Genetic monitoring as a promising tool for conservation and management. Trends in Ecology and Evolution. 2007;22:25–33. doi: 10.1016/j.tree.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Slaney TL, Hyatt KD, Northcote TG, Fielden RJ. Status of anadromous salmon and trout in British Columbia and Yukon. Fisheries. 1996;21:20–32. [Google Scholar]
- Slatkin M. Gene flow in natural populations. Annual Review of Ecology and Systematics. 1985;16:393–430. [Google Scholar]
- Small MP, Currens K, Johnson TH, Frye AE, Von Bargen JF. Impacts of supplementation: genetic diversity in supplemented and unsupplemented populations of summer chum salmon (Oncorhynchus keta) in Puget Sound (Washington, USA) Canadian Journal of Fisheries and Aquatic Sciences. 2009;66:1216–1229. [Google Scholar]
- Smith BD, Ward BR. Trends in wild adult steelhead (Oncorhynchus mykiss) abundance for coastal regions of British Columbia support the variable marine survival hypothesis. Canadian Journal of Fisheries and Aquatic Sciences. 2000;57:271–284. [Google Scholar]
- Stickney RR. Aquaculture in the United States: A Historical Survey. New York: John Wiley and Sons; 1996. [Google Scholar]
- Taberlet PS, Griffin B, Goossens S, Questiau V, Manceau N, Escaravage N, Waits LP, et al. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Research. 1996;24:3189–3194. doi: 10.1093/nar/24.16.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlet PS, Waits LP, Luikart G. Noninvasive genetic sampling: look before you leap. Trends in Evolution and Ecology. 1999;14:323–327. doi: 10.1016/s0169-5347(99)01637-7. [DOI] [PubMed] [Google Scholar]
- Tamkee P, Parkinson E, Taylor EB. The influence of Wisconsinan glaciation and contemporary stream hydrology on microsatellite DNA variation in rainbow trout (Oncorhynchus mykiss. Canadian Journal of Fisheries and Aquatic Sciences. 2010;67:919–935. [Google Scholar]
- Taylor EB, Tamkee P, Sterling G, Hughson W. Microsatellite DNA analysis of rainbow trout (Oncorhynchus mykiss) from western Alberta, Canada: native status and evolutionary distinctiveness of “Athabasca” rainbow trout. Conservation Genetics. 2007;8:1–15. [Google Scholar]
- Tessier N, Bernatchez L. Stability of population structure and genetic diversity across generations assessed by microsatellites among sympatric populations of landlocked Atlantic salmon (Salmo salar L.) Molecular Ecology. 1999;8:169–179. [Google Scholar]
- Tessier N, Bernatchez L, Wright JM. Population structure and impact of supportive breeding inferred from mitochondrial and microsatellite DNA analyses in land-locked Atlantic salmon Salmo salar L. Molecular Ecology. 1997;6:735–750. [Google Scholar]
- Utter F. Genetic problems of hatchery-reared progeny released into the wild and how to deal with them. Bulletin of Marine Science. 1998;62:623–640. [Google Scholar]
- Van Doornik DM, Waples RS, Baird MC, Moran P, Berntson EA. Genetic monitoring reveals genetic Stability within and among threatened Chinook salmon populations in the Salmon River, Idaho. North American Journal of Fisheries Management. 2011;31:96–105. [Google Scholar]
- Wang J. A new method for estimating effective population sizes from a single sample of multilocus genotypes. Molecular Ecology. 2009;18:2148–2164. doi: 10.1111/j.1365-294X.2009.04175.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Ryman N. Genetic effects of multiple generations of supportive breeding. Conservation Biology. 2001;15:1619–1631. [Google Scholar]
- Waples RS. A generalized approach for estimating effective populations size from temporal changes in allele frequency. Genetics. 1989;121:379–391. doi: 10.1093/genetics/121.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples RS. Conservation genetics of Pacific salmon. II. Effective population size and the rate of loss of genetic variability. Journal of Heredity. 1990;81:267–276. [Google Scholar]
- Waples RS. Salmonid insights into effective population size. In: Hendry AP, Stearns SC, editors. Evolution Illuminated: Salmon and Their Relatives. Oxford: Oxford University Press; 2004. pp. 295–314. [Google Scholar]
- Waples RS. Genetic estimates of contemporary effective population size: to what time periods do the estimates apply? Molecular Ecology. 2005;14:3335–3352. doi: 10.1111/j.1365-294X.2005.02673.x. [DOI] [PubMed] [Google Scholar]
- Waples RS, Do C. ldne: a program for estimating effective population size from data on linkage disequilibrium. Molecular Ecology Resources. 2008;8:753–756. doi: 10.1111/j.1755-0998.2007.02061.x. [DOI] [PubMed] [Google Scholar]
- Waples RS, Masuda M, Pella J. SalmonNb: a program for computing cohort-specific effective population sizes (Nb) in Pacific salmon and other semelparous species using the temporal method. Molecular Ecology Notes. 2007;7:21–24. [Google Scholar]
- Ward BR. 2006. The case for wild steelhead recovery without artificial fish culture intervention. Report prepared for the BC Ministry of Environment.
- Ward BR, Slaney PA. Egg-to-smolt survival and fry-to-smolt density dependence of Keogh River steelhead. In: Gibson RJ, Cutting RE, editors. Production of Juvenile Atlantic Salmon, Salmo Salar, in Natural Waters. 1993. pp. 209–217. Can. Spec. Publ. Fish. Aquat. Sci. 118. [Google Scholar]
- Wattier R, Engel CR, Saumitou-Laprade P, Valero M. Short allele dominance as a source of heterozygote deficiency at microsatellite loci: experimental evidence at the dinucleotide locus GvICT in Gracilaria gracilis (Rhodophyta) Molecular Ecology. 1998;7:1569–1573. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Williamson KS, May B. Homogenization of fall-run chinook salmon gene pools in the Central Valley of California, USA. North American Journal of Fisheries Management. 2005;25:993–1009. [Google Scholar]
- Withler IL. Variability in life history characteristics of steelhead trout (Salmo gairdneri) along the Pacific coast of North America. Journal of the Fisheries Research Board of Canada. 1966;23:365–393. [Google Scholar]
- Young KA. Toward evolutionary management: lessons from salmonids. In: Hendry AP, Stearns SC, editors. Evolution Illuminated: Salmon and Their Relatives. Oxford: Oxford University Press; 2003. pp. 358–376. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


