Abstract
Human-induced land use changes are causing extensive habitat fragmentation. As a result, many species are not able to shift their ranges in response to climate change and will likely need to adapt in situ to changing climate conditions. Consequently, a prudent strategy to maintain the ability of populations to adapt is to focus conservation efforts on areas where levels of intraspecific variation are high. By doing so, the potential for an evolutionary response to environmental change is maximized. Here, we use modeling approaches in conjunction with environmental variables to model species distributions and patterns of genetic and morphological variation in seven Ecuadorian amphibian, bird, and mammal species. We then used reserve selection software to prioritize areas for conservation based on intraspecific variation or species-level diversity. Reserves selected using species richness and complementarity showed little overlap with those based on genetic and morphological variation. Priority areas for intraspecific variation were mainly located along the slopes of the Andes and were largely concordant among species, but were not well represented in existing reserves. Our results imply that in order to maximize representation of intraspecific variation in reserves, genetic and morphological variation should be included in conservation prioritization.
Keywords: Andes, conservation prioritization, ecological modeling, Ecuador, evolutionary process, generalized dissimilarity modeling, landscape genetics, species distribution
Introduction
Human-induced land use changes are leading to rapid habitat fragmentation. As a result, local populations are increasingly unable to track ongoing climate change (Travis 2003) and will have to adapt to the new environmental conditions or go extinct. The ability to adapt genetically depends on the amount of heritable variation currently present in a population. Thus, one way to mitigate the potential impacts of climate change on biodiversity is to ensure that the evolutionary processes underlying the generation and maintenance of such variation are preserved (Smith et al. 1993). Evolutionary processes have long been regarded of major importance in reserve design (e.g. Frankel 1974; Moritz 2002; Gregory et al. 2006; Smith and Grether 2008), but in practice have seen limited use in conservation planning. Nevertheless, given the current rate of anthropogenic climate change, there is an urgent need for approaches that enable conservation biologists to take into account evolutionary processes in prioritization schemes.
Recently, we proposed a novel, integrative approach to incorporate evolutionary processes in conservation prioritization based on spatially explicit models of intraspecific genetic and phenotypic variation, in combination with species-level data (Thomassen et al. 2010). The approach is based on the assumption that retaining the highest possible levels of adaptive variation will be a useful strategy to maximize a species’ adaptive potential (e.g. Vandergast et al. 2008; Tymchuk et al. 2010; Sgrò et al. 2011). In contrast to species richness, adaptive intraspecific variation is likely to be the result of relatively recent microevolutionary processes and represents options for an evolutionary response to current and future environmental conditions. To accomplish its inclusion in conservation prioritization, ecological modeling approaches used to map intraspecific variation should be integrated with existing conservation strategies. To this end, we suggested a framework that includes the modeling of species distributions and corresponding environmentally associated intraspecific variation using population-level genetic and phenotypic variation and the subsequent integration of the resulting models with species-level information and socio-economic data to finalize prioritization (Thomassen et al. 2010). As a result of recent advances in spatially explicit modeling methods and the availability of climate and high-resolution remotely sensed environmental information, such an integrative approach is now feasible. Climate and remote sensing data capture many habitat properties that are important to characterize a species’ ecological niche and to identify selective processes that result in adaptive variation. Because environmental variables often cover most of the global land surface, it is possible to predict environmentally associated intraspecific genetic and phenotypic variation to areas that have not been previously sampled.
Here, we extend our model to include multiple amphibian, bird, and mammal species. Our aim is to assess the utility of environmentally associated intraspecific variation in conservation prioritization. We focus on the association between intraspecific variation and environmental heterogeneity, because it is reasonable to suspect that this association is the combined result of direct selective forces on linked genes and of decreased gene flow because of reduced dispersal ability caused by habitat differences. Thus, intraspecific variation insofar explained by spatial heterogeneity in environmental variables may represent adaptive variation, even if the markers examined are evolving neutrally. Moreover, the morphological characters studied are fitness-related in many species. We investigate two important issues. First, if historical evolutionary processes that have resulted in speciation remain important today, traditional methods of reserve design based on species-level data may also capture relevant intraspecific variation. However, if this is not the case, valuable intraspecific variation may not be conserved with traditional methods. Second, because it is not feasible to gather data for all species present in a region under consideration for conservation, reserve design must rely on a limited subset of species that may or may not be representative of the entire community. Our objectives in this paper are to (i) model the distributions of a set of target species; (ii) model genetic and morphological variation in those species; (iii) design reserves based on patterns of intraspecific variation; (iv) design reserves based on species-level data; (v) compare the overlap between priority areas based on intraspecific variation and species-level data; and (vi) assess whether priority areas based on intraspecific variation are consistent among species. Our goal is not to produce a reserve plan, which would require the inclusion of socio-economic, cost, and threat criteria, as well as the input of stakeholders. Rather, we aim to examine the utility of our proposed strategy from a purely biological perspective, focusing on maximizing different aspects of biodiversity. Our framework is also largely independent of the choice of modeling and reserve selection software, and the methods that we use here might readily be replaced by others developed in the future.
We conducted our research in continental Ecuador, a region that has received considerable conservation focus, because of its high species richness, endemism, and level of threat (e.g. Orme et al. 2005; Finer et al. 2010). Ecuador is geographically highly diverse, harboring steep environmental gradients along the slopes of the Andes, with the Pacific coastal region on the west, and the Amazon basin on the east. The topographic and climatologic heterogeneity of the country has likely resulted in diversifying selection, and divergence through isolation, resulting in high biodiversity (e.g. Graham et al. 2004; Chaves et al. 2007; Milá et al. 2009). Two previously defined biodiversity hotspots occur in Ecuador: the Chocó/Darien/West Ecuador and the Tropical Andes ecoregions (Myers et al. 2000). Lowland, eastern Ecuador is part of the Amazon basin ecoregion. Ecuador is highly suitable for our study, because our results can be compared to previous prioritization plans and existing protected areas. Further, its variety of environmental gradients and opportunities for isolation make it an ideal test of our approach for complex regions harboring much biodiversity.
Materials and methods
An overview of the workflow used in this study is provided in Fig. 1. First, geographic distribution models for each of seven target species were created using Maxent (Phillips et al. 2006) in conjunction with species-presence point localities and environmental variables. Second, using the modeled species distributions to define the spatial extent, we predicted genetic and morphological intraspecific variation in a generalized dissimilarity modeling framework (GDM; Ferrier et al. 2007). GDM modeled beta-diversity by evaluating how genetic and morphological variation vary as a function of geographic distance and environmental heterogeneity. For each trait, these analyses resulted in a GIS layer with values representing 50 classes of similarity in the respective traits, each class representing an equal amount of trait variation. These classes were separated as individual layers in ArcGIS (version 9.3) and subsequently used in reserve selection software (ResNet; Sarkar et al. 2009) that aimed at representing each class at a set target to prioritize areas for conservation based on genetic and morphological variation. In addition, we identified areas harboring the highest levels of intraspecific variation, thus taking into account both alpha- and beta-diversity. We also prioritized areas for conservation based on species richness using species distributions of birds, mammals, and amphibians, available from public databases.
Figure 1.
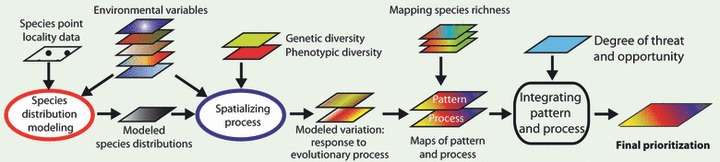
Schematic of our workflow for this paper. (i) Species distribution models for the seven target taxa were created using Maxent (Phillips et al. 2006), species-presence point localities, and a set of environmental variables. The modeled species distributions served to delimit our study area for modeling intraspecific variation. (ii) We predicted the patterns of genetic and morphological diversity using in situ data and the environmental variables in a generalized dissimilarity modeling framework (Ferrier et al. 2007). For each genetic or morphological trait, these analyses resulted in a GIS layer with values representing 50 similarity classes. These classes were separated as individual layers in ArcGIS (version 9.3) and used in (iii) subsequent ResNet reserve selection software (Sarkar et al. 2009) to prioritize areas for conservation based on genetic and morphological variation. (iv) We also used species distributions of birds, mammals, and amphibians, available from public databases, to prioritize areas for conservation based on species richness and complementarity. Finally, we combined the results from steps (iii) and (iv) and compared all results with each other and with currently protected areas.
Environmental data
We used a set of moderately high-resolution (∼1 km) climate and satellite remote sensing variables to characterize the often sharp habitat transitions in Ecuador (Data S1; Table S1). A detailed description of these variables can be found elsewhere (Buermann et al. 2008; see the Supporting Information in Thomassen et al. 2010 for maps of the variables used). Briefly, we used bioclimatic metrics (WorldClim; Hijmans et al. 2005), which are derived from meteorological station data (1950–2000), and capture variations in the annual mean, extreme, and seasonality of temperature and precipitation. To characterize ecosystem function and structure, we used optical passive and microwave active satellite data and derived products. The optical data were derived from MODIS (Justice et al. 1998) and include Leaf Area Index (LAI; Myneni et al. 2002) and percent tree cover (Hansen et al. 2002). Microwave data include QuikScat (QSCAT; Long et al. 2001) and Shuttle Radar Topography Mission (SRTM) elevation data. All raw satellite data were collected during 2001, and information on corresponding ecological inference can be found in Table S1.
Species-level data
Species distributions of birds (949 species; Ridgely et al. 2007) and mammals (336 species; Patterson et al. 2007) (Data S1) were obtained from the NatureServe database. Amphibian distribution maps were obtained from the Global Amphibian Assessment, accessed through the IUCN Red List database (467 species; IUCN, Conservation International, and NatureServe, 2008). These data were compiled based on field surveys and expert opinion and have seen widespread use in conservation planning (e.g. Hess et al. 2006; Darst et al. 2009). Because our study was aimed at terrestrial species only, coastal and pelagic species were not included in our analyses. Downloaded data consisted of GIS feature layers for each species, which were clipped to the extent of continental Ecuador, resulting in a total of 1752 species for further analyses, subdivided as follows: 467 amphibians, 336 mammals, and 949 birds. Feature layers were subsequently converted to raster grid files in ASCII format at a 2-km spatial resolution, which was found to result in files that were small enough for computational handling in the ResNet reserve selection software. Data layers were processed using ArcGIS 9.3 (ESRI, Redlands, CA, USA).
Genetic and morphological data
To map intraspecific variation, we focused on seven species: three bird species: wedge-billed woodcreeper (Glyphorynchus spirurus), masked flowerpiercer (Diglossa cyanea), and streak-necked flycatcher (Mionectes striaticollis); three mammal species: silky, chestnut, and Seba's short-tailed bats (Carollia brevicauda, C. castanea, and C. perspicillata, respectively), and one amphibian species [the frog Pristimantis (formerly Eleutherodactylus) w-nigrum]. These species were chosen, because they are abundant, easily sampled, and represent a range of different vagilities and niches. They have different habitat preferences with respect to elevation and vegetation cover, occupy a range of vegetation strata, and show a range of food habits, including insectivores, frugivores, and nectarinivores. Genetic data were available for all species, except for two of the three bat species (C. brevicauda and C. castanea).
Amplified fragment length polymorphism (AFLP) loci were used for the wedge-billed woodcreeper (136 loci, 178 individuals, 15 sites) and for the bat C. perspicillata (311 loci, 83 individuals, 9 sites). For the flowerpiercer and streak-necked flycatcher, we selected 10 microsatellite loci each and genotyped them for 102 individuals among 12 sampling locations and 106 individuals from nine sampling locations, respectively. For the frog P. w-nigrum, we sequenced two anonymous loci and an 840-bp region of the recombination activating gene 1 (RAG1) (n = 76–127 per locus, nine sampling sites). We calculated appropriate genetic population pairwise distances (Nei's D, Fst, and Φst) for subsequent use in GDM. Whereas the genetic markers used are different across taxa and may evolve at different rates, it seems justified to assume that the variation at this intraspecific level is the result of relatively recent evolutionary processes, which are the ones most likely to be relevant given future environmental change.
Morphological variation among populations was assessed through the measurement of fitness-related traits. For birds, these include wing, tail, tarsus, and bill lengths (the latter is defined as exposed culmen or the length of the bill from tip to the onset where the feathers start); bill width; and bill depth (wedge-billed woodcreeper n = 195, 15 sites; flowerpiercer n = 90, 10 sites; flycatcher n = 122, eight sites). For the bats, the following measurements were taken or calculated from landmarks of skulls using geometric morphometrics or inter-landmark measurements from the body on a total of 413 individuals (C. brevicauda: n = 167, 43 sites; C. castanea: n = 86, 25 sites; C. perspicillata: n = 160, 44 sites): centroid size for the size of the skull (hereafter referred to as skull size), angle of curvature of the zygomatic arch (hereafter referred to as zygomatic arch), and forearm length (Jarrín-V. et al. 2010). For P. w-nigrum, we measured the following morphological variables in 224 adult males among 16 sites: snout-vent length (SVL), gape width and the lengths of metacarpal phalanges (from the distal margin of the carpals to the tip of the third finger), radio-ulna, metatarsal phalanges (from the distal margin of the tarsal to the tip of the fourth toe), tarsal, tibio-fibula, femur (from the distal margin of the femur to the anterior margin of the urostyle), and lower jaw (from the posterior margin of the lower jaw to the tip of the snout). We calculated pairwise Euclidean distances from site averages as a measure of dissimilarity between sampling sites and divided these by the sum of the standard deviations within each site to take into account within-population variation. Further details on the genetic and morphological data used can be found in the Data S1.
Modeling of species distributions and intraspecific variation
Modeling morphological and genetic variation of the target species across Ecuador first required the delineation of each species’ geographic range. Maps of continuous habitat suitability for each species were generated using Maxent 3.0 (Phillips et al. 2006), using known occurrences from throughout their ranges (Table 1), with particular focus on Ecuador. Rather than relying on both presence and absence data points, Maxent uses presence-only data. The input data consist of a set of environmental layers for the study region and the observed species-presence localities within that region. The program then uses these data to estimate the environmental niche space that most accurately describes the observed occurrences. To test for the importance of environmental predictor variables in determining the species’ range, we used the variable jackknifing approach implemented in Maxent. Using a subset of the original variable set, new models were computed and their performance as measured by the regularized training gain (the average log probability of the presence samples, corrected for a uniform distribution with gain = 0) compared to that of the full model (Phillips et al. 2006). We tested whether models were significantly better than random predictions using a one-tailed binomial test on the proportion of test sites falling outside the prediction resulting from a model that used 60% of the data for training and 40% for testing (Data S1) (Anderson et al. 2002). In addition, we used the area under the receiver operator curve (AUC) to evaluate model performance (Phillips et al. 2006). To convert the continuous Maxent predictions into presence–absence maps defining the study area for subsequent GDM analyses (see below), we used thresholds for habitat suitability, which were centered within the range of a number of optimized Maxent thresholds, including ‘Equal training sensitivity and specificity’ and ‘balance threshold’. Criteria for choosing this threshold level included verification whether the Ecuadorian field sampling sites were included in the suitable area and comparisons to published range estimates in field guides.
Table 1.
Results from Maxent species distribution models
| Most important variables | ||||||
|---|---|---|---|---|---|---|
| Species | No. sites | AUC | Test AUC | Test P | By themselves | When omitted |
| WBWC | 71 | 0.898* | 0.899* | 0.001 | QSCATmean, Bio18* | Bio18, QSCATmean* |
| MFP | 103 | 0.997 | 0.995 | <0.000 | SRTM, Bio4 | Bio4, SRTM |
| SNFC | 34 | 0.987 | 0.983 | <0.000 | SRTM, SRTMstd | SRTM |
| Cb | 31 | 0.949 | 0.786 | 0.009 | Bio16, 12, 18 | LAImax, -range, SRTMstd |
| Cc | 20 | 0.935 | 0.790 | 0.002 | Bio18, 16 | LAImax |
| Cp | 36 | 0.895 | 0.744 | 0.007 | Bio16, 18 | ∼Equal for all variables |
| Pw-n | 135 | 0.990 | 0.983 | 0.000 | Bio1, 5, 6, SRTM | ∼Equal for all variables |
Shown are the number of georeferenced sites used to build the distribution model; two measures of model performance: (i) the area under the receiver operator curve (AUC) for full models and for test data sets (40% of the full data set), and (ii) the P-value from a one-tailed binomial test on the Maxent extrinsic omission rate and proportional predicted area (Anderson et al. 2002), where significant P-values indicate high model performance; the most important environmental predictor variables as assessed by a jackknifing procedure, where the variable either is used on its own or is the only one omitted from the data set. See Table S1 for information on the environmental variables.
WBWC, wedge-billed woodcreeper; MFP, masked flowerpiercer; SNFC, streak-necked flycatcher; Cb, Carollia brevicauda; Cc, Carollia castanea; Cp, Carollia perspicillata; Pw-n, Pristimantis w-nigrum; SRTM, Shuttle Radar Topography Mission.
From Buermann et al. (2008).
To predict the distribution of environmentally associated genetic and phenotypic variation across the landscape, we used GDM (Ferrier et al. 2007), a matrix regression technique that predicts biotic dissimilarity (i.e. beta-diversity) between sites based upon environmental dissimilarity and geographic distance. GDM explicitly considers the influence of geographic distance on explaining biological variation. This is carried out in a two-step method: first, dissimilarities of a set of environmental predictor variables are fitted to the pairwise genetic (e.g. Fst, Nei's D, or Фst) or phenotypic dissimilarities (Euclidean distance), the response variables. The contributions of predictor variables to explaining the observed response variation are tested by permutation, and only those variables that are significant are retained in the final model. The relative importance of predictor variables in a GDM can be assessed by means of response curves. These procedures result in a function that describes the relationship between environmental and response variables. Second, using the function resulting from the first step, a spatial prediction is made of the response variable patterns. The output is a map of classes, in our case 50, that summarize the variation in different phenotypes or genotypes. These classes were used in subsequent reserve design algorithms as a way to identify areas of high conservation priority.
Geographic distance may not represent the distance an individual might travel among localities, because of variation in habitat suitability, or the presence of barriers, such as wide rivers or high mountain ranges. We therefore also included least-cost-paths (LCP; PathMatrix 1.1; Ray 2005) and resistance distances (RD; Circuitscape 2.2, McRae 2006) in our models (Data S1), which take into account the permeability of the habitat matrix. LCP and RD were computed using friction surfaces in which unsuitable habitat (defined as those areas predicted to be unsuitable by a species distribution model, after applying a threshold value for suitability) was assumed to be 10 times as difficult to penetrate as suitable habitat.
To further evaluate the extent to which distance is potentially correlated with environmental differences, for each region and for each dependent variable we ran independent tests with the following sets of predictor variables: (i) environmental variables and distance (geographic, LCP, or RD); (ii) only distance (geographic, LCP, or RD); (iii) only environmental variables. Comparison of the results from these three runs provided an indication of the correlation between geographic distance and environmental differences.
In order to assess the significance of the level of variation that was explained by our models, we ran additional models in which the environmental layers were substituted by layers with random values for each grid cell. The resulting percentage of variation explained was compared to that of the full model. We considered the performance of the full model not significant if it explained an equal amount or less of the total variation than a model with random environmental variables.
To identify areas harboring the highest amounts of genetic and phenotypic alpha-diversity, where reserves could most efficiently protect a large percentage of the intraspecific variation, we calculated the standard deviation from GDM predictions in an area of 3 × 3 grid cells at a spatial resolution of 1 km. Thus, we first identified variation that is related to environmental variables, and which is likely to be representative of adaptive variation, and subsequently mapped areas showing high levels of such genetic and morphological variation within a 3 × 3 km area.
Reserve selection
We designed conservation areas to cover two types of measures for overall biodiversity: (i) species occurrences and (ii) intraspecific genetic and morphological variation. The same area prioritization procedure was used for both data types: a rarity-complementarity algorithm implemented in the ResNet software package (Sarkar et al. 2009). The design of a set of areas to protect features of biodiversity is typically formulated as a constrained optimization problem in which the objective is to establish protected areas that meet the representation targets for the features while taking up as little land as possible (Sarkar et al. 2006). A representation target of 10% of the total habitat suitable for a species has seen widespread use in conservation planning exercises and is included in the Convention on Biological Diversity (Justus et al. 2008). We examined a range of conservation targets for the species and genetic and phenotypic classes, specified as a single occurrence (one site of 2 × 2 km), and 5%, 10%, and 20% of the total suitable habitat.
The input data for ResNet consist of a set of sites and a list of the elements of biodiversity in each site. Using this information, ResNet employs a heuristic algorithm to prioritize areas for conservation. The iterative algorithm first selects the site that contains the rarest species or class of genetic or phenotypic similarity. ResNet breaks ties in rarity by selecting the site with the greatest complementarity, which is the one containing the largest number of elements of biodiversity that are not currently represented adequately to meet their targets. ResNet stops selecting sites when all elements of biodiversity have met their targets in the selected areas. Because of the large number of grid cells in combination with the amount of species or classes of genetic and morphological similarity, we used a heuristic algorithm implemented in ResNet, which has been shown to find near-optimal solutions to large conservation planning problems (Sarkar et al. 2009). We utilized ResNet because the large number of grid cells in combination with the amount of species or classes of genetic and morphological similarity was tractable only with a heuristic algorithm. Further, the rarity-complementarity algorithm is more effective for the rapid analysis of large-scale data sets than other techniques that have been used to select reserves such as simulated annealing or integer programming (Kelley et al. 2002; Sarkar et al. 2004).
To avoid putting specific weight on either species-level data or intraspecific variation, we chose not to combine these two types of data into a single ResNet run, but instead made a composite map of areas harboring high levels of intraspecific alpha-diversity, reserves based on species-level data and those based on classes of similar morphology and genotype identified using GDM. To create this composite map, we started by mapping areas comprising the highest levels of genetic and morphological alpha-diversity for each species (see above). This was carried out in ArcGIS using the following three steps: (i) for each species we calculated the highest 10% of variation in each trait (alpha-diversity) and compiled all traits in a single map; (ii) the resulting GIS layer was reclassified such that areas comprising the highest 10% of variation were coded 1, and all other areas 0; (iii) the layers from the previous step were added across species, resulting in a layer with values 0–7, with increasing values indicating more cross-species overlap in high levels of variation. On top of this map, we then superimposed reserves selected by ResNet using intraspecific variation or species-level data, because the algorithm capitalizes on unique variation that may not be captured in areas harboring high levels of intraspecific alpha-diversity. The composite map is thus likely to be the most comprehensive representation of variation at all levels.
Results
Species distribution models
Threshold-dependent one-tailed binomial tests on the Maxent extrinsic omission rate and proportional predicted area of the species distribution models (Fig. S1) were highly significant (Table 1), suggesting that the models performed significantly better than random. Evaluation of the AUC values also suggested high model performance (AUC values ≥0.895 for all models; test AUC values ≥0.744; Table 1). The variable importance for each of the models is shown in Table 1. The most important variable in defining distribution models for the streak-necked flycatcher and flowerpiercer was elevation, whereas precipitation variables were important for defining the ranges of the wedge-billed woodcreeper (see also Buermann et al. 2008) and the three bat species (Table 1). Temperature variables were particularly informative in defining the range of the frog species. The wedge-billed woodcreeper and the three bat species were predicted to be present across much of the Ecuadorian lowland areas west and east of the Andes (Fig. S1). The masked flowerpiercer was restricted to mid to high elevations. Likewise, the streak-necked flycatcher was predicted to occur from mid to high elevations, but with the highest probability in the mid-elevations, whereas the frog species occurred along a fairly narrow band at mid-elevations. The models corresponded well with published range maps (Lynch 1979; Eisenberg and Redford 2000; Ridgely and Greenfield 2001) and assessments based upon personal observations in the field (TBS, BM, PJ, CMK, CJS).
Models of intraspecific variation
Generalized dissimilarity models varied in their performance across species and across genetic and morphological traits. A total of 36 out of 60 models were found to perform better than random (Table 2). For the majority of these 36 models, environmental variables far outperformed measures of geographic distance as predictors of intraspecific variation (Table 2), suggesting that isolation-by-distance was not an important driver of diversification in these traits. However, for some of the models, geographic distance explained a considerable proportion of the total intraspecific variation, which suggests that this part of the observed variation is potentially attributable to isolation-by-distance. In these cases, environmental heterogeneity and geographic distance are partially correlated, and it is not possible to distinguish between the effects of the two on patterns of variation. In the Data S1, we describe the models for each species, and predictive maps of the models are shown in Fig. S2 (for the wedge-billed woodcreeper, see Thomassen et al. 2010). In summary, differentiation in the flowerpiercer is driven by moisture levels and elevation differences and located along elevational gradients on both sides of the Andes. In the flycatcher, vegetation and temperature variables were most important in explaining the observed genetic and morphological variation. Differences were most apparent between the eastern and western sides of the Andes, as well as along the western elevational gradient for microsatellites and tarsus length, and the eastern elevational gradient for tail length. In the bats, variation in the zygomatic arch was mostly related to seasonal measures of precipitation and vegetation characteristics. Forearm length was best explained by vegetation variables (including QSCAT, which distinguishes different forest types), which is potentially interpreted as adaptive given the importance of this trait in maneuverability in relation to vegetation density. Skull size was related to climate and vegetation variables, but also to geographic distance. Differentiation on the east side of the Andes was most pronounced between the lowlands and mid-elevations, but for some variables also along a latitudinal gradient (Fig. S2). Similarly, the pattern of variation in the bats on the west side of the Andes was most apparent along a latitudinal axis, with additional differentiation between the lowlands and mid-elevations. Finally, temperature and vegetation variables were most important in explaining the observed genetic and morphological variation in frogs. The resulting patterns of variation are located along the elevational gradients on both sides of the Andes, with additional variation located along a latitudinal gradient (Fig. S2).
Table 2.
Generalized dissimilarity modeling results for significant models
| Species/region | Trait | % Explained* | Variables selected† |
|---|---|---|---|
| WBWC/West | AFLP | 98.5 / 98.4 / 50.8 / 19.4 | 18, 6, 8, 4, 7, 1, 16 |
| Tarsus length | 60.7 / 60.7 / 0.0 / 34.3 | 6, 1, 8 | |
| Wing length | 91.7 / 91.7 / 6.1 / 34.3 | 7, 13, 6, 14 | |
| Tail length | 82.4 / 81.5 / 6.1 / 24.6 | 7, 13, 9, 1, 3 | |
| Bill depth | 92.5 / 91.5 / 22.7 / 11.4 | 11, 12, 1, 5, 8, 9 | |
| Bill length | 63.9 / 63.9 / 0.0 / 0.0 | 5, 8, 14, 6, 4 | |
| WBWC/East | AFLP | 72.2 / 71.5 / 8.8 / 28.0 | 12, 17, 9, 6, 1, 16, 5, 11, 14 |
| Tarsus length | 70.5 / 50.5 / 10.9 / 13.8 | 14, 5, 6, 4, 9 | |
| Bill depth | 27.2 / 23.1 / 10.4 / 2.4 | 8, 1, 5, 16, 17, 9 | |
| MFP/Ecuador | Msat | 51.0 / 51.0 / 2.4 / 11.2 | 6, 17, 16, 9, 11, 18, 8 |
| Tarsus length | 43.2 / 42.9 / 8.7 / 20.0 | 3, 14, 8, 5, 1, 12 | |
| Bill length | 70.8 / 70.8 / 0.0 / 8.1 | 17, 5, 11, 9, 12, 13, 4 | |
| SNFC/Ecuador | Msat | 42.0 / 40.6 / 12.0 / 2.4 | 9, 16, 18, 1, 4, 17, 11 |
| Tarsus length | 73.8 / 73.8 / 0.0 / 45.8 | 13, 12, 14, 4 | |
| Tail length | 90.8 / 90.8 / 0.0 / 84.3 | 8, 12, 9, 5, 16, 18 | |
| Cb/West | Zygomatic arch | 33.1 / 33.1 / 0.1 / 25.7 | 13, 7, 5, 6, 15, 18, 9 |
| Forearm length | 68.9 / 68.9 / 0.1 / 9.6 | 8, 16, 9, 7, 1 | |
| Cb/East | Forearm length | 16.1 / 16.0 / 3.7 / 0.7 | 7, 16, 13, 12, 6, 4, 1, 9 |
| Cc/West | Zygomatic arch | 40.0 / 40.0 / 0.8 / 35.8 | 17, 15, 10, 8 |
| Cc/East | Zygomatic arch | 20.0 / 18.2 / 3.4 / 10.9 | 9, 5, 6, 4, 1, 7 |
| Forearm length | 60.2 / 60.2 / 15.8 / 3.6 | 7, 4, 8, 11, 9, 6 | |
| Skull size | 42.4 / 42.4 / 23.1 / 8.4 | 4, 8, 6, 5, 4, 17, 11, 12 | |
| Cp/West | Zygomatic arch | 78.5 / 78.5 / 1.3 / 39.5 | 18, 16, 14, 4, 17, 9, 8, 3, 10 |
| Forearm length | 33.7 / 32.3 / 21.4 / 1.1 | 5, 11, 15, 1, 9, 17 | |
| Skull size | 61.6 / 61.2 / 39.6 / 5.7 | 16, 1, 18, 14, 3, 12, 4, 7 | |
| Cp/East | AFLP | 34.6 / 34.6 / 0.0 / 11.1 | 17, 18, 5 |
| Zygomatic arch | 19.0 / 18.5 / 2.2 / 1.0 | 6, 1, 9, 4, 12, 11, 14, 5, 16 | |
| Forearm length | 12.8 / 12.8 / 0.0 / 0.2 | 5, 6, 7, 16 | |
| Skull size | 22.3 / 19.2 / 6.2 / 1.3 | 1, 8, 11, 5, 6, 15, 7, 9, 18 | |
| Pw-n | nDNA | 79.2 / 76.7 / 49.5 / 4.1 | 18, 15, 12, 1, 16, 11, 4 |
| Gape width | 54.1 / 53.8 / 0.05 / 11.5 | 11, 8, 17, 16, 13, 6, 1, 9, 7 | |
| Jaw length | 34.1 / 33.6 / 0.8 / 17.4 | 8, 15, 13, 5, 3, 1, 6, 17, 11, 12 | |
| Met-car | 34.9 / 34.6 / 0.0 / 1.9 | 9, 4, 17, 6, 11, 12, 1, 18 | |
| Phalanges | 55.9 / 55.9 / 0.0 / 4.5 | 9, 4, 7, 11, 12, 8, 3, 17 | |
| Radio-ulna | 36.9 / 36.9 / 0.8 / 18.6 | 8, 11, 6, 15, 1, 16, 3, 4, 10, 17 | |
| Femur length | 40.9 / 40.9 / 0.0 / 4.3 | 11, 9, 7, 5, 17, 8 |
WBWC, wedge-billed woodcreeper; MFP, masked flowerpiercer; SNFC, streak-necked flycatcher; Cb, Carollia brevicauda; Cc, Carollia castanea; Cp, Carollia perspicillata; Pw-n, Pristimantis w-nigrum; AFLP, amplified fragment length polymorphism; SRTM, Shuttle Radar Topography Mission.
Percentages of total variation explained are shown for models with the following predictor variables: distance and environment (full model)/only environment/only distance/random layers.
Variables are shown in decreasing order of importance. 1 geographic distance; 2 Andean barrier; 3 elevation (SRTM); 4 elevation std (SRTMstd); 5 QSCATMean; 6 QSCATStd; 7 Treecover; 8 LAImax; 9 LAIrange; 10 Bio1; 11 Bio2; 12 Bio4; 13 Bio5; 14 Bio6; 15 Bio12; 16 Bio15; 17 Bio16; 18 Bio17.
Levels of intraspecific variation
To identify areas harboring the highest levels of intraspecific variation per unit area (alpha-diversity), we mapped the top 10% of standard deviations for GDM classes in a 3 × 3 km area (Fig. 2). The highest levels of alpha-diversity in morphological and genetic traits were found mostly along the slopes of the Andes (Fig. 2). These results are in contrast with the location of areas comprising the highest levels of species richness, which are in the Amazon basin (Fig. S3). Thus, the spatial patterns of species-level data and intraspecific variation are different, suggesting that the former may not be a suitable representation of the latter, and that, if the goal is to maximize intraspecific variation, it should be considered in conservation prioritization. Even though the wedge-billed woodcreeper and the bats C. brevicauda, C. castanea, and C. perspicillata also showed higher levels of genetic and morphological alpha-diversity within the Amazon basin, as well as along the Andes, the lowland areas capture only a fraction of the intraspecific variation. Interestingly, areas of high intraspecific alpha-diversity are at least partly concordant across the species examined here, suggesting that a subset of species might accurately represent a broad array of taxa.
Figure 2.
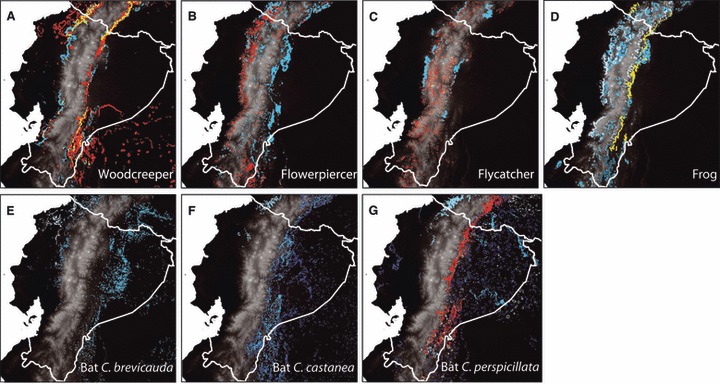
Maps indicating the highest 10% of variation per unit area (3 × 3 km) (alpha-diversity) in genetic and morphological variation in the seven target species. (A) wedge-billed woodcreeper Glyphorynchus spirurus; (B) masked flowerpiercer Diglossa cyanea; (C) streak-necked flycatcher Mionectes striaticollis; (D) the frog species Pristimantis w-nigrum; (E) silky short-tailed bat Carollia brevicauda; (F) chestnut short-tailed bat Carollia castanea; and (G) Seba's short-tailed bat Carollia perspicillata. Colors indicate the types of variation examined: red: genetic data; blue: morphological data (different shades of blue indicate different morphological traits); yellow: overlapping regions of high levels of alpha-diversity in both genetic and morphological data. Grey scale indicates elevation, with low elevations in black and high elevations in white.
Reserve selection
To generate maps of conservation priority areas based on species richness and complementarity of amphibians, birds, mammals, all species, and based on genetic and morphological variation, we performed ResNet runs at representation targets of one occurrence and 5%, 10%, and 20% of the total area occupied by each species or each GDM class of similar genotypes or phenotypes. The percentage of total land area selected for conservation increased linearly with higher targets to a maximum of 22.7% at a 20% target for birds (Table 3). The area selected for conservation based on genetic and morphological variation was up to 5.2% smaller than that based on biodiversity pattern, represented by species-level data. The overlap between priority areas for each taxonomic group was relatively low, ranging from 10.4% to 12.8% at a 10% representation target (Table 4). Interestingly, priority areas at the 10% representation target based on all species captured only 17.8–32.8% of those based on each individual taxonomic group. These results suggest that none of the three taxonomic groups (amphibians, birds, and mammals) is a good surrogate for any of the others and that only their combined analysis can ensure good representation of all species in priority areas. In addition, the overlap between genetic/morphological variation and species-level data at a 10% representation target was also low (7.6–12.4%; Table 4), suggesting that reserves based on species-level data do not effectively represent genetic and morphological variation, or vice versa. The existing reserve network covers about 16% of continental Ecuador and is overlapping with a higher percentage of selected areas based on genetic and morphological variation than based on species-level information (Table 3). Overall, the percentage of priority areas that were located in existing reserves ranged between 11.1% and 23.8%.
Table 3.
Comparison of areas selected by ResNet to total area of Ecuador and percentages located in existing reserves
| Measure of biodiversity | Target* | % Area of Ecuador | % In reserves |
|---|---|---|---|
| Reserves | 16.0 | ||
| Species | |||
| Amphibians | 1 | 0.1 | 18.1 |
| 5% | 4.7 | 16.6 | |
| 10% | 9.5 | 16.1 | |
| 20% | 18.9 | 16.1 | |
| Birds | 1 | 0.0 | 18.2 |
| 5% | 5.7 | 15.9 | |
| 10% | 11.4 | 17.7 | |
| 20% | 22.7 | 18.0 | |
| Mammals | 1 | 0.0 | 11.1 |
| 5% | 5.8 | 15.7 | |
| 10% | 11.4 | 17.7 | |
| 20% | 22.1 | 17.6 | |
| All species | 1 | 0.1 | 14.1 |
| 5% | 5.8 | 15.2 | |
| 10% | 11.3 | 16.5 | |
| 20% | 22.4 | 15.9 | |
| Intraspecific variation | |||
| Gen/morph | 1 | 0.0 | 23.8 |
| 5% | 4.3 | 23.3 | |
| 10% | 8.6 | 21.1 | |
| 20% | 17.5 | 19.7 | |
Figures are provided for reserves based on each of the three different taxonomic groups (amphibians, birds, mammals, and the three groups combined) for which species-level data were available, as well as for intraspecific variation (gen/morph).
The set of selected sites was required to represent at least a certain target percentage of the total area occupied by a species, or by a generalized dissimilarity modeling class of similar genotypes or phenotypes. We specified targets of one occurrence (one site of 2 × 2 km), 5%, 10%, and 20%.
Table 4.
Percent overlap between ResNet solutions for different levels of information at targets of 10% and 20%
| Process | Pattern | ||||
|---|---|---|---|---|---|
| Gen/morph | All species | Amphibians | Birds | Mammals | |
| Target = 10 | |||||
| Gen/morph | 100.0 | 10.0 | 9.9 | 11.5 | 12.4 |
| All species | 7.6 | 100.0 | 27.5 | 30.4 | 17.9 |
| Amphibians | 9.0 | 32.8 | 100.0 | 12.4 | 12.8 |
| Birds | 8.7 | 30.3 | 10.4 | 100.0 | 11.6 |
| Mammals | 9.4 | 17.8 | 10.7 | 11.6 | 100.0 |
| Target = 20 | |||||
| Gen/morph | 100.0 | 19.4 | 18.3 | 22.1 | 22.7 |
| All species | 15.2 | 100.0 | 35.1 | 41.9 | 28.3 |
| Amphibians | 17.0 | 41.6 | 100.0 | 22.6 | 23.8 |
| Birds | 17.1 | 41.2 | 18.8 | 100.0 | 20.4 |
| Mammals | 18.0 | 28.5 | 20.3 | 20.9 | 100.0 |
Figures are provided for overlap between reserves based on each of the three different taxonomic groups (amphibians, birds, mammals, and the three groups combined) for which species-level data were available and based on intraspecific variation (gen/morph). Results are shown for representation targets of 10% and 20% of the total area occupied by each species or by each class of similar genotypes or phenotypes. The table reads as: percentage of ‘row’ overlapping with ‘column’.
The composite map of reserves using algorithm-based selection of species richness and intraspecific variation, and regions harboring the highest levels of intraspecific alpha-diversity, suggests that high priority should be given to regions located along the western and eastern slopes of the Andes (Fig. 3). Although the areas of highest intraspecific alpha-diversity are in the Andean foothills (Figs 2 and 3), when we selected areas to represent intraspecific variation with ResNet (Fig. 3), some sites in the Amazon lowlands were also selected. We hypothesized that the Amazon lowlands were selected because there was high intraspecific alpha-diversity in bat species in these lowland areas (Fig. 2E–G), as well as unique variation not present elsewhere. We tested these hypotheses by removing bats from the analysis and repeating the ResNet prioritization. This resulted in a more pronounced concentration of selected sites in the Andean foothills (Fig. S4). Nevertheless, areas in the Amazon basin continued to be selected, suggesting that unique genetic and morphological variation in wedge-billed woodcreepers from the Amazon basin contributed to its importance in the ResNet solutions. To determine which sites were regarded most important by the ResNet algorithm, we plotted the order of site selection of the map of Ecuador (Fig. S4), which suggested that sites in the Andes were selected first. This gave us confidence that the Andean foothills were indeed the most important in reserve selection.
Figure 3.
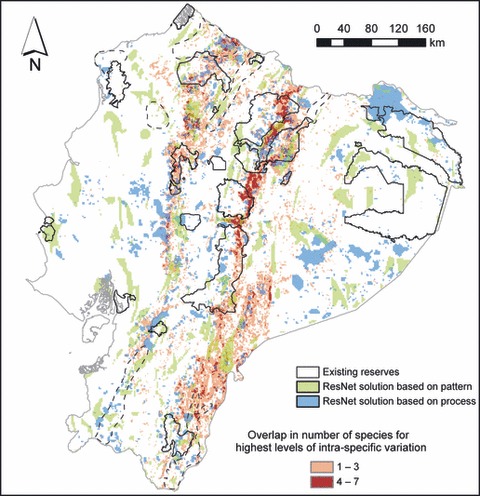
Composite map of selected reserves at a 10% representation target using species-level data (green) and genetic and morphological data (blue) in ResNet and areas harboring high levels of intraspecific alpha-diversity (orange and red). Areas delimited with solid black lines indicate existing reserves. Areas indicated with dashed lines are those where one or more of the generalized dissimilarity models show high uncertainty, because the associated environmental conditions are outside the range of those encountered at sampled locations.
Discussion
Climate change poses an urgent and significant threat to biodiversity (Fischlin et al. 2007; Sinervo et al. 2010). Species may not be able to respond to climate change unless they have the capacity to shift their ranges. However, range shifts may often not be possible for many reasons, including disparities in vagility between mutualistic species, isolation of populations because of habitat fragmentation, new biotic interactions resulting from a different species composition, or elevational constraints. Hence, many populations will have no choice but to adapt to the changing climate conditions in situ. A prudent conservation strategy therefore is to design reserves that maximize intraspecific variation. Our results provide the first, country-wide assessments of the utility of intraspecific genetic and morphological variation for conservation prioritization. Future work is planned to predict future morphological changes under climate change based on current spatial relationships between morphology and environment, but this is beyond the scope of this paper.
In considering evolutionary processes in reserve design, conservation planners have relied on a number of different measures of biodiversity as surrogates for evolutionary process. First, the phylogenetic species definition has been used to identify evolutionarily significant units (ESU) that represent different (genetic) lineages for conservation prioritization (Moritz 1994; Peterson and Navarro-Sigüenza 1998). An advantage of this approach is that some level of sub-specific variation is likely to be preserved. However, it is probably biased toward broad-scale vicariant events and unable to capture the finer-scale adaptive variation we identified with our method. Waples et al. (2001) used another approach to define ESUs based on a combination of habitat characteristics, genetic data, and life history traits. Although this approach may capture adaptive variation in the study species, its use has been limited to a single species complex, where detailed studies of life history were available, which is unlikely to be the case for the vast majority of species. A second approach to preserve evolutionary processes is the protection of habitat variability, primarily along environmental gradients (e.g. Cowling and Pressey 2001; Smith et al. 2001, 2005; Rouget et al. 2006; Klein et al. 2009). However, it is unclear to what extent total adaptive diversity is protected by focusing exclusively on gradients. Finally, phylogenetic diversity (PD) – as measured by the branch lengths in a phylogenetic tree – has served as a measure of genetic variation (e.g. Vane-Wright et al. 1991; Faith 1992; Forest et al. 2007). Such species-level information is useful in determining the phylogenetic diversity of communities, but may fail to capture extant population-level variation and adaptive diversity. It will likely miss very recent episodes of local adaptation and genetic isolation where sufficient time has not elapsed for species-level phylogenetic trees to show distinct genetic partitions. To help alleviate this issue, several phylogeographic studies have focused on intraspecific PD (Moritz and Faith 1998; Rissler et al. 2006). However, the genetic markers used in these studies typically evolve slowly, and the timescale of differentiation between lineages may be unsuitable for the purposes of preserving extant evolutionary processes. Moreover, patterns of genetic lineages may be the result of neutral rather than selective processes and consequently of less relevance to understanding adaptive variation that might be germane to climate change.
In contrast to the approaches discussed earlier, our spatially explicit models focus on the relationship between environmental heterogeneity and genetic and phenotypic variation and emphasize local adaptation and genetic turnover on recent timescales. The combined use of two types of data – genetic and morphological – is likely a benefit to our approach (Crandall et al. 2000). The genetic markers used may or may not be linked to adaptive variation, but the fact that a large proportion of the variation was explained by environmental variables, and not by geographic distance, suggests that the markers may be a suitable proxy for adaptive variation. In addition, although there is no direct evidence for the adaptive context of the morphological traits in the species studied, there is evidence in other species that these traits may be fitness-related (e.g. Grant et al. 1976; Wassersug and Sperry 1977; Arnold and Wade 1984; Smith 1990; Norberg 1994; Sedinger et al. 1995; Heinen and Hammond 1997; Bardwell et al. 2001; Benkman 2003; Jensen et al. 2004; Milá et al. 2008; Sol 2008; Nogueira et al. 2009). Thus, these traits are likely to represent adaptive variation. The combined use of morphological and genetic markers may provide more confidence in the resulting maps of priority areas than either alone.
Between-species comparisons
Because reserve selection is often scale-dependent and our focal species comprise distributions of varying sizes and coverage, we did not select reserves for each focal species to compare specific priority areas, but instead focused on the areas with the highest levels of intraspecific alpha-diversity to assess whether these priority areas are concordant among different species. Such areas are expected to be the ones where a given reserve could most likely harbor as much intraspecific variation as possible. All seven target species showed high levels of intraspecific alpha-diversity along the slopes of the Andes, and some in the Amazon basin, most notably the three bat species (Fig. 2). This result is perhaps not surprising given that the steep elevational gradient from lowlands to highlands is correlated with steep gradients in many other environmental variables. Interestingly, however, the highest levels of genetic and morphological alpha-diversity are mostly located in the mid-elevations from approximately 1500–2500 m, extending down to about 700 m in the southeast. In addition, areas harboring high levels of genetic variation do not in all species overlap with those comprising high morphological alpha-diversity. This may be explained by the fact that it is unlikely that the genetic markers used are linked to the morphological traits measured, and the two data types may thus represent different levels of diversity. In addition, variation in the phenotypic traits may in part be the result of a plastic response, without underlying genetic differences. Plasticity in itself is likely important to the versatility of populations to respond to climate change. Thus, the reduction of levels of plasticity because of habitat loss may have negative impacts on the persistence of populations and species under changing environmental conditions. As a consequence, considering phenotypic trait variation in conservation prioritization is important irrespective of whether the variation is because of underlying genetic differences or because of plasticity.
Comparison between reserves based on species richness versus intraspecific variation
Priority areas based on species-level data were only partly concordant with those based on intraspecific variation (Table 4) and may thus not be sufficiently effective in preserving either species richness or intraspecific variation given ongoing and future climate change. This result suggests that the reverse is also true: reserves based on intraspecific variation do not adequately capture species-level variation. A comparison of reserves based on intraspecific variation using either the ResNet rarity-complementarity algorithm or the criterion that reserves should harbor high levels of intraspecific alpha-diversity also reveals discordant patterns (Fig. 3). This can partly be explained by the distribution of unique variation not found elsewhere and by the way ResNet selects sites important for conservation. Sites comprising high levels of variation are selected first, because these represent the observed intraspecific variation very efficiently (Fig. S4). Thus, the target for the full range of genetic and morphological variation found in areas with high intraspecific alpha-diversity across species is quickly met in relatively small areas (the small, scattered blue areas along the Andes in Fig. 3). However, areas harboring unique variation, but little overall intraspecific diversity, have at that point not been selected yet, and the targets for the unique variation have not been met. In subsequent iterations of the algorithm, these are the areas selected next, but as overlap in unique intraspecific variation among species decreases, larger areas will need to be selected in order to meet the respective targets of representation. Thus, the reserves in areas harboring high levels of variation are smaller than those in areas of low variation, because of the higher efficiency of protecting intraspecific variation in the former.
Taken together, these results suggest that the spatial patterns of species richness are not concordant with those of intraspecific variation and emphasize the need for both to be taken into account in reserve design. Our results also suggest that the evolutionary processes underlying observed patterns of species richness may occur on different spatial scales or have been differently distributed geographically than those causing current levels of intraspecific variation. For example, intraspecific variation may evolve in response to habitat heterogeneity on a smaller spatial scale than that necessary for speciation (Losos and Schluter 2000).
Comparison with Ecuador's existing reserves
The existing reserve network in Ecuador does not effectively capture species richness or intraspecific variation, despite the fact that a relatively large proportion of the total landmass is protected (∼16%). In addition, large areas of high intraspecific or species diversity remain unprotected by the existing reserves (Fig. 3). This discrepancy may not be surprising given the fact that reserves are often established using socio-economic and political criteria rather than biological ones (Pressey and Tully 1994; Pressey et al. 1994; Bass et al. 2010). In fact, a similar ineffectiveness of the existing reserves was found in previous studies using ecosystem-level data across Ecuador (Sierra et al. 2002), species-level data for the equatorial Pacific region (Peralvo et al. 2007), and a combination of the representation of ecosystems and Red List species in the Tropical Andes and Chocó (Sarkar et al. 2009). Our study thus supports the conclusions of previous studies regarding the inefficacy of the existing reserve network in Ecuador, but is novel in showing that this is also true when using intraspecific variation as a measure of biodiversity.
Comparison to other proposed reserves
One of the most comprehensive conservation prioritization studies for the entire country of Ecuador was carried out using differences in habitat type as a measure of diversity (Sierra et al. 2002). Our results agree with Sierra et al. (2002) in placing high priority on the Andean slopes in regions that are largely unprotected in existing reserves. In addition, our study placed low to medium priority on some areas in the Amazon basin (blue areas in Fig. 3), which corresponds to the prioritization by Sierra et al. (2002). However, only our study identifies these areas as having unique intraspecific variation found nowhere else in Ecuador. Consequently, even though the Amazon basin may not harbor high levels of intraspecific alpha-diversity, a prioritization scheme that aims to protect the entire range of intraspecific variation in the species studied should also include areas in the Amazon basin. Moreover, the Amazon basin comprises the highest species richness per unit area found in Ecuador (Fig. S3).
In contrast to our study, the prioritization scheme by Sierra et al. (2002) put more emphasis on the far northwestern and central coasts, likely as a result of the presence of unique ecosystems, comprised of coastal lowland evergreen forests and dry shrub, found nowhere else in the country. Two additional studies that prioritized areas for conservation in Ecuador did so only for parts of Ecuador (Peralvo et al. 2007; Sarkar et al. 2009). Because of this difference in scale, and because of likely scale dependence in reserve selection (e.g. Sierra et al. 2002; Warman et al. 2004), we refrain from making comparisons with our study.
Advantages and limitations
A potential limitation of our study is that the range maps used as input for the species-level analyses are at a broad scale and do not capture finer-scale nuances of the species distributions, which may influence the results of prioritization software. In addition, it would be desirable to add intraspecific variation of additional species, most notable plant and insect species, which may improve the representation of intraspecific variation of the entire community. Nevertheless, we are confident in concluding that reserves based on species-level data are not concordant with those based on intraspecific variation and that there is a need for both in conservation prioritization.
Over- or underprediction of the species’ ranges may have consequences for the models of intraspecific variation. Over- and underprediction is likely to occur at the extremes of the suitable range of environmental conditions. Thus, prediction of a species’ range into areas where it is not present may result in the false prediction of unique intraspecific variation. In contrast, failure to predict the full species range may result in neglecting unique intraspecific variation that might be important to conserve. Nevertheless, based on expert knowledge and our own experience in the field with the seven target species, we are confident about the predicted species distribution. While our target species are unequally distributed across Ecuador, the combined distributions of the target species span almost the entire country. In addition, it is important to note that the patterns of high intraspecific alpha-diversity are concordant among target species (see above and Figs 2 and 3).
Several questions remain with regard to the implementation of the approach proposed here. For instance, the intraspecific genetic and morphological variation considered may not be equally important to fitness. Thus, when such data are available, weighting of the traits may improve the relevance of the prioritization scheme to maximizing the evolutionary potential of populations in the face of climate change. In addition, the consequence of adding or removing traits is unknown. In general, adding species or traits will likely result in down weighting the relative influence of individual traits, which may be beneficial to the utility of the prioritization scheme for as many species as possible. We demonstrated that removing the three bat species affected the resulting prioritization scheme, but not in a way that it significantly altered the locations of the top priority reserves.
Finally, the methods presented here appear to be generally applicable to other areas insofar as there is reasonable overlap between the ranges of species targeted for modeling intraspecific variation. Conservation prioritization is scale-dependent, and our approach is no exception. Careful consideration of the size of the study area is therefore warranted. The framework presented here could be tailored to any region subject to data availability. The data required to use this framework include the longitude and latitude of species’ occurrences along with data on intraspecific phenotypic variation and population structure inferred from genetic markers. The software and GIS layers utilized in this study are freely available on the internet and could be deployed to develop conservation plans for anywhere in the world. As for Ecuador, it should be emphasized that we did not include socio-economic criteria, levels of threat, and opportunities, all critical elements in reserve design (Fig. 1) (reviewed in Moffett et al. 2006). Future work will attempt to incorporate this information in order to develop a final prioritization strategy for Ecuador.
Conclusions
We have shown that the locations of the highest levels of intraspecific genetic and morphological variation among a diverse group of vertebrate taxa are broadly concordant. This suggests that a subset of taxa could potentially serve as suitable representatives for community-wide levels of intraspecific variation. We also demonstrate that priority areas for conservation-based species-level data are not concordant with those based on intraspecific genetic and morphological variation. Because maximizing the level of environmentally associated genetic and morphological variation is likely to benefit populations in the face of climate change, our results emphasize the need for incorporating intraspecific variation in conservation prioritization. Finally, we have developed a new framework for considering multiple types of diversity for conservation planning. While we used Ecuador as a test case for these methods, they are broadly applicable to other regions and species and should serve to improve conservation planning targeted at maintaining all levels of diversity.
Acknowledgments
For help in the field we thank Fernando Alaya, Diego Almeida-Reinoso, Italo Tapia, Juan Fernando Freile, Tatiana Santander, Jaime Chaves, Gabriela Castañeda, Brandt T. Ryder, Daniela Gross, Juan Diego Ortiz, Orfa Rodríguez, María Fernanda Salazar, Suzanne Tomassi, John McCormack, Brenda Larison, Luis Carrasco, Marcelo Tobar, Jordan Karubian, and the Timpe family. We also thank Martín R. Bustamante, Santiago R. Ron, and Juan Manuel Guayasamin for frog specimen identification and locality data; Elisa Bonaccorso for facilitating molecular laboratory work on the frogs; Néstor Acosta-Buenaño and Pablo Menéndez-Guerrero for frog locality data; Mónica Páez and Andrea Terán for assistance in frog identification; Luis A. Coloma (Museo de Zoología at Pontifica Universidad Catolíca del Ecuador (PUCE)) for tissue loans; and the Kansas University of Natural History for access to the collection. We are grateful to Dr. M. Kinnison and two anonymous reviewers for thoughtful comments on a previous version of this manuscript. For assistance on woodcreeper work in the laboratory, we thank Navi Timber and Daniel Greenfield. Funding was provided by grants from NSF (IRCEB9977072 to TBS, RKW, and CJS) and NASA (IDS/03-0169-0347 to TBS, RKW, and CJS; NNG05GB37G to CHG).
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Data S1.Supporting materials and methods.
Figure S1. Predicted species distributionsbased on Maxent (Phillips et al. 2006) models for the followingspecies: (A) wedge-billed woodcreeper G. spirurus; (B)streak-necked flycatcher M. striaticollis; (C) maskedflowerpiercer D. cyanea; (D) the frog species P.w-nigrum; (E) silky short-tailed bat C. brevicauda; (F)chestnut short-tailed bat C. castanea; and (G) seba'sshort-tailed bat C. perspicillata. Colors indicate thepredicted logistic probability of occurrence (see color bar).
Figure S2. Maps of the generalized dissimilarity models that performed better than random.
Figure S3. Total species richness maps for birds, mammals, and amphibians in Ecuador.
Figure S4. ResNet solutions for intraspecific variation at 10% representation targets.
Table S1. List of environmental variables used in our analyses.
Table S2. Elevation and measures of geneticdiversity per site for M. striaticollis microsatellite data.
Table S3. Elevation and measures of geneticdiversity per site for D. cyanea microsatellite data.
Table S4. Elevation and measures of geneticdiversity per site for C. perspicillata AFLP data.
Table S5. Elevation and measures of geneticdiversity per site for P. w-nigrum nuclear DNA data.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Literature cited
- Anderson RP, Gomez-Laverde M, Peterson AT. Geographical distributions of spiny pocket mice in South America: insights from predictive models. Global Ecology and Biogeography. 2002;11:131–141. [Google Scholar]
- Arnold SJ, Wade MJ. On the measurement of natural and sexual selection: applications. Evolution. 1984;38:720–734. doi: 10.1111/j.1558-5646.1984.tb00345.x. [DOI] [PubMed] [Google Scholar]
- Bardwell E, Benkman CW, Gould WR. Adaptive geographic variation in western scrub-jays. Ecology. 2001;82:2617–2627. [Google Scholar]
- Bass MS, Finer M, Jenkins CN, Kreft H, Cisneros-Heredia DF, McCracken SF, Pitman NCA, et al. Global conservation significance of Ecuador's Yasuni National Park. PLoS ONE. 2010;5:e8767. doi: 10.1371/journal.pone.0008767. doi: 10.1371/journal.pone.0008767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkman CW. Divergent selection drives the adaptive radiation of crossbills. Evolution. 2003;57:1176–1181. doi: 10.1111/j.0014-3820.2003.tb00326.x. [DOI] [PubMed] [Google Scholar]
- Buermann W, Saatchi S, Graham CH, Zutta BR, Chaves JA, Milá B, Smith TB. Predicting species distributions across the Amazonian and Andean regions using remote sensing data. Journal of Biogeography. 2008;35:1160–1176. [Google Scholar]
- Chaves JA, Pollinger JP, Smith TB, LeBuhn G. The role of geography and ecology in shaping the phylogeography of the speckled hummingbird (Adelomyia melanogenys) in Ecuador. Molecular Phylogenetics and Evolution. 2007;43:795–807. doi: 10.1016/j.ympev.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Cowling RM, Pressey RL. Rapid plant diversification: planning for an evolutionary future. Proceedings of the National Academy of Sciences of the USA. 2001;98:5452–5457. doi: 10.1073/pnas.101093498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crandall KA, Bininda-Emonds ORP, Mace GM, Wayne RK. Considering evolutionary processes in conservation biology. Trends in Ecology and Evolution. 2000;15:290–295. doi: 10.1016/s0169-5347(00)01876-0. [DOI] [PubMed] [Google Scholar]
- Darst CR, Huffman KA, Jarvis J. Conservation significance of America's newest system of protected areas: national landscape conservation system. Natural Areas Journal. 2009;29:224–254. [Google Scholar]
- Eisenberg JF, Redford KH. Mammals of the Neotropics, Volume 3: Ecuador, Bolivia, Brazil. Chicago, IL: University of Chicago Press; 2000. [Google Scholar]
- Faith DP. Conservation evaluation and phylogenetic diversity. Biological Conservation. 1992;61:1–10. [Google Scholar]
- Ferrier S, Manion G, Elith J, Richardson K. Using generalized dissimilarity modelling to analyse and predict patterns of beta diversity in regional biodiversity assessment. Diversity and Distributions. 2007;13:252–264. [Google Scholar]
- Finer M, Moncel R, Jenkins CN. Leaving the oil under the Amazon: Ecuador's Yasuni-ITT Initiative. Biotropica. 2010;42:63–66. [Google Scholar]
- Fischlin A, Midgley GF, Price JT, Leemans R, Gopal B, Turley C, Rounsevell MDA. Ecosystems, their properties, goods, and services. In: Parry ML, Canziani OF, Palutikof JP, van der Linden PJ, Hanson CE, et al., editors. Climate Change 2007: Impacts, Adaptation and Vulnerability. Contribution of Working Group II to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge: Cambridge University Press; 2007. pp. 211–272. [Google Scholar]
- Forest E, Grenyer R, Rouget M, Davies TJ, Cowling RM, Faith DP, Balmford A, et al. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature. 2007;445:757–760. doi: 10.1038/nature05587. [DOI] [PubMed] [Google Scholar]
- Frankel OH. Genetic conservation: our evolutionary responsibility. Genetics. 1974;78:53–65. doi: 10.1093/genetics/78.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham CH, Ron SR, Santos JC, Schneider CJ, Moritz C. Integrating phylogenetics and environmental niche models to explore speciation mechanisms in dendrobatid frogs. Evolution. 2004;58:1781–1793. doi: 10.1111/j.0014-3820.2004.tb00461.x. [DOI] [PubMed] [Google Scholar]
- Grant PR, Grant BR, Smith JNM, Abbott I, Abbott LK. Darwin's finches: population variation and natural selection. Proceedings of the National Academy of Sciences. 1976;73:257–261. doi: 10.1073/pnas.73.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory A, Burke T, Ferris R, Robson J, Smithers R, Whitlock R. 2006. The Conservation of Genetic Diversity: Science and Policy Needs in a Changing World. JNCC report, No. 383.
- Hansen MC, DeFries RS, Townshend JRG, Sohlberg RA, Dimicelli C, Carroll M. Towards an operational MODIS continuous field of percent tree cover algorithm: examples using AVHRR and MODIS data. Remote Sensing of the Environment. 2002;83:303–319. [Google Scholar]
- Heinen JT, Hammond G. Antipredator behaviors of newly metamorphosed green frogs (Rana clamitans) and leopard frogs (R. pipiens) in encounters with eastern garter snakes (Thamnophis s-sirtalis. American Midland Naturalist. 1997;137:136–144. [Google Scholar]
- Hess GR, Bartel RA, Leidner AK, Rosenfeld KM, Rubino MJ, Snider SB, Ricketts TH. Effectiveness of biodiversity indicators varies with extent, grain, and region. Biological Conservation. 2006;132:448–457. [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology. 2005;25:1965–1978. [Google Scholar]
- IUCN, Conservation International, and NatureServe. 2008. An analysis of amphibians on the 2008 IUCN Red List http://www.iucnredlist.org/amphibians (accessed on 12 November 2008)
- Jarrín-V P, Flores-C C, Salcedo-Q J. Morphological variation in the Short-tailed Fruit Bat (Carollia) in Ecuador, with comments on the practical and philosophical aspects of boundaries among species. Integrative Zoology. 2010;5:226–240. doi: 10.1111/j.1749-4877.2010.00208.x. [DOI] [PubMed] [Google Scholar]
- Jensen H, Sæther B-E, Ringsby TH, Tufto J, Griffith SC, Ellegren H. Lifetime reproductive success in relation to morphology in the house sparrow Passer domesticus. Journal of Animal Ecology. 2004;73:599–611. [Google Scholar]
- Justice CO, Vermote E, Townsend JRG, DeFries R, Roy DP, Hall DK, Salomonson VV, et al. The Moderate Resolution Imaging Spectroradiometer (MODIS): land remote sensing for global change research. IEEE Transactions on Geoscience and Remote Sensing. 1998;36:1228–1249. [Google Scholar]
- Justus J, Fuller T, Sarkar S. Influence of representation targets on the total area of conservation-area networks. Conservation Biology. 2008;22:673–682. doi: 10.1111/j.1523-1739.2008.00928.x. [DOI] [PubMed] [Google Scholar]
- Kelley C, Garson J, Aggarwal A, Sarkar S. Place prioritization for biodiversity reserve network design: a comparison of the SITES and ResNet software packages for coverage and efficiency. Diversity and Distributions. 2002;8:297–306. [Google Scholar]
- Klein C, Wilson K, Watss M, Stein J, Berry S, Carwardine J, Stafford Smith M, et al. Incorporating ecological and evolutionary processes into continental-scale conservation planning. Ecological Applications. 2009;19:206–217. doi: 10.1890/07-1684.1. [DOI] [PubMed] [Google Scholar]
- Long DG, Drinkwater MR, Holt B, Saatchi S, Bertoia C. Global ice and land climate studies using scatterometer image data. EOS Transactions AGU. 2001;82:503. [Google Scholar]
- Losos JB, Schluter D. Analysis of an evolutionary species–area relationship. Nature. 2000;408:847–850. doi: 10.1038/35048558. [DOI] [PubMed] [Google Scholar]
- Lynch JD. A new frog species of the Eleutherodactylus fitzingeri group from the Pacific Andean versant in Ecuador. Herpetologica. 1979;35:228–233. [Google Scholar]
- McRae BH. Isolation by resistance. Evolution. 2006;60:1551–1561. [PubMed] [Google Scholar]
- Milá B, Wayne RK, Smith TB. Ecomorphology of migratory and sedentary populations of the yellow-rumped warblers (Dendroica coronata. Condor. 2008;110:335–344. [Google Scholar]
- Milá B, Wayne RK, Fitze P, Smith TB. Divergence with gene flow and fine-scale phylogeographic structure in the wedge-billed woodcreeper Glyphorynchus spirurus, a Neotropical rainforest bird. Molecular Ecology. 2009;18:2879–2995. doi: 10.1111/j.1365-294X.2009.04251.x. [DOI] [PubMed] [Google Scholar]
- Moffett A, Dyer J, Sarkar S. Integrating biodiversity representation with multiple criteria in North-Central Namibia using non-dominated alternatives and a modified Analytic Hierarchy Process. Biological Conservation. 2006;129:181–191. [Google Scholar]
- Moritz C. Defining ‘evolutionarily significant units’ for conservation. Trends in Ecology and Evolution. 1994;9:373–375. doi: 10.1016/0169-5347(94)90057-4. [DOI] [PubMed] [Google Scholar]
- Moritz C. Strategies to protect biological diversity and the evolutionary processes that sustain it. Systematic Biology. 2002;51:238–254. doi: 10.1080/10635150252899752. [DOI] [PubMed] [Google Scholar]
- Moritz C, Faith D. Comparative phylogeography and the identification of genetically divergent areas for conservation. Molecular Ecology. 1998;7:419–429. [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Myneni RB, Hoffman S, Knyazikhin Y, Privette JL, Glassy J, Tian Y, Wang Y, et al. Global products of vegetation leaf area and fraction absorbed PAR from year one of MODIS data. Remote Sensing of the Environment. 2002;83:214–231. [Google Scholar]
- Nogueira MR, Peracchi AL, Monteiro LR. Morphological correlates of bite force and diet in the skull and mandible of phyllostomid bats. Functional Ecology. 2009;23:715–723. [Google Scholar]
- Norberg UM. Wing design, flight performance and habitat use in bats. In: Wainwright PC, Reilly SM, editors. Ecological Morphology: Integrative Organismal Biology. Chicago: University of Chicago Press; 1994. pp. 205–239. [Google Scholar]
- Orme CDL, Davies RG, Burgess M, Eigenbrod F, Pickup N, Olson VA, Webster AJ, et al. Global hotspots of species richness are not congruent with endemism or threat. Nature. 2005;436:1016–1019. doi: 10.1038/nature03850. [DOI] [PubMed] [Google Scholar]
- Patterson BD, Ceballos G, Sechrest W, Tognelli MF, Brooks T, Luna L, Ortega P, et al. Digital Distribution Maps of the Mammals of the Western Hemisphere, version 3.0. Arlington, VA, USA: NatureServe; 2007. [Google Scholar]
- Peralvo M, Sierra R, Young KR, Ulloa-Ulloa C. Identification of biodiversity conservation priorities using predictive modeling: an application for the equatorial pacific region of South America. Biodiversity Conservation. 2007;16:2649–2675. [Google Scholar]
- Peterson AT, Navarro-Sigüenza AG. Alternate species concepts as bases for determining priority conservation areas. Conservation Biology. 1998;13:427–431. [Google Scholar]
- Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecological Modelling. 2006;190:231–259. [Google Scholar]
- Pressey RL, Tully SL. The cost of ad hoc conservation: a case study in western New South Wales. Australian Journal of Ecology. 1994;19:375–384. [Google Scholar]
- Pressey RL, Johnson IR, Wilson PD. Shades of irreplaceability: towards a measure of the contribution of sites to a reservation goal. Biodiversity Conservation. 1994;3:242–262. [Google Scholar]
- Ray N. PATHMATRIX: a geographical information system tool to compute effective distances among samples. Molecular Ecology Notes. 2005;5:177–180. [Google Scholar]
- Ridgely RS, Greenfield PJ. The Birds of Ecuador. Volume II: A Field Guide. London: Christopher Helm; 2001. [Google Scholar]
- Ridgely RS, Allnutt TF, Brooks T, McNicol DK, Mehlman DW, Young BE, Zook JR. Digital Distribution Maps of the Birds of the Western Hemisphere, version 3.0. Arlington, VA, USA: NatureServe; 2007. [Google Scholar]
- Rissler LJ, Hijmans RJ, Graham CH, Moritz C, Wake DB. Phylogeographic lineages and species comparisons in conservation analysis: a case study of California herpetofauna. American Naturalist. 2006;167:655–666. doi: 10.1086/503332. [DOI] [PubMed] [Google Scholar]
- Rouget M, Cowling RM, Lombard AT, Knight AT, Kerley GIH. Designing large-scale conservation corridors for pattern and process. Conservation Biology. 2006;20:549–561. doi: 10.1111/j.1523-1739.2006.00297.x. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Pappas C, Garson J, Aggarwal A, Cameron S. Place prioritization for biodiversity conservation using probabilistic surrogate distribution data. Diversity and Distributions. 2004;10:125–133. [Google Scholar]
- Sarkar S, Pressey RL, Faith DP, Margules CR, Fuller T, Stoms DM, Moffett A, et al. Biodiversity conservation planning tools: present status and challenges for the future. Annual Review of Environment and Resources. 2006;31:123–159. [Google Scholar]
- Sarkar S, Fuller T, Aggarwal A, Moffett A, Kelley CD. The ConsNet software platform for systematic conservation planning. In: Moilanen A, Wilson KA, Possingham HP, editors. Spatial Conservation Prioritization: Quantitative Methods and Computational Tools. New York: Oxford University Press; 2009. pp. 235–248. [Google Scholar]
- Sedinger JS, Flint PL, Lindberg MS. Environmental influence on life-history traits – growth, survival, and fecundity in Black Brant (Branta bernicla. Ecology. 1995;76:2404–2414. [Google Scholar]
- Sgrò CM, Lowe AJ, Hoffman AA. Building evolutionary resilience for conserving biodiversity under climate change. Evolutionary Applications. 2011;4:326–337. doi: 10.1111/j.1752-4571.2010.00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra R, Campos F, Chamberlin J. Assessing biodiversity conservation priorities: ecosystem risk and representativeness in continental Ecuador. Landscape and Urban Planning. 2002;59:95–110. [Google Scholar]
- Sinervo B, Méndez-de-la-Cruz F, Mile DB, Heulin B, Bastiaans E, Villagrán-Santa Cruz M, Lara-Resediz R, et al. Erosion of lizard diversity by climate change and altered thermal niches. Science. 2010;328:894–899. doi: 10.1126/science.1184695. [DOI] [PubMed] [Google Scholar]
- Smith TB. Resource use by bill morphs of an African finch: evidence for intraspecific competition. Ecology. 1990;71:1246–1257. [Google Scholar]
- Smith TB, Grether G. The importance of conserving evolutionary processes. In: Carroll SP, Fox CW, editors. Conservation Biology: Evolution in Action. Oxford, UK: Oxford University Press; 2008. pp. 85–98. [Google Scholar]
- Smith TB, Bruford MW, Wayne RK. The preservation of process: the missing element of conservation programs. Biodiversity Letters. 1993;1:164–167. [Google Scholar]
- Smith TB, Kark S, Schneider CJ, Wayne RK. Biodiversity hotspots and beyond: the need for preserving environmental transitions. Trends in Ecology and Evolution. 2001;16:431. [Google Scholar]
- Smith TB, Saatchi S, Graham CH, Slabbekoorn H, Spicer G. Putting process on the map: why ecotones are important for conserving biodiversity. In: Purvis A, Gittleman J, Brooks T, editors. Phylogeny and Conservation. Cambridge, UK: Cambridge University Press; 2005. pp. 166–197. [Google Scholar]
- Sol D. Artificial selection, naturalization, and fitness: Darwin's pigeons revisited. Biological Journal of the Linnean Society. 2008;93:657–665. [Google Scholar]
- Thomassen HA, Buermann W, Milá B, Graham CH, Cameron SE, Schneider CJ, Pollinger JP, et al. Modeling environmentally associated morphological and genetic variation in a rainforest bird, and its application to conservation prioritization. Evolutionary Applications. 2010;3:1–16. doi: 10.1111/j.1752-4571.2009.00093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis JMJ. Climate change and habitat destruction: a deadly anthropogenic cocktail. Proceedings of Biological Sciences. 2003;270:467–473. doi: 10.1098/rspb.2002.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymchuk WV, O'Reilly P, Bittman J, MacDonald D, Schulte P. Conservation genomics of Atlantic salmon: variation in gene expression between and within regions of the Bay of Fundy. Molecular Ecology. 2010;19:1842–1859. doi: 10.1111/j.1365-294X.2010.04596.x. [DOI] [PubMed] [Google Scholar]
- Vandergast AG, Bohonak AJ, Hathaway SA, Boys J, Fisher RN. Are hotspots of evolutionary potential adequately protected in southern California? Biological Conservation. 2008;141:1648–1664. [Google Scholar]
- Vane-Wright RI, Humphries CJ, Williams PH. What to protect? Systematics and the agony of choice. Biological Conservation. 1991;55:235–254. [Google Scholar]
- Waples RS, Gustafson RG, Weitkamp LA, Myers JM, Johnson OW, Busby PJ, Hard JJ, et al. Characterizing diversity in salmon from the Pacific Northwest. Journal of Fish Biology. 2001;59(Suppl. A):1–41. [Google Scholar]
- Warman LD, Sinclair ARE, Scudder GGE, Klinkenberg B, Pressey RL. Sensitivity of systematic reserve selection to decision about scale, biological data, and targets: case study from Southern British Columbia. Conservation Biology. 2004;18:655–666. [Google Scholar]
- Wassersug RJ, Sperry DG. The relationships of locomotion to differential predation on Pseudacris triseriata (Anura: Hylidae) Ecology. 1977;58:830–839. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


