Abstract
Evolution occurs rapidly and is an ongoing process in our environments. Evolutionary principles need to be built into conservation efforts, particularly given the stressful conditions organisms are increasingly likely to experience because of climate change and ongoing habitat fragmentation. The concept of evolutionary resilience is a way of emphasizing evolutionary processes in conservation and landscape planning. From an evolutionary perspective, landscapes need to allow in situ selection and capture high levels of genetic variation essential for responding to the direct and indirect effects of climate change. We summarize ideas that need to be considered in planning for evolutionary resilience and suggest how they might be incorporated into policy and management to ensure that resilience is maintained in the face of environmental degradation.
Keywords: adaptive potential, biodiversity, climate change, conservation, evolution, evolutionary resilience, genetic diversity
Introduction
Natural populations are responding to global climate change both through altered timing of life history traits, geographical shifts in species ranges and potentially altered ecosystem interactions (Root et al. 2003; Parmesan 2006). Climate change is of particular concern for ecological communities restricted to montane areas and local moist pockets, because populations of species in these communities cannot readily move when conditions become warmer and drier. This leads to high predicted rates of extinction in animals and plants from some local regions (Thomas et al. 2004). However, if populations change genetically and can evolve and adapt to these predicted environmental changes, then species extinction risks because of climate change might be substantially reduced.
Ecologists now generally recognize that rapid rates of evolution are possible within species, with consequences for species abundance and distribution. There is already evidence for rapid evolution in response to climate change in several short-lived species (Reusch and Wood 2007), suggesting that many organisms have the capacity to respond to climate change within a time frame of tens of years (Bradshaw and Holzapfel 2008). These responses depend on the presence of genetic variation in populations. In the absence of genetic variation, there is now strong evidence for an increased risk of extinction in wild populations (Spielman et al. 2004). If genetic diversity for adaptive evolution can be conserved, and restoration practices put in place that help promote in situ adaptive processes, the long-term implications go well beyond the persistence of species, with potential impacts on biodiversity and ecosystem function (Bailey et al. 2009) as well as resilience in response to climate extremes (Reusch et al. 2005).
However, managers currently tend to ignore evolutionary processes when devising ways to protect biodiversity under climate change and developing criteria for conservation (Mace and Purvis 2008; Crandall 2009; Gebremedhin et al. 2009). We argue that management plans for species and habitats should aim at developing resilient landscapes where the evolutionary potential of species and populations can be conserved. This can be achieved by explicit consideration of genetic diversity and the processes that support ongoing, in situ evolutionary processes in biodiversity management and planning.
We build on the ecological definition of resilience (e.g., Gunderson 2000; Thrush et al. 2009) by explicitly including the role played by genetic diversity and evolutionary processes, not only on the persistence of populations and species but also in influencing community ecology and ecosystem function. In this sense, evolutionary resilience refers both to the ability of populations to persist in their current state (analogous to resistance in the ecological literature) and to undergo evolutionary adaptation in response to changing environmental conditions (analogous to the transition between multiple stability domains in response to perturbation) (Gunderson 2000; Thrush et al. 2009). This definition of evolutionary resilience explicitly recognizes that ongoing evolutionary change is the norm in nature and that it is one of the dynamic processes that generate and maintain biodiversity patterns and processes. By using this definition, we place evolutionary considerations at the centre of biodiversity conservation and management (Hendry et al. 2010).
Measuring genetic diversity and evolutionary potential
Genetic diversity can be divided into two categories. Adaptive genetic diversity underpins the way an organism adapts to a new environment. In contrast, neutral genetic diversity involves parts of the genome that are not under natural selection, and this diversity reflects population dynamics and evolutionary forces such as genetic drift, mutation and migration. Conservation biologists have largely focussed on the latter, even though adaptive genetic diversity is needed for organisms to evolve and persist in changing environments.
While neutral diversity is commonly used to infer the potential of populations to evolve, this connection tends to be weak (e.g., Reed and Frankham 2001; McKay and Latta 2002). Instead, this potential should ideally be investigated by directly understanding variation in genes that are involved in an adaptive response (Hoffmann and Willi 2008). If these genes are unknown, adaptive variation can be assessed by directly measuring the extent to which traits under selection are genetically determined and variable (heritability and evolvability). This requires quantitative traits to be measured across multiple generations of an organism (typically parents and their offspring) ideally under similar environmental conditions, such as through the use of common garden experiments. Although time-consuming and not always possible for the focal species, common garden experiments have been used to assess the levels of adaptive genetic diversity in a range of taxa, including plants (Dorman et al. 2009; Ramirez-Valiente et al. 2009), insects (Crozier 2004; Klemme and Hanski 2009) and vertebrates (Johansson et al. 2007). Long-term studies of wild animal populations under field conditions can also be used to assess adaptive genetic variation and the extent of adaptation to environmental change (Grueber and Jamieson 2008; Kruuk et al. 2008; Charmantier et al. 2009; Ozgul et al. 2009) although this approach has limitations (Hadfield et al. 2010).
Because of the difficulty (real and perceived) of measuring adaptive genetic diversity, this form of diversity has not been considered a priority in conservation planning and management, with the exception of revegetation and captive breeding programs, which we address later. However, advances in genotyping techniques combined with more sophisticated statistical methods provide the means by which adaptive (and neutral) genetic diversity can be estimated more easily in a range of organisms in the absence of any prior information on either molecular or quantitative trait variation (Beaumont and Balding 2004; Storz 2005). In one of the first empirical tests of this approach, Bonin et al. (2007) characterized amplified fragment length polymorphisms to identify neutral and selected loci in six populations of the widespread common frog, Rana temporaria, and seven populations of the threatened and restricted plant Austrian dragonhead, Dracocephalum austriacum. This information was used to asses four different conservation strategies aimed at maximizing the genetic diversity (neutral and adaptive) for both species. In doing so, the authors developed a population adaptive index to account for the adaptive value of a particular population. In both species, the neutral and adaptive diversities within and among populations were not correlated, so conservation strategies based on one type of index would not select the same populations for protection.
In another study, Joost et al. (2007) combined molecular data with geographical information systems (GIS) and environmental variables to detect regions of the genome under natural selection using a spatial analysis method. They examined a species of pine weevil (Hylobius abietis) with a large geographical range, and 57 breeds of sheep originating from European and Middle Eastern countries. There were strong signals associating loci with environmental variables, such that these loci did not behave in a neutral manner. This approach holds promise in being able to take advantage of genome-wide scans of molecular markers across large geographic scales to detect genomic regions under selection and identifying likely instances of adaptive divergence across species’ ranges.
Finally, new information is continually emerging on candidate genes that underlie adaptive differences between populations and species. While not yet readily available as a broadly applicable tool, such information may make it possible in the future to measure adaptive genetic diversity directly rather than relying on multiple generation or indirect measurements (Burdon and Wilcox 2007; Hoffmann and Willi 2008; Gebremedhin et al. 2009).
Maintaining genetic variation and evolutionary potential: population size and beyond
An important component of the evolutionary resilience of individual populations is to maintain them at a large enough size to maintain genetic variation and allow ongoing evolution (Table 1– Aim A, B). A number of models have been applied to predict the likelihood of populations evolving under climate change. The simplest model involves the breeder's equation (R = h2S), which predicts the size of the selection response (R) given a certain selection pressure (S) and trait heritability (h2). This model explicitly links evolutionary responses to environmental change in the presence of adaptive genetic variation (h2).
Table 1.
Aims and approaches/outcomes for developing evolutionary resilience in populations and landscapes against climate change
| Aim | Scale where applied | Approach/Outcome | Comments/Limitations |
|---|---|---|---|
| A. Increase population size and genetic variation generally | Population | Increased census size | Needs to be related to effective size, which depends on life history and environmental variability |
| Increased effective size | Can be enhanced by population connectedness and breeding systems | ||
| Maintenance/increase in mtDNA/nuclear DNA variation (neutral) | Can be increased by including individuals from different populations (translocation) as well as through population size | ||
| B. Maintain adaptive potential in target genes and traits | Population | Identification and maintenance of genetic variation in candidate genes for adaptation | Focus of candidate gene work is on model species, but increasingly being applied to nonmodel systems |
| Identification/maintenance of variation in key quantitative traits (heritability/evolvability) | Potentially could be used to assess selection response potential but still fairly rarely measured | ||
| C. Identify species with little adaptive potential = low diversity in key ecological traits | Multiple populations of one species | Measure and identify traits involved in maintaining distribution with low heritability/evolvability or other constraints limiting directional evolution | Requires substantial genetic information on target species unless ecological correlates can be identified |
| D. Identify and protect evolutionary refugia | Multiple populations of multiple species within a landscape | Identify hotspots with high levels of mtDNA/nuclear DNA variation (neutral) | Depends on the accumulation of data across multiple species |
| Identify mtDNA/nuclear DNA uniqueness across regions | Depends on the accumulation of data across multiple species, could be applied at higher taxonomic levels to preserve evolutionary uniqueness | ||
| E. Increase connectedness and gene flow across environmental gradients | Multiple populations in a landscape | Movement of genes within landscape | Involves gene flow rather than just migration of individuals |
| Allow in situ selection across heterogeneous areas and climatic gradients | Needs large populations to ensure effective selection of high fitness genotypes | ||
| F. Increase adaptability to future environments by translocation | Population | Introduction of genetic material from provenances that match likely future climate at a site | Genotypes can be matched to likely future environments, but approach still rarely applied outside of deliberate introductions of species |
Models have also examined the combined effects of heritability and population size on the ability of populations to undergo adaptive evolutionary change and ‘keep up’ with climate change (Lynch and Lande 1993; Burger and Lynch 1995). These highlight the importance of population sizes on adaptive potential; large effective population sizes are required for maintaining genetic variation and evolutionary potential – typically a thousand rather than a hundred breeding individuals are required (Fig. 1). At small effective population sizes, demographic and environmental stochasticity will have a much larger impact on extinction probabilities than genetic variation (Willi and Hoffmann 2008). There is thus a direct association between the heritability of a trait and extinction risk as long as climatic effects on growth rate are not too severe (Fig. 1). Models have also recently been extended to include plasticity, relaxing conditions under which extinction is inevitable unless the costs of plasticity are high (Chevin et al. 2010).
Figure 1.
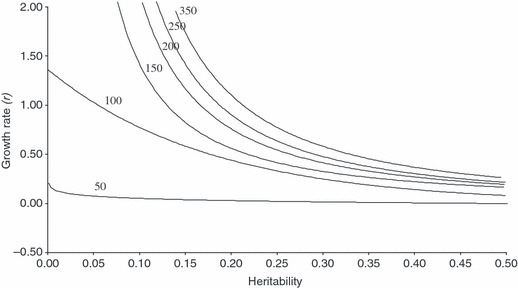
Contour plots presenting median number of generations to extinction as a function of the narrow-sense heritability of the trait under selection and mean intrinsic rate of natural increase (r) for a population of size 1000. The rate of environmental change (k) in this case was set at 0.1, and the width of the fitness function (VW) was 20. Populations of 1000 flies were predicted to persist well within the approximate ranges of r > 0.5 and h2 > 0.29. Modified from (Willi and Hoffmann 2008).
Given that rates of evolution increase with population size up to at least a few thousand individuals from a randomly mating population, how well do conservation and restoration efforts currently preserve this genetic diversity and evolutionary potential? The answer to this question would seem to be ‘not very well’. When dealing with highly threatened species, there is often little opportunity to increase population size although breeding programs can maximize effective size (Frankham et al. 2002). In addition, there is also adaptation to conditions of captivity resulting in reduced fitness of populations for eventual release into the wild (Frankham 2008).
Reserve systems typically should aim at conserving several hundred and preferably several thousand individuals if populations are isolated. This will help ensure that evolutionary potential is maintained for adapting to climate change. Otherwise, genetic diversity will be lost, a process that might take only a few years in threatened populations (Mitrovski et al. 2008). Adaptive genetic variation is expected to decrease alongside neutral genetic variation but follow a different trajectory depending on the patterns of selection acting on traits (Willi et al. 2006).
Genetic translocations: insuring against extinction and increasing adaptive potential
Most conservation efforts focus on threatened species where genetic diversity has been lost, usually as a consequence of small population size resulting from habitat loss and fragmentation. Such populations face high risk of extinction, and the option of assisted migration to alleviate these threats has been the focus of increasing debate (Hunter 2007; McLachlan et al. 2007; Grueber and Jamieson 2008; Hoegh-Guldberg et al. 2008; Menges 2008; Ricciardi and Simberloff 2009; Richardson et al. 2009; Swarts and Dixon 2009). Much of the recent debate concerns the movement of species beyond their current range (Hoegh-Guldberg et al. 2008; Ricciardi and Simberloff 2009; Richardson et al. 2009), following suggestions that intentional movement of species outside of their natural (current) range should be considered as an option for species at immediate risk of extinction (Hoegh-Guldberg et al. 2008). While the translocation of individuals to areas outside their current range has been used as a last resort for some highly threatened species, particularly in New Zealand (Jamieson et al. 2006; Miskelly et al. 2009), the issue remains highly controversial.
Moving individuals from one population to another (genetic translocation or assisted migration) within a species’ current range as a way of enabling gene flow and conserving or enhancing the adaptive potential of species has also been suggested as a conservation tool. There are two circumstances under which genetic translocation could be considered in the context of evolutionary resilience and climate change. The first concerns populations of threatened species that have suffered severe reductions in genetic diversity and where dispersal processes have been disrupted by habitat fragmentation. The genetic translocation of a few individuals per generation is likely to be enough to reduce the detrimental consequences of inbreeding, while minimizing the risks of outbreeding depression that may follow the introduction of genes from populations that have adapted to different environmental conditions than the recipient population (Lopez et al. 2009). The risk of outbreeding depression should be carefully weighed against the risk that ongoing loss of genetic diversity poses to the long-term persistence of populations (Edmands 2007; Lopez et al. 2009). Common garden or field-based experiments, where individuals from different populations are crossed, will assist in assessing the risk of genetic translocations and subsequent outbreeding depression. Combined with the estimates of genetic divergence using neutral genetic markers, this type of information can help inform decisions about the feasibility of genetic translocations (Holmes et al. 2008).
The second set of circumstances under which genetic translocations should be considered in the context of evolutionary resilience and climate change involves cases where there is strong local adaptation (ecotype differentiation). Moving individuals from warm-adapted populations to colder locations may increase the probability of adaptation, and thus, persistence and resilience of cold-adapted populations under a warming environment. Such approaches could be applied to species that display very wide altitudinal or latitudinal ranges and that have been shown to display genetically based clines in performance under different thermal or aridity gradients. Examples of clinal patterns along such gradients exist for terrestrial invertebrates (Hoffmann and Weeks 2007), vertebrates (Cheviron and Brumfield 2009), plants (Viveros-Viveros et al. 2009) and marine organisms (Berkelmans and van Oppen 2006). The ecological risks of such genetic translocations are likely to be minimal (Hoegh-Guldberg et al. 2008; Lopez et al. 2009). The long-term aim of genetic translocations is populations that harbour the adaptive genetic diversity to enable ongoing adaptation to the environmental changes caused by climate change and other threats. This helps to obviate the need for ongoing intervention and management, ensuring evolutionary resilient populations.
Geographic variation: moving beyond local provenances
The issue of genetic translocation to maximize population adaptability and evolutionary resilience under climate change is pertinent to ongoing landscape restoration (Menges 2008; Jones and Monaco 2009). Large-scale revegetation is widely carried out to restore degraded landscapes. Because local adaptation is recognized as being commonplace (Hereford 2009), the focus has been on local provenance when making decisions about which seed to source for restoration and reintroduction programmes (Callaham 1964; Keller et al. 2000; McKay et al. 2005; O'Brien et al. 2007). This is because it is widely assumed that local adaptation will always result in a fitness trade-off between local and nonlocal environments. However, such fitness trade-offs are not ubiquitous and when present, they are weak (Hereford 2009). Despite this evidence, an emphasis on local provenance prevails. A ‘local is best’ sourcing practice misses two important points, which may seriously impact restoration or reintroduction outcomes in the face of future climatic changes (Table 1– Aims A, B, F).
The first potential problem with ‘local is best’ recommendations is that there is a risk of encouraging the establishment of populations that do not harbour sufficient genetic variation and evolutionary potential [i.e., establish genetic ghettos (Schneider et al. 1999)]. In addition, strict adherence to ‘local is best’ protocols may encourage the selection of inbred or genetically depauperate seed sources (Broadhurst et al. 2008), when genetically healthier sources further afield may produce a more efficacious restoration result (C. Navarro, S. Cavers and A. Lowe, unpublished data). This may serve to perpetuate the number of small inbred populations across highly degraded landscapes that are unlikely to persist in the long term (Broadhurst et al. 2008).
The second issue is that particular environmental conditions driving local adaptation can change very rapidly. The environment is continually changing at different rates and scales (Wilkinson 2001) particularly when anthropogenic influences can rapidly change selection pressures (e.g., increased salinity, irrigation, and heavy metal deposition). In many regions of the world, conditions under which a 200-year-old tree established are likely to be quite different to those existing today. Source material from more distant (geographically and ecologically) populations may often harbour adaptations that more closely match the environment of the focal restoration site today.
Identifying highly adapted genotypes has long been a central issue in forestry and crop breeding but is just as applicable for natural populations. For instance, genotypes of river redgum, Eucalyptus camuldulensis, from arid environments show a strong fitness advantage under dry conditions over genotypes from humid environments because the arid genotypes allocate resources to roots under dry conditions (Gibson et al. 1995). These types of interactions between genotype performance and the environment (‘GE’ interactions) are extremely common and have been documented for hundreds of species.
In recommending new provenance or reintroduction practices, it is instructive to consider the strength of GE interactions across current and future environments (Fig. 2) and simulate natural gene flow dynamics that facilitate the redistribution of genetic variation within a species (Fig. 3). For long-lived species like most trees, where strong GE interactions exist, and in areas where large changes in climate are predicted, provenances likely to be the most suitable in the future should be selected (predictive provenancing, B in Fig. 2). Climatic matching can be used to locate such provenances at least in widespread species (Rice and Emery 2003). The extent to which sourcing other provenances can help mitigate against climate change will depend on the strength of GE interactions. Where GE interactions appear weak or have not been tested, it may be sensible to simulate leptokurtic gene flow dynamics, where most propagules disperse proximally, but with a significant proportion moving over longer distances (Fig. 3). Such a restoration practice would mix locally sourced material, taken from genetically healthy stock, with proximate and ecogeographically matched sources. In addition, a smaller proportion of material, depending on the natural gene flow dynamics of the focal species, should be comprised of material from much further afield to increase genetic variation and promote adaptation (Fig. 3). This practice is defined as composite provenancing by Broadhurst et al. (2008) and represents a cautionary strategy that might also be appropriate for species where GE interactions are strong but the predicted changes in climate are small or unknown (A in Fig. 2).
Figure 2.
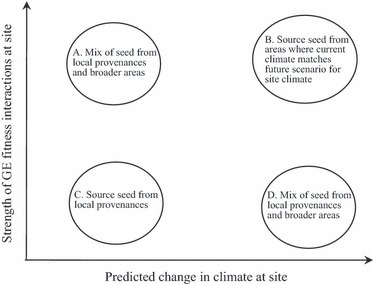
Seed sources future scenarios depend on whether climatic conditions at a site are expected to change and/or whether there are strong genotype–environment interactions for fitness.
Figure 3.
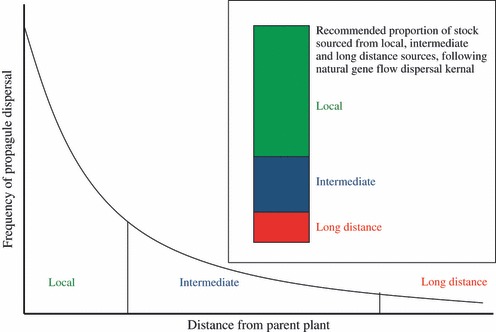
Recommendations for provenancing based on dispersal distance from parental plant. For most species, dispersal distance can be described by an exponential decay curve, with the majority of dispersal occurring locally, a smaller proportion occurring at an intermediate distance and a much lower proportion occurring over long distances. The practice of composite provenancing mimics this dispersal kernel in the proportion of locally sourced material (high proportion), intermediate distance–sourced material that is ecologically matched (medium proportion) and material introduced from distant populations that are ecologically diverse (low proportion). Such a strategy retains a core of locally adapted material. However, the intermediate sourcing will mimic medium-range gene flow that may bring in additional adaptive or beneficial genes. The long-distance provenancing allows an opportunity for novel in situ adaptation at the restoration site either through admixture or recombination. This latter material may be maladapted to the planting site and so an increase in sowing or stocking rate may be required, but the ultimate advantage will be the increased adaptive potential of restoration plantings employing this strategy.
Even when genetic data are not available, it may be possible to identify provenances for introduction based on ecological data. In alpine ash, Eucalyptus delegatensis, variation in average growth performance of 68 populations in a series of common gardens was used to identify patterns of genetic variation; these patterns were then linked back to features of the environment from where the populations originated to identify surrogates of genetic variability (Garnier-Gere and Ades 2001).
Finally, genetic information (both molecular and quantitative) can be used for an increasing number of species to identify the extent of adaptive divergence across species’ geographic ranges, which can help in the choice of source populations (Broadhurst et al. 2006). When combined with GIS environmental data across species’ distributions (Lipow et al. 2007), such approaches enable much more sophisticated insight into the factors underlying phenotypic divergence both within and between species (Kozak et al. 2008).While used to the greatest extent so far with tree species of high value to forestry (Lipow et al. 2004, 2007)), such approaches should become part of restoration practices more generally (Broadhurst et al. 2006, 2008; Butcher et al. 2009).
Susceptible species and evolutionary refugia
Distribution models suggest that many species will be threatened under climate change because they can only tolerate minor changes in temperature and other conditions. For example, tropical lizards (Williams et al. 2003) and other ectotherms (Deutsch et al. 2008) might be particularly threatened by global warming because their optimal temperature is close to their thermal maximum. In these cases, plastic changes and evolutionary adaptation may be insufficient to counter the effects of climate change, and protection is required.
Susceptible species with a low evolutionary potential can be identified if heritable variation has been characterized for key traits limiting distributions (Table 1– Aim C). Where heritabilities are very low, there is likely to be limited evolutionary potential to adapt, as in the case of the response of rainforest Drosophila to low humidity conditions (Kellermann et al. 2006, 2009). Even where genetic variation is present, adaptive shifts may be limited by interactions among traits (Etterson and Shaw 2001; Hellmann and Pineda-Krch 2007). It is possible to identify these limits experimentally – such as by transplanting populations to a variety of habitats that might even extend outside their current range (Crozier 2004; Pelini et al. 2009).
Evolutionary processes need to be considered in prioritizing regions for protection (Table 1– Aim D). When recovering from mass extinction and responding to climate change, refugia play a critical role. Evolutionary refugia represent areas where species persist under specific optimal conditions, generally representing a small fraction of their original range. Migration and/or dispersal to more suitable habitat in response to climate change will not be possible for species restricted to such refugia (Schneider et al. 1999), so they should become priorities for protection.
Refugia can be identified through ecological criteria. Mountain top areas are refugia for many species unable to persist under the warmer conditions of lowland regions. These areas act as refugia in the tropics and in temperate regions. For instance, one of the predictions of climate change in the Victorian Alps in Australia is an altitudinally encroaching treeline (Wearne and Morgan 2001). Because these mountains are flat regions, there is no upper area where alpine meadow plants and animals can move. The meadows in effect are already an evolutionary refuge, and the only option for long-term survival of these organisms is to actively maintain and protect the area as a refuge.
Refugia can also be identified by their genetic uniqueness. Phylogenetic comparisons (Crozier 1997) and phylogenetic diversity metrics (Moritz et al. 2009) can identify the uniqueness of taxa in specific regions. In addition, measures of uniqueness based on comparative phylogeographic data can be used to identify unique refugia across multiple taxa (Moritz 2002). Areas identified by such studies should have a high ranking for reserves. Genetic markers can also indicate populations with high levels of genetic diversity that might have priority for conservation. Again this type of information can be accumulated across taxa to develop general patterns of biodiversity hotspots (Davis et al. 2008; Vandergast et al. 2008).
Conservation planning, in situ evolution and climate change
Systematic conservation planning is often about spatial planning, and traditional conservation reserves and methods of designing them are static, with the implicit assumption that threats to biodiversity are themselves static. Protected areas play a central role in the conservation of biodiversity, but they are geographically fixed and increasingly isolated by habitat fragmentation (Hannah et al. 2007). Furthermore, current conservation practices are based on an implicit assumption of a relatively stable climate. However, range shifts have been a predominant response to past climate change, with each species tracking its preferred climatic conditions (Hannah 2010). Although range shifts in response to current climate change have also been observed, the ability of species to track climate change will be affected by ongoing habitat loss and fragmentation. Moving beyond single-species approaches to planning, it is increasingly recognized that ecological processes must also be conserved on biologically relevant scales (Hannah 2010), which may not fit within the fixed boundaries of protected areas as they now stand. However, evolutionary considerations are still lacking from these discussions about protected area planning under climate change. Overall, protected areas as they currently stand are poorly suited to accommodating in situ evolution in response to climate change.
While evolutionary processes have been acknowledged by some as central to the maintenance of biodiversity in reserves and in the maintenance of species borders (Cowling and Pressey 2001; Moritz 2002; Rouget et al. 2006; Taylor and Figgis 2007; Mace and Purvis 2008), they are yet to be explicitly incorporated into conservation planning schemes and approaches. Reserves need to be interconnected across landscapes. This will help to increase population size, protect against ecological catastrophes and provide links to refuge areas (Hannah 2010). Yet, reserve selection can be based on factors that extend beyond interconnectedness (which has been the traditional argument for such strategies): they can facilitate ongoing in situ evolution by encompassing a range of habitats where specific genotypes can be selected (Dunlop and Brown 2008). These habitats might include steep ecological gradients and areas with recent geological or climatic change (Cowling and Pressey 2001; Davis et al. 2008).
From an evolutionary perspective, the development of connectedness in landscapes can help ensure the movement of individuals and genes along corridors linking environments and increase evolutionary resilience (Table 1– Aim E). When populations are interconnected along climatic gradients, there is the potential for in situ adaptive evolution (e.g., Balanya et al. 2003; Umina et al. 2005).
Environmental gradients and refugia within landscapes have been included in conservation planning in the Cape Floristic Region (Cowling and Pressey 2001; Hannah et al. 2005; Pressey et al. 2007) and the Thicket Biome of South Africa (Rouget et al. 2006). Here, reserve design targeted regions that included riverine corridors crossing mountain ranges, allowing for dispersal and providing climatic refugia that included environmental/climatic gradients. This shift in thinking around planning and design is also reflected in the recognition of the need for the restoration of ecological connectivity to prepare for climate change within Australia (Soule et al. 2004; Mackey et al. 2007), where large-scale landscape restoration and connectivity programs have also been initiated (Mackey et al. 2007; Mansergh and Cheal 2007).
Finally, there is still a need to improve our understanding of what constitutes a permeable landscape for species migrations. Although continuous and intact native habitat is, in the majority of cases, the optimal solution for biodiversity outcomes, many refuge areas are separated by an economically or socially important matrix (e.g., farmland or cities). In such cases, alternative ‘corridor’ strategies need to be considered, such as stepping stones and narrow linear routes (e.g., road-side verges). To establish ‘ecosystem corridors’, we will also need an understanding of which species can migrate through different corridor and matrix types. If movement can occur between different refuge and conserved areas, levels of genetic variation in quantitative traits in populations can be increased (Whitlock 1999). Landscape planning, informed by species’ dispersal/migration and establishment characteristics, is critical to this endeavour. Recent advancements in the modelling of connectivity of populations across landscapes mean that population genetic data can explicitly be used to inform about how organisms move through different landscape configurations (McRae and Beier 2007; Fortuna et al. 2009; Pavlacky et al. 2009). We are now at the stage where we can explicitly plan for and conserve the evolutionary processes that underpin species and ecosystem responses to climate change, to plan for evolutionary resilience.
Conclusions: planning for evolutionary resilient landscapes under climate change
While the need to include evolutionary processes that maintain genetic diversity and adaptive potential in protected area planning and management has been advocated for some time (Pressey et al. 2007; Mace and Purvis 2008), there is as yet little guidance about how this might occur. With new emerging genomic tools, and an increased understanding of the genetic basis of adaptive responses to environmental change, more broadly, we have the opportunity to seriously consider and include evolutionary processes in conservation planning.
Traditionally, conservation efforts have focussed on the species level. However, intra-specific genetic variation needs to be considered in prioritization for conservation purposes (Frankham et al. 2002; Moritz 2002). Loss of genetic diversity within populations can be associated with inbreeding depression, which in turn results in lowered fitness and increased risk of extinction. Genetic variation is essential for adaptation to environmental change and evolution over the longer term. Importantly, intra-specific genetic diversity favours species richness in plant communities (Whitham et al. 2006; Bailey et al. 2009) and contributes to ecosystem functioning and resilience (Reusch et al. 2005). This recognition requires a shift in how ‘units’ of conservation are defined and considered, from a species-orientated approach to one that includes diversity at both inter- and intra-specific levels. It also requires a shift in how genetic diversity is thought about in the conservation literature.
We have developed a checklist that provides guidelines for the development of evolutionary resilient landscapes that will help to promote biodiversity at a time of climate change (Table 2, but see also Table 1). They outline how evolutionary resilience might be constructed and maintained and how it can be incorporated into policy and planning to ensure that species resilience is maintained in the face of a looming mass extinction.
Table 2.
A checklist for evolutionary resilience
| Maintaining population sizes |
| Management and conservation programs should aim at conserving population sizes of one to several thousand rather than tens to several hundred individuals to maintain high levels of variation for adaptation. The maintenance of genetic diversity must also be considered in captive breeding programs, sourcing of seed for revegetation programs and landscape restoration |
| Ranking areas for conservation by incorporating evolutionary processes |
| Interconnected reserves must include environmental gradients across landscapes, in particular, steep ecological gradients and areas with recent geological or climatic change. This will increase their long-term ability to sustain large populations, allow for migration and maximize the opportunity for in situ adaptation. Refugia will be essential where genetic variation cannot be maintained, and the potential for evolution is decreased. When choosing areas for reserves or refugia, it is important to assess genetic uniqueness and genetic diversity across taxa. Increasing the connectivity of refuge and conservation areas will not only allow for migration but, depending on corridor design, also increase genetic connectivity and population sizes |
| Incorporating genetic diversity when restoring degraded landscapes |
| Seed material for restoration should maximize genetic diversity and adaptedness. Local provenance collections should be supplemented by a smaller proportion of material from regions with different climates, where there has been evolutionary divergence and local adaptation, to promote evolutionary potential (composite provenancing). Climate matching for the future, predictive provenancing, should also be considered for source populations particularly where organisms are long-lived. For instance, programs could begin by determining future climate scenarios for area(s) of concern in 2050. If climate scenarios fall outside the current climate envelope of target species, ample adaptive genetic diversity might still allow in situ evolution and persistence |
| High-priority species – maximizing evolution for species survival and persistence |
| For highly threatened and endangered species, it may not be possible to maintain populations of thousands, and programs should aim at minimizing the probability of extinction. This can be performed by monitoring populations/species for genetic variation using neutral genetic markers and undertaking management decisions that maximize genetic diversity and the probability of persistence and survival (see Table 1). When modelling changes in species abundance and distribution, evolutionary considerations should be included. This requires the estimation of appropriate parameters but can be performed within a spatial context (Kearney et al. 2009). When predictions suggest that species and populations face extinction even with evolutionary change, direct intervention through translocation or ex situ conservation (e.g., seed banks, zoos or aquaria) should be considered |
Acknowledgments
We thank the Australian Research Council and the Commonwealth Environment Research Facility for funding and Andrew Weeks for the many discussions and insight.
Literature cited
- Bailey J, Schweitzer J, Ubeda F, Koricheva J, LeRoy C, Madritch M, Rehill B, et al. From genes to ecosystems: a synthesis of the effects of plant genetic factors across levels of organization. Philosophical Transactions of the Royal Society B. 2009;362:1607–1616. doi: 10.1098/rstb.2008.0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balanya J, Serra L, Gilchrist GW, Huey RB, Pascual M, Mestres F, Sole E. Evolutionary pace of chromosomal polymorphism in colonizing populations of Drosophila subobscura: an evolutionary time series. Evolution. 2003;57:1837–1845. doi: 10.1111/j.0014-3820.2003.tb00591.x. [DOI] [PubMed] [Google Scholar]
- Beaumont M, Balding D. Identifying adaptive genetic divergence among populations from genome scans. Molecular Ecology. 2004;13:969–980. doi: 10.1111/j.1365-294x.2004.02125.x. [DOI] [PubMed] [Google Scholar]
- Berkelmans R, van Oppen MJH. The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget’ of hope for coral reefs in an era of climate change. Proceedings of the Royal Society B. 2006;273:2305–2312. doi: 10.1098/rspb.2006.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin A, Nicole F, Pompanon F, Miaud C, Taberlet P. Population adaptive index: a new method to help measure intraspecific geneytic diversity and prioritize populations for conservation. Conservation Biology. 2007;21:697–708. doi: 10.1111/j.1523-1739.2007.00685.x. [DOI] [PubMed] [Google Scholar]
- Bradshaw WE, Holzapfel CM. Genetic response to rapid climate change: it's seasonal timing that matters. Molecular Ecology. 2008;17:157–166. doi: 10.1111/j.1365-294X.2007.03509.x. [DOI] [PubMed] [Google Scholar]
- Broadhurst L, Young A, Thrall P, Murray B. Sourcing seed for Acacia acinacea, a key revegetation species in south eastern Australia. Conservation Genetics. 2006;7:749–763. [Google Scholar]
- Broadhurst LM, Lowe A, Coates DJ, Cunningham SA, McDonald M, Vesk PA, Yates C. Seed supply for broadscale restoration: maximising evolutionary potential. Evolutionary Applications. 2008;1:587–597. doi: 10.1111/j.1752-4571.2008.00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon R, Wilcox P. Population management:potential impacts of advances in genomics. New Forests. 2007;34:187–206. [Google Scholar]
- Burger R, Lynch M. Evolution and extinction in a changing environment – a quantitative-genetic analysis. Evolution. 1995;49:151–163. doi: 10.1111/j.1558-5646.1995.tb05967.x. [DOI] [PubMed] [Google Scholar]
- Butcher P, McDonald M, Bell J. Congruence between environmental parameters, morphology and genetic structure in Australia's most widely distributed eucalypt, Eucalyptus camaldulensis. Tree Genomics & Genomes. 2009;5:189–210. [Google Scholar]
- Callaham R. Provenance research: investigation of genetic diversity associated with geography. Unasylva. 1964;18:40–50. [Google Scholar]
- Charmantier A, McCleery R, Cole L, Perrins C, Kruuk L, Sheldon B. Adaptive phenotpyic plasticity in response to climate change in a wild bird population. Science. 2009;320:800–803. doi: 10.1126/science.1157174. [DOI] [PubMed] [Google Scholar]
- Chevin L-M, Lande R, Mace GM. Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. PLoS Biology. 2010;8:e1000357. doi: 10.1371/journal.pbio.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheviron Z, Brumfield R. Migration-selection balance and local adaptation of mitochondrial haplotypes in rufoucollared sparrows (Zonotrichia capensis) along an elevational gradient. Evolution. 2009;63:1593–1605. doi: 10.1111/j.1558-5646.2009.00644.x. [DOI] [PubMed] [Google Scholar]
- Cowling RM, Pressey RL. Rapid plant diversification: planning for an evolutionary future. Proceedings of the National Academy of Sciences USA. 2001;98:5452–5457. doi: 10.1073/pnas.101093498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crandall K. A multifacted approach to species conservation. Animal Conservation. 2009;12:105–106. [Google Scholar]
- Crozier RH. Preserving the information content of species: genetic diversity, phylogeny, and conservation worth. Annual Review of Ecology and Systematics. 1997;28:243–268. [Google Scholar]
- Crozier L. Field transplants reveal summer constraints on a butterfly range expansion. Oecologia. 2004;141:148–157. doi: 10.1007/s00442-004-1634-z. [DOI] [PubMed] [Google Scholar]
- Davis EB, Koo MS, Conroy C, Patton JL, Moritz C. The California Hotspots Project: identifying regions of rapid diversification of mammals. Molecular Ecology. 2008;17:120–138. doi: 10.1111/j.1365-294X.2007.03469.x. [DOI] [PubMed] [Google Scholar]
- Deutsch CA, Tewksbury JJ, Huey RB, Sheldon KS, Ghalambour CK, Haak DC, Martin PR. Impacts of climate warming on terrestrial ectotherms across latitude. Proceedings of the National Academy of Sciences USA. 2008;105:6668–6672. doi: 10.1073/pnas.0709472105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman M, Sapir Y, Volis S. Local adaptation in four Iris species tsted in a common garden experiment. Biological Journal of the Linnean Society. 2009;98:267–277. [Google Scholar]
- Dunlop M, Brown PR. Implications of climate change for Australia's National Reserve System: A Preliminary Assessment. A Report to the Department of Climate Change, February 2008. Canberra, Australia: Department of Climate Change; 2008. [Google Scholar]
- Edmands S. Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Molecular Ecology. 2007;16:463–475. doi: 10.1111/j.1365-294X.2006.03148.x. [DOI] [PubMed] [Google Scholar]
- Etterson JR, Shaw RG. Constraint to adaptive evolution in response to global warming. Science. 2001;294:151–154. doi: 10.1126/science.1063656. [DOI] [PubMed] [Google Scholar]
- Fortuna M, Albaladejo R, Fernandez L, Aparicio A, Bascompte J. Networks of spatial genetic variation across species. Proceedings of the National Academy of Sciences USA. 2009;10:19044–19049. doi: 10.1073/pnas.0907704106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. Genetic adaptation to captivity in species conservation programs. Molecular Ecology. 2008;17:325–333. doi: 10.1111/j.1365-294X.2007.03399.x. [DOI] [PubMed] [Google Scholar]
- Frankham R, Ballou JD, Briscoe DA. Introduction to Conservation Genetics. Cambridge: Cambridge University Press; 2002. [Google Scholar]
- Garnier-Gere PH, Ades PK. Environmental surrogates for predicting and conserving adaptive genetic variability in tree species. Conservation Biology. 2001;15:1632–1644. [Google Scholar]
- Gebremedhin B, Ficetola G, Naderi S, Rezaei H-R, Maudet C, Rioux D, Luikart G, et al. Frontiers in developing conservation units: from neutral markers to adaptive genetic variation. Animal Conservation. 2009;12:107–109. [Google Scholar]
- Gibson A, Bachelard EP, Hubick KT. Relationship between climate and provenance variation in Eucalyptus camaldulensis Dehnh. Australian Journal of Plant Physiology. 1995;22:453–460. [Google Scholar]
- Grueber CE, Jamieson IG. Quantifying and managing the loss of genetic variation in a free-ranging population of takahe through the use of pedigrees. Conservation Genetics. 2008;9:645–651. [Google Scholar]
- Gunderson L. Ecological resilience – in theory and application. Annual Review of Ecology and Systematics. 2000;31:425–439. [Google Scholar]
- Hadfield JD, Wilson AJ, Garant D, Sheldon BC, Kruuk LEB. The misuse of BLUP in ecology and evolution. American Naturalist. 2010;175:116–125. doi: 10.1086/648604. [DOI] [PubMed] [Google Scholar]
- Hannah L. A global conservation system for climate change adaptation. Conservation Biology. 2010;24:70–77. doi: 10.1111/j.1523-1739.2009.01405.x. [DOI] [PubMed] [Google Scholar]
- Hannah L, Midgley G, Hughes G, Bomhard B. The view from the cape. Extinction risk, protected areas, and climate change. BioScience. 2005;55:231–242. [Google Scholar]
- Hannah L, Midgley G, Andelman S, Araujo M, Hughes G, Martinez-Meyer E, Pearson R, et al. Protected area needs in a changing climate. Frontiers in Ecology and the Environment. 2007;5:131–138. [Google Scholar]
- Hellmann J, Pineda-Krch M. Constraints and reinforcement on adaptation under climate change: selection of genetically correlated traits. Biological Conservation. 2007;137:599–609. [Google Scholar]
- Hendry AP, Lohmann LG, Conti E, Cracraft J, Crandall KA, Faith DP, Hauser C, et al. Evolutionary biology in biodiversity science, conservation, and policy: a call to action. Evolution. 2010;64:1517–1528. doi: 10.1111/j.1558-5646.2010.00947.x. [DOI] [PubMed] [Google Scholar]
- Hereford J. A quantiative survey of local adaptation and fitness trade-offs. The American Naturalist. 2009;173:579–588. doi: 10.1086/597611. [DOI] [PubMed] [Google Scholar]
- Hoegh-Guldberg O, Hughes L, McIntyre S, Lindenmayer DB, Parmesan C, Possingham HP, Thomas CD. Assisted colonization and rapid climate change. Science. 2008;321:345–346. doi: 10.1126/science.1157897. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Weeks AR. Climatic selection on genes and traits after a 100 year-old invasion: a critical look at the temperate-tropical clines in Drosophila melanogaster from eastern Australia. Genetica. 2007;129:133–147. doi: 10.1007/s10709-006-9010-z. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Willi Y. Detecting genetic responses to environmental change. Nature Reviews Genetics. 2008;9:1–12. doi: 10.1038/nrg2339. [DOI] [PubMed] [Google Scholar]
- Holmes GD, James EA, Hoffmann AA. Limitations to reproductive output and genetic rescue in populations of the rare shrub Grevillea repens (Proteaceae) Annals of Botany. 2008;102:1031–1041. doi: 10.1093/aob/mcn195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter ML. Climate change and moving species: furthering the debate on assisted colonization. Conservation Biology. 2007;21:1356–1358. doi: 10.1111/j.1523-1739.2007.00780.x. [DOI] [PubMed] [Google Scholar]
- Jamieson I, Wallis G, Briske J. Inbreeding and endangered species management: is New Zealand out of step with teh rest of the world? Conservation Biology. 2006;20:38–47. doi: 10.1111/j.1523-1739.2005.00282.x. [DOI] [PubMed] [Google Scholar]
- Johansson M, Primmer C, Merila J. Does habitat fragmentation reduce fitness and adaptability? A case study of the common frog (Rana temporaria. Molecular Ecology. 2007;16:2693–2700. doi: 10.1111/j.1365-294X.2007.03357.x. [DOI] [PubMed] [Google Scholar]
- Jones TA, Monaco TA. A role for assisted evolution in designing native plant materials for domesticated landscapes. Frontiers in Ecology and the Environment. 2009;7:541–547. [Google Scholar]
- Joost S, Bonin A, Bruford MW, Despres L, Conord C, Erhadrt G, Taberlet P. A spatial analysis method (SAM) to detect candidate loci for selection: towards a landscape genomics approach to adaptation. Molecular Ecology. 2007;16:3955–3969. doi: 10.1111/j.1365-294X.2007.03442.x. [DOI] [PubMed] [Google Scholar]
- Kearney M, Porter WP, Williams C, Ritchie S, Hoffmann AA. Integrating biophysical models and evolutionary theory to predict climatic impacts on species’ ranges: the dengue mosquito Aedes aegypti in Australia. Functional Ecology. 2009;23:528–538. [Google Scholar]
- Keller M, Kollman J, Edwards P. Genetic introgression from distant provenances reduces fitness in local weed populations. Journal of Applied Ecology. 2000;37:647–659. [Google Scholar]
- Kellermann VM, van Heerwaarden B, Hoffmann AA, Sgro CM. Very low additive genetic variance and evolutionary potential in multiple populations of two rainforest Drosophila species. Evolution. 2006;60:1104–1108. doi: 10.1554/05-710.1. [DOI] [PubMed] [Google Scholar]
- Kellermann V, van Heerwaarden B, Sgro C, Hoffmann A. Fundamental evolutionary limits in ecological traits drive Drosophila species distributions. Science. 2009;325:1244–1246. doi: 10.1126/science.1175443. [DOI] [PubMed] [Google Scholar]
- Klemme I, Hanski I. Heritability of and strong single gene (Pgi) effects on life history traits in the Glanville fritillary butterfly. Journal of Evolutionary Biology. 2009;22:1944–1953. doi: 10.1111/j.1420-9101.2009.01807.x. [DOI] [PubMed] [Google Scholar]
- Kozak K, Graham C, Wiens J. Integrating GIS-based environmental data into evolutionary biology. Trends in Ecology & Evolution. 2008;23:141–148. doi: 10.1016/j.tree.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Kruuk L, Slate J, Wilson A. New answers for old questions: the evolutionary quantitative genetics of wild animal populations. Annual Review of Ecology, Evolution and Systematics. 2008;39:525–548. [Google Scholar]
- Lipow SR, Vance-Borland K, St Clair JB, Henderson J, McCain C. Gap analysis of conserved genetic resources for forest trees. Conservation Biology. 2004;18:412–423. [Google Scholar]
- Lipow S, Vance-Borland K, St Clair J, Henderson J, McCain C. In situ gene conservation of six conifers in western Washington and Oregon. Western Journal of Applied Forestry. 2007;22:176–187. [Google Scholar]
- Lopez S, Rousset F, Shaw F, Shaw R, Ronce O. Joint effects of inbreeding and local adaptation on the evolution of genetic load after fragmentation. Conservation Biology. 2009;23:1618–1627. doi: 10.1111/j.1523-1739.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- Lynch M, Lande R. Evolution and extinction in response to environmental change. In: Kareiva PM, Kingsolver JG, Huey RB, editors. Biotic Interactions and Global Change. Sunderland, Massachusetts: Sinauer Associates Inc; 1993. pp. 234–250. [Google Scholar]
- Mace GM, Purvis A. Evolutionary biology and practical conservation: bridging a widening gap. Molecular Ecology. 2008;17:9–19. doi: 10.1111/j.1365-294X.2007.03455.x. [DOI] [PubMed] [Google Scholar]
- Mackey BG, Soule ME, Nix HA, Recher HF, Lesslie RG, Williams JE, Woinarski JCZ. Towards a scientific framework for the Wild Country project. In: Wu J, Hobbs RJ, et al., editors. Key Topics and Perspectives in Landscape Ecology. Cambridge: Cambridge University Press; 2007. pp. 192–208. [Google Scholar]
- Mansergh I, Cheal D. Protected Area Planning and Management for Eastern Australian Temperate Forests and Woodland Ecosystems. Protected Areas: Buffering Nature Against Climate Change, Sydney, Australia. Sydney: WWF-Australia; 2007. [Google Scholar]
- McKay JK, Latta RG. Adaptive population divergence: markers, QTL and traits. Trends in Ecology & Evolution. 2002;17:285–291. [Google Scholar]
- McKay J, Christian C, Harrison S, Rice K. “How local is local?”– a review of practical and conceptual issues in the genetics of restoration. Restoration Ecology. 2005;13:432–440. [Google Scholar]
- McLachlan JS, Hellmann JJ, Schwartz MW. A frmework for debate of assisted migration in an era of climate change. Conservation Biology. 2007;21:297–302. doi: 10.1111/j.1523-1739.2007.00676.x. [DOI] [PubMed] [Google Scholar]
- McRae BH, Beier P. Circuit theory predicts gene flow in plant and animal populations. Proceedings of the National Academy of Sciences USA. 2007;104:19885–19890. doi: 10.1073/pnas.0706568104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges ES. Restoration demography and genetics of plants: when is a translocation successful? Australian Journal of Botany. 2008;56:187–196. [Google Scholar]
- Miskelly C, Taylor G, Gummer H, Williams R. Translocations of eight species of burrow-nesting seabirds (genera PterodromoaPelecanoidesPachyptila and Puffinus: Family Procellariidae) Biological Conservation. 2009;142:1965–1980. [Google Scholar]
- Mitrovski P, Hoffmann A, Heinze D, Weeks A. Rapid loss of genetic variation in an endangered possum. Biology Letters. 2008;4:134–138. doi: 10.1098/rsbl.2007.0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz C. Strategies to protect biological diversity and the evolutionary processes that sustain it. Systematic Biology. 2002;51:238–254. doi: 10.1080/10635150252899752. [DOI] [PubMed] [Google Scholar]
- Moritz C, Hoskin C, Graham CH, Hugall A, Moussalli A. Historical biogeography, diversity and conservation of Australia's tropical rainforest herpetofauna. In: Purvis A, Gittleman JL, Brooks T, editors. Phylogeny and Conservation. Cambridge: Cambridge University Press; 2009. pp. 243–267. [Google Scholar]
- O'Brien E, Mazanec R, Krauss S. Provenance variation of ecologically important traits of forest trees: implications for restoration. Journal of Applied Ecology. 2007;44:583–595. [Google Scholar]
- Ozgul A, Tuljapurkar S, Benton T, Pemberton J, Clutton-Brock T, Coulson T. The dynamics of phenotypic change and teh shrinking sheep of St. Kilda. Science. 2009;325:464–467. doi: 10.1126/science.1173668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmesan C. Ecological and evolutionary responses to recent climate change. Annual Review of Ecology Evolution and Systematics. 2006;37:637–669. [Google Scholar]
- Pavlacky D, Goldizen A, Prentis P, Nicholls J, Lowe A. A landscape genetics approach for quantifying the relative influence of historic and contemporary habitat heterogeneity on the genetic connectivity of a rainforest bird. Molecular Ecology. 2009;18:2945–2960. doi: 10.1111/j.1365-294X.2009.04226.x. [DOI] [PubMed] [Google Scholar]
- Pelini SL, Dzurisin JDK, Prior KM, Williams CM, Marsico TD, Sinclair BJ, Hellmann JJ. Translocation experiments with butterflies reveal limits to enhancement of poleward populations under climate change. Proceedings of the National Academy of Sciences USA. 2009;106:11160–11165. doi: 10.1073/pnas.0900284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pressey RL, Cabeza M, Watts ME, Cowling RM, Wilson KA. Conservation planning in a changing world. Trends in Ecology and Evolution. 2007;22:583–592. doi: 10.1016/j.tree.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Ramirez-Valiente J, Lorenzo Z, Soto A, Valladares F, Gil L, Aranda I. Elucidating the role of genetic drift and natural selection in cork oak differentiation regarding drought tolerance. Molecular Ecology. 2009;18:3803–3815. doi: 10.1111/j.1365-294X.2009.04317.x. [DOI] [PubMed] [Google Scholar]
- Reed DH, Frankham R. How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evolution. 2001;55:1095–1103. doi: 10.1111/j.0014-3820.2001.tb00629.x. [DOI] [PubMed] [Google Scholar]
- Reusch T, Wood T. Molecular ecology of global change. Molecular Ecology. 2007;16:3973–3992. doi: 10.1111/j.1365-294X.2007.03454.x. [DOI] [PubMed] [Google Scholar]
- Reusch TB, Ehlers A, Hammerli A, Worm B. Ecosystem recovery after climatic extremes enhanced by genotypic diversity. Proceedings of the National Academy of Sciences USA. 2005;102:2826–2831. doi: 10.1073/pnas.0500008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciardi A, Simberloff D. Assisted colonization is not a viable conservation strategy. Trends in Ecology & Evolution. 2009;24:248–253. doi: 10.1016/j.tree.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Rice K, Emery NC. Managing microevolution: restoration in the face of global change. Frontiers in Ecology and the Environment. 2003;1:469–478. [Google Scholar]
- Richardson DM, Hellmann JJ, McLachlan JS, Sax DF, Schwartz MW, Gonzalez P, Brennan EJ, et al. Multidimensional evaluation of managed relocation. Proceedings of the National Academy of Sciences USA. 2009;106:9721–9724. doi: 10.1073/pnas.0902327106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root TL, Price JT, Hall KR, Schneider SH, Rosenzweig C, Pounds JA. Fingerprints of global warming on wild animals and plants. Nature. 2003;421:57–60. doi: 10.1038/nature01333. [DOI] [PubMed] [Google Scholar]
- Rouget M, Cowling RM, Lombard AT, Knight AT, Kerley GH. Designing large-scale conservation corridors for pattern and process. Conservation Biology. 2006;20:549–561. doi: 10.1111/j.1523-1739.2006.00297.x. [DOI] [PubMed] [Google Scholar]
- Schneider CJ, Smith TB, Larison B, Moritz C. A test of alternative models of diversification in tropical rainforests: ecological gradient vs. rainforest refugia. Proceedings of the National Academy of Sciences USA. 1999;96:13869–13873. doi: 10.1073/pnas.96.24.13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soule ME, Mackey BG, Recher HF, Williams JE, Woinarski JCZR, Driscol D, Dennison WC, et al. The role of connectivity in Australian conservation. Pacific Conservation Biology. 2004;10:266–279. [Google Scholar]
- Spielman D, Brook BW, Frankham R. Most species are not driven to extinction before genetic factors impact them. Proceedings of the National Academy of Sciences USA. 2004;101:15261–15264. doi: 10.1073/pnas.0403809101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz J. Using genome scans of DNA polymorphisms to infer adaptive population divergence. Molecular Ecology. 2005;14:671–688. doi: 10.1111/j.1365-294X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Swarts ND, Dixon KW. Terrestrial orchid conservation in the age of extinction. Annals of Botany. 2009;104:543–556. doi: 10.1093/aob/mcp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Figgis P. Protected Areas: Buffering Nature Against Climate Change, Sydney, Australia. Sydney: WWF-Australia; 2007. [Google Scholar]
- Thomas CD, Cameron A, Green RE, Bakkenes M, Beaumont LJ, Collingham YC, Erasmus BFN, et al. Extinction risk from climate change. Nature. 2004;427:145–148. doi: 10.1038/nature02121. [DOI] [PubMed] [Google Scholar]
- Thrush SF, Hewitt JE, Dayton PK, Coco G, Lohrer AM, Norkko A, Norkko J, et al. Forecasting the limits of resilience: integrating empirical research with theory. Proceedings of the Royal Society B-Biological Sciences. 2009;276:3209–3217. doi: 10.1098/rspb.2009.0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umina PA, Weeks AR, Kearney MR, McKechnie SW, Hoffmann AA. A rapid shift in a classic clinal pattern in Drosophila reflecting climate change. Science. 2005;308:691–693. doi: 10.1126/science.1109523. [DOI] [PubMed] [Google Scholar]
- Vandergast AG, Bohonak AJ, Hathaway SA, Boys J, Fisher RN. Are hotspots of evolutionary potential adequately protected in southern California? Biological Conservation. 2008;141:1648–1664. [Google Scholar]
- Viveros-Viveros H, Saenz-Romeroa C, Vargas-Hernandez J, Lopez-Upton J, Ramirez-Valverde G, Santacruz-Varela A. Altitudinal variation in Pinus hartwegii Lindl. I: height growth, shoot phenology and frost damage in seedlings. Forest Ecology and Management. 2009;257:836–842. [Google Scholar]
- Wearne LJ, Morgan JW. Recent forest encroachment into subalpine grasslands near Mount Hotham, Victoria, Australia. Arctic Antarctic and Alpine Research. 2001;33:369–377. [Google Scholar]
- Whitham TG, Bailey JK, Schweitzer JA, Shuster SM, Bangert RK, LeRoy CJ, Lonsdorf EV, et al. A framework for community and ecosystem genetics: from genes to ecosystems. Nature Reviews Genetics. 2006;7:510–523. doi: 10.1038/nrg1877. [DOI] [PubMed] [Google Scholar]
- Whitlock M. Neutral additive genetic variance in a metapopulation. Genetical Research. 1999;74:215–221. doi: 10.1017/s0016672399004127. [DOI] [PubMed] [Google Scholar]
- Wilkinson D. Is local provenance important in habitat creation? Journal of Applied Ecology. 2001;38:1371–1373. [Google Scholar]
- Willi Y, Hoffmann AA. Demographic factors and genetic variation influence population persistence under environmental change. Journal of Evolutionary Biology. 2008;22:124–133. doi: 10.1111/j.1420-9101.2008.01631.x. [DOI] [PubMed] [Google Scholar]
- Willi Y, van Buskirk J, Hoffmann AA. Limits to the adaptive potential of small populations. Annual Review of Ecology, Evolution and Systematics. 2006;37:433–478. [Google Scholar]
- Williams SE, Bolitho EE, Fox S. Climate change in Australian tropical forests: an impending environmental catastrophe. Proceedings of the Royal Society, London, Series B: Biological Sciences. 2003;270:1887–1892. doi: 10.1098/rspb.2003.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]


